Abstract
In Xenopus, the germline is specified by the inheritance of germ-plasm components synthesized at the beginning of oogenesis. Only the cells in the early embryo that receive germ plasm, the primordial germ cells (PGCs), are competent to give rise to the gametes. Thus, germ-plasm components continue the totipotent potential exhibited by the oocyte into the developing embryo at a time when most cells are preprogrammed for somatic differentiation as dictated by localized maternal determinants. When zygotic transcription begins at the mid-blastula transition, the maternally set program for somatic differentiation is realized. At this time, genetic control is ceded to the zygotic genome, and developmental potential gradually becomes more restricted within the primary germ layers. PGCs are a notable exception to this paradigm and remain transcriptionally silent until the late gastrula. How the germ-cell lineage retains full potential while somatic cells become fate restricted is a tale of translational repression, selective degradation of somatic maternal determinants, and delayed activation of zygotic transcription.
1. INTRODUCTION
Primordial germ cells (PGCs), sole precursors of the gametes in the adult animal, are able to differentiate into all cell lineages, including themselves. Thus, PGCs are considered the “stem cells” of the species. PGC specification is an early event in embryonic development and relies on a unique set of germ-cell determinants. In mammals, germ-cell determinants are induced within a small population of cells prior to gastrulation by BMP signaling (reviewed in Saitou & Yamaji, 2012). In species including worm, fly, fish, and frog, these germ-cell determinants are synthesized during early oogenesis and aggregate into germ plasm (reviewed in King, 2014; Seydoux & Braun, 2006). After fertilization, the germ plasm distributes to just a few blastomeres and those cells acquire the PGC fate. Although initiation of germline development can occur through these two distinct mechanisms, many mammalian PGC-specific genes are components of the maternally synthesized germ plasm in other species. Interestingly, a number of these genes encode RNA-binding proteins that function in regulating zygotic genome activity, totipotency, proliferation, differentiation, and migration of PGCs. They do so through controlling translation and mRNA stability as discussed in this chapter.
In classic studies, Nieuwkoop and Faber (1956) described “cytoplasmic inclusions” or germ plasm at the vegetal pole of eggs and early embryos of Xenopus laevis that segregated exclusively into the germline. Both loss- and gain-of-function experiments supported the conclusion that germ plasm is necessary for PGC formation (Nieuwkoop & Sutasurya, 1976; Smith, 1966; Wakahara, 1977, 1978). Recent compelling experiments have shown that germ plasm is both required and sufficient to specify the germline in Xenopus. Germ plasm introduced into animal pole blastomeres caused them to enter the germ-cell lineage and give rise to functioning gametes, albeit, in small numbers (Tada, Mochii, Orii, & Watanabe, 2012). The only requirement for proper migration was that these animal pole blastomeres be placed back into the endodermal region.
Most of oogenesis is spent in G2 of the first meiotic prophase. At this time, high rates of transcription from all four copies of each chromosome achieve maximum transcript production. The products of these maternal transcripts are sufficient to meet the requirements of the embryo through early development in the absence of zygotic transcription. Fully-grown oocytes are arrested at this meiotic stage and can remain quiescent for a long period of time. In response to the hormone progesterone, oocytes are released from G2 prophase arrest, undergo maturation events including germinal vesicle breakdown (GVBD), and the completion of meiosis I. Matured oocytes now arrest at the second meiotic metaphase and, after passage through the oviducts, are capable of being fertilized. Sperm entry triggers completion of meiosis II. The female pronucleus then fuses with the male pronucleus, leading to the formation of the zygote and initiation of embryonic development (reviewed in Ferrell, 1999; Heasman, 2006).
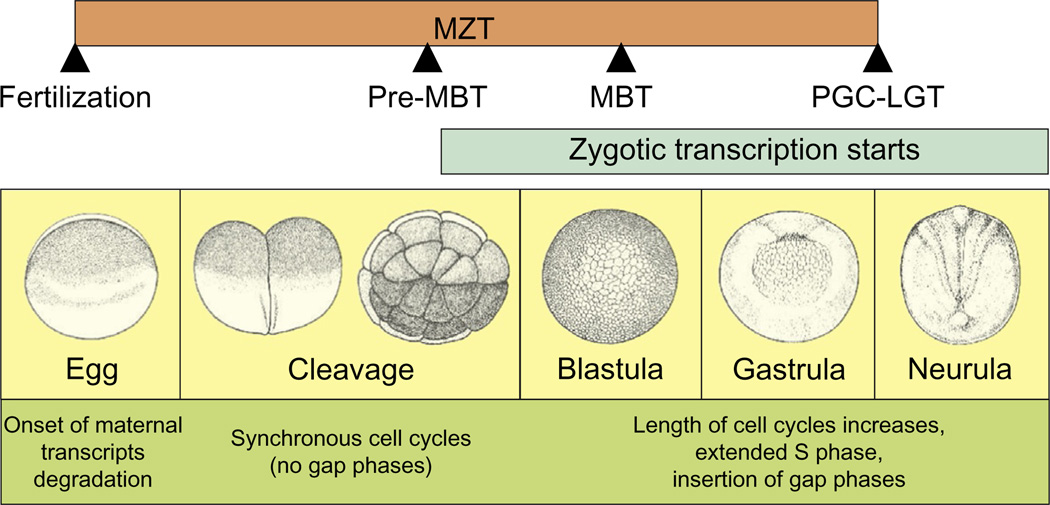
Although the diploid genome is reconstituted at fertilization, in Xenopus laevis, the major activation of the zygotic genome occurs ~6 h later, midway through the blastula stage. This critical time is known as the mid-blastula transition (MBT) and it is distinguished by a shift in the cell cycle as well as de novo RNA synthesis. For the 12 divisions prior to the MBT, the cell cycle is synchronous, and occurs fairly rapidly, every 25 min. To achieve such rates, the gap phases are skipped and the cell cycle goes from mitosis to DNA synthesis to mitosis. At the MBT, the embryo is comprised of some 4000 cells, cell divisions slow and the gap phases enter the cycle. All major developmental decisions up until this point have been made from maternal transcripts. These decisions include the specification of the three major axes (animal/vegetal, dorsal-anterior/ventral, right/left), the three primary germ layers (ectoderm, mesoderm, and endoderm), and the germ-cell lineage (reviewed in Heasman, 2006; King, 2014; White & Heasman, 2008). However, the transition from maternal to zygotic genetic control of development begins before the MBT, at fertilization, when degradation of maternal messages begins and ends during gastrulation when zygotic transcripts are required for further embryonic development. Thus the maternal-to-zygotic transition (MZT) encompasses a longer developmental time period than the MBT (Langley, Smith, Stemple, & Harvey, 2014; Tadros & Lipshitz, 2009; Fig. 1).
Figure 1.
The maternal-to-zygotic transition (MZT). MZT is a period that occurs very early during embryonic development, when degradation of maternal transcripts is initiated and is complete with the first morphological change caused by zygotic transcription, gastrulation. After the 12 mitotic divisions, the synchronous cell cycles are followed by asynchronous cleavages, with the introduction of gap phases during a developmental event called the “mid-blastula transition” (MBT). Most zygotic genes are silent until large-scale zygotic gene activation (ZGA) that, in Xenopus, coincides with the MBT. Several genes required for embryo patterning are transcribed before the MBT (pre-MBT) starting at the 32-cell stage. PGC transcription begins later, during the transition between gastrula and neurula (primordial germ cell late-gastrula transition, PGC-LGT). Images of Xenopus embryos are shown to highlight the different developmental stages.
The cleavage stages before the MBT accomplish the important task of generating enough cells to begin the process of regional differentiation according to germ layer identity. Not surprisingly, these early-stage blastomeres are maintained in a pluripotent state by the expression of the maternal factors, Oct 60 and Oct 25, orthologs of mammalian Oct 3/4 (Cao et al., 2004; Cao, Siegel, & Knochel, 2006; Cao, Siegel, Oswald, & Knochel, 2008; Hinkley, Martin, Leibham, & Perry, 1992; Whitfield, Heasman, & Wylie, 1993). Post-MBT, these factors gradually decline and are replaced by lineage-specific transcription factors such as Xsox17, Bix4, GATAs, and Xnrs, many of which are activated by the maternal transcription factor, VegT, at the MBT (reviewed in Heasman, 2006; White & Heasman, 2008). Thus, as development proceeds and genetic control is ceded to the zygotic genome, developmental potential gradually becomes more restricted within the primary germ layers. The germline is a notable exception to this paradigm and raises the fundamental question as to how this lineage retains the potential for totipotency while the somatic cells surrounding them become fate restricted. There is strong evidence for overlapping “antidifferentiation” mechanisms involving repression at both transcriptional and translational levels that operate to preserve the germline through the MZT (Leatherman & Jongens, 2003; Venkatarama et al., 2010). In this chapter, we will examine the MZT within the context of germline specification.
2. GERM-PLASM RNAs AND CYTOSKELETAL DYNAMICS: STAGE VI OOCYTE
In Stage VI Xenopus oocytes, germ plasm is organized into numerous small islands. At the ultrastructural level, each of these germ-plasm islands contains mitochondria, endoplasmic reticulum, membraneless electron-dense materials and matrix. The electron-dense material or germinal granules, can be fibrillar or round-shaped and is a hallmark of germ plasm (Heasman, Quarmby, & Wylie, 1984). RNAs within the germ plasm are found with distinct localization patterns (Fig. 2). Germinal granules contain nanos RNA, while Xpat and DeadSouth are peripherally associated with granules. Xdazl, Xlsirts, Xwnt11, DEADSouth, and fatvg are found within the matrix as ribonucleoprotein particles (RNPs) (Kloc et al., 2002). The significance of this organization of germ-plasm mRNAs in relation to when they may be translated after fertilization is not known. In full-grown oocytes, the germ-plasm islands are finely dispersed in the subcortex at the vegetal pole, located between the cortical granules and yolk. This geographic location and the structural organization of the germ plasm are likely maintained by a highly organized cytoskeletal network, which is established during early stages of oogenesis (Gard, Cha, & King, 1997). At least, two types of intermediate filament are closely associated with germ plasm. These include a large amount of vimentin, which is colocalized with germ plasm (Godsave, Anderton, Heasman, & Wylie, 1984; Torpey, Heasman, & Wylie, 1990; Wylie, Brown, Godsave, Quarmby, & Heasman, 1985; Wylie, Heasman, Parke, Anderton, & Tang, 1986), and a well-developed network of cytokeratin filaments that can be easily detected in the vegetal cortex and subcortex (Gard, 1999; Kloc, Bilinski, & Dougherty, 2007; Kloc et al., 2005; Torpey, Heasman, & Wylie, 1992). Ultrastructural analysis reveals that some germ-plasm islands are surrounded and penetrated by the long cytokeratin filaments (Kloc et al., 2007). Although actin filaments (Gard, 1999) and a small number of microtubules (Gard, 1991, 1994; Pfeiffer & Gard, 1999) are present in the vegetal subcortex region, it is unclear whether they play a direct role in maintaining germ-plasm islands there. Recently, it has been reported that depletion of Vangl2 and aPKC, which results in a reduction in the acetylated microtubule cytoskeleton, disrupts vegetal localization of VegT and Wnt11 mRNAs (Cha, Tadjuidje, Wylie, & Heasman, 2011). These results suggest that the microtubule cytoskeleton plays a role in maintaining at least some RNA components at the vegetal pole.
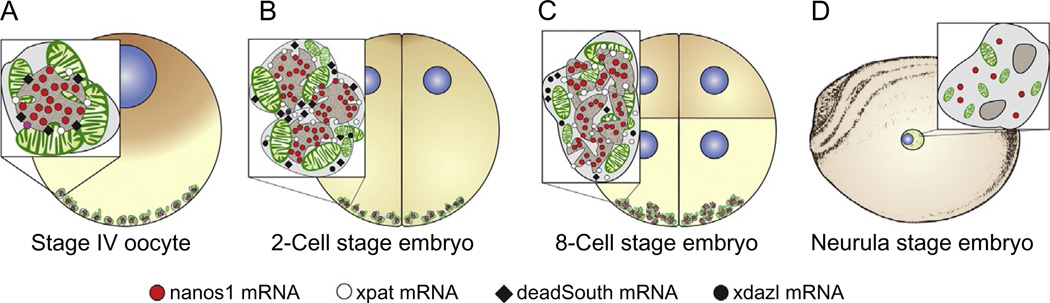
Figure 2.
