Abstract
Due to the abundant presence of alkylating agents in living cells and the environment, DNA alkylation is generally unavoidable. Among the alkylated DNA lesions, O4-alkylthymidine (O4-alkyldT) are known to be highly mutagenic and persistent in mammalian tissues. Not much is known about how the structures of the alkyl group affect the repair and replicative bypass of the O4-alkyldT lesions, or how the latter process is modulated by translesion synthesis polymerases. Herein, we synthesized oligodeoxyribonucleotides harboring eight site-specifically inserted O4-alkyldT lesions and examined their impact on DNA replication in Escherichia coli cells. We showed that the replication past all the O4-alkyldT lesions except (S)- and (R)-sBudT was highly efficient, and these lesions directed very high frequencies of dGMP misincorporation in E. coli cells. While SOS-induced DNA polymerases play redundant roles in bypassing most of the O4-alkyldT lesions, the bypass of (S)- and (R)-sBudT necessitated Pol V. Moreover, Ada was not involved in the repair of any O4-alkyldT lesions, Ogt was able to repair O4-MedT and, to a lesser extent, O4-EtdT and O4-nPrdT, but not other O4-alkyldT lesions. Together, our study provided important new knowledge about the repair of the O4-alkyldT lesions and their recognition by the E. coli replication machinery.
INTRODUCTION
The genomic integrity is constantly challenged by endogenous metabolism and environmental exposure, leading to a diverse array of damage products in DNA (1,2). Alkylation of DNA is generally unavoidable owing to the ubiquitous presence of alkylating agents in the environment and in living cells, generating adducts at multiple sites on nucleobases as well as the phosphate backbone (3–6). Depending on the nature of the alkylating agents involved, the size of alkyl groups adducted to DNA varies from a simple methyl group to complex alkyl functionalities (6,7). Despite the cytotoxic, teratogenic and carcinogenic effects, alkylating agents also constitute a major class of cancer chemotherapeutic drugs (8).
Among all DNA alkylation adducts, O6-alkyl-2′-deoxyguanosine (O6-alkyldG) and O4-alkylthymidine (O4-alkyldT) are known to be highly mutagenic (6,9–11). Although O6-alkyldG is induced more efficiently than O4-alkyldT (6) and can be detected both in vivo and in vitro (12), O4-alkyldT was found to accumulate at higher levels than O6-alkyldG in cellular and tissue DNA (13–16), suggesting that O4-alkyldT may be a more or equally important DNA alkylation adduct. Thus, it is important to understand how the O4-alkyldT lesions perturb the efficiency and accuracy of DNA replication, and how they are repaired.
Several shuttle vector studies with the use of site-specifically incorporated DNA lesions have been conducted for assessing how O4-alkyldT lesions compromise DNA replication in Escherichia coli and mammalian cells. In this vein, it was observed that O4-methyl- and O4-ethyl-dT (O4-MedT and O4-EtdT, Scheme 1) could direct high frequencies of dGMP misincorporation in E. coli cells (17–20). Likewise, O4-MedT, O4-EtdT and O4-n-propryl-dT (O4-nPrdT, Scheme 1) were found to be highly mutagenic during replication in mammalian cells (21–24), with the former two inducing T→C transition mutations at higher frequencies (∼20%) than O4-nPrdT (∼12%) (23). The relatively low mutation frequency of O4-nPrdT was attributed to the more efficient repair of this lesion, presumably by the nucleotide excision repair pathway (23). The miscoding properties of these lesions revealed from the in vivo replication experiments are in keeping with the findings made from in vitro replication studies, where O4-MedT, O4-EtdT and O4-iso-propyl-dT (O4-iPrdT, Scheme 1) were shown to direct preferential misincorporation of dGMP by purified DNA polymerases (25–27). However, very little is known about the roles of translesion synthesis DNA polymerases in bypassing these lesions in cells (20).
Scheme 1.
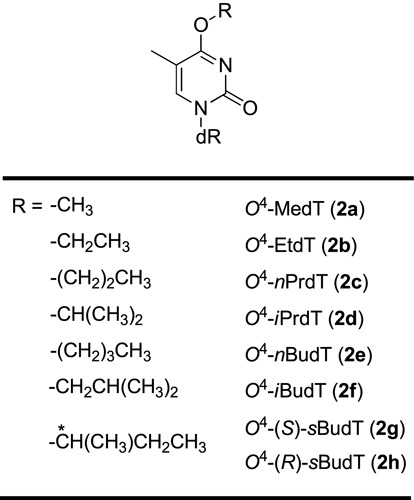
Structures of the O4-alkyldT lesions examined in the present study.
Previous studies showed that O6-alkylguanine-DNA alkyltransferase (AGT) was relatively inefficient in repairing the O4-alkyldT lesions (28–32). In vitro biochemical assays revealed that purified E. coli Ada repairs O4-MedT 10 000 times slower than O6-MedG, and Ogt repairs O4-MedT at a ∼84-fold higher efficiency than Ada (29,30). The lack of involvement of Ada in repairing O4-MedT was also manifested by the observation that the mutation frequency of O4-MedT, when situated in a single-stranded plasmid, was not altered by the depletion of this repair protein in E. coli cells (17). In addition, a recent study showed that the T:A→C:G transition mutations induced by ethylating agents and propylating agents in E. coli cells could be efficiently reduced by Ogt, suggesting the involvement of Ogt in repairing O4-EtdT and O4-nPrdT (32). Considering that these alkylating agents could also induce alkylation at adenine and at other nucleophilic sites (i.e., O2 and N3) on thymine, it is not clear whether the reduction in T:A→C:G transition mutations is primarily attributed to the repair of O4-alkyldT lesions. Thus, it is important to assess systematically the AGT-mediated repair of O4-alkyldT lesions and the modulation of this repair by the structure of the alkyl group.
Although site-specific mutagenesis studies have been carried out for several O4-alkyldT lesions, the studies were conducted with the use of somewhat disparate shuttle vector systems relying on colony picking/counting and sequencing to estimate the degrees to which these lesions impede DNA replication and induce mutations, respectively. In the present study, we set out to assess comprehensively, by using a highly quantitative and accurate shuttle vector-based assay (33,34), how the size (from methyl to butyl), branching (iPr, iBu and sBu versus their straight chain counterparts) and stereochemsitry (R versus S diastereomers of sBu) of the alkyl group incorporated to the major-groove O4 position of thymine affect the fidelity and efficiency of DNA replication, and how replication past these lesions is affected by AGT proteins and SOS-induced DNA polymerases.
