Abstract
In Drosophila, color vision and wavelength-selective behaviors are mediated by the compound eye’s narrow-spectrum photoreceptors, R7 and R8, and their downstream neurons, Tm5a/b/c and Tm20, in the second optic neuropil, or medulla. These chromatic Tm neurons project axons to a deeper optic neuropil, the lobula, which in insects has been implicated in processing and relaying color information to the central brain. The synaptic targets of the chromatic Tm neurons in the lobula are not known, however. Using a modified GRASP (GFP reconstitution across synaptic partners) method to probe connections between the chromatic Tm neurons and 28 known and novel types of lobula neurons, we identified anatomically the visual projection neurons LT11 and LC14, and the lobula intrinsic neurons Li3 and Li4, as synaptic targets of the chromatic Tm neurons. Single-cell GRASP analyses revealed that Li4 receives synaptic contacts from over 90% of all four types of chromatic Tm neurons while LT11 is postsynaptic to the chromatic Tm neurons with only modest selectivity and at a lower frequency and density. To visualize synaptic contacts at the ultrastructural level, we developed and applied a “two-tag” double labeling method to label LT11’s dendrites and the mitochondria in Tm5c’s presynaptic terminals. Serial electron microscopic reconstruction confirmed that LT11 receives direct contacts from Tm5c. This method would be generally applicable to map the connections of large complex neurons in Drosophila and other animals.
Keywords: Color vision, GRASP, lobula, retina, phototaxis
Graphical Abstract
Using trans-synaptic tracing and ultrastructural “two-tag” double-labeling approaches for EM, the authors identify synaptic targets of first-order chromatic interneurons in the deep visual processing center, the lobula. Lobula intrinsic neuron Li4 and projection neuron LT11 receive parallel chromatic inputs but with differential synaptic densities.
INTRODUCTION
The fly’s visual system collects spectrally encoded images of the world through its paired compound eyes, each an array of about 750 unit eyes or ommatidia (Ready et al., 1976). Each ommatidium accommodates two photoreceptor types: rod-like R1-R6 that innervate the first neuropil layer, the lamina, and cone-like R7/R8 that innervate the second neuropil, or medulla (Fischbach and Dittrich, 1989). The visual pathways arising from these inputs are arranged in structural modules, lamina cartridges and medulla columns. Ultraviolet- (R7) or blue/green- (R8) sensitive (Mikeladze-Dvali et al., 2005) photoreceptors each express one of two rhodopsins (Morante and Desplan, 2008). Each photoreceptor type thus has two subtypes and all four subtypes are spectrally distinct (Hardie et al., 1979; Hardie and Kirschfeld, 1983). R7, R8 cell pairs coordinately express a particular rhodopsin to construct one of two types of ommatidial rhodopsin partnerships, pale or yellow (Franceschini et al.,1981;, Mazzoni, et al., 2008). In the ommatidia of the “pale” type, the R7 cell expresses the Rh3 opsin, which is most sensitive to ultraviolet (UV) light, while R8 expresses the Rh5 opsin, sensitive in the blue; whereas in “yellow” ommatidia, the R7 cell expresses the long-UV sensitive Rh4 opsin and the underlying R8 expresses the green-sensitive Rh6 opsin. The two types pattern the eye randomly (Bell et al., 2007) and presumably extend the spectral range of the retina (Morante and Desplan, 2008). Both R7 and R8 are required functionally to enable spectral preference and color vision (Gao et al., 2008; Schnaitmann et al., 2010, 2013; Yamaguchi et al., 2010; Melnattur et al., 2014).
Significant progress has recently been made in identifying the chromatic circuits of R7 and R8 in the medulla (Gao et al., 2008; Takemura et al., 2013; Karuppudurai et al., 2014). Among more than 60 morphologically distinct medulla neurons, at least four types of medulla projection (Tm) neurons, Tm5a/b/c and Tm20, and one type of amacrine neuron, Dm8, express histamine-gated chloride channels and receive direct synaptic inputs from R7 and/or R8 photoreceptors (Gao et al., 2008). In particular, the medulla projection neurons Tm5a/b/c and Tm20 relay chromatic information to the deeper visual center, the lobula, suggesting that they are anatomically comparable to vertebrate retinal ganglion cells (Cajal and Sánchez, 1915; Sanes and Zipursky, 2010). Genetically inactivating all of these four types of medulla projection neurons abolishes color learning, indicating that as a collective they are functionally required for hue discrimination (Melnattur et al., 2014). However, inactivating subsets of these neurons is insufficient to block color learning, suggesting that true color vision is mediated by multiple redundant pathways. In contrast, innate ultraviolet (UV) preference is dominated by a single pathway, from UV-sensing R7 photoreceptors to the amacrine neuron Dm8 and to the projection neuron Tm5c (Karuppudurai et al., 2014). Tm5a receives photoreceptor input exclusively from yellow-type photoreceptors while the other three Tm types do not appear to differentiate yellow from pale photoreceptor subtypes. Using the Tango-Trace method, four TmY types of projection neurons have been identified as the targets of specific pale or yellow photoreceptor subtypes but their contribution to color-driven behaviors has yet to be demonstrated (Jagadish et al., 2014). The behaviorally relevant Tm5a/b/c and Tm20 medulla neurons project axons to innervate the lobula’s deep strata (Lo4–6 of the total of six strata), however their synaptic targets in the lobula are entirely unknown.