Schemes of germ-plasm structure during Xenopus laevis embryogenesis. At oocyte Stage VI, germ-plasm islands are associated with the vegetal cortex (A). Between fertilization and the 8-cell stage, germinal granules (GGs) undergo ultrastructural changes thought to represent a permissive state for translation. After fertilization, the germ plasm contains a large number of small (250 nm) spherical GGs (B). During early cleavage, GGs coalesce into progressively more complex aggregates at the apex of vegetal pole blastomeres (C). Germ plasm at the 8-cell stage contains a few irregularly shaped large GG(about 2000 nm) that, in cross section, show complex and variable morphology (C). Between the gastrula and neurula stages, large GGs disappear and primordial germ cells from the neurula stage contain only a few small (300 nm) GGs (D).
The cytokeratin network at the vegetal pole has been studied extensively. Evidence supports the cytokeratin network as a scaffold, allowing some vegetally localized maternal RNAs to associate with it. These include the noncoding RNAs Xlsirts (Kloc & Etkin, 1994) and VegT mRNA, which encodes the master regulator for Xenopus germ layer specification (Zhang et al., 1998). Both Xlsirts and VegT RNPs in Stage VI oocytes are found associated with the cytokeratin network (Kloc et al., 2005). Strikingly, both Xlsirts and VegT mRNAs play important structural roles in maintaining the architecture of the cytokeratin network in the vegetal cortex. Depletion of these RNAs disrupts the network in a transcript-specific manner and impairs the proper formation of the germinal granules and subsequent development of the germline (Kloc et al., 2007, 2005). In the case of VegT, Heasman and colleagues have provided convincing evidence that it is the VegT mRNA, not the VegT protein, which is required for maintaining the structure of the cytokeratin network. In Xlsirts- or VegT-depleted oocytes, vegetal localization of a number of maternal RNAs is disrupted, likely due to the defective cytokeratin network (Heasman, Wessely, Langland, Craig, & Kessler, 2001; Kloc & Etkin, 1994; Kloc et al., 2005).
3. GERM PLASM AND CYTOSKELETAL DYNAMICS: OOCYTE MATURATION/FERTILIZATION
During oocyte maturation and subsequent fertilization, the cytoskeleton in the vegetal pole undergoes a series of dramatic changes. While vimentin remains colocalized with germ plasm throughout these transitions (Godsave et al., 1984), other cytoskeletal architectures are largely erased during the process of oocyte maturation. For example, the cytokeratin network is disrupted nearly completely after GVBD, with only some cytokeratin punctuate foci remaining within the vegetal cortex (Kloc et al., 2005). In addition, actin filaments and microtubules become barely detectable. It is likely that disassembly of the existing cytoarchitecture in the fully-grown quiescent oocyte prepares the oocyte for fertilization. The fertilized egg quickly builds a new cytoskeletal system, which facilitates the rapid coalescence of germ-plasm islands found widely distributed throughout the vegetal subcortex before egg activation (Ressom & Dixon, 1988).
In addition to cytoskeletal dynamics, a few other changes associated with oocyte maturation have been observed. For example, endogenous aPKC protein, which is homogenously distributed in the oocyte, is translocated to the animal hemisphere of the egg and can no longer be detected at the vegetal pole (Cha et al., 2011; Nakaya et al., 2000). Hermes protein, a germ-plasm RNA-binding protein highly expressed in oocytes, is markedly downregulated after GVBD, and cannot be detected during early stages of embryonic development (Song et al., 2007; Zearfoss, Chan, Wu, Kloc, & Etkin, 2004). Interestingly, Hermes protein colocalizes with Xvelo, the homologue of zebrafish Bucky ball, which regulates assembly of the Balbiani body (Bb) in oocytes (Marlow & Mullins, 2008) and organizes germ plasm after fertilization (Bontems et al., 2009; Heim et al., 2014). Although the significance of these molecular events remains unclear, it is highly likely that some of these molecular changes trigger remodeling of the germ plasm that is required for the initiation of germline development after fertilization.
3.1 Postfertilization
To initiate PGC development, the germ-plasm islands (>500) that are widely dispersed in the oocyte’s vegetal subcortex must coalesce into larger aggregates for segregation into a small number of blastomeres. This process begins at fertilization and with each division small aggregates are pushed into larger aggregates as the cleavage furrow advances. By the 32-cell stage, there are now on average 36 islands of germ plasm (Savage & Danilchik, 1993). During the first two Xenopus embryonic cell cycles, cleavage furrows are initiated in the animal hemisphere and move toward the vegetal pole. As a consequence of the first two embryonic cell divisions, germ-plasm aggregates become concentrated into the vegetal corner of each blastomere. In zebrafish embryos, germ-plasm RNPs first move toward the animal pole together with the cytoplasm during cytoplasmic streaming. Once streaming is completed, the blastoderm is created on top of the yolky mass. The large volume of yolk prevents the cleavage furrows from reaching the vegetal pole. For this reason, four large germ-plasm aggregates form at the periphery of the blastoderm as the zebrafish embryo divides. Interestingly, large germ-plasm aggregates can form in artificially activated eggs. Notably, it took 3–4 h for germ-plasm islands to coalesce in activated eggs, whereas the same process is largely completed by around 2 h postfertilization in normal embryos (Robb, Heasman, Raats, & Wylie, 1996). Considering that artificially activated eggs do not undergo cleavage, cleavage may not be absolutely required for coalescence of germ plasm.
A growing body of evidence suggests that formation of germ-plasm aggregates relies heavily on cytoskeletal dynamics after fertilization. Upon egg activation during fertilization, several cytoskeleton networks are established very rapidly in Xenopus. This includes a complex arrangement of cytokeratin network after egg activation (Kloc et al., 2005). Also, shortly after fertilization, a dense microtubule network is established in the entire fertilized egg. These microtubules, nucleated by the sperm centrosome and egg pronuclear-associated components, extend radially into the entire embryo. Treatments that block microtubule polymerization after egg activation (Ressom & Dixon, 1988; Savage & Danilchik, 1993), or that deplete the maternal kinesin-like protein, Xklp1, impair the aggregation process of the germ plasm. These studies have highlighted the importance of the microtubule network in germ-plasm aggregation.
It is worth mentioning that, shortly after fertilization, a unique microtubule network forms in the vegetal cortex. These microtubule arrays are organized into numerous parallel bundles and drive cortical rotation, a process that is essential for axis specification. This microtubule network exists only transiently and for a short period of time, and is disassembled prior to the beginning of the first embryonic division (Houliston & Elinson, 1991; Schroeder & Gard, 1992). In Xenopus, these microtubules are continuous with microtubules extending from the animal hemisphere. Polymerization of microtubules into the vegetal cortex is regulated by vegetally localized maternal factors such as Trim36 (Cuykendall & Houston, 2009; Olson, Oh, & Houston, 2015) and Dead-end (Dnd1) (Mei et al., 2013), both of which are actually components of the germ plasm. There is no evidence to suggest that these vegetal cortical microtubules are essential for germ-plasm aggregation. Recently, the zebrafish maternal-effect mutant, Hecate, has been characterized, which carries mutations in the glutamate receptor-interacting protein 2a (grip2a) gene. Hecate embryos show defects in the alignment and bundling of microtubules at the vegetal cortex. Such bundling abnormalities result in defective initiation of Wnt signaling during cleavage and subsequent axis specification. In contrast, the aggregation of germ plasm occurs normally in hecate mutants at the 4-cell stage. Even in severely ventralized hecate embryos, the average number of PGCs is unaffected at 24 h postfertilization (Ge et al., 2014). It is also worth pointing out that the defects in hecate embryos, which are maternal-effect mutant, does not necessarily rule out the possibility that zygotic Grip2 may be involved in germ-cell development. In Xenopus, injection of morpholino antisense oligos that block Grip2.1 translation, or overexpression of a dominant negative Grip2.1, reduces average PGC number and impairs the proper anterior-posterior positioning of PGCs in embryos (Tarbashevich, Koebernick, & Pieler, 2007). Nonetheless, the observation that germ-plasm coalesces as normal into four large masses in hecate embryos, which are defective in vegetal cortical microtubules, suggests that these aligned vegetal cortical microtubule arrays are unlikely to be required for germ-plasm aggregation.
Our current knowledge regarding how cleavage coordinates germ-plasm aggregation comes largely from studies in zebrafish. In zebrafish, germ-plasm RNPs exist as single particles before fertilization. After fertilization, these particles begin to form aggregates. This process is accelerated as the first embryonic cell cycle begins. As the astral microtubules extend from the sperm aster toward the periphery of the blastodisc, germ-plasm RNPs and associated actin move outwardly. As a result, germ-plasm RNPs are displaced to the outer edge of the blastodisc and the center of the blastodisc becomes free from germ-plasm RNPs. During cytokinesis, through the action of astral microtubules of the bipolar spindle, some germ-plasm RNPs are recruited to the cleavage furrow, forming two rod-like structures. These rod-like structures are pushed in opposite directions as the cleavage furrow advances outwardly from the center of the blastodisc, leading to distal compaction of germ-plasm RNP aggregates at both ends of the mature cleavage furrow. This is reminiscent of astral microtubule-dependent germ-plasm transportation during Drosophila germ-cell development (Lerit & Gavis, 2011). Subsequently, in zebrafish, similar processes occur repeatedly during the second and third cytokinesis. Ultimately, the majority of germ-plasm RNPs is packaged into four compact masses located at the periphery of the embryo (Eno & Pelegri, 2013; Nair et al., 2013; Theusch, Brown, & Pelegri, 2006). These large germ plasm aggregates are inherited by a few blastomeres, leading to the specification of the germline.
Several zebrafish maternal-effect mutants show defects in the aggregation of germ-plasm RNPs. These include cellular island, which carries mutation in Aurora B Kinase that is critical for cleavage furrow formation during cytokinesis (Yabe et al., 2009), and motley, a maternal-effect mutant deficient in the chromosomal passenger protein Birc5b/Survivin. Birc5b/Survivin regulates chromosomal passenger complex function, astral microtubule remodeling in the initiation of cytokinesis furrow ingression, and cortical microfilament dynamics (Nair et al., 2013). In addition, overexpression of Bucky ball, which encodes a RNA-binding protein important for Bb formation (Marlow & Mullins, 2008), induces abnormal germ-plasm aggregation, and generates ectopic PGCs in the zebrafish embryo (Bontems et al., 2009).
After coalescence of the germ plasm is completed, the location of zebrafish germ plasm remains unchanged in the embryo until morphogenesis begins. In Xenopus, until the 16-cell stage, germ plasm aggregates are located in the subcortex at the vegetal pole. As cleavage proceed, germ plasm changes its location from the vegetal subcortical regions to deep within the yolky endodermal mass (Ressom & Dixon, 1988; Savage & Danilchik, 1993). Interestingly, this movement can occur in artificially activated eggs and is independent of microtubules, microfilaments, or protein synthesis, although it takes longer in activated eggs as compared to fertilized embryos. This again suggests that cleavage may not be absolutely required for the germ-plasm dynamics in Xenopus embryos. In activated eggs, it seems that germ plasm is carried by cytoplasmic flow from the region of the vegetal pole toward the interior (Ressom & Dixon, 1988). By the blastula stage, four to five PGCs with concentrated germ plasm can be found in the endodermal mass. There, PGCs later divide a few times and eventually migrate out of the endoderm. At the tadpole stage, they migrate into the gonad where they proliferate and differentiate into gametes.