MATERIALS AND METHODS
Material
All chemicals, unless otherwise specified, were purchased from Sigma-Aldrich (St Louis, MO, USA) or EMD Millipore (Billerica, MA, USA). 1,1,1,3,3,3-Hexafluoro-2-propanol (HFIP) was obtained from Oakwood Products Inc. (West Columbia, SC, USA). Common reagents for solid-phase DNA synthesis were obtained from Glen Research Co. (Sterling, VA, USA) and unmodified oligodeoxyribonucleotides (ODNs) were from Integrated DNA Technologies (Coralville, IA, USA). [γ-32P]ATP was obtained from Perkin Elmer (Piscataway, NJ, USA). Shrimp alkaline phosphatase was purchased from USB Corporation (Cleveland, OH, USA) and all other enzymes were obtained from New England Biolabs (Ipswich, MA, USA).
M13mp7(L2), wild-type AB1157 and C215 E. coli strains, and Ada- and Ogt-deficient E. coli strains (as FC215 derivatives), including C216 (Δogt::kan), C217 (Δada ΔalkB::cam) and C218 (Δogt::kan Δada ΔalkB::cam), were kindly provided by Prof. John M. Essigmann (35). Polymerase-deficient AB1157 strains [Δpol B1::spec (Pol II-deficient), ΔdinB (Pol IV-deficient), ΔumuC::kan (Pol V-deficient) and Δpol B1::specΔdinB ΔumuC::kan(Pol II, Pol IV, Pol V-triple knockout)] were generously provided by Prof. Graham C. Walker (36).
Chemical syntheses
Eight O4-alkyldT derivatives were synthesized (Scheme 1). The synthetic route for the phosphoramidite building blocks of the O4-alkyldT lesions was adapted from previously published procedures (Scheme 2) (37).
Scheme 2.
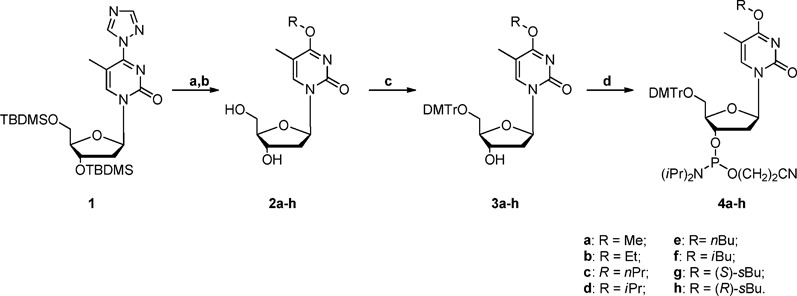
Syntheses of phosphoramidite building blocks of O4-alkylthymidineα. αReagents and conditions: (a) ROH, DBU, r.t., 10 h; (b) TBAF, THF, r.t., 1 h; (c) DMTr-Cl, DMAP, pyridine, r.t., 10 h; (d) 2-cyanoethyl-N,N-diisopropyl chlorophosphoramidite, DIEA, CH2Cl2, 1 h.
General procedures for the syntheses of O4-alkylthymidine
Compound 1 (300 mg, 0.58 mmol), prepared at an overall yield of 81% according to the published procedures (37), was dissolved in 6 ml of anhydrous acetonitrile at 0°C, to which solution were added 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU, 0.17 ml, 1.16 mmol) and the corresponding alcohol (1 ml). After 30 min, the resulting mixture was allowed to warm to r.t. and the solution was stirred overnight. The solution was neutralized by addition of 0.5 M aqueous solution of KH2PO4 (pH 6.5) and the product was extracted into chloroform (40 ml). The organic layer was then washed with brine (3 × 20 ml) and dried over anhydrous Na2SO4. The filtrate was concentrated and the residue was dissolved in 4 ml of anhydrous tetrahydrofuran (THF), to which 0.26 ml of tetrabutylammonium fluoride (TBAF) in THF (1.0 M) was added. The resulting mixture was stirred for another 2 h and concentrated. The residue was purified by silica gel column chromatography with a step gradient of methanol (0–7%) in methylene chloride to afford the desired product 2a-h. In this context, the stereospecific syntheses of O4-(S)-sBudT and O4-(R)-sBudT were accomplished by employing the commercially available (S)-sBuOH and (R)-sBuOH, respectively.
General procedures for DMTr protection
To 100 mg of compound 2a-h, which was dried three times by evaporation with pyridine and dissolved in anhydrous pyridine (10 ml) on an ice bath, were added 4-dimethylaminopyridine (DMAP, 0.5% mol) and 4,4′-dimethoxytrityl chloride (DMTr-Cl, 1.2 eq.). The resulting solution was stirred at room temperature for 10 h. The reaction was then quenched with methanol (0.5 ml), and the solvent was removed in vacuo. The residue was purified by silica gel column chromatography with ethyl acetate as the mobile phase to yield the desired product 3a-h.
General procedures for phosphoramidite synthesis
To a round bottom flask, which was stirred in an ice bath and contained a solution of compounds 3a-h (60 mg) in anhydrous methylene chloride (3.0 ml), were added N,N-diisopropylethylamine (DIEA, 2.2 eq.) and 2-cyanoethyl-N,N-diisopropyl chlorophosphoramidite (1.2 eq.). The mixture was then stirred at room temperature for 1 h under an argon atmosphere. The reaction was quenched by cooling the mixture in an ice bath followed by slow addition of methanol (0.20 ml). The solution was quickly diluted with ethyl acetate (8.0 ml). The organic layer was washed sequentially with saturated NaHCO3 (4.0 ml) and brine (4.0 ml), and dried over anhydrous Na2SO4. The solvent was evaporated under reduced pressure to yield 4a-h in a foam that was used directly for ODN synthesis.
The reaction yields and spectroscopic characterizations of the above-synthesized products are provided in the online Supplementary Materials. The nuclear magnetic resonance spectra for these compounds are shown in Supplementary Figures S1–S18.
ODN synthesis
The 12-mer lesion-containing ODNs 5′-ATGGCGXGCTAT-3′ (‘X’ represents the O4-alkyldT lesions) were synthesized on a Beckman Oligo 1000S DNA synthesizer (Fullerton, CA) at 1 μmol scale. The synthesized phosphoramidite building block was dissolved in anhydrous acetonitrile at a concentration of 0.067 M. Commercially available phosphoramidite building blocks (ultramild) were employed for the incorporation of the unmodified nucleotides (Glen Research Co., Sterling, VA, USA) following the standard ODN assembly protocol. The ODNs were cleaved from the controlled pore glass (CPG) support and deprotected with concentrated ammonium hydroxide at r.t. for 1 h. The solvents were evaporated, and the residues were redissolved in water and purified by high-performance liquid chromatography (HPLC).