Anatomically, the deep lobula strata bridge between the optic lobe and the central brain: their lobula neurons receive retinotopic inputs from the medulla and connect with the protocerebrum, including the visual processing glomeruli (Strausfeld and Okamura, 2007). By screening Gal4 expression patterns, Ito and colleagues have identified over 14 classes of visual projection neurons (VPN) that connect the lobula to the central brain (Otsuna and Ito, 2006). These include the columnar type (LC) VPNs, which have small receptive fields, and the large tree-like type (LT) VPNs, which elaborate large dendritic trees in distinct strata of the lobula. Despite these extensive anatomical findings, little is known about either the functions or connections of the VPNs. LT11, a pair of bilateral hemivisual-field VPNs, have been shown to play a role in spectrum-specific phototaxis (Otsuna et al., 2014). In bees, electrophysiological studies have demonstrated that neurons innervating the deep lobula strata (Lo5–6) are color sensitive and in general have a larger receptive field than the corresponding medulla neurons (Hertel, 1980; Paulk et al., 2008). Given that the central brain targets of lobula output neurons (VPN) lack retinotopic organization, the lobula seems likely to transform images into feature-based information, such as associating color and motion visual attributes with objects. The connections between lobula neurons and medulla Tm neurons are not known and their identification would be an important step in understanding the computations carried out by the lobula.
In this study, we identified the lobula neurons that receive synaptic inputs from chromatic Tm neurons. We combined the GRASP (GFP reconstitution across synaptic partners) method with presynaptic labeling to visualize potential synaptic contacts between candidate lobula neurons and their chromatic Tm neuron inputs. We further used single-cell analyses to estimate the synaptic numbers for different types of chromatic Tm neurons. Finally, we developed a novel double-labeling method to label both presynaptic terminals and postsynaptic dendrites for electron microscopic analyses. Using this novel method, we examined the contacts between a selected pair of Tm and lobula neurons at the ultrastructural level.
MATERIALS AND METHODS
Fly strains
Flies were maintained on standard Drosophila medium at 23~25°C under a 12–12 hour light-dark cycle. Fly stocks used in this study were as follows: (1) ortC1a-LexA::VP16; (2) ortC1a-LexADBD; (3) OK371-dVP16AD; (4) ET18k–dVP16AD; (5) ET24g–dVP16AD; (6) ET24b–dVP16AD; (7) 13XLexAop2-IVS-FRTstopFRT- spGFP11::CD4::HA-T2A–BrpD3::mCherry in attP2 (Karuppudurai et al., 2014); (8) UAS-spGFP1–10::CD4 (Gordon and Scott, 2009); (9) UAS-mCD8::GFP; (10) 8XLexAop2-FLPL; (11) LexAop-HRP::CD2; (12) UAS>CD2,y+>mCD8::GFP; (13) hsFLP122; (14) hsFLP; (15) Janelia Farm FlyLight Gal4 lines (Bloomington Drosophila Stock Center, BDSC); (16) NP Gal4 Lines (Kyoto DGRC); (17) brp-FSF-GFP in attP2 (Chen et al., 2014); (18) 13XLexAop2-mito::APX::HA (this study); and (19) UAS-HRP::DsRed::GPI (Han et al., 2014). The lobula Gal4 lines are listed in Table 1. The detailed genotypes of animals used in this study are listed in Table 2.
Table 1.