4. MATERNAL DETERMINANTS FOR TWO LINEAGES INHERITED BY THE SAME BLASTOMERES: A DILEMMA
While the overwhelming majority of mRNAs are distributed uniformly throughout the Xenopus oocyte, several hundred are found concentrated specifically at the vegetal pole. As alluded to earlier, many of these localized maternal RNAs are highly enriched in a cytokeratin-containing fraction, a characteristic that allowed their isolation (Mosquera, Forristall, Zhou, & King, 1993; Pondel & King, 1988). RNAs appear to localize using two different pathways during oogenesis while some use both pathways. In general, germ-plasm RNAs accumulate within the Bb along with mitochondria during the earliest oocyte stages. All RNAs identified thus far that localize into the Bb have functions in the germline (reviewed in King, Messitt, & Mowry, 2005). The Bb was shown to expand toward the vegetal pole, pushing germ plasm into the subcortex in Stage I to II oocytes (Wilk, Bilinski, Dougherty, & Kloc, 2005). RNAs encoding factors that function only in the soma such as Vg1 and VegT, localize within large particles transported along microtubules (reviewed in Pratt & Mowry, 2013). Localization of these RNAs occurs after germ-plasm RNAs have arrived at the vegetal cortex and for this reason, has been called the late pathway. However, not all RNAs that become restricted to PGCs during development are localized within the Bb germ plasm. A set of RNAs with dual functions in both the oocyte and PGCs (Dead-end) or in PGCs and the soma (Fatvg) is localized during the late pathway. In summary, by the end of oogenesis, maternal RNAs required to specify both germline and somatic lineages become localized to the oocyte vegetal cortex. The latter include VegT and Vg1 responsible for endoderm/mesoderm identity (Birsoy, Kofron, Schaible, Wylie, & Heasman, 2006; Zhang & King, 1996) and Xwnt11 and Trim36 for dorsal/ventral patterning (Cuykendall & Houston, 2009; Tao et al., 2005).
Such colocalization of determinants for different lineages presents a dilemma that must be resolved before the MBT. As the fertilized egg divides, germ plasm is asymmetrically segregated into only a few cells, while localized somatic RNAs are activated for translation and are passively transmitted into all vegetal cells, including PGCs (King et al., 2005; Lai, Singh, & King, 2012; Venkatarama et al., 2010). Thus, well before zygotic gene transcription begins 12 divisions later at the MBT (~4000 cells), determinants, including VegT, are available to initiate somatic gene-expression programs in PGCs (Fig. 3; Venkatarama et al., 2010). VegT RNA is actually translated earlier, after maturation, so its protein is available from the earliest stages of development. It is likely preloaded into nuclei and ready to initiate somatic gene programs at the MBT (Stennard, Zorn, Ryan, Garrett, & Gurdon, 1999). In spite of the presence of VegT, however, PGCs do not activate the endoderm gene-expression program, which includes the transcription factors, Xsox17α and Bix4 (Casey et al., 1999; Hudson, Clements, Friday, Stott, & Woodland, 1997). Instead, they remain competent to form all cell types. How are PGCs protected from somatic fates at this time? What prevents VegT protein from activating the endoderm genetic program in PGCs at the MBT?
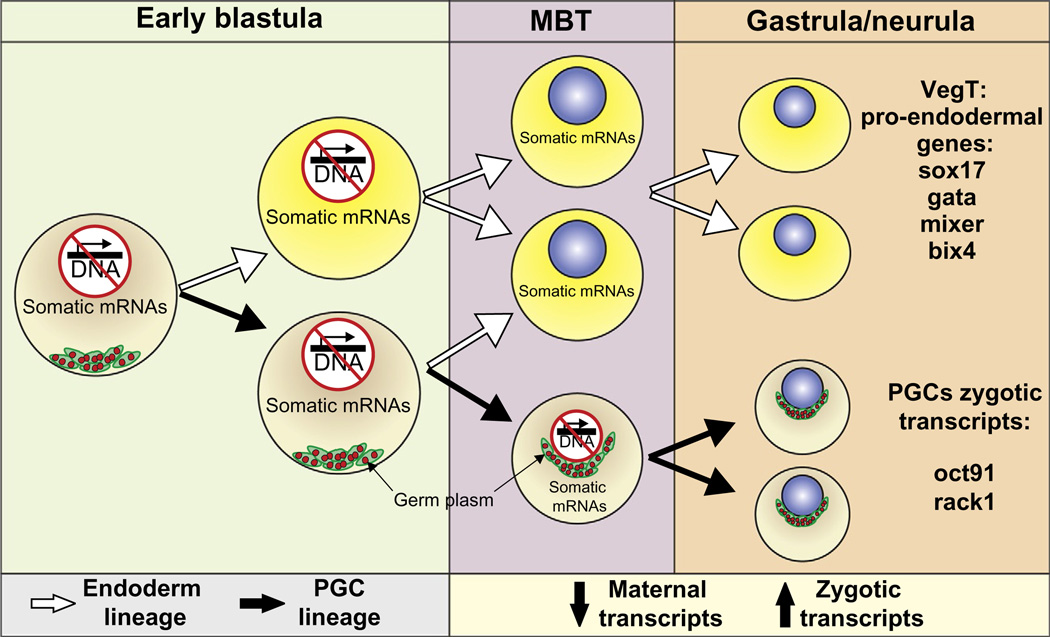
Figure 3.
Scheme of primordial germ cell (PGC) segregation from soma. In Xenopus, the germline is specified through the inheritance of germ plasm formed during oogenesis and asymmetrically segregated into the future germ-cell lineage (black arrows). Endodermal somatic fate is established by the inheritance of maternal somatic transcripts, including VegT (white arrows). Germ cells repress somatic maternal mRNAs. PGCs delay starting mRNA transcription to ensure that somatic differentiation programs remain inactive at the time that zygotic transcription is initiated in the rest of the embryo.
5. MECHANISMS FOR PROTECTING THE GERMLINE DURING THE MZT
Two critical activities appear to be required in Xenopus PGCs to protect them from VegT activity and other somatic differentiation signals: (1) translational repression of VegT by Pumilio and Nanos, and (2) transient genome-wide suppression of mRNA transcription at the MBT. The latter activity ensures that somatic differentiation programs remain inactive when zygotic transcription is initiated in the rest of the embryo (Fig. 3). Nanos-family members are distinguished throughout the animal kingdom by a conserved CCHC Zinc-finger motif that is required for RNA binding (Hashimoto et al., 2010). Alone, Nanos has little RNA-sequence-specific binding activity, but in all species studied it acts as a translational repressor (reviewed in Lai & King, 2013; Lai, Zhou, Luo, Fox, & King, 2011). Like Nanos, Pumilio (PUM) is also a member of a conserved family of proteins found in all eukaryotes. The conserved C-terminal region contains eight tandem repeats that defines its RNA-binding domain and confers specificity in RNA recognition for binding. PUM specifically binds an 8-nt canonical sequence within the 3′UTRs (UGUANAUA) of targeted RNAs for repression (Jaruzelska et al., 2003; Lai et al., 2012; Wang, Opperman, Wickens, & Hall, 2009; Wharton, Sonoda, Lee, Patterson, & Murata, 1998; Zamore, Williamson, & Lehmann, 1997; Zhang et al., 1997); hence, it is called the Pumilio Binding Element (PBE; formerly Nanos Response Element). The first four nucleotides (UGUA) are absolutely required and N could be A, U, or C. One change in the last three nucleotides (AUA) is also tolerated and those PBEs are referred to as noncanonical (ncPBE). The last three nucleotides most common in noncanonical PBEs are AUG and AAA (Table 1).
Table 1.
Candidate RNAs for Pumilio/Nanos Repression
| PGC Transcripts |
Canonical PBE: UGUANAUA |
Noncanonical PBE: UGUANAUAa |
Function |
|---|---|---|---|
| ddx25 (DeadSouth) |
0 | 2 | RNA helicase |
| Cdk9A | 3 | 3 | CTD-kinase, RNA synthesis |
| Cyclin B1b | 1 | 0 | Cell cycle regulator |
| Vg1 | 1 | 2 | Growth factor, endoderm patterning, mesoderm induction |
| otx1 | 1 | 4 | Transcription factor |
| pumilio1 | 1 | 7 | Translation |
| rbpms2 (hermes) |
0 | 2 | Translation |
| sox7 | 2 | 5 | Transcription factor |
| VegTb | 1 | 2 | Transcription factor, endoderm determinant |
| sybu | 0 | 5 | D/V axis |
| trim36 | 1 | 1 | D/V axis |
| wnt11bb | 1 | 5 | D/V axis |
| xdazl | 0 | 4 | Translation (activator) |
| xpat | 0 | 2 | Germ-plasm development |
| xvelo | 2 | 2 | Cell polarity |
Noncanonical PBE: One change occurs in the last three nucleotides (AUA), most frequently AAA or AUG.
Validated in vivo as repressed by Pumilio/Nanos or Pumilio.
Although Nanos activity mediates repression and PUM selects the RNA target, there are reports that other regions of PUM can mediate repression alone (Padmanabhan & Richter, 2006; Weidmann & Goldstrohm, 2012). Two Pumilio family proteins have been characterized in Xenopus oocytes that differ significantly in their N-terminal regions, while their RNA-binding domains are virtually identical and are expected to bind the same set of RNAs (Ota, Kotani, & Yamashita, 2011). PUM has many targets, associates with proteins other than Nanos, including Dazl (Fox, Urano, & Reijo Pera, 2005; Padmanabhan & Richter, 2006), and is part of complexes in both somatic and germ cells. Thus, the challenge is to identify PGC RNAs that bind PUM, but which require either Nanos for repression or Dazl for activation. Recently, a set of RNAs involved in meiosis and repressed by Nanos2 has been identified in mouse male gonads by immunoprecipitation with Nanos antibody (Suzuki, Igarashi, Aisaki, Kanno, & Saga, 2010; Suzuki & Saga, 2008). While Xenopus has only one nanos gene, the mouse has three. Interestingly, the Nanos2-interacting proteins identified in this study include the CCR4-NOT deadenylation complex. These findings suggest that Nanos represses expression of these RNAs and leads to their degradation.
Compared to control ornithine decarboxylases1 (ODC), both maternal VegT and survivin (Xsurv) RNA levels at Stage 8 are significantly lower in PGCs than in somatic cells and they are not detected by Stage 10 (Venkatarama et al., 2010). Maternally expressed Xsurv is a positive regulator of cell-cycle progression localized to the mitotic spindle (Murphy, Sabel, Sandler, & Dagle, 2002; Yan et al., 2005). Loss of Xsurv may reflect the difference in cell-cycle regulation in PGCs (Zust & Dixon, 1975). These differences in RNA levels cannot be accounted for by zygotic expression of VegT RNA in the embryo sample. Specific antisense depletion of maternal VegT shows that most of VegT at Stage 10 is maternal RNA (Zhang et al., 1998). Therefore, it seems that VegT and Xsurv are regulated differently (less stable) in PGCs than in somatic cells. PGCs depleted of Nanos activity misexpress the downstream targets of VegT, Xsox17, and Bix4 (Lai et al., 2012). The phenotype can be rescued by injection of Nanos RNA. These results suggest that Nanos normally represses translation of the maternal VegT mRNA within the germline. Indeed, the 3′UTR of VegT contains one canonical PBE that binds PUM and is required for VegT repression (Table 1; Lai et al., 2012). Further, sometime between MBT and gastrulation, VegT is dramatically reduced in PGCs relative to the whole embryo, but not in nanos-depleted embryos (Lai et al., 2012). The elimination of somatic determinants from PGCs is part of the MZT for the germline. Thus, RNA degradation does indeed play a role in Nanos/PUM translational repression in Xenopus early embryos. Nanos functions to repress RNAs that normally specify endoderm and promote apoptosis, both of which are critical for preserving the germline.
What other germline RNAs may be regulated in this manner? Cyclin B1, a central regulator of the cell-cycle, is a prime example also targeted for repression by PUM/Nanos in other species (Lai et al., 2011). Pumilio also binds and represses Xwnt11 but in this case does not require Nanos (Lai & King, 2013). Scores of other RNAs found within the germ plasm contain PBEs, but without direct testing for repressive activity, no conclusions can be drawn (see candidates in Table 1). Of interest is Vg1 RNA, a somatic determinant involved in endoderm and mesoderm patterning (Birsoy et al., 2006). Vg1 RNA contains 1 PBE and 2 ncPBEs and may be regulated as VegT is regulated. PUM mRNA itself has the highest number with 1 PBE and 6 ncPBEs, suggesting that a negative autoregulatory loop may operate in the germline. That is, PUM may be allowed to only reach a certain concentration within PGCs before its mRNA is repressed and degraded. Transcripts encoding Nanos, Dead-end, and all helicases except DeadSouth, do not have PBEs within their 3′UTRs.