HPLC
HPLC separations were conducted on an Agilent 1100 HPLC system with a Kinetex XB-C18 column (4.60 × 150 mm, 5 μm in particle size and 100 Å in pore size; Phenomenex Inc., Torrance, CA, USA). For the purification of ODNs, a triethylammonium acetate buffer (50 mM, pH 6.8, Solution A) and a mixture of solution A and acetonitrile (70/30, v/v, Solution B) were employed as mobile phases. The flow rate was 0.8 ml/min and the gradient profile was 5–25% B in 5 min followed by 25–55% B in 60 min. The HPLC traces for the purifications of the 12-mer lesion-containing ODNs are shown in Supplementary Figure S19 and the electrospray ionization-mass spectrometry (ESI-MS) and tandem MS (MS/MS) of the purified lesion-containing ODNs are displayed in Supplementary Figures S20–S26.
Preparation of the lesion-carrying 22-mer ODNs
The 12-mer O4-alkyldT-containing ODNs were 5′-phosphorylated and ligated individually with a 10-mer ODN (5′-AGTGGAAGAC-3′) in the presence of a template in the ligation buffer with T4 DNA ligase and ATP at 16°C for 8 h. The resulting 22-mer ODNs were purified by denaturing PAGE.
Construction of single-stranded lesion-containing and lesion-free competitor M13 genomes
The lesion-containing and lesion-free M13mp7(L2) genomes were prepared following the previously reported procedures (Supplementary Figure S27) (38). First, 20 pmol of single-stranded M13 genome was digested with 40 U EcoRI at 23°C for 8 h to linearize the vector. The resulting linearized vector was then mixed with two scaffolds, 5′-CTTCCACTCACTGAATCATGGTCATAGCTTTC-3′ and 5′-AAAACGACGGCCAGTGAATTATAGC-3′ (25 pmol), each spanning one end of the linearized vector. To the mixture a 30 pmol of the 5′-phosphorylated 22-mer O4-alkyldT-bearing ODN or the competitor ODN (25-mer, 5′-GCAGGATGTCATGGCGATAAGCTAT-3′) was subsequently added, and the DNA was annealed. The resulting mixture was treated with T4 DNA ligase at 16°C for 8 h, followed by incubation with T4 DNA polymerase (22.5 U) at 37°C for 4 h to degrade the excess scaffolds and the unligated vector. The lesion-containing and the lesion-free M13 genomes were purified from the solution by using Cycle Pure Kit (Omega). The constructed lesion-containing genomes were normalized against the lesion-free competitor genome following published procedures (38).
Transfection of control, lesion-containing and competitor M13 genomes into E. coli cells
The control lesion-free M13 genome was mixed with competitor genome at a molar ratio of 1:1, the M13 genomes containing the O4-alkyldT lesions with the alkyl group being Me, Et, nPr and nBu were mixed individually with the competitor genome at a molar ratio of 2:1, and those with the alkyl group being iPr, iBu, (S)-sBu and (R)-sBu were mixed separately with the competitor genome at a molar ratio of 5:1 (25 fmol each of competitor genome was used). The mixtures were transfected into SOS-induced, electrocompetent wild-type AB1157 E. coli cells and the isogenic E. coli cells that are deficient in Pol II, Pol IV, Pol V or all three polymerases, as well as wild-type E. coli cells and isogenic strains that are deficient in Ogt, Ada or both, following the previously published procedures (38). The SOS induction was achieved by irradiating the E. coli cells with 254 nm light at a dose of 45 J/m2 (36). The E. coli cells were subsequently grown in lysogeny broth (LB) medium at 37°C for 6 h. The phage was recovered from the supernatant by centrifugation at 13 000 r.p.m. for 5 min and further amplified in SCS110 E. coli cells to increase the progeny/lesion-genome ratio. The amplified phage was finally purified using the QIAprep Spin M13 kit (Qiagen) to obtain the single-stranded M13 DNA template for polymerase chain reaction (PCR) amplification.
Quantification of bypass efficiencies and mutation frequencies
We employed a modified version of the competitive replication and adduct bypass (CRAB) assay to assess the bypass efficiencies and mutation frequencies of the O4-alkyldT lesions upon replication in E. coli cells (Supplementary Figure S28) (33,34,38). The PCR amplification was carried out with the use of Phusion high-fidelity DNA polymerase for the sequence region of interest in the single-stranded M13 DNA template. The primers were 5′-YCAGCTATGACCATGATTCAGTGAGTGGA-3′ and 5′-YTCGGTGCGGGCCTCTTCGCTATTAC-3′ (‘Y’ represents a 5′-amino modifier, i.e., H2N(CH2)6-, added to the 5′ phosphate group of the ODNs). The amplification cycles were 30, with each cycle consisting of 10 s at 98°C, 30 s at 65°C and 15 s at 72°C, along with a final extension at 72°C for 5 min. The PCR products were purified by using Cycle Pure Kit (Omega). For the determination of bypass efficiency, a portion of the above PCR products was digested with 10 U BbsI restriction endonuclease and 1 U shrimp alkaline phosphatase in 10 μl New England Biolabs (NEB) CutSmart buffer at 37°C for 30 min, followed by heating at 80°C for 20 min to deactivate the phosphatase. To the above mixture were subsequently added 5 mM DTT, 1 μM ATP (premixed with 1.66 pmol [γ-32P]ATP) and 10 U T4 polynucleotide kinase in a 15 μl solution and the mixture was incubated at 37°C for 30 min, followed by heating at 65°C for 20 min to deactivate the T4 polynucleotide kinase. To the resulting solution was added 10 U MluCI restriction endonuclease, and the mixture was incubated at 37°C for 30 min, followed by quenching with 15 μl formamide gel loading buffer containing xylene cyanol FF and bromophenol blue dyes. The mixture was separated by 30% native polyacrylamide gel (acrylamide:bis-acrylamide = 19:1). The DNA bands were quantified using a Typhoon 9410 variable mode Imager.
The above restriction digestion of the PCR products gave rise to a short duplex, d(p*GGCGMGCTAT)/d(AATTATAGCN), where ‘M’ represents the nucleobase incorporated at the original lesion site during DNA replication in vivo, ‘N’ is the complementary base of ‘M’ in the opposite strand and ‘p*’ designates the [5′-32P]-labeled phosphate (Supplementary Figure S28). The bypass efficiency was determined by using the following formula:
Bypass efficiency (%) = (lesion signal/competitor signal)/(non-lesion control signal/competitor signal) × 100%.