List of GAL4 strains used in this study
| Gal4 strain | Reference | |
|---|---|---|
| VPNs | ||
| LC4 | NP7281, NP7365, NP7476 | (1) |
| LC6 | NP6250, NP6813, NP7067, R41C07 | (1), (3) |
| LC9 | NP6250, NP6813, NP7067 | (1) |
| LC10 | NP6250, NP6813, NP7067, R22D06 | (1), (3) |
| LC11 | NP3045, NP7226, R12H12 | (1), (3) |
| LC13 | NP6502 | (1) |
| LC14 | NP1011, NP1320, NP6558, ato-GAL4 | (1) |
| LC151 | R74B04 | (3) |
| LC161 | R89B12 | (3) |
| LT1 | NP1195, NP2331 | (1) |
| LT10 | NP1035, NP5006, NP7121 | (1) |
| LT11 | NP1035, NP1047, NP6099 | (1) |
| LT12 | NP7233, NP7365 | (1) |
| LT32 | NP2450 | (1) |
| Complex neurons | ||
| CT1 | NP2400 | (2) |
| CT02 | NP7318 | (2) |
| CT03 | NP2397 | (2) |
| CT31 | NP2302, NP2404 | (2) |
| CT33 | NP2784 | (2) |
| CT34 | NP2364, NP7233 | (2) |
| CT35 | NP0722, NP1049, NP1149 | (2) |
| CC1 | NP0528, NP3337, NP7281 | (2) |
| CC2 | NP6645, | (2) |
| CC61 | NP0302, NP1194, NP5279 | (2) |
| Lobula intrinsic | ||
| Li3 | R43E05 | (3) |
| Li4 | R89D06, NP1582 | (3) |
| Li51 | R84C012 | (3) |
| Uncharacterized | ||
| R76G07 | (3) |
Tentatively named
R84C01 also labels additional unidentified lobula neurons
Shinomiya et al., 2010 (Flybrain Neuron Database)
This study
TABLE 2.
Summary of Experimental Genotypes
| Figures | Genotype |
|---|---|
| 1B–B1 | hsFLP122; ortC1aLexA::VP16/+; 13XLexAop2-FSF-spGFP11::HA-T2A–BrpD3::mCherry/+ |
| 1C–C1 | yw; ortC1aLexA::VP16/8XLexAop2-FLPL; brp-FSF-GFP/LexAop-HRP::CD2 |
| 2A | yw/w1118; UAS-mCD8GFP/+; R43E05-GAL4/UAS-mCD8GFP |
| 2B–C1 | hsFLP122/w1118; UAS-D2Y+ -mCD8GFP/+; R43E05-GAL4/+ |
| 2D | yw; UAS-mCD8GFP/+; NP1582-GAL4/UAS-mCD8GFP |
| 2E–F1 | hsFLP122/w1118; NP1582-GAL4/UAS-D2Y+ -mCD8GFP |
| 3A | yw/w1118; UAS-mCD8GFP/+; R43E05-GAL4/UAS-mCD8GFP |
| 3A1-A2 |
hsFLP122/w1118; ortC1aLexA::VP16/+; R43E05-GAL4/13XLexAop2-FSF-spGFP11::HA-T2A- BrpD3::mCherry |
| 3B | yw; UAS-mCD8GFP/+; NP1582-GAL4/UAS-mCD8GFP |
| 3B1-B2 |
hsFLP122/yw; ortC1aLexA::VP16/+; NP1582-GAL4/13XLexAop2-FSF-spGFP11::HA-T2A- BrpD3::mCherry |
| 3C | yw; UAS-mCD8GFP/+; NP7476-GAL4/UAS-mCD8GFP |
| 3C1-C2 |
hsFLP122/yw; ortC1aLexA::VP16/+; NP7476-GAL4/13XLexAop2-FSF-spGFP11::HA-T2A- BrpD3::mCherry |
| 4A | NP6099-GAL4/yw; UAS-mCD8GFP/+; UAS-mCD8GFP/+ |
| 4A1-A2 |
hsFLP122/NP6099-GAL4; ortC1aLexA::VP16/+;13XLexAop2-FSF-spGFP11::HA-T2A- BrpD3::mCherry/+ |
| 4B | yw; UAS-mCD8GFP/+; ato-GAL4/UAS-mCD8GFP |
| 4B1-B2 |
hsFLP122/yw; ortC1aLexA::VP16/+; ato-GAL4/13XLexAop2-FSF-spGFP11::HA-T2A- BrpD3::mCherry |
| 5B–B2 |
hsFLP1/yw; ortC1a-LexADBD/+; NP1582-GAL4, ET7B1dVP16AD/13XLexAop2-FSF- spGFP11::CD4::HA-T2A–BrpD3::mCherry, UAS-spGFP1-10::CD4, UAS-HRP::CD2 |
| 5C–C2 |
hsFLP1/yw; ortC1a-LexADBD/+; NP1582-GAL4, ET18kdVP16AD/13XLexAop2-FSF- spGFP11::CD4::HA-T2A–BrpD3::mCherry, UAS-spGFP1-10::CD4, UAS-HRP::CD2 |
| 5D–D2 |
hsFLP1/yw; ortC1a-LexADBD, OK371-dVP16AD/+; NP1582-GAL4/13XLexAop2-FSF- spGFP11::CD4::HA-T2A–BrpD3::mCherry, UAS-spGFP1-10::CD4, UAS-HRP::CD2 |