6. PGCs ARE TRANSCRIPTIONALLY REPRESSED AT THE MBT
The second level of protection for PGCs during the MZT acts to prevent translated somatic determinants from causing inappropriate transcription. While somatic cells enter the MBT, PGCs are transcriptionally repressed and therefore unable to respond to maternally inherited signals like VegT. Similar to the Drosophila and Caenorhabditis elegans germline, the absence of new transcription at the MBT correlates with the unphosphorylated state of the carboxy-terminal domain in RNA polymerase II that must be phosphorylated for transcriptional elongation steps to occur. Zygotic transcription in PGCs does not occur until late gastrula stages, well after somatic endoderm cells have become fate restricted (Venkatarama et al., 2010; Wylie, Snape, Heasman, & Smith, 1987). What accounts for this delay? In nanos loss-of-function mutants in Drosophila, C. elegans, and Xenopus, the delay in transcription is lost and PGCs are transcriptionally active at the same time as somatic cells are at the MBT (Deshpande, Calhoun, Jinks, Polydorides, & Schedl, 2005; Lai et al., 2012; Schaner, Deshpande, Schedl, & Kelly, 2003). How does Nanos’ function as a translational repressor in PGCs play a role in the transient genome-wide suppression of mRNA transcription at the MBT?
7. HOW DOES THE GERMLINE ESCAPE THE MBT?
It seems unlikely that PGCs have a different timing mechanism from somatic cells for initiating zygotic transcription. Global regulation of transcription must follow very conserved rules. It seems more likely that the mechanism governing the delay in transcription for both the soma and PGCs will be related. Studies on the regulation of MBT onset suggest that the first step is that genomic DNA is released from a repressed state. This event is correlated with the nuclear-to-cytoplasmic (N/C) ratio and depletion of a putative repressor (reviewed in Langley et al., 2014). Cell cycle regulation, therefore, is one component, driving changes in the N/C ratio although nuclear size also plays a role (Jevtic & Levy, 2015). As mentioned, a target of Nanos/PUM repression, apparently conserved across species, is cyclin B1. Cyclin B1 regulates the cell cycle in a concentration-dependent fashion (Kotani, Yasuda, Ota, & Yamashita, 2013; Mendez & Richter, 2001). By repressing the cyclin B1 mRNA, the levels of Cyclin B1 protein would be expected to decline in PGCs. Consistent with this observation, whereas endodermal cells can divide every 40–75 min, PGCs undergo cell divisions less frequently, dividing only during three discrete time periods by the time they exit the endoderm at tadpole stages. A second requirement for correct initiation of zygotic transcription depends on de novo translation and accumulation of key maternal transcription factors, which are in limited supply. In Xenopus, VegT was identified as being one of these critical factors (Skirkanich, Luxardi, Yang, Kodjabachian, & Klein, 2011). Translational repression of VegT, cyclinB1, and other maternal factors in PGCs by Nanos/PUM may contribute to the observed delay in transcription; sufficient levels of core factors are not achieved. In PGCs, VegT is translationally repressed early on and degraded by gastrulation (Lai et al., 2012). In zebrafish, the pluripotency factors, Oct 91 and Sox, were also found to be key maternal factors important in this regard (Harvey et al., 2013; Lee et al., 2013). Interestingly, maternal Sox7 is found in the germ plasm, like VegT. It, too, is a potential target for Nanos/PUM repression as the Sox7 3′UTR contains no less than 7 PBEs (Table 1).
PGCs have an open chromatin structure early that becomes condensed later. Global silencing of mRNA transcription could be explained at the level of chromatin structure. Dimethylated lysine 4 of histone H3 (H3K4) or hyperacetylated Histone H4 (Penta) are markers of transcriptionally active chromatin; methylation of lysine 9 of histone H3 (H3K9) marks inactive chromatin. When PGCs and somatic cells were isolated from embryos at MZT stages (Fig. 1) and compared for these histone modifications, no significant differences were found (Venkatarama et al., 2010). All cells stained positively for H4Penta and H3meK4, indicating an active chromatin structure. Interestingly, H1c linker protein, which restricts chromatin remodeling, is not expressed in PGCs while it is in the endoderm. PGCs may retain the more permissive oocyte linker variant, B4 (Venkatarama et al., 2010). For Xenopus PGCs, global transcriptional repression does not involve a dramatic alteration of chromatin architecture. Indeed, the mechanism involved prevents transcription in spite of “permissive” chromatin.
8. RNA DEGRADATION DURING THE MZT
Two important events occur during the MZT: turnover of maternal gene products and activation of the zygotic genome. In some species, for example mammals, these two events are accomplished in a very narrow time window shortly after fertilization. In mice, interfering with either degradation of maternal gene products or zygotic genome activation (ZGA) leads to developmental arrest around the 2-cell stage, suggesting that these two events are tightly coupled. Interestingly, degradation of maternal gene products and activation of the zygotic genome are not tightly coupled in many other species. In Drosophila, zebrafish, and Xenopus, degradation of maternal mRNAs lasts a long time. While some RNAs are degraded relatively rapidly, a large proportion of maternal mRNAs remains in the embryo even by the end of gastrulation (Bashirullah et al., 1999; Ferg et al., 2007; Mathavan et al., 2005). In these species, high-level ZGA occurs abruptly at the MBT, which corresponds to the 14th, 12th, and 10th embryonic cell division in Drosophila, Xenopus, and zebrafish, respectively. In Drosophila, turnover of maternal mRNAs is essential for ZGA. Mutation of Smaug, an RNA-binding protein essential for the destabilization of a large subset of the maternal mRNAs, causes a significant reduction in zygotic transcription at 2–3-h postfertilization (Benoit et al., 2009). In zebrafish, turnover of maternal RNAs does not seem to be necessary for ZGA. It has been reported that maternal-zygotic Dicer-mutant zebrafish embryos, which do not process precursor miRNAs into mature miRNAs, display mild defects during gastrulation, brain formation, somitogenesis, and heart development (Giraldez et al., 2005). Considering that inhibition of ZGA leads to the death of the embryo during gastrulation in both zebrafish and Xenopus (Kane et al., 1996; Sible, Anderson, Lewellyn, & Maller, 1997; Stack & Newport, 1997), the finding that maternal-zygotic Dicer mutants can survive for a number of days argues that interfering with maternal RNA degradation has no, or only a minimal, effect on ZGA in zebrafish. Thus, whether degradation of maternal gene products is a prerequisite for ZGA may be species specific.
Many maternal mRNAs are synthesized very early during oogenesis. In fact, in most species, more than 50% of protein-coding genes are expressed in the oocyte. In fully-grown oocytes, these maternal RNAs are very stable. After fertilization, a significant fraction of the maternal RNA pool is actively degraded. Existing evidence in the literature suggests that a significant amount of maternal mRNA turnover events relies on zygotic transcription, especially zygotically transcribed miRNAs (Bushati, Stark, Brennecke, & Cohen, 2008; Giraldez et al., 2006; Lund, Liu, Hartley, Sheets, & Dahlberg, 2009). The pioneering work of Newport and Kirschner elegantly demonstrated that, prior to the MBT, the zygotic genome is globally silenced. Large-scale zygotic transcription occurs at the MBT. Intriguingly, even though the major ZGA occurs at the MBT, the transcriptional machinery is present in pre-MBT stage embryos (Newport & Kirschner, 1982; Prioleau, Huet, Sentenac, & Mechali, 1994). In at least two independent studies in Xenopus, phosphorylated Ser2 in the CTD of RNA polymerase II was detected during early cleavage stages and indicated that Pol II-dependent transcription elongation could occur (Blythe, Cha, Tadjuidje, Heasman, & Klein, 2010; Collart, Ramis, Down, & Smith, 2009). These results suggested that at least a low level of RNA polymerase II-dependent transcription may occur pre-MBT. Indeed, it has been observed that a small number of zygotic genes are transcribed during pre-MBT stages when the zygotic genome is globally silenced in both Drosophila (Ali-Murthy, Lott, Eisen, & Kornberg, 2013; Edgar & Schubiger, 1986; Harrison, Botchan, & Cline, 2010; Harrison, Li, Kaplan, Botchan, & Eisen, 2011; Karr, Weir, Ali, & Kornberg, 1989; Liang et al., 2008; ten Bosch, Benavides, & Cline, 2006) and Xenopus (Blythe et al., 2010; Nakakura, Miura, Yamana, Ito, & Shiokawa, 1987; Shiokawa et al., 1989; Skirkanich et al., 2011; Suzuki, Ueno, & Hemmati-Brivanlou, 1997; Tan et al., 2013; Yang, Tan, Darken, Wilson, & Klein, 2002). The pre-MBT zygotic transcription is unlikely to be “transcriptional noise.” Among 151 genes that are transcribed prior to the Xenopus MBT, many are associated with cellular events such as cell division, apoptosis, signal transduction, and phosphorylation. The pluripotency-associated gene, Oct 25, is also on the list of pre-MBT genes (Tan et al., 2013). Interestingly, some of the pre-MBT transcriptional events are precisely regulated by signaling pathways that are essential for early embryonic patterning. For example, at least a subset of pre-MBT transcription in Xenopus embryos relies on Wnt/β-catenin signaling (Blythe et al., 2010; Skirkanich et al., 2011; Yang et al., 2002). Inhibition of β-catenin-dependent pre-MBT transcription impairs axis specification, leading to ventralized embryos (Yang et al., 2002). Interestingly, we recently found that transcription of Xenopus miR-427, the homologue of miR-430, which plays key roles during the zebrafish MZT, can be detected as early as the 16-cell stage. Given that miR-427 is essential for deadenylation of maternal mRNAs during the Xenopus MZT (Lund et al., 2009), it seems likely that the low level of transcription occurring prior to the MBT may be important to initiate and coordinate the turnover of maternal RNAs. Later on the degradation machinery is accelerated by the major ZGA, leading to a rapid clearance of maternal products during gastrulation (Fig. 4).
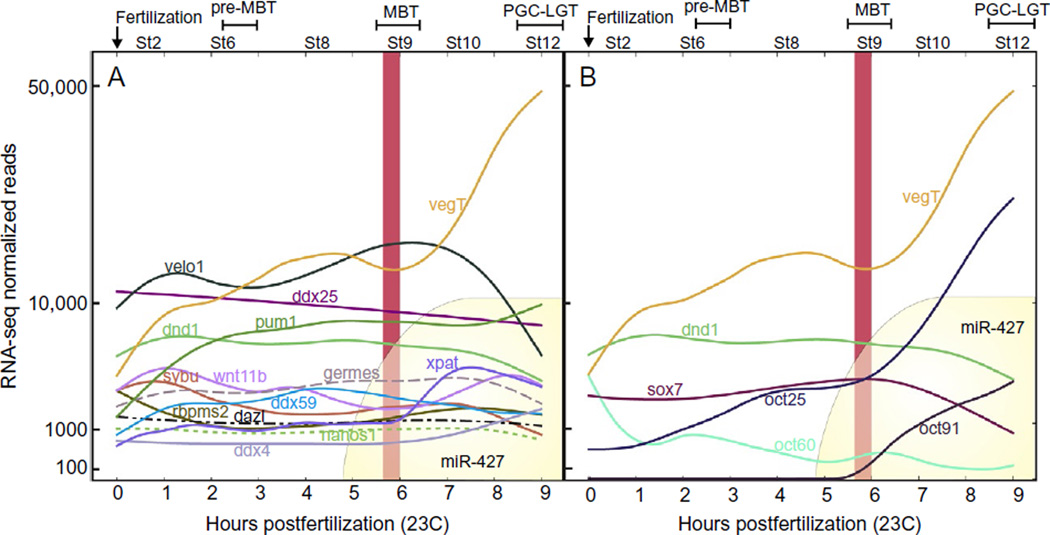
Figure 4.
Transcriptional profiling in Xenopus early development. Representation of gene-expression patterns of (A) PGC-related genes and (B) pluripotency factors between fertilization and the onset of gastrulation. Before the MBT, positive/negative changes in reads are because of polyadenylation and deadenylation rather than degradation while, after the MBT, miR-427-dependent degradation occurs (Collart et al., 2014). Polyadenylation of maternally deposited transcripts occurs immediately after fertilization and is required for zygotic transcription. Two waves of zygotic genome activation occur: At the 32/64-cell stage (pre-MBT), ~150 new transcripts are synthesized while the bulk of transcription occurs hours later, at the MBT (Tan et al., 2013). The maternal determinant VegT is degraded in PGCs while it is polyadenlyated and translated in the soma and zygotically transcribed at the MBT (included in both graphics as a reference). miR-427 is essential for deadenylation of maternal mRNAs during the MZT (yellow curves in A and B). (A) Germline RNAs appear to be polyadenylated soon after fertilization and remain stable through the MBT. (B) Pluripotency factors generally decline after the MBT except for Oct 91, which is newly transcribed in the PGCs. Modified from Collart et al. (2014).