Identification of mutagenic products by LC-MS/MS
The PCR products were digested with 50 U each of BbsI and MluCI, along with 20 U shrimp alkaline phosphatase in 250 μl NEB CutSmart buffer at 37°C for 2 h, followed by deactivation of enzymes at 80°C for 20 min. The resulting solution was extracted once with phenol/chloroform/isoamyl alcohol (25:24:1, v/v). The aqueous layer was subsequently dried in a Speed-vac, desalted with HPLC and dissolved in 20 μl water. A 10-μl aliquot was injected for LC-MS/MS analysis and an Agilent Zorbax SB-C18 column (0.5 × 250 mm, 5 μm in particle size) was used. The gradient for LC-MS/MS analysis was 5 min of 5–20% methanol followed by 35 min of 20–50% methanol in 400 mM HFIP (pH was adjusted to 7.0 with triethylamine). The temperature for the ion-transport tube was maintained at 300°C. The LTQ linear ion trap mass spectrometer (Thermo Electron, San Jose, CA, USA) was set up for monitoring the fragmentation of the [M-3H]3− ions of the 10-mer d(GGCGMGCTAT) and d(AATTATAGCN), with ‘M’ and ‘N’ being ‘A’, ‘T’, ‘C’ or ‘G’. The fragment ions detected in the MS/MS were manually assigned.
RESULTS
The major objectives of this study were to investigate how the O4-alkyldT lesions with varying structures of the alkyl group compromise DNA replication and to define the respective roles of SOS-induced DNA polymerases and O6-alkylguanine-DNA alkyltransferases in bypassing and repairing these lesions in E. coli cells. To this end, we first synthesized lesion-carrying ODNs with a site-specifically incorporated O4-alkyldT (Scheme 1) and characterized these ODNs by ESI-MS and MS/MS analyses (Supplementary Figures S20–S26). The LC-MS results revealed no detectable level of degradation of O4-alkyldT to dT in these modified ODNs.
To assess the bypass efficiencies and mutation frequencies of the O4-alkyldT lesions, we ligated the aforementioned lesion-containing ODNs into single-stranded M13 genome and performed a modified version of the CRAB and restriction endonuclease and post-labeling (REAP) assays to examine how these lesions inhibit DNA replication and induce mutations in E. coli cells (Supplementary Figure S28). We employed two restriction enzymes, i.e. MluCI and BbsI, to digest the PCR products of the progeny genome, affording 10mer ODN fragment(s) from the lesion-containing or lesion-free genome and a 13mer fragment from the competitor genome (Figure 1). The released ODNs were subjected to LC-MS/MS and native PAGE analyses to identify the replication products (Supplementary Figures S29–S39), as described elsewhere (20,39). As illustrated in Figure 1, by switching the order of the two restriction enzyme digestion, we can selectively incorporate a 32P-labeled phosphate to the 5′ ends of lesion-situated strand, i.e. d(p*GGCGMGCTAT) or its complementary strand, i.e. d(p*AATTATAGCN). When BbsI was first added, we could resolve, with 30% native PAGE, the [5′-32P]-labeled d(p*GGCGTGCTAT) (non-mutagenic product, 10mer-T) from the products carrying a T→A or T→G mutation, i.e. d(p*GGCGAGCTAT) (10mer-A) and d(p*GGCGGGCTAT) (10mer-G). However, the corresponding product harboring a T→C mutation at the lesion site, i.e. d(p*GGCGCGCTAT) (10mer-C), could not be separated from the non-mutagenic 10mer-T (Figure 1C and Supplementary Figures S33–S39). On the other hand, the respective radiolabeled complementary strand, i.e. d(p*AATTATAGCN), with ‘N’ being ‘A’ or ‘G’, obtained from sequential digestion with MluCI and BbsI, could be readily resolved from each other (Figure 1B and Supplementary Figures S33–S39). Thus, with the combination of the two enzyme digestion procedures, we were able to distinguish unequivocally the four potential types of replication products, and by monitoring the products from the strand of d(p*AATTATAGCN), we could quantify the frequencies of T→C mutation. It is worth noting that our assay is also capable of detecting putative −1 or −2 frame-shift mutation products; such putative frameshift mutations emanating from the replication of any of the O4-alkyldT-bearing genomes were, however, below the detection limit of our method. In this vein, it is of note that large deletions resulting in the loss of MluCI and/or BbsI recognition site will not give rise to the restriction fragments used for this assay, and are thus treated as lack of replicative bypass.
Figure 1.
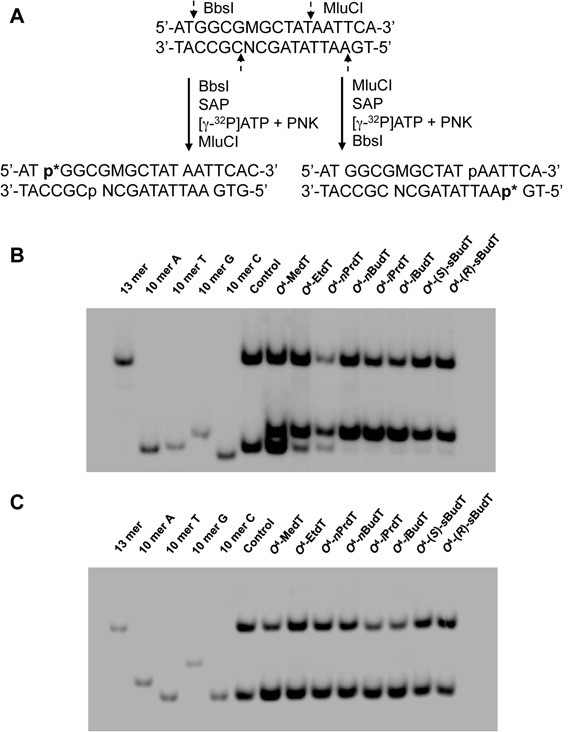
Native PAGE (30%) for monitoring the bypass efficiencies and mutation frequencies of O4-alkyldT in SOS-induced wild-type (WT) AB1157 Escherichia coli cells. (A) Sequential restriction enzyme digestion for the selective labeling of the strand initially bearing the lesion or the complementary strand. ‘SAP’ and ‘PNK’ designate shrimp alkaline phosphatase and T4 polynucleotide kinase, respectively. (B) Gel image showing the 13-mer and 10-mer products released from the bottom-strand (opposite to lesion-containing strand) of the PCR products of the progeny of the competitor genome and the control or lesion-carrying genome, where 10mer A, 10mer C, 10mer G and 10mer T represent the [5′-32P]-labeled standard ODNs 5′-AATTATAGCN-3′, with ‘N’ being A, C, G and T, respectively. (C) Gel image showing the 13-mer and 10-mer products released from the top-strand (lesion-containing strand) of the PCR products of the progeny of the competitor genome and the control or lesion-carrying genome, where 10mer A, 10mer C, 10mer G and 10mer T represent the [5′-32P]-labeled standard ODNs 5′-GGCGMGCTAT-3′, with ‘M’ being A, C, G and T, respectively.