| 5E–E2 |
hsFLP1/yw; ortC1a-LexADBD/+; NP1582-GAL4, ET24bdVP16AD/13XLexAop2-FSF- spGFP11::CD4::HA-T2A–BrpD3::mCherry, UAS-spGFP1-10::CD4, UAS-HRP::CD2 |
|
5F–F2 5G–G2 |
NP6099-GAL4/yw; ortC1a-LexADBD/hsFLP; ET18kdVP16AD/13XLexAop2-FSF- spGFP11::CD4::HA-T2A–BrpD3::mCherry, UAS-spGFP1-10::CD4, UAS-HRP::CD2 |
| 5H–H2 |
NP6099-GAL4/yw; ortC1a-LexADBD, OK371-dVP16AD/hsFLP; 13XLexAop2-FSF- spGFP11::CD4::HA-T2A–BrpD3::mCherry, UAS-spGFP1-10::CD4, UAS-HRP::CD2/+ |
| 5I–I2 |
NP6099-GAL4/hsFLP122; ortC1a-LexA::VP16/+; ET18kdVP16AD/13XLexAop2-FSF- spGFP11::CD4::HA-T2A–BrpD3::mCherry, UAS-spGFP1-10::CD4, UAS-HRP::CD2 |
| 6A–A1 | yw; ortC1a-LexADBD, OK371-dVP16AD/+; 13XLexAop2-mito-APX41F-HA/+ |
| 6B–E |
NP1582-GAL4/yw; ortC1a-LexADBD, OK371-dVP16AD/+; 13XLexAop2-mito-APX41F- HA/UAS-HRP::DsRed::GPI |
GRASP
To screen candidate neurons postsynaptic to chromatic Tm neurons, hsFLP122; ortC1a-LexA::VP16/CyO; LexAop2-IVS-FRTstopFRT-spGFP11::CD4::HA-T2A–BrpD3::mCherry, UAS-spGFP1–10::CD4, UAS-HRP::CD2/TM2 was crossed with various lobula Gal4 lines (either from FlyLight or NP Gal4 lines). To remove the FRT-stop-FRT cassette and express both the split-GFP (spGFP11::CD4::HA) and active zone marker (BrpD3::mCherry) in presynaptic neurons, newly eclosed adults were exposed to a 30°C heat shock for 3 days to activate flippase expression before dissection. Under this condition, more than 90% of presynaptic neurons expressed both reporters as expected. Meanwhile, postsynaptic lobula neurons expressed the other split-GFP (spGFP1–10::CD4), and functional GFP (GRASP) was reconstituted at the contact sites between these two groups of neurons. Colocalization of the active zone marker and GRASP to generate fluorescent puncta indicated possible synapses, suggesting candidate lobula targets for chromatic pathways. For single-cell GRASP, the experiment was carried out as described previously (Karuppudurai et al., 2014).
Immunohistochemistry and Confocal Microscopy
Standard methods were performed as described previously (Ting et al., 2007). In brief, brain samples were dissected and fixed in fresh prepared 4% paraformaldehyde for 90 min. Samples were then washed extensively with PBT (0.5 % TritonX-100 in PBS) and blocked by 10% normal goat serum for 30 min. Primary and corresponding secondary antibodies were used for the immunolabeling. Primary antibodies used in this study are listed in Table 3. The secondary antibodies including goat anti-mouse, rabbit, or rat conjugated with Alexa Fluoro 488, 568, or 647 (Life technologies) were used at a 1:400 dilution. Confocal images were obtained using either a Zeiss LSM510 Meta or a Zeiss LSM780 microscope. Images were deconvolved to remove z-distortion using Huygens Professional software (Scientific Volume Imaging). The colocalization of two fluorescent signals was defined using the coloc module of Imaris software (Bitplane; RRID: nif-0000–00314).
TABLE 3.