9. STABILITY OF GERMLINE RNAs AND PROTEINS
Like all other maternal RNAs, germ-plasm RNAs are degraded during embryonic development. Unfortunately, in Xenopus, it is largely unclear when germ-plasm RNAs are degraded and how their turnover is regulated. So far, only a few studies have addressed these important questions. These studies suggest that germ-plasm RNAs are degraded through at least two different mechanisms. The first wave of germ-plasm RNA turnover occurs during gastrulation, and is most likely focused on the clearance of germ-plasm RNAs from the soma. Within PGCs, these maternal germ-plasm RNAs are protected and turnover is very slow. Little is known about the temporal control of germ-plasm RNAs in terms of translation and decay. However, based upon limited experimental evidence in the literature, we suggest a few general themes and highlight important questions that remain to be addressed.
The stability of germ-plasm RNAs must be regulated differently in the soma than in the germline as germ-plasm components can be detrimental to establishing somatic cell fates, but essential for PGC development. A good example is Nanos whose expression in the soma leads to abnormal development but its loss from germ plasm results in the failure of the germline while the soma is unaffected (Lai et al., 2012; Luo, Nerlick, An, & King, 2011). During early cleavage stages in zebrafish, nanos and other germ-plasm RNAs are also present in the soma and must be degraded there (Kedde et al., 2007). However, it is not clear whether that situation holds for Xenopus. The early history of germ-plasm RNAs is well established from whole mount in situ hybridization (WISH) analyses, but precise quantitation of these RNAs within germ plasm or within the ooplasm or soma has not been determined as this would require a separation of the two. For example, we know that in the oocyte, germ-plasm RNAs are either sequestered in the germinal granules or associated with the germ-plasm matrix. As described above, by the 8-cell stage, they coalesce into a few large aggregates that are inherited later by a few blastomeres, which consequently become PGCs. During these processes, germ-plasm RNAs are transported into the PGCs along with other components of the germ plasm. Once these processes are complete, highly concentrated germ-plasm RNAs, including nanos1 (Forristall, Pondel, Chen, & King, 1995), Xpat (Hudson & Woodland, 1998), Xdazl (Houston, Zhang, Maines, Wasserman, & King, 1998), DEADSouth (MacArthur, Houston, Bubunenko, Mosquera, & King, 2000), and dnd1 (Horvay, Claussen, Katzer, Landgrebe, & Pieler, 2006), can be detected in a few blastomeres by WISH. However, detection of highly concentrated germ-plasm RNAs in PGCs by WISH does not rule out the possibility that a certain amount of these RNAs fails to be transported into PGCs and is left in somatic cells. How efficient the process is at shuttling germline RNAs exclusively into PGCs is not known. Studies in C. elegans have revealed that, during segregation of the germline from soma, a certain amount of germ plasm is indeed left in the somatic cells and these RNAs are rapidly degraded (Zhang et al., 2009; Zhao, Tian, & Zhang, 2009). In Xenopus embryos, mechanisms capable of actively degrading germ-plasm RNAs in the soma have recently been identified (see below for details). Future work will have to establish in a more quantitative way the distribution and turnover of germ-plasm RNAs. Only then will it be possible to establish whether different mechanisms operate to control the stability of germ-plasm RNAs in the soma versus the germline.
Although only a few studies address how germ-plasm RNAs are degraded, the temporal and spatial expression patterns of many germ-plasm RNAs have been studied. RNA-seq analysis of RNAs isolated at different stages from Xenopus tropicalis, offers some insights (Collart et al., 2014). In this study, polyadenylated and nonpolyadenylated RNAs were quantitated. If we look at germ-plasm RNAs with VegT as a comparison, germline RNAs appear polyadenylated soon after fertilization and remain stable through the MBT (Fig. 4A). A general picture that emerges from these studies is that germ-plasm RNAs are degraded dramatically during gastrulation. For example, degradation of Xpat (Hudson & Woodland, 1998), Xdazl (Houston et al., 1998), DEADSouth (MacArthur et al., 2000), dnd1 (Horvay et al., 2006), and nanos1 (Lai et al., 2011), was detected easily by Northern Blot or RT-PCR analysis of whole gastrulae RNA. Interestingly, persistent expression of these germ-plasm RNAs can be detected in PGCs by WISH even as late as the tadpole stages (Kataoka et al., 2006). The significance of this degradation event during gastrulation is not fully understood. In our opinion, it is critically important to clarify in which cell lineages this RNA turnover event occurs. If degradation of germ-plasm RNAs during gastrulation occurs mainly in PGCs, it would suggest that this turnover event is a critical step during the MZT within the germline. If it happens in somatic cells, it would suggest that this turnover event is more likely responsible for removal of germ-plasm components from the somatic tissues, which ensures proper segregation of the germline from soma. In principle, these two possibilities are not mutually exclusive.
Some recent studies have demonstrated that miR-427 and miR-18 play important roles in controlling the turnover of DeadSouth and dnd1, respectively. Xenopus DEADSouth, a homologue of mammalian DDX25, is a DEAD-box RNA helicase that is specifically localized in the germ plasm. Its function during PGC development can be substituted by VASA/ DDX4 (Yamaguchi, Taguchi, Watanabe, & Orii, 2013). The 3′UTR of DEADSouth mRNA contains three miR-427 targeting sequences (Yamaguchi, Kataoka, Watanabe, & Orii, 2014). When the 3′UTR of DEADSouth is fused to a fluorescent protein, Venus, the reporter construct undergoes rapid turnover during gastrulation. By the tadpole stage, the reporter RNA is degraded completely in somatic tissues and Venus can be detected only in PGCs (Kataoka et al., 2006). Mutation in these miR-427 targeting sequences or knockdown of miR-427 interferes with clearance of the reporter RNA in somatic cells (Yamaguchi et al., 2014). Interestingly, the expression of miR-427 could not be detected in isolated PGCs (Yamaguchi et al., 2014). This raises the intriguing possibility that the miR-427-dependent turnover, which begins during gastrulation, does not occur in the PGCs. It is solely responsible for removal of DEADSouth mRNA from the somatic tissues. Therefore, it is unlikely that miR-427-dependent turnover plays a role during the MZT within the germline. In the case of dnd1, a regulatory element within the 3′UTR, which regulates vegetal localization of dnd1 in the oocyte, plays an essential role in somatic dnd1 mRNA clearance. This dnd1 mRNA turnover event is mediated by miR-18, which binds to the GCACUU(U) sequence in this regulatory element of the dnd1 3′UTR. Interfering with the binding of miR-18 to dnd1 significantly increases the level of endogenous dnd1 RNA in somatic tissues at the tadpole stage (Koebernick, Loeber, Arthur, Tarbashevich, & Pieler, 2010). As expected, when the dnd1 3′UTR is fused to GFP, it is competent to direct PGC-specific GFP expression. However, the PGC-specific GFP expression can be observed only when this miR-18-dependent somatic clearance event is complete (Dzementsei, Schneider, Janshoff, & Pieler, 2013; Koebernick et al., 2010). Taken together, these two lines of evidence support the idea that the dramatic decrease in the level of germ-plasm RNAs during gastrulation is a consequence of somatic RNA clearance.
The important question remains, how are maternal germ-plasm RNAs degraded within PGCs during germline development? Taking advantage of the X. laevis and borealis hybrid embryos, it was found that after the dramatic RNA degradation events during gastrulation, nanos1 RNA, presumably located in PGCs, remained stable at least until the end of the tailbud stage. Zygotic transcription of nanos1 does not occur in early-stage embryos (Lai et al., 2011). In the case of DEADSouth, maternal messages are removed nearly completely by the tailbud stage. Around the same time, embryos start to express zygotic DEADSouth (Yamaguchi et al., 2014). Other than these two RNAs, we do not know how other maternal germ-plasm RNAs are regulated during PGC development. Another issue is the half-life of the proteins encoded by germ-plasm RNAs. Nanos is present in germ plasm until PGCs leave the endoderm at Stage 39, well past when its RNA is detected. Xdazl protein has been detected even as late as tadpole Stage 52 and within germ cells of the developing gonads (Kataoka et al., 2006). Clearly, more analysis needs to be done to assess the gene-expression profiles within PGCs during germline development. Such information would lay the foundation to study how ZGA and removal of maternal germ-plasm RNAs are coordinated during germline development.
10. THE PGC LATE-GASTRULA TRANSITION
Germ plasm can be thought of as a mechanism to continue the potential for totipotency exhibited by the oocyte, forward into development. Germ plasm then, functions as an “escape hatch” for totipotency from the oocyte. Interestingly, some of the germ-plasm components that are passed on to PGCs as untranslated RNAs, are translated in the oocyte. These include the RNA-binding proteins Dead-end, DeadSouth, Xdazl, and RINGO/Spy. These proteins play a role in translational regulation or in regulating meiotic events. After maturation and fertilization, are these proteins degraded while being preserved as RNAs within the germ plasm? Is this one mechanism for carrying gametic identity forward through embryogenesis? What is the fate of oocyte proteins in the soma? We simply do not know the answers to these questions in Xenopus.
Segregation of the germline from the somatic endoderm occurs at gastrulation when the germ plasm moves to a perinuclear location; subsequent divisions result in both daughter cells receiving germ plasm. Before complete separation from the soma, PGCs are presumptive PGCs or pPGCs. Only after separation and during gastrulation do PGCs initiate their zygotic transcription program, or the PGC Late-Gastrula Transition (Fig. 3). Which maternal factor(s) activate the PGC program remains unknown. Importantly, Xenopus Oct 25, Oct 60, and Oct 91 are in the same POU subclass as the human pluripotency factor, Oct 3/4 (Hinkley et al., 1992), and Oct 3/4 acts as a functional homolog for Oct 25 or Oct 60 in rescue experiments (Cao et al., 2006). Maternal Oct 60 is expressed in isolated pPGCs prior to gastrulation and Oct 91 is expressed de novo in PGCs after they segregate from the endoderm (Hinkley et al., 1992; Venkatarama et al., 2010). The RNA-seq results from X. tropicalis for the Oct and Sox pluripotent factors show they generally decline after the MBT except for Oct 91, which is newly transcribed in PGCs (Fig. 4B; Collart et al., 2014; Tan et al., 2013).
Xenopus Oct 60 plays a key role in maintaining pluripotency by suppressing signaling in differentiation pathways including Activin/Nodal, BMP, and WNT (Cao et al., 2004, 2006, 2008; Whitfield et al., 1993). Oct 25 is not expressed in PGCs, while Oct 60 expression in pPGCs persists until gastrulation. Importantly, zygotic Oct 91 is subsequently expressed de novo in PGCs (Venkatarama et al., 2010). Oct 60 expression and function have primarily been investigated in somatic cells and its role in PGCs remains unknown. The maternally expressed transcription factor, Sox7, localizes to the vegetal pole and may have a role in PGC fate in addition to its known role in somatic cells (Zhang, Basta, Fawcett, & Klymkowsky, 2005; Zhang, Basta, & Klymkowsky, 2005). Interestingly, maternal expression of Oct 60 depends on an Octamer-Sox-binding motif in its promoter region and Oct 60 has been shown to bind this motif (Morichika, Sugimoto, Yasuda, & Kinoshita, 2014). Perhaps, Oct 60 and Sox proteins promote PGC transcription during the PCG-LGT. Consistent with this proposal, TRANSFAC promoter analysis identifies putative Oct and Sox binding motifs within the Oct 91 promoter region.
What gene programs are likely to be initiated at the PGC-LGT? Beyond keeping competency for totipotency, a zygotic differentiation program must be activated that confers the ability to migrate. Directed migration is one characteristic that PGCs acquire, which is missing from somatic endoderm. Because PGCs originate outside the gonads within embryonic endoderm, PGCs must migrate through somatic tissues and reach the developing gonads. Once inside the presumptive gonad, the PGCs divide, enter meiosis, and differentiate into definitive gametes.