The identities of the above restriction digestion products were also confirmed by LC-MS/MS analyses. In particular, we monitored the higher-resolution ‘ultra-zoom scan’ MS for the [M-3H]3− ions of d(GGCGMGCTAT) and d(AATTATAGCN) and their MS/MS (Supplementary Figures S29–S32), where ‘M’ and ‘N’ designate the nucleotides inserted at the initial damage site and the opposing nucleotide in the complementary strand, respectively. Unlike the promiscuous nucleotide misinsertions directed by the minor-groove O2-alkyldT lesions (39), our results revealed that the eight O4-alkyldT lesions exclusively induced T→C transition mutation, which is consistent with the previously published results (23). Along this line, our previous study showed that the sensitivity of our LC-MS- and MS/MS-based assay can allow for detection of a mutation frequency that is as low as 0.2% (33).
The bypass efficiencies of the O4-alkyldT lesions were then calculated from the ratio of the combined intensities of bands observed for the 10-mer products from the lesion-containing genome over the intensity of the 13-mer product from the competitor genome, with the consideration of the molar ratio of the lesion over competitor genomes employed in the initial transfection. The bypass efficiencies for the lesion-carrying genomes were then normalized against that for the control lesion-free genome as described in the ‘Materials and Methods’ section. It turned out that, except for the two diastereomers of O4-sBudT, whose replication bypass efficiencies are ∼5%, other O4-alkyldT lesions are not strong impediments to DNA replication in wild-type AB1157 cells (Figure 2A). In addition, the bypass efficiencies for the two diastereomers of O4-sBudT are very similar in wild-type AB1157 cells (Figure 2A).
Figure 2.
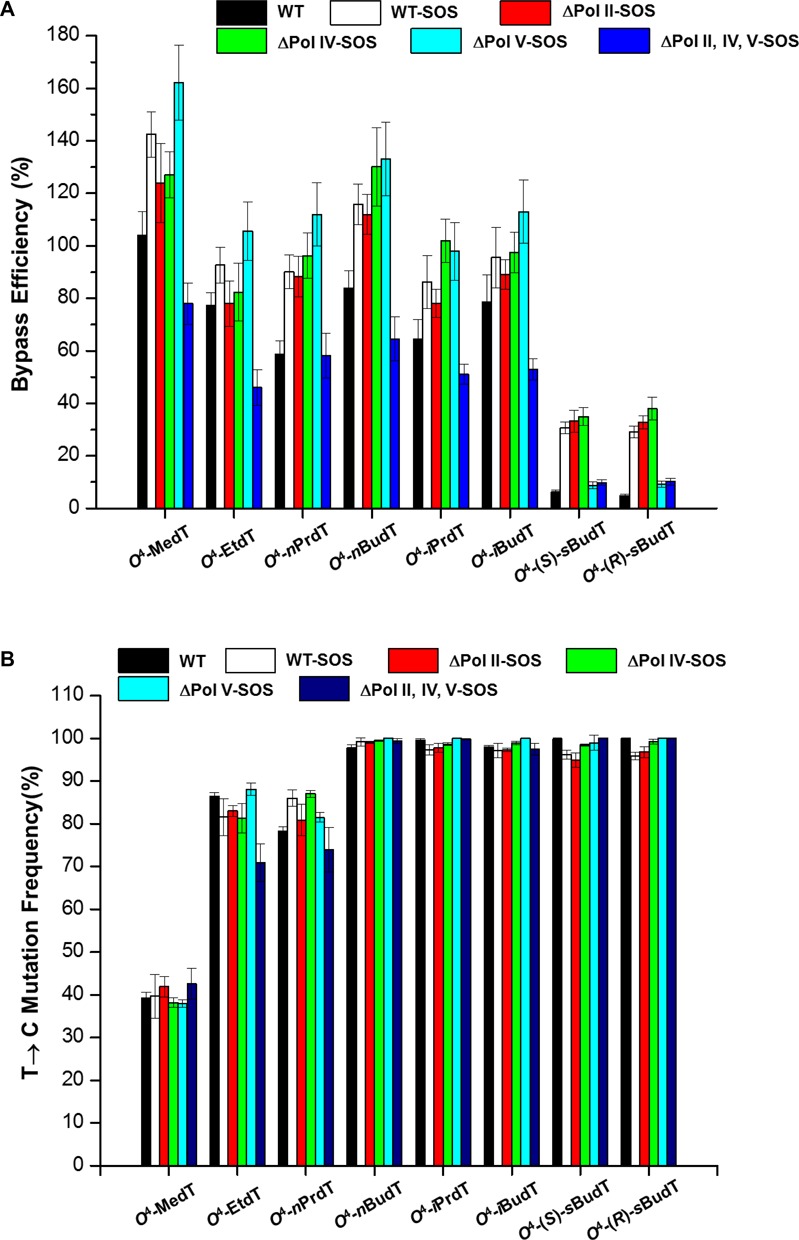
The bypass efficiencies (A) and mutation frequencies (B) of the O4-alkyldT lesions in AB1157 Escherichia coli strains that are proficient in translesion synthesis or deficient in Pol II, Pol IV, Pol V or all three SOS-induced DNA polymerases. The data represent the means and standard deviations of results from three independent replication experiments.
We also examined the roles of the SOS-induced DNA polymerases in bypassing the O4-alkyldT lesions by conducting the replication experiments in E. coli strains deficient in these DNA polymerases. Our results showed that, similar to what we found previously for O4-EtdT (20), no significant differences in bypass efficiency were observed for all O4-alkyldT lesions when any of the three SOS-induced polymerases were individually depleted. However, a marked reduction in bypass efficiency was observed for the triple knockout cells (Figure 2A), indicating that Pol II, Pol IV and Pol V play somewhat redundant roles in bypassing the O4-alkyldT lesions in E. coli cells.
The results from native PAGE analysis also allowed us to measure the mutation frequencies of O4-alkyldT lesions in wild-type and DNA polymerase-deficient AB1157 E. coli strains (Figure 2B). Our results revealed that all O4-alkyldT lesions are highly mutagenic, resulting exclusively in T→C mutation at frequencies of up to nearly 100%. Interestingly, the mere increase in the size of the alkyl group from methyl to ethyl led to a two-fold elevation in the T→C mutation frequency (i.e. from ∼40 to 80%). It is also worth noting that single or even triple depletion of these three polymerases did not confer any significant change in mutation frequencies, suggesting that the miscoding property of the O4-alkyldT lesions is independent of the nature of polymerases involved.