Antibodies used, their immunogens and sources.
| Antigen | Immunogen | Source, species, type, catalog number |
Dilution | Reference, RRID |
|---|---|---|---|---|
| Green fluorescent protein (GFP) |
GFP isolated directly from Aequorea victoria |
Life Technologies, A-11120 Mouse monoclonal (IgG2a) |
1:400 | (Ting et al., 2014) RRID: AB_10055152 |
| Green fluorescent protein (GFP) |
full-length GFP | Life Technologies, G10362 Rabbit monoclonal |
1:400 | (Ting et al., 2014) RRID: AB_10565179 |
| Green fluorescent protein (GFP) |
recombinant full length GFP protein |
Abcam, ab13970 Chicken polyclonal |
1:1000 | (Ting et al., 2014) RRID: AB_300798 |
| HA | synthetic peptide (residues 76- 111 of X47 hemaglutinin 1) |
Roche, 11867423001 Rat monoclonal (IgG1) |
1:400 | (Karuppudurai et al., 2014) RRID: AB_390918 |
| Horseradish peroxidase(HRP) |
purified peroxidase from horseradish |
Abcam, ab10183 Mouse monoclonal (IgG2b) |
1:2000 | (Karuppudurai et al., 2014) RRID: AB_296913 |
| FasIII | nerve cord and primary neuronal membrane lysate (10- 13 hr development) |
DSHB, Iowa City, IA Mouse monoclonal (IgG2a) |
1:50 | (Snow et al., 1989) RRID: AB_528238 |
| Connectin |
Drosophila Connectin protein expressed in pGEX vector |
DSHB, Iowa City, IA Mouse monoclonal (IgG1) |
1:50 | (Meadows et al., 1994) RRID: AB_10660830 |
Antibody Characterization
Primary antibodies against green fluorescent proteins (GFP) were used in our previous studies. No immunopositive signal was seen in animals that lacked the expression of fluorescent or fusion proteins. The labeling specificity of all three antibodies in the Drosophila brain has been demonstrated in single-neuron analysis (Ting et al., 2011; Ting et al., 2014; Karuppudurai et al., 2014). In immunohistochemistry assays, expression signals stained and detected by anti-GFP mouse IgG2a (3E6; Life Technologies, #A11120) or anti-GFP ABfinity™ recombinant rabbit monoclonal antibody (Life Technologies, #G10362) colocalized with dim GFP fluorescence from the same protein in culture cells (manufacturer's technical information). Chicken anti-GFP antibody was raised against recombinant full length GFP protein. Its specificity was confirmed by immunohistochemistry and Western blotting in various tissues of GFP transgenic animal and transfected culture cells (manufacturer's technical information), and is lacking in those that lack GFP expression.
Monoclonal antibodies against epitope tags (hemagglutinin [HA] or horseradish peroxidase [HRP]) of fusion proteins were examined in previous studies (Ting et al., 2011; Karuppudurai et al., 2014). Both antibodies lacked detectable signals in wild-type fly brain tissue. Anti-HA (3F10) antibody was raised against a synthetic peptide (residues 76–111 of X47 hemaglutinin 1) conjugated to KLH. Anti-HRP (2H11) antibody was raised against an immunogen of purified peroxidase from horseradish.
Both anti-FasIII (Snow et al., 1989) and anti-Connectin (Meadows et al., 1994) antibodies were obtained from Developmental Studies Hybridoma Bank. Both immunolabeled distinct medulla and lobula strata in the Drosophila optic lobe (Gao et al., 2008; Karuppudurai et al., 2014). For anti-FasIII antibody, its specificity was confirmed by the Western blotting of an 80 kDa immunosignal in S2 cells expressing fasIII cDNA (Snow et al. 1989). For anti-Connectin, its specificity was confirmed by the Western blotting of a 61 kDa immunosignal in 6- to 18-hour embryos and S2 cells expressing of connectin cDNA (Meadows et al., 1994).
Construction of 13XLexAop2-IVS-mito::APXW41F::HA
To target APX to mitochondria, we used the mitochondrial targeting sequence that contains 29 amino acids of human Cox subunit 8a (MSVLTPLLLRGLTGSARRLPVPRAKIHSL). The peptide sequence of mutant pea ascorbate peroxidase (APXW41F) is as reported previously (Martell et al., 2012). APXW41F forms a constitutive homodimer and therefore exhibits greater enzyme activity than the wild-type APX. Codon optimized mito::APXW41F::HA cDNA was synthesized and subcloned to the pUC57 vector (GenScript, Piscataway, NJ). The mito::APX fragment was subcloned to replace the myr::GFP fragment from NotI and XbaI digested pJFRC19 (13XLexAop2-IVS-myr::GFP) plasmid (Addgene, Cambridge, MA).