Mutational analysis has identified three different signaling systems that affect PGC migration: CXCR4/SDF1, Notch/Delta2 (Morichika et al., 2010), and KIF13B-PIP3-GRIP2.1. PGCs begin their migration out of the endoderm and toward the presumptive gonads as early as tailbud (Stage 24), 10–12 h after PGC transcription begins (Terayama et al., 2013; Venkatarama et al., 2010). At this time, they are found clustered at the midline in the posterior endoderm. Early midline clustering is likely a passive step as CXCR4 is not expressed (RNA or protein) at this stage but it is during lateral and dorsal movements (Nishiumi, Komiya, & Ikenishi, 2005). Six hours later, PGCs are found spread more laterally and express CXCR4 on their surface (Takeuchi, Tanigawa, Minamide, Ikenishi, & Komiya, 2010) while its ligand, SDF-1 is expressed in the dorsal mesentery (Braun et al., 2002). Also around this time, a decreased attachment to ECM components like fibronectin, reduced adhesion to somatic endodermal cells, and downregulation of E-cadherin occurs in isolated PGCs but not in endodermal cells (Dzementsei et al., 2013).
The cellular movements that underpin these migration steps—blebbing and persistent blebbing at the leading edge—and which are required for directional migration, all occur in isolated PGCs cultured in a serum-free medium with a fibronectin substrate. As expected, these cellular behaviors require F-actin, myosin II activity, and RhoA/Rho-associated protein kinase (ROCK) signaling. ROCK was identified as an early zygotic transcript in PGCs (Venkatarama et al., 2010). Antisense depletion of either maternal Xdazl or Dead-end RNA prevents this step from occurring and PGCs are lost soon afterward (Horvay et al., 2006; Houston & King, 2000). Both of these RNA-binding proteins play a role in regulating translation, suggesting that maternal control of translation may extend to zygotically synthesized transcripts in the PGCs.
Finally, PGCs will give rise to the gametes and only they are competent to execute the reductional divisions during meiosis. How is this ability carried forward in PGCs through the MZT, but not in the soma? What maternal and/or zygotic factors are required to maintain competence for meiosis? These are questions that remain to be answered.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Amanda Butler and Ms. Dawn Owens (Dept. of Cell Biology, University of Miami, Miller School of Medicine, Miami, FL, USA) who helped to assemble the data shown in Table 1. The authors would like to acknowledge support from NIH to M.L.K. (grant R01GM102397) and to J.Y. (grant R01GM111816).
REFERENCES
- Ali-Murthy Z, Lott SE, Eisen MB, Kornberg TB. An essential role for zygotic expression in the pre-cellular Drosophila embryo. PLoS Genetics. 2013;9:e1003428. doi: 10.1371/journal.pgen.1003428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirullah A, Halsell SR, Cooperstock RL, Kloc M, Karaiskakis A, Fisher WW, et al. Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. The EMBO Journal. 1999;18:2610–2620. doi: 10.1093/emboj/18.9.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit B, He CH, Zhang F, Votruba SM, Tadros W, Westwood JT, et al. An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development. 2009;136:923–932. doi: 10.1242/dev.031815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy B, Kofron M, Schaible K, Wylie C, Heasman J. Vg 1 is an essential signaling molecule in Xenopus development. Development. 2006;133:15–20. doi: 10.1242/dev.02144. [DOI] [PubMed] [Google Scholar]
- Blythe SA, Cha SW, Tadjuidje E, Heasman J, Klein PS. beta-Catenin primes organizer gene expression by recruiting a histone H3 arginine 8 methyltransferase, Prmt2. Developmental Cell. 2010;19:220–231. doi: 10.1016/j.devcel.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontems F, Stein A, Marlow F, Lyautey J, Gupta T, Mullins MC, et al. Bucky ball organizes germ plasm assembly in zebrafish. Current Biology. 2009;19:414–422. doi: 10.1016/j.cub.2009.01.038. [DOI] [PubMed] [Google Scholar]
- Braun M, Wunderlin M, Spieth K, Knochel W, Gierschik P, Moepps B. Xenopus laevis stromal cell-derived factor 1: Conservation of structure and function during vertebrate development. Journal of Immunology. 2002;168:2340–2347. doi: 10.4049/jimmunol.168.5.2340. [DOI] [PubMed] [Google Scholar]
- Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Current Biology. 2008;18:501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- Cao Y, Knochel S, Donow C, Miethe J, Kaufmann E, Knochel W. The POU factor Oct-25 regulates the Xvent-2B gene and counteracts terminal differentiation in Xenopus embryos. The Journal of Biological Chemistry. 2004;279:43735–43743. doi: 10.1074/jbc.M407544200. [DOI] [PubMed] [Google Scholar]
- Cao Y, Siegel D, Knochel W. Xenopus POU factors of subclass V inhibit activin/nodal signaling during gastrulation. Mechanisms of Development. 2006;123:614–625. doi: 10.1016/j.mod.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Cao Y, Siegel D, Oswald F, Knochel W. Oct25 represses transcription of nodal/activin target genes by interaction with signal transducers during Xenopus gastrulation. The Journal of Biological Chemistry. 2008;283:34168–34177. doi: 10.1074/jbc.M803532200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey ES, Tada M, Fairclough L, Wylie CC, Heasman J, Smith JC. Bix4 is activated directly by VegT and mediates endoderm formation in Xenopus development. Development. 1999;126:4193–4200. doi: 10.1242/dev.126.19.4193. [DOI] [PubMed] [Google Scholar]
- Cha SW, Tadjuidje E, Wylie C, Heasman J. The roles of maternal Vangl2 and aPKC in Xenopus oocyte and embryo patterning. Development. 2011;138:3989–4000. doi: 10.1242/dev.068866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart C, Owens ND, Bhaw-Rosun L, Cooper B, De Domenico E, Patrushev I, et al. High-resolution analysis of gene activity during the Xenopus mid-blastula transition. Development. 2014;141:1927–1939. doi: 10.1242/dev.102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart C, Ramis JM, Down TA, Smith JC. Smicl is required for phosphorylation of RNA polymerase II and affects 3′-end processing of RNA at the midblastula transition in Xenopus. Development. 2009;136:3451–3461. doi: 10.1242/dev.027714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuykendall TN, Houston DW. Vegetally localized Xenopus trim36 regulates cortical rotation and dorsal axis formation. Development. 2009;136:3057–3065. doi: 10.1242/dev.036855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G, Calhoun G, Jinks TM, Polydorides AD, Schedl P. Nanos downregulates transcription and modulates CTD phosphorylation in the soma of early Drosophila embryos. Mechanisms of Development. 2005;122:645–657. doi: 10.1016/j.mod.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Dzementsei A, Schneider D, Janshoff A, Pieler T. Migratory and adhesive properties of Xenopus laevis primordial germ cells in vitro. Biology Open. 2013;2:1279–1287. doi: 10.1242/bio.20135140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Schubiger G. Parameters controlling transcriptional activation during early Drosophila development. Cell. 1986;44:871–877. doi: 10.1016/0092-8674(86)90009-7. [DOI] [PubMed] [Google Scholar]
- Eno C, Pelegri F. Gradual recruitment and selective clearing generate germ plasm aggregates in the zebrafish embryo. Bioarchitecture. 2013;3:125–132. doi: 10.4161/bioa.26538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferg M, Sanges R, Gehrig J, Kiss J, Bauer M, Lovas A, et al. The TATA-binding protein regulates maternal mRNA degradation and differential zygotic transcription in zebrafish. The EMBO Journal. 2007;26:3945–3956. doi: 10.1038/sj.emboj.7601821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell JE., Jr Xenopus oocyte maturation: new lessons from a good egg. Bioessays. 1999;21:833–842. doi: 10.1002/(SICI)1521-1878(199910)21:10<833::AID-BIES5>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Forristall C, Pondel M, Chen L, King ML. Patterns of localization and cyto-skeletal association of two vegetally localized RNAs, Vg1 and Xcat-2. Development. 1995;121:201–208. doi: 10.1242/dev.121.1.201. [DOI] [PubMed] [Google Scholar]
- Fox M, Urano J, Reijo Pera RA. Identification and characterization of RNA sequences to which human PUMILIO-2 (PUM2) and deleted in Azoospermia-like (DAZL) bind. Genomics. 2005;85:92–105. doi: 10.1016/j.ygeno.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Gard DL. Organization, nucleation, and acetylation of microtubules in Xenopus laevis oocytes: A study by confocal immunofluorescence microscopy. Developmental Biology. 1991;143:346–362. doi: 10.1016/0012-1606(91)90085-h. [DOI] [PubMed] [Google Scholar]
- Gard DL. Gamma-tubulin is asymmetrically distributed in the cortex of Xenopus oocytes. Developmental Biology. 1994;161:131–140. doi: 10.1006/dbio.1994.1015. [DOI] [PubMed] [Google Scholar]
- Gard DL. Confocal microscopy and 3-D reconstruction of the cytoskeleton of Xenopus oocytes. Microscopy Research and Technique. 1999;44:388–414. doi: 10.1002/(SICI)1097-0029(19990315)44:6<388::AID-JEMT2>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Gard DL, Cha BJ, King E. The organization and animal-vegetal asymmetry of cytokeratin filaments in stage VI Xenopus oocytes is dependent upon F-actin and microtubules. Developmental Biology. 1997;184:95–114. doi: 10.1006/dbio.1997.8508. [DOI] [PubMed] [Google Scholar]
- Ge X, Grotjahn D, Welch E, Lyman-Gingerich J, Holguin C, Dimitrova E, et al. Hecate/Grip2a acts to reorganize the cytoskeleton in the symmetry-breaking event of embryonic axis induction. PLoS Genetics. 2014;10:e1004422. doi: 10.1371/journal.pgen.1004422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Godsave SF, Anderton BH, Heasman J, Wylie CC. Oocytes and early embryos of Xenopus laevis contain intermediate filaments which react with anti-mammalian vimentin antibodies. Journal of Embryology and Experimental Morphology. 1984;83:169–187. [PubMed] [Google Scholar]
- Harrison MM, Botchan MR, Cline TW. Grainyhead and Zelda compete for binding to the promoters of the earliest-expressed Drosophila genes. Developmental Biology. 2010;345:248–255. doi: 10.1016/j.ydbio.2010.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Li XY, Kaplan T, Botchan MR, Eisen MB. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genetics. 2011;7:e1002266. doi: 10.1371/journal.pgen.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey SA, Sealy I, Kettleborough R, Fenyes F, White R, Stemple D, et al. Identification of the zebrafish maternal and paternal transcriptomes. Development. 2013;140:2703–2710. doi: 10.1242/dev.095091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Hara K, Hishiki A, Kawaguchi S, Shichijo N, Nakamura K, et al. Crystal structure of zinc-finger domain of Nanos and its functional implications. EMBO Reports. 2010;11:848–853. doi: 10.1038/embor.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J. Patterning the early Xenopus embryo. Development. 2006;133:1205–1217. doi: 10.1242/dev.02304. [DOI] [PubMed] [Google Scholar]
- Heasman J, Quarmby J, Wylie CC. The mitochondrial cloud of Xenopus oocytes: The source of germinal granule material. Developmental Biology. 1984;105:458–469. doi: 10.1016/0012-1606(84)90303-8. [DOI] [PubMed] [Google Scholar]
- Heasman J, Wessely O, Langland R, Craig EJ, Kessler DS. Vegetal localization of maternal mRNAs is disrupted by VegT depletion. Developmental Biology. 2001;240:377–386. doi: 10.1006/dbio.2001.0495. [DOI] [PubMed] [Google Scholar]
- Heim AE, Hartung O, Rothhamel S, Ferreira E, Jenny A, Marlow FL. Oocyte polarity requires a Bucky ball-dependent feedback amplification loop. Development. 2014;141:842–854. doi: 10.1242/dev.090449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkley CS, Martin JF, Leibham D, Perry M. Sequential expression of multiple POU proteins during amphibian early development. Molecular and Cellular Biology. 1992;12:638–649. doi: 10.1128/mcb.12.2.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvay K, Claussen M, Katzer M, Landgrebe J, Pieler T. Xenopus Dead end mRNA is a localized maternal determinant that serves a conserved function in germ cell development. Developmental Biology. 2006;291:1–11. doi: 10.1016/j.ydbio.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Houliston E, Elinson RP. Patterns of microtubule polymerization relating to cortical rotation in Xenopus laevis eggs. Development. 1991;112:107–117. doi: 10.1242/dev.112.1.107. [DOI] [PubMed] [Google Scholar]