The above replication experiments showed that mutation frequencies for the O4-alkyldT lesions with small straight-chain alkyl groups (i.e., O4-MedT, O4-EtdT and O4-nPrdT) were not as high as the other lesions investigated. This might be attributed to the differential repair of the O4-alkyldT lesions. To test this, we exploited the involvement of O6-alkylguanine-DNA alkyltransferases in the repair of O4-MedT, O4-EtdT, O4-nPrdT and O4-nBudT lesions by conducting the replication experiments in isogenic E. coli cells that are proficient or deficient in Ogt and/or Ada (Figure 3). We found that the bypass efficiencies for the four O4-alkyldT lesions were not significantly altered upon the depletion of these DNA repair factors (Figure 3A). However, the removal of Ogt in the wild-type or Ada/AlkB-double knockout background gave rise to elevations in T→C mutation frequencies of O4-MedT, O4-EtdT and O4-nPrdT to nearly 100%, supporting the involvement of Ogt in the repair of these lesions (Figure 3B). Dual depletion of Ada and AlkB in wild-type or Ogt-deficient background, however, did not result in any appreciable increase in mutation frequencies for any of the O4-alkyldT lesions, suggesting the lack of involvement of Ada in their repair. The lower mutation frequency of O4-MedT than O4-EtdT and O4-nPrdT in wild-type cells also reveals that the Ogt-mediated repair of O4-MedT is more efficient than O4-EtdT and O4-nPrdT.
Figure 3.
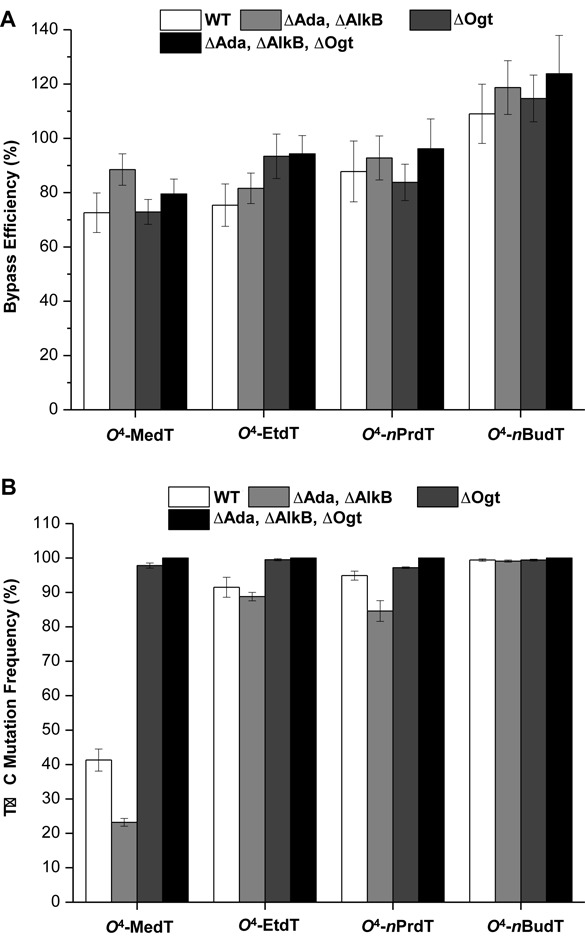
The bypass efficiencies (A) and and mutation frequencies (B) of the O4-alkyldT lesions in SOS-induced Escherichia coli cells that are proficient in O6-alkylguanine-DNA alkyltransferases or deficient in Ogt (ΔOgt), both Ada and Ogt (ΔAda, ΔOgt), or Ada, AlkB and Ogt (ΔAda, ΔAlkB, ΔOgt). The data represent the means and standard deviations of results from three independent replication experiments.
DISCUSSION
We systematically investigated the cytotoxic and mutagenic properties of the O4-alkyldT lesions in E. coli cells and our results led to several important findings. First, we observed that all O4-alkyldT lesions except for (S)- and (R)-O4-sBudT do not strongly inhibit DNA replication in E. coli cells. However, the two diastereomers of O4-sBudT are still bypassed more efficiently than simple O2-MedT (39), suggesting that the major-groove pyrimidine lesions are much better tolerated than the minor-groove counterparts by the E. coli DNA replication machinery.
Second, our results demonstrated that the nucleotide insertion opposite O4-alkyldT lesions is specific, where only T→C mutation was observed for all the eight O4-alkyldT lesions and the mutation frequencies of the O4-alkyldT lesions were not affected by depletion of Pol II, Pol IV, Pol V or all three polymerases. The mutation frequencies were also not perturbed by SOS induction in wild-type AB 1157 cells, indicating that the selective dGMP misinsertion may be attributed to the distinct chemical properties of the O4-alkyldT lesions. In this vein, the addition of a methyl group to O4 position of thymine is known to facilitate its favorable base pairing with guanine (9,40).
Third, our results revealed that the three SOS-induced DNA polymerases play somewhat redundant roles in bypassing the O4-alkyldT lesions with the exception of (S)- and (R)-O4-sBudT, as reflected by the observation that the bypass efficiencies of these lesions dropped substantially only upon depletion of all three polymerases. Efficient bypass of O4-sBudT lesions, however, necessitates Pol V. In this connection, Yuan et al. (41) observed that Pol V also plays an important role in the bypass of O4-carboxymethyl-dT in E. coli cells.
Fourth, we found that Ogt, but not Ada, is involved in repairing O4-alkyldT with small straight-chain alkyl functionality, with O4-MedT being much more efficiently repaired than O4-EtdT and O4-nPrdT. The nearly 100% T→C mutation frequency observed for O4-iPrdT, O4-nBudT, O4-iBudT or O4-sBudT suggest the lack of repair of these lesions in E. coli cells. This observation is in line with the previous observations that Ada is not involved in repairing O4-alkyldT and that Ogt is slightly better than Ada in tolerating large O6-alkyldG adducts (17,29,30). Likewise, all the eight O4-alkyldT lesions were completely mutagenic in Ogt-deficient E. coli cells, suggesting the absence of other DNA repair mechanisms in the removal of these DNA lesions when they are situated in single-stranded M13 genome. In this context, it is important to note that the replication of these lesions in the single-stranded M13 genome differs from the replication of these lesions in double-stranded chromosomal DNA in E. coli cells. In particular, some DNA repair machinery (e.g. nucleotide excision repair) necessitates double-stranded DNA substrates; thus, the modulation of mutation frequencies of these lesions by some DNA repair activities may not be revealed from the replication experiments with the use of single-stranded plasmid.
Different from the minor-groove O2-alkyldT lesions (39), the bypass efficiencies of the major-groove O4-alkyldT do not exhibit a clear trend with the sizes of alkyl groups attached to the O4 position of thymine, which could be attributed to the lack of perturbation of base pairing between O4-alkyldT and dG by the substituent groups on the O4 of thymine (9,40). The failure to observe a marked decrease in bypass efficiency with the increase of the size of the alkyl group also suggests that the active sites of polymerases are spacious enough to accommodate the majority of the alkyl groups situated on the O4 position of thymine. However, the two diastereomers of O4-sBudT significantly blocked DNA replication, suggesting that a branched ethyl group, but not a methyl group (as in O4-iPrdT), located at the α-carbon of the O4 of thymine may render it difficult for the modified thymine to fit into the active sites of DNA polymerases.