EM sample preparation and image processing
EM samples were prepared as previously reported (Edwards and Meinertzhagen, 2009) with the following modifications. The fly’s brain was dissected from the head capsule in phosphate-buffered saline (PBS), then fixed with 4% formaldehyde and 0.3% glutaraldehyde in 0.1M phosphate buffer (PB) for 1 hour at 4°C. After washing with 0.1M PB, the brain was embedded in 7% agarose and sectioned at 150 µm thickness in a horizontal plane with a Leica VT1200 S vibrating microtome. Specimen slices were pre-incubated in DAB solution for 40 minutes then incubated in the solution with H2O2 for 6 hours at 4°C to generate the oxidation reaction product and were finally osmicated. Serial 50 nm EM sections were cut and images captured using an FEI Tecnai T12 electron microscope, aligned using the TrakEM2 plug-in (Cardona et al., 2012; RRID:nlx_151924) for Fiji (http://fiji.sc/; RRID:SciRes_000137) and processed as previously reported (Shinomiya et al., 2014).
RESULTS
Four types of chromatic Tm neurons are presynaptic in the lobula’s deep strata
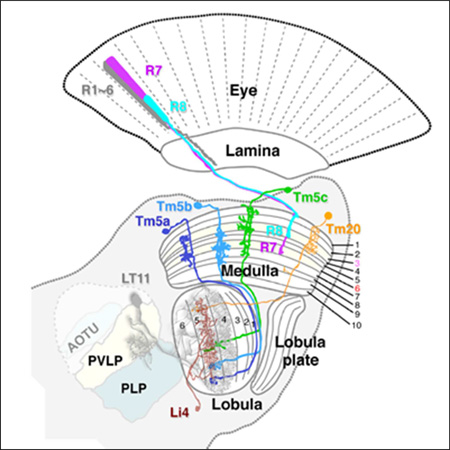
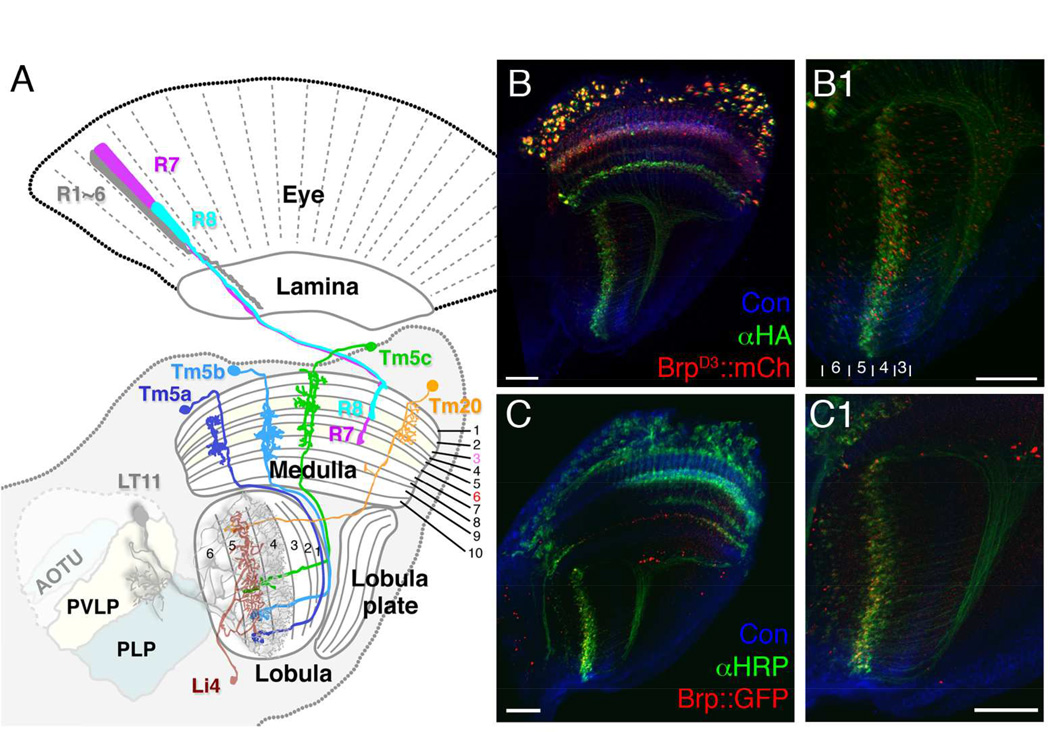
We have previously identified four types of transmedulla neurons (Tm5a, Tm5b, Tm5c and Tm20) that receive inputs from narrow-spectrum photoreceptors R7 and R8 (Figure 1A) and are involved in generating visual behaviors driven by specific wavelengths (Karuppudurai et al., 2014, Melnattur et al., 2014). To understand how color information is processed in the deep visual centers, we set out to identify the synaptic targets of these chromatic Tm neurons. First, we used the ortC1a-LexAVP16 driver to express an active-zone marker, BrpD3::mCherry, in all four types of Tm neurons, to mark their presynaptic sites (Christiansen et al., 2011; Karuppudurai et al., 2014; Ting et al., 2014; Melnattur et al., 2014). We found that the presynaptic sites of these neurons were localized to the axon terminals in the deep lobula strata, Lo4–6 (Fischbach and Dittrich, 1989; Figure 1B1), as well as their dendrites in the medulla (stratum M8; Figure 1B). BrpD3::mCherry expression was also found in the cell bodies, likely due to marker overexpression. We further confirmed the locations of presynaptic sites using the STaR (synaptic tagging with recombination) method (Figures 1C, 1C1), which expresses Brp::GFP at the endogenous level (Chen et al., 2014). In summary, the four types of chromatic Tm neurons are presynaptic in lobula strata 4–6, consistent with previous studies (Gao et al., 2008; Karuppudurai et al., 2014).