- Houston DW, King ML. A critical role for Xdazl, a germ plasm-localized RNA, in the differentiation of primordial germ cells in Xenopus. Development. 2000;127:447–456. doi: 10.1242/dev.127.3.447. [DOI] [PubMed] [Google Scholar]
- Houston DW, Zhang J, Maines JZ, Wasserman SA, King ML. A Xenopus DAZ-like gene encodes an RNA component of germ plasm and is a functional homologue of Drosophila boule. Development. 1998;125:171–180. doi: 10.1242/dev.125.2.171. [DOI] [PubMed] [Google Scholar]
- Hudson C, Clements D, Friday RV, Stott D, Woodland HR. Xsox17alpha and -beta mediate endoderm formation in Xenopus. Cell. 1997;91:397–405. doi: 10.1016/s0092-8674(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Hudson C, Woodland HR. Xpat, a gene expressed specifically in germ plasm and primordial germ cells of Xenopus laevis. Mechanisms of Development. 1998;73:159–168. doi: 10.1016/s0925-4773(98)00047-1. [DOI] [PubMed] [Google Scholar]
- Jaruzelska J, Kotecki M, Kusz K, Spik A, Firpo M, Reijo Pera RA. Conservation of a Pumilio-Nanos complex from Drosophila germ plasm to human germ cells. Development Genes and Evolution. 2003;213:120–126. doi: 10.1007/s00427-003-0303-2. [DOI] [PubMed] [Google Scholar]
- Jevtic P, Levy DL. Nuclear size scaling during Xenopus early development contributes to midblastula transition timing. Current Biology. 2015;25:45–52. doi: 10.1016/j.cub.2014.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane DA, Hammerschmidt M, Mullins MC, Maischein HM, Brand M, van Eeden FJ, et al. The zebrafish epiboly mutants. Development. 1996;123:47–55. doi: 10.1242/dev.123.1.47. [DOI] [PubMed] [Google Scholar]
- Karr TL, Weir MP, Ali Z, Kornberg T. Patterns of engrailed protein in early Drosophila embryos. Development. 1989;105:605–612. doi: 10.1242/dev.105.3.605. [DOI] [PubMed] [Google Scholar]
- Kataoka K, Yamaguchi T, Orii H, Tazaki A, Watanabe K, Mochii M. Visualization of the Xenopus primordial germ cells using a green fluorescent protein controlled by cis elements of the 3′ untranslated region of the DEADSouth gene. Mechanisms of Development. 2006;123:746–760. doi: 10.1016/j.mod.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- King ML. Germ cell specification in Xenopus. In: Kloc M, Kubiak JZ, editors. Xenopus development. Hoboken, New Jersey: John Wiley & Sons, Inc.; 2014. pp. 75–100. [Google Scholar]
- King ML, Messitt TJ, Mowry KL. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biology of the Cell. 2005;97:19–33. doi: 10.1042/BC20040067. [DOI] [PubMed] [Google Scholar]
- Kloc M, Bilinski S, Dougherty MT. Organization of cytokeratin cytoskeleton and germ plasm in the vegetal cortex of Xenopus laevis oocytes depends on coding and non-coding RNAs: Three-dimensional and ultrastructural analysis. Experimental Cell Research. 2007;313:1639–1651. doi: 10.1016/j.yexcr.2007.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M, Dougherty MT, Bilinski S, Chan AP, Brey E, King ML, et al. Three-dimensional ultrastructural analysis of RNA distribution within germinal granules of Xenopus. Developmental Biology. 2002;241:79–93. doi: 10.1006/dbio.2001.0488. [DOI] [PubMed] [Google Scholar]
- Kloc M, Etkin LD. Delocalization of Vg1 mRNA from the vegetal cortex in Xenopus oocytes after destruction of Xlsirt RNA. Science. 1994;265:1101–1103. doi: 10.1126/science.7520603. [DOI] [PubMed] [Google Scholar]
- Kloc M, Wilk K, Vargas D, Shirato Y, Bilinski S, Etkin LD. Potential structural role of non-coding and coding RNAs in the organization of the cytoskeleton at the vegetal cortex of Xenopus oocytes. Development. 2005;132:3445–3457. doi: 10.1242/dev.01919. [DOI] [PubMed] [Google Scholar]
- Koebernick K, Loeber J, Arthur PK, Tarbashevich K, Pieler T. Elr-type proteins protect Xenopus Dead end mRNA from miR-18-mediated clearance in the soma. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16148–16153. doi: 10.1073/pnas.1004401107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani T, Yasuda K, Ota R, Yamashita M. Cyclin B1 mRNA translation is temporally controlled through formation and disassembly of RNA granules. The Journal of Cell Biology. 2013;202:1041–1055. doi: 10.1083/jcb.201302139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, King ML. Repressive translational control in germ cells. Molecular Reproduction and Development. 2013;80:665–676. doi: 10.1002/mrd.22161. [DOI] [PubMed] [Google Scholar]
- Lai F, Singh A, King ML. Xenopus Nanos1 is required to prevent endoderm gene expression and apoptosis in primordial germ cells. Development. 2012;139:1476–1486. doi: 10.1242/dev.079608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Zhou Y, Luo X, Fox J, King ML. Nanos1 functions as a translational repressor in the Xenopus germline. Mechanisms of Development. 2011;128:153–163. doi: 10.1016/j.mod.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley AR, Smith JC, Stemple DL, Harvey SA. New insights into the maternal to zygotic transition. Development. 2014;141:3834–3841. doi: 10.1242/dev.102368. [DOI] [PubMed] [Google Scholar]
- Leatherman JL, Jongens TA. Transcriptional silencing and translational control: Key features of early germline development. Bioessays. 2003;25:326–335. doi: 10.1002/bies.10247. [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, et al. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013;503:360–364. doi: 10.1038/nature12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerit DA, Gavis ER. Transport of germ plasm on astral microtubules directs germ cell development in Drosophila. Current Biology. 2011;21:439–448. doi: 10.1016/j.cub.2011.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Liu M, Hartley RS, Sheets MD, Dahlberg JE. Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. RNA. 2009;15:2351–2363. doi: 10.1261/rna.1882009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Nerlick S, An W, King ML. Xenopus germline nanos1 is translationally repressed by a novel structure-based mechanism. Development. 2011;138:589–598. doi: 10.1242/dev.056705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur H, Houston DW, Bubunenko M, Mosquera L, King ML. DEADSouth is a germ plasm specific DEAD-box RNA helicase in Xenopus related to eIF4A. Mechanisms of Development. 2000;95:291–295. doi: 10.1016/s0925-4773(00)00357-9. [DOI] [PubMed] [Google Scholar]
- Marlow FL, Mullins MC. Bucky ball functions in Balbiani body assembly and animal-vegetal polarity in the oocyte and follicle cell layer in zebrafish. Developmental Biology. 2008;321:40–50. doi: 10.1016/j.ydbio.2008.05.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathavan S, Lee SG, Mak A, Miller LD, Murthy KR, Govindarajan KR, et al. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genetics. 2005;1:260–276. doi: 10.1371/journal.pgen.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei W, Jin Z, Lai F, Schwend T, Houston DW, King ML, et al. Maternal Dead-End1 is required for vegetal cortical microtubule assembly during Xenopus axis specification. Development. 2013;140:2334–2344. doi: 10.1242/dev.094748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Richter JD. Translational control by CPEB: A means to the end. Nature Reviews. Molecular Cell Biology. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- Morichika K, Kataoka K, Terayama K, Tazaki A, Kinoshita T, Watanabe K, et al. Perturbation of Notch/Suppressor of Hairless pathway disturbs migration of primordial germ cells in Xenopus embryo. Development, Growth & Differentiation. 2010;52:235–244. doi: 10.1111/j.1440-169X.2009.01164.x. [DOI] [PubMed] [Google Scholar]
- Morichika K, Sugimoto M, Yasuda K, Kinoshita T. Possible regulation of Oct60 transcription by a positive feedback loop in Xenopus oocytes. Zygote. 2014;22:266–274. doi: 10.1017/S0967199412000536. [DOI] [PubMed] [Google Scholar]
- Mosquera L, Forristall C, Zhou Y, King ML. A mRNA localized to the vegetal cortex of Xenopus oocytes encodes a protein with a nanos-like zinc finger domain. Development. 1993;117:377–386. doi: 10.1242/dev.117.1.377. [DOI] [PubMed] [Google Scholar]
- Murphy CR, Sabel JL, Sandler AD, Dagle JM. Survivin mRNA is down-regulated during early Xenopus laevis embryogenesis. Developmental Dynamics. 2002;225:597–601. doi: 10.1002/dvdy.10194. [DOI] [PubMed] [Google Scholar]
- Nair S, Marlow F, Abrams E, Kapp L, Mullins MC, Pelegri F. The chromosomal passenger protein birc5b organizes microfilaments and germ plasm in the zebrafish embryo. PLoS Genetics. 2013;9:e1003448. doi: 10.1371/journal.pgen.1003448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakakura N, Miura T, Yamana K, Ito A, Shiokawa K. Synthesis of heterogeneous mRNA-like RNA and low-molecular-weight RNA before the midblastula transition in embryos of Xenopus laevis. Developmental Biology. 1987;123:421–429. doi: 10.1016/0012-1606(87)90400-3. [DOI] [PubMed] [Google Scholar]
- Nakaya M, Fukui A, Izumi Y, Akimoto K, Asashima M, Ohno S. Meiotic maturation induces animal-vegetal asymmetric distribution of aPKC and ASIP/PAR-3 in Xenopus oocytes. Development. 2000;127:5021–5031. doi: 10.1242/dev.127.23.5021. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin): A systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. Amsterdam: North-Holland Publ. Co.; 1956. [Google Scholar]
- Nieuwkoop PD, Sutasurya LA. Embryological evidence for a possible poly-phyletic origin of the recent amphibians. Journal of Embryology and Experimental Morphology. 1976;35:159–167. [PubMed] [Google Scholar]
- Nishiumi F, Komiya T, Ikenishi K. The mode and molecular mechanisms of the migration of presumptive PGC in the endoderm cell mass of Xenopus embryos. Development, Growth & Differentiation. 2005;47:37–48. doi: 10.1111/j.1440-169x.2004.00777.x. [DOI] [PubMed] [Google Scholar]
- Olson DJ, Oh D, Houston DW. The dynamics of plus end polarization and microtubule assembly during Xenopus cortical rotation. Developmental Biology. 2015;401:249–263. doi: 10.1016/j.ydbio.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota R, Kotani T, Yamashita M. Biochemical characterization of Pumilio1 and Pumilio2 in Xenopus oocytes. The Journal of Biological Chemistry. 2011;286:2853–2863. doi: 10.1074/jbc.M110.155523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan K, Richter JD. Regulated Pumilio-2 binding controls RINGO/Spy mRNA translation and CPEB activation. Genes & Development. 2006;20:199–209. doi: 10.1101/gad.1383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer DC, Gard DL. Microtubules in Xenopus oocytes are oriented with their minus-ends towards the cortex. Cell Motility and the Cytoskeleton. 1999;44:34–43. doi: 10.1002/(SICI)1097-0169(199909)44:1<34::AID-CM3>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Pondel MD, King ML. Localized maternal mRNA related to transforming growth factor β mRNA is concentrated in a cytokeratin-enriched fraction from Xenopus oocytes. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:7612–7616. doi: 10.1073/pnas.85.20.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt CA, Mowry KL. Taking a cellular road-trip: mRNA transport and anchoring. Current Opinion in Cell Biology. 2013;25:99–106. doi: 10.1016/j.ceb.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioleau MN, Huet J, Sentenac A, Mechali M. Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell. 1994;77:439–449. doi: 10.1016/0092-8674(94)90158-9. [DOI] [PubMed] [Google Scholar]
- Ressom RE, Dixon KE. Relocation and reorganization of germ plasm in Xenopus embryos after fertilization. Development. 1988;103:507–518. doi: 10.1242/dev.103.3.507. [DOI] [PubMed] [Google Scholar]
- Robb DL, Heasman J, Raats J, Wylie C. A kinesin-like protein is required for germ plasm aggregation in Xenopus. Cell. 1996;87:823–831. doi: 10.1016/s0092-8674(00)81990-x. [DOI] [PubMed] [Google Scholar]