In summary, our systematic shuttle vector-based study of a group of structurally defined O4-alkyldT lesions provided important new insights into the impact of this group of DNA lesions on the efficiency and accuracy of DNA replication and the repair of these lesions in vivo. Our results also indicated that the carcinogenic potentials of these lesions may arise, at least in part, from their high bypass efficiencies and strong mutagenic effects. It will be important to assess how the O4-alkyldT lesions compromise DNA replication and how they are repaired in mammalian cells in the future.
Supplementary Material
Acknowledgments
The authors would like to thank Prof. Graham C. Walker for providing the E. coli strains and Prof. John M. Essigmann for providing the initial M13 vector.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01 ES025121, T32 ES018827]. Funding for open access charge: NIH [R01 ES025121].
Conflict of interest statement. None declared.
REFERENCES
- 1.Friedberg E.C., Walker G.C., Siede W., Wood R.D., Schultz R.A., Ellenberger T. DNA Repair and Mutagenesis. Washington, D.C: ASM Press; 2006. [Google Scholar]
- 2.Fu D., Calvo J.A., Samson L.D. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer. 2012;12:104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballschmiter K. Pattern and sources of naturally produced organohalogens in the marine environment: biogenic formation of organohalogens. Chemosphere. 2003;52:313–324. doi: 10.1016/S0045-6535(03)00211-X. [DOI] [PubMed] [Google Scholar]
- 4.Hecht S.S. DNA adduct formation from tobacco-specific N-nitrosamines. Mutat. Res. 1999;424:127–142. doi: 10.1016/s0027-5107(99)00014-7. [DOI] [PubMed] [Google Scholar]
- 5.Taverna P., Sedgwick B. Generation of an endogenous DNA-methylating agent by nitrosation in Escherichia coli. J. Bacteriol. 1996;178:5105–5111. doi: 10.1128/jb.178.17.5105-5111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shrivastav N., Li D., Essigmann J.M. Chemical biology of mutagenesis and DNA repair: cellular responses to DNA alkylation. Carcinogenesis. 2010;31:59–70. doi: 10.1093/carcin/bgp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drablos F., Feyzi E., Aas P.A., Vaagbo C.B., Kavli B., Bratlie M.S., Pena-Diaz J., Otterlei M., Slupphaug G., Krokan H.E. Alkylation damage in DNA and RNA - repair mechanisms and medical significance. DNA Repair (Amst). 2004;3:1389–1407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Kufe D.W., Pollock R.E., Weichselbaum R.R., Bast R.C., Gansler T.S., Holland J.F., Frei E. Cancer Medicine. 6th edn. Hamilton: BC Decker; 2003. [Google Scholar]
- 9.Swann P.F. Why do O6-alkylguanine and O4-alkylthymine miscode - the relationship between the structure of DNA containing O6-alkylguanine and O4-alkylthymine and the mutagenic properties of these bases. Mutat. Res. 1990;233:81–94. doi: 10.1016/0027-5107(90)90153-u. [DOI] [PubMed] [Google Scholar]
- 10.Singer B. O-alkyl pyrimidines in mutagenesis and carcinogenesis - occurrence and significance. Cancer Res. 1986;46:4879–4885. [PubMed] [Google Scholar]
- 11.Singer B., Dosanjh M.K. Site-directed mutagenesis for quatitation of base base interactions at defined sites. Mutat. Res. 1990;233:45–51. doi: 10.1016/0027-5107(90)90150-3. [DOI] [PubMed] [Google Scholar]
- 12.Singer B., Grunberger D. Molecular Biology of Mutagens & Carcinogens. NY: Plenum Press; 1983. [Google Scholar]
- 13.Bedell M.A., Lewis J.G., Billings K.C., Swenberg J.A. Cell specificity in hepatocarcinogenesis - preferential accumulation of O6-methylguanine in targest cell DNA during continuous exposure of rats to 1,2-dimethylhydrazine. Cancer Res. 1982;42:3079–3083. [PubMed] [Google Scholar]
- 14.Swenberg J.A., Dyroff M.C., Bedell M.A., Popp J.A., Huh N., Kirstein U., Rajewsky M.F. O4-ethyldeoxythymidine, but not O6-ethyldeoxyguanosine, accumulates in hepatocyte DNA of rats exposed continuously to diethylnitrosamine. Proc. Natl. Acad. Sci. U.S.A. 1984;81:1692–1695. doi: 10.1073/pnas.81.6.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang H., Konishi C., Kuroki T., Huh N. Detection of O6-methylguanine, O4-methylthymine and O4-ethylthymine in human liver and peripheral-blood leukocyte DNA. Carcinogenesis. 1995;16:1277–1280. doi: 10.1093/carcin/16.6.1277. [DOI] [PubMed] [Google Scholar]
- 16.Sassanfar M., Dosanjh M.K., Essigmann J.M., Samson L. Relative efficiencies of the bacterial, yeast, and human DNA methyltransferases for the repair of O6-methylguanine and O4-methylthymine - suggestive evidence for O4-methylthymine repair by eukaryotic methyltransferases. J. Biol. Chem. 1991;266:2767–2771. [PubMed] [Google Scholar]
- 17.Dosanjh M.K., Singer B., Essigmann J.M. Comparative mutagenesis of O6-methylguanine and O4-methylthymine in Escherichia coli. Biochemistry. 1991;30:7027–7033. doi: 10.1021/bi00242a031. [DOI] [PubMed] [Google Scholar]
- 18.Pauly G.T., Hughes S.H., Moschel R.C. Comparison of mutagenesis by O6-methyl- and O6-ethylguanine and O4-methylthymine in Escherichia coli using double-stranded and gapped plasmids. Carcinogenesis. 1998;19:457–461. doi: 10.1093/carcin/19.3.457. [DOI] [PubMed] [Google Scholar]
- 19.Preston B.D., Singer B., Loeb L.A. Mutagenic potential of O4-methylthymine in vivo determined by an enzymatic approach to site-specific mutahenesis. Proc. Natl. Acad. Sci. U.S.A. 1986;83:8501–8505. doi: 10.1073/pnas.83.22.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhai Q., Wang P., Wang Y. Cytotoxic and mutagenic properties of regioisomeric O2-, N3- and O4-ethylthymidines in bacterial cells. Carcinogenesis. 2014;35:2002–2006. doi: 10.1093/carcin/bgu085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altshuler K.B., Hodes C.S., Essigmann J.M. Intrachromosomal probes for mutagenesis by alkylated DNA bases replicated in mammalian cells: a comparison of the mutagenicities of O4-methylthymine and O6-methylguanine in cells with different DNA repair backgrounds. Chem. Res. Toxicol. 1996;9:980–987. doi: 10.1021/tx960062w. [DOI] [PubMed] [Google Scholar]