Figure 1.
The axons of chromatic Tm neurons, Tm5a/b/c and Tm20, form terminals in deep lobula strata. (A) Schematic illustration of the Drosophila visual system, including the compound eye’s retina, and successive optic neuropils, the lamina, medulla and lobula, with central brain regions. R1–6 innervate the lamina, and R7, R8 the medulla. A single lobula projection neuron LT11 (gray) receives chromatic inputs from strata Lo4 and Lo5 strata and projects an axon to optic glomeruli in the posterior ventrolateral protocerebrum (PVLP). A representative lobula intrinsic neuron (Li4, brown) elaborates web-like processes in lobula stratum Lo5. Its cell body is located primarily in the posterior lateral cell body region (LCBR). AOTU, anterior optic tubercle; PLP, posterior lateral protocerebrum. (B, B1) ortC1a-LexA::VP16 drives expression of a HA-tagged membrane marker (αHA, green) and an active zone marker, Brp::mCherry (red) in the chromatic Tm neurons. Presynaptic sites of Tm5a/b/c and Tm20 are located in the deep lobula strata (Lo4, Lo5, and sparsely in Lo6). (C,C1) The presynaptic sites of chromatic Tm neurons marked using the STaR method, which expresses GFP-tagged Brp (pseudo-colored red) from the endogenous brp promoter (see main text for details), showing a pattern of presynaptic sites similar to (B1) in the lobula. B1 and C1 enlarged from B and C, respectively. Scale bar, 20 µm for B-C1.
We next took an anatomical approach to identify the lobula neurons that receive direct synaptic inputs from the chromatic Tm neurons. First, we collected and characterized Gal4 lines that express in lobula neurons. Second, we used the GRASP (GFP reconstitution across synaptic partners) method (Feinberg et al., 2008) to examine contacts between the axon terminals of chromatic Tm neurons and the dendrites of their candidate lobula neuron targets. To differentiate genuine synapses from mere membrane contacts, we marked the presynaptic sites of the chromatic Tm neurons and examined the juxtapositions between GRASP signals and presynaptic markers. Third, to characterize more completely the patterns of synaptic connections between candidate partner neurons we employed a flip-out GRASP approach to visualize and probe synaptic contacts at single-neuron resolution (Karuppudurai et al., 2014). Finally, we developed and used a novel double-labeling method to examine those contacts in selected neuron pairs at the ultrastructural level, using targeted peroxidase markers.
Identification of novel lobula neuron drivers
To collect lobula-specific GAL4 drivers, we examined two collections of Gal4 strains. The first is a previously reported library of GAL4 enhancer-trap strains (Osutna and Ito, 2006; Shinomiya et al., 2011; http://flybrain-ndb.iam.u-tokyo.ac.jp). These include 14 pathways bridging between the lobula and central brain reported previously (Osutna and Ito, 2006). The second is the Janelia FlyLight GAL4 collection, which contains approximately 7000 lines made using small enhancer fragments (Jenett et al., 2012). We sought drivers that selectively express in the lobula neuropil by screening entire image stacks of fly brains (http://flweb.janelia.org/cgi-bin/flew.cgi). By restricting anatomical expression criteria to the optic lobes, we first narrowed down our targets to 2048 lines which we then inspected more closely. Among 59 lobula driver lines identified, we focused on eight lines that showed restricted arborizations in the deep lobula strata, and subjected these to further anatomical analysis.