- Saitou M, Yamaji M. Primordial germ cells in mice. Cold Spring Harbor Perspectives in Biology. 2012;4:1–20. doi: 10.1101/cshperspect.a008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage RM, Danilchik MV. Dynamics of germ plasm localization and its inhibition by ultraviolet irradiation in early cleavage Xenopus embryos. Developmental Biology. 1993;157:371–382. doi: 10.1006/dbio.1993.1142. [DOI] [PubMed] [Google Scholar]
- Schaner CE, Deshpande G, Schedl PD, Kelly WG. A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Developmental Cell. 2003;5:747–757. doi: 10.1016/s1534-5807(03)00327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder MM, Gard DL. Organization and regulation of cortical microtubules during the first cell cycle of Xenopus eggs. Development. 1992;114:699–709. doi: 10.1242/dev.114.3.699. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Braun RE. Pathway to totipotency: Lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Shiokawa K, Misumi Y, Tashiro K, Nakakura N, Yamana K, Oh-uchida M. Changes in the patterns of RNA synthesis in early embryogenesis of Xenopus laevis. Cell Differentiation and Development. 1989;28:17–25. doi: 10.1016/0922-3371(89)90019-1. [DOI] [PubMed] [Google Scholar]
- Sible JC, Anderson JA, Lewellyn AL, Maller JL. Zygotic transcription is required to block a maternal program of apoptosis in Xenopus embryos. Developmental Biology. 1997;189:335–346. doi: 10.1006/dbio.1997.8683. [DOI] [PubMed] [Google Scholar]
- Skirkanich J, Luxardi G, Yang J, Kodjabachian L, Klein PS. An essential role for transcription before the MBT in Xenopus laevis. Developmental Biology. 2011;357:478–491. doi: 10.1016/j.ydbio.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LD. The role of a “germinal plasm” in the formation of primordial germ cells in Rana pipiens. Developmental Biology. 1966;14:330–347. doi: 10.1016/0012-1606(66)90019-4. [DOI] [PubMed] [Google Scholar]
- Song HW, Cauffman K, Chan AP, Zhou Y, King ML, Etkin LD, et al. Hermes RNA-binding protein targets RNAs-encoding proteins involved in meiotic maturation, early cleavage, and germline development. Differentiation. 2007;75:519–528. doi: 10.1111/j.1432-0436.2006.00155.x. [DOI] [PubMed] [Google Scholar]
- Stack JH, Newport JW. Developmentally regulated activation of apoptosis early in Xenopus gastrulation results in cyclin A degradation during interphase of the cell cycle. Development. 1997;124:3185–3195. doi: 10.1242/dev.124.16.3185. [DOI] [PubMed] [Google Scholar]
- Stennard F, Zorn AM, Ryan K, Garrett N, Gurdon JB. Differential expression of VegT and Antipodean protein isoforms in Xenopus. Mechanisms of Development. 1999;86:87–98. doi: 10.1016/s0925-4773(99)00119-7. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Igarashi K, Aisaki K, Kanno J, Saga Y. NANOS2 interacts with the CCR4-NOT deadenylation complex and leads to suppression of specific RNAs. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3594–3599. doi: 10.1073/pnas.0908664107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Saga Y. Nanos2 suppresses meiosis and promotes male germ cell differentiation. Genes & Development. 2008;22:430–435. doi: 10.1101/gad.1612708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Ueno N, Hemmati-Brivanlou A. Xenopus msx1 mediates epidermal induction and neural inhibition by BMP4. Development. 1997;124:3037–3044. doi: 10.1242/dev.124.16.3037. [DOI] [PubMed] [Google Scholar]
- Tada H, Mochii M, Orii H, Watanabe K. Ectopic formation of primordial germ cells by transplantation of the germ plasm: Direct evidence for germ cell determinant in Xenopus. Developmental Biology. 2012;371:86–93. doi: 10.1016/j.ydbio.2012.08.014. [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: A play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Tanigawa Y, Minamide R, Ikenishi K, Komiya T. Analysis of SDF-1/CXCR4 signaling in primordial germ cell migration and survival or differentiation in Xenopus laevis. Mechanisms of Development. 2010;127:146–158. doi: 10.1016/j.mod.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Tan MH, Au KF, Yablonovitch AL, Wills AE, Chuang J, Baker JC, et al. RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Research. 2013;23:201–216. doi: 10.1101/gr.141424.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q, Yokota C, Puck H, Kofron M, Birsoy B, Yan D, et al. Maternal wnt11 activates the canonical wnt signaling pathway required for axis formation in Xenopus embryos. Cell. 2005;120:857–871. doi: 10.1016/j.cell.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Tarbashevich K, Koebernick K, Pieler T. XGRIP2.1 is encoded by a vegetally localizing, maternal mRNA and functions in germ cell development and anteroposterior PGC positioning in Xenopus laevis. Developmental Biology. 2007;311:554–565. doi: 10.1016/j.ydbio.2007.09.012. [DOI] [PubMed] [Google Scholar]
- ten Bosch JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development. 2006;133:1967–1977. doi: 10.1242/dev.02373. [DOI] [PubMed] [Google Scholar]
- Terayama K, Kataoka K, Morichika K, Orii H, Watanabe K, Mochii M. Developmental regulation of locomotive activity in Xenopus primordial germ cells. Development, Growth & Differentiation. 2013;55:217–228. doi: 10.1111/dgd.12018. [DOI] [PubMed] [Google Scholar]
- Theusch EV, Brown KJ, Pelegri F. Separate pathways of RNA recruitment lead to the compartmentalization of the zebrafish germ plasm. Developmental Biology. 2006;292:129–141. doi: 10.1016/j.ydbio.2005.12.045. [DOI] [PubMed] [Google Scholar]
- Torpey NP, Heasman J, Wylie CC. Identification of vimentin and novel vimentin-related proteins in Xenopus oocytes and early embryos. Development. 1990;110:1185–1195. doi: 10.1242/dev.110.4.1185. [DOI] [PubMed] [Google Scholar]
- Torpey NP, Heasman J, Wylie CC. Distinct distribution of vimentin and cytokeratin in Xenopus oocytes and early embryos. Journal of Cell Science. 1992;101(Pt. 1):151–160. doi: 10.1242/jcs.101.1.151. [DOI] [PubMed] [Google Scholar]
- Venkatarama T, Lai F, Luo X, Zhou Y, Newman K, King ML. Repression of zygotic gene expression in the Xenopus germline. Development. 2010;137:651–660. doi: 10.1242/dev.038554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakahara M. Partial characterization of “primordial germ cell-forming activity” localized in vegetal pole cytoplasm in anuran eggs. Journal of Embryology and Experimental Morphology. 1977;39:221–233. [PubMed] [Google Scholar]
- Wakahara M. Induction of supernumerary primordial germ cells by injecting vegetal pole cytoplasm into Xenopus eggs. The Journal of Experimental Zoology. 1978;203:159–164. [Google Scholar]
- Wang Y, Opperman L, Wickens M, Hall TM. Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20186–20191. doi: 10.1073/pnas.0812076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann CA, Goldstrohm AC. Drosophila Pumilio protein contains multiple autonomous repression domains that regulate mRNAs independently of Nanos and brain tumor. Molecular and Cellular Biology. 2012;32:527–540. doi: 10.1128/MCB.06052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton RP, Sonoda J, Lee T, Patterson M, Murata Y. The Pumilio RNA-binding domain is also a translational regulator. Molecular Cell. 1998;1:863–872. doi: 10.1016/s1097-2765(00)80085-4. [DOI] [PubMed] [Google Scholar]
- White JA, Heasman J. Maternal control of pattern formation in Xenopus laevis. Journal of Experimental Zoology. Part B, Molecular and Developmental Evolution. 2008;310:73–84. doi: 10.1002/jez.b.21153. [DOI] [PubMed] [Google Scholar]
- Whitfield T, Heasman J, Wylie C. XLPOU-60, a Xenopus POU-domain mRNA, is oocyte-specific from very early stages of oogenesis, and localised to presumptive mesoderm and ectoderm in the blastula. Developmental Biology. 1993;155:361–370. doi: 10.1006/dbio.1993.1035. [DOI] [PubMed] [Google Scholar]
- Wilk K, Bilinski S, Dougherty MT, Kloc M. Delivery of germinal granules and localized RNAs via the messenger transport organizer pathway to the vegetal cortex of Xenopus oocytes occurs through directional expansion of the mitochondrial cloud. The International Journal of Developmental Biology. 2005;49:17–21. doi: 10.1387/ijdb.041906kw. [DOI] [PubMed] [Google Scholar]
- Wylie CC, Brown D, Godsave SF, Quarmby J, Heasman J. The cytoskeleton of Xenopus oocytes and its role in development. Journal of Embryology and Experimental Morphology. 1985;89(Suppl.):1–15. [PubMed] [Google Scholar]
- Wylie CC, Heasman J, Parke JM, Anderton B, Tang P. Cytoskeletal changes during oogenesis and early development of Xenopus laevis. Journal of Cell Science. 1986;5(Supplement):329–341. doi: 10.1242/jcs.1986.supplement_5.21. [DOI] [PubMed] [Google Scholar]
- Wylie CC, Snape A, Heasman J, Smith JC. Vegetal pole cells and commitment to form endoderm in Xenopus laevis. Developmental Biology. 1987;119:496–502. doi: 10.1016/0012-1606(87)90052-2. [DOI] [PubMed] [Google Scholar]
- Yabe T, Ge X, Lindeman R, Nair S, Runke G, Mullins MC, et al. The maternal-effect gene cellular island encodes aurora B kinase and is essential for furrow formation in the early zebrafish embryo. PLoS Genetics. 2009;5:e1000518. doi: 10.1371/journal.pgen.1000518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Kataoka K, Watanabe K, Orii H. Restriction of the Xenopus DEADSouth mRNA to the primordial germ cells is ensured by multiple mechanisms. Mechanisms of Development. 2014;131:15–23. doi: 10.1016/j.mod.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Taguchi A, Watanabe K, Orii H. DEADSouth protein localizes to germ plasm and is required for the development of primordial germ cells in Xenopus laevis. Biology Open. 2013;2:191–199. doi: 10.1242/bio.20123111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Cao L, Li Q, Wu Y, Zhang H, Saiyin H, et al. Aurora C is directly associated with Survivin and required for cytokinesis. Genes to Cells. 2005;10:617–626. doi: 10.1111/j.1365-2443.2005.00863.x. [DOI] [PubMed] [Google Scholar]
- Yang J, Tan C, Darken RS, Wilson PA, Klein PS. Beta-catenin/Tcf-regulated transcription prior to the midblastula transition. Development. 2002;129:5743–5752. doi: 10.1242/dev.00150. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Williamson JR, Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA. 1997;3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- Zearfoss NR, Chan AP, Wu CF, Kloc M, Etkin LD. Hermes is a localized factor regulating cleavage of vegetal blastomeres in Xenopus laevis. Developmental Biology. 2004;267:60–71. doi: 10.1016/j.ydbio.2003.10.032. [DOI] [PubMed] [Google Scholar]
- Zhang C, Basta T, Fawcett SR, Klymkowsky MW. SOX7 is an immediate-early target of VegT and regulates Nodal-related gene expression in Xenopus. Developmental Biology. 2005;278:526–541. doi: 10.1016/j.ydbio.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Zhang C, Basta T, Klymkowsky MW. SOX7 and SOX18 are essential for cardiogenesis in Xenopus. Developmental Dynamics. 2005;234:878–891. doi: 10.1002/dvdy.20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, et al. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- Zhang J, Houston DW, King ML, Payne C, Wylie C, Heasman J. The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell. 1998;94:515–524. doi: 10.1016/s0092-8674(00)81592-5. [DOI] [PubMed] [Google Scholar]
- Zhang J, King ML. Xenopus VegT RNA is localized to the vegetal cortex during oogenesis and encodes a novel T-box transcription factor involved in mesodermal patterning. Development. 1996;122:4119–4129. doi: 10.1242/dev.122.12.4119. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yan L, Zhou Z, Yang P, Tian E, Zhang K, et al. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 2009;136:308–321. doi: 10.1016/j.cell.2008.12.022. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Tian E, Zhang H. Selective autophagic degradation of maternally-loaded germline P granule components in somatic cells during C. elegans embryogenesis. Autophagy. 2009;5:717–719. doi: 10.4161/auto.5.5.8552. [DOI] [PubMed] [Google Scholar]
- Zust B, Dixon KE. The effect of u.v. irradiation of the vegetal pole of Xenopus laevis eggs on the presumptive primordial germ cells. Journal of Embryology and Experimental Morphology. 1975;34:209–220. [PubMed] [Google Scholar]