- 22.Klein J.C., Bleeker M.J., Lutgerink J.T., van Dijk W.J., Brugghe H.F., van den Elst H., van der Marel G.A., van Boom J.H., Westra J.G., Berns A.J., et al. Use of shuttle vectors to study the molecular processing of defined carcinogen-induced DNA damage: mutagenicity of single O4-ethylthymine adducts in HeLa cells. Nucleic Acids Res. 1990;18:4131–4137. doi: 10.1093/nar/18.14.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein J.C., Bleeker M.J., Roelen H., Rafferty J.A., Margison G.P., Brugghe H.F., Vandenelst H., Vandermarel G.A., Vanboom J.H., Kriek E., et al. Role of nucleotide excision repair in processing of O4-alkylthymines in human cells. J. Biol. Chem. 1994;269:25521–25528. [PubMed] [Google Scholar]
- 24.Pauly G.T., Moschel R.C. Mutagenesis by O6-methyl-, O6-ethyl-, and O6-benzylguanine and O4-methylthymine in human cells: effects of O6-alkylguanine-DNA alkyltransferase and mismatch repair. Chem. Res. Toxicol. 2001;14:894–900. doi: 10.1021/tx010032f. [DOI] [PubMed] [Google Scholar]
- 25.Dosanjh M.K., Menichini P., Eritja R., Singer B. Both O4-methylthymine and O4-ethylthymine preferentially form alkyl T-G pairs that do not block in-vitro replication in a defined sequence. Carcinogenesis. 1993;14:1915–1919. doi: 10.1093/carcin/14.9.1915. [DOI] [PubMed] [Google Scholar]
- 26.Singer B., Spengler S.J., Fraenkelconrat H., Kusmierek J.T. O4-methyl, O4-ethyl, or O4-isopropyl substituents on thymidine in poly(dA-dT) all lead to transitions upon replication. Proc. Natl. Acad. Sci. U.S.A. 1986;83:28–32. doi: 10.1073/pnas.83.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen N., Wang P., Wang Y. Replication across regioisomeric ethylated thymidine lesions by purified DNA polymerases. Chem. Res. Toxicol. 2013;26:1730–1738. doi: 10.1021/tx4002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brent T.P., Dolan M.E., Fraenkel-Conrat H., Hall J., Karran P., Laval L., Margison G.P., Montesano R., Pegg A.E., Potter P.M., et al. Repair of O-alkylpyrimidines in mammalian cells: a present consensus. Proc. Natl. Acad. Sci. U.S.A. 1988;85:1759–1762. doi: 10.1073/pnas.85.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkinson M.C., Potter P.M., Cawkwell L., Georgiadis P., Patel D., Swann P.F., Margison G.P. Purification of the E. coli ogt gene product to homogeneity and its rate of action on O6-methylguanine, O6-ethylguanine and O4-methylthymine in dodecadeoxyribonucleotides. Nucleic Acids Res. 1989;17:8475–8484. doi: 10.1093/nar/17.21.8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graves R.J., Li B.F., Swann P.F. Repair of O6-methylguanine, O6-ethylguanine, O6-isopropylguanine and O4-methylthymine in synthetic oligodeoxynucleotides by Escherichia coli ada gene O6-alkylguanine-DNA-alkyltransferase. Carcinogenesis. 1989;10:661–666. doi: 10.1093/carcin/10.4.661. [DOI] [PubMed] [Google Scholar]
- 31.Bronstein S.M., Skopek T.R., Swenberg J.A. Efficient repair of O6-ethylguanine, but not O4-ethylthymine or O2-ethylthymine, is dependent upon O6-alkylguanine-DNA alkyltransferase and nucleotide excision repair activities in human cells. Cancer Res. 1992;52:2008–2011. [PubMed] [Google Scholar]
- 32.Fang Q., Kanugula S., Tubbs J.L., Tainer J.A., Pegg A.E. Repair of O4-alkylthymine by O6-alkylguanine-DNA alkyltransferases. J. Biol. Chem. 2010;285:8185–8195. doi: 10.1074/jbc.M109.045518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong H., Cao H., Wang Y. Formation and genotoxicity of a guaninecytosine intrastrand cross-link lesion in vivo. Nucleic Acids Res. 2007;35:7118–7127. doi: 10.1093/nar/gkm851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan B., Cao H., Jiang Y., Hong H., Wang Y. Efficient and accurate bypass of N2-(1-carboxyethyl)-2’-deoxyguanosine by DinB DNA polymerase in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 2008;105:8679–8684. doi: 10.1073/pnas.0711546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rye P.T., Delaney J.C., Netirojjanakul C., Sun D.X., Liu J.Z., Essigmann J.M. Mismatch repair proteins collaborate with methyltransferases in the repair of O6-methylguanine. DNA Repair (Amst). 2008;7:170–176. doi: 10.1016/j.dnarep.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neeley W.L., Delaney S., Alekseyev Y.O., Jarosz D.F., Delaney J.C., Walker G.C., Essigmann J.M. DNA polymerase V allows bypass of toxic guanine oxidation products in vivo. J. Biol. Chem. 2007;282:12741–12748. doi: 10.1074/jbc.M700575200. [DOI] [PubMed] [Google Scholar]
- 37.Xu Y.Z., Swann P.F. A simple method for the solid-phase synthesis of oligodeoxynucleotides containing O4-alkylthymine. Nucleic Acids Res. 1990;18:4061–4065. doi: 10.1093/nar/18.14.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delaney J.C., Essigmann J.M. Assays for determining lesion bypass efficiency and mutagenicity of site-specific DNA lesions in vivo. Methods Enzymol. 2006;408:1–15. doi: 10.1016/S0076-6879(06)08001-3. [DOI] [PubMed] [Google Scholar]
- 39.Zhai Q., Wang P., Cai Q., Wang Y. Syntheses and characterizations of the in vivo replicative bypass and mutagenic properties of the minor-groove O2-alkylthymidine lesions. Nucleic Acids Res. 2014;42:10529–10537. doi: 10.1093/nar/gku748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brennan R.G., Pyzalska D., Blonski W.J.P., Hruska F.E., Sundaralingam M. Crystal structure of the promutagen O4-methylthymidine - importance of the anticonformation of the O4 methoxy group and possible mispairing of O4-methylthymidine with guanine. Biochemistry. 1986;25:1181–1185. doi: 10.1021/bi00353a036. [DOI] [PubMed] [Google Scholar]
- 41.Yuan B., Wang J., Cao H., Sun R., Wang Y. High-throughput analysis of the mutagenic and cytotoxic properties of DNA lesions by next-generation sequencing. Nucleic Acids Res. 2011;39:5945–5954. doi: 10.1093/nar/gkr159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