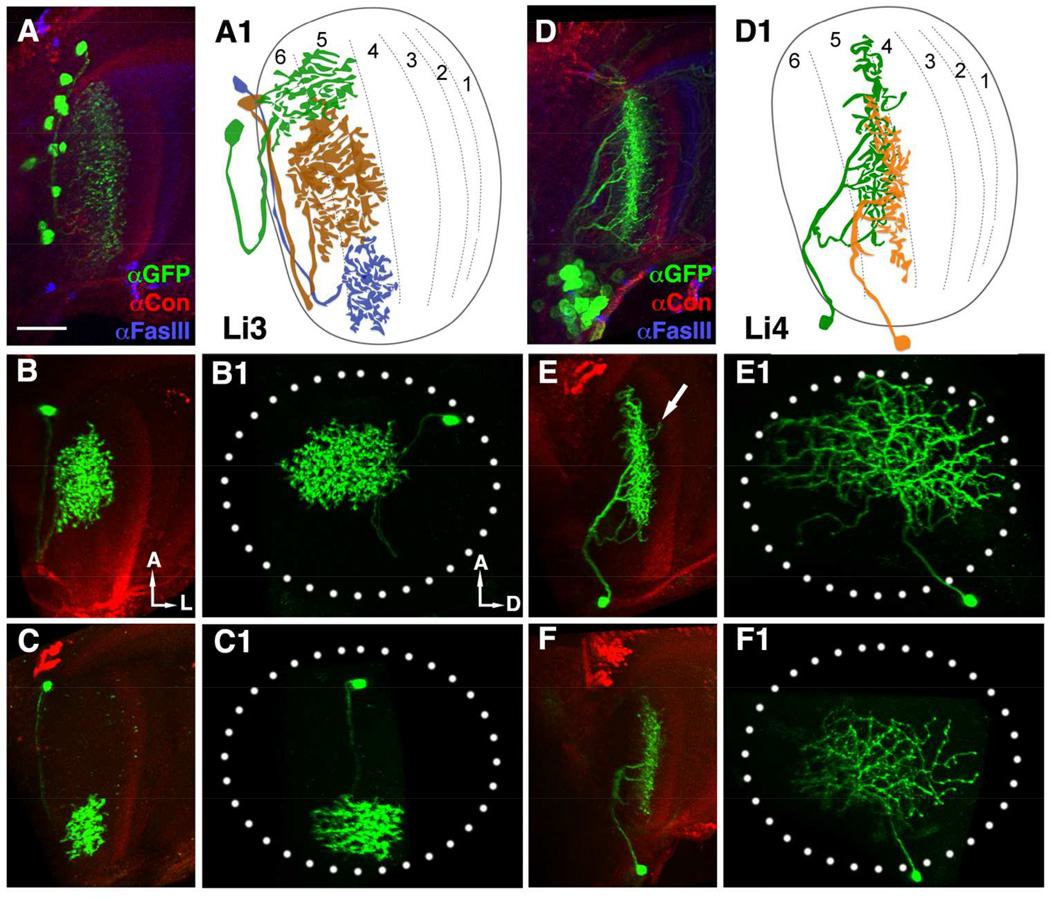
GFP expression driven by specific lobula drivers revealed neurons with distinct morphological patterns in some of the lobula’s six strata (Figure 2 and data not shown). Based on the locations of their dendritic arborizations and cell bodies, we found three drivers, R12H12, R22D06, and R41C07, that label neurons resembling previously reported lines LC11, LC10, and LC6, respectively (Table 1 and data not shown). We identified additional drivers for two novel types of lobula intrinsic neurons, Li3 and Li4, as well as two LC types of lobula columnar neurons, tentatively identified as LC15 and LC16 (Table 1). Li3-Gal4 (R43E05) labels 11–13 cell bodies dispersed in each lobe (n=5 brains), located mostly in the anterior lobula with dendritic arbors in lobula strata 5 and 6 (Figure 2A). Using the flip-out genetic mosaic method (Wong et al., 2002), we analyzed the morphologies of single Li3 neurons. The “bush-like” arborization of each Li3 covered a small region of the lobula neuropil’s retinotopic field (about 11–20%: Figures 2B1 and C1). In contrast, Li4-Gal4 (NP1582 and R89D06) labels a cluster of 26–32 neurons (n=9 brains), each of which arborizes over a large area of lobula neuropil (about 45–78%: Figures 2E1 and F1). While most neurites arborized in lobula stratum 5, they also extended small branches that invaded lobula stratum 4 (Figure 2E). These morphological features indicate that Li3 and Li4 are two different types of lobula intrinsic neurons.
Figure 2.
Lobula intrinsic neurons, Li3 and Li4, extend dendritic arbors in deep strata of the lobula. (A, D) Li3- and Li4-specific Gal4 drivers drive expression of a membrane-tethered GFP marker (mCD8::GFP, green) to reveal the dendritic arbors of these cells. Anti-Fas III (blue) and anti-Connectin (magenta) immunolabeling were used to mark lobula strata 1, 2, 4, 5 and 3, respectively (Gao et al., 2008). In each optic lobe, about 12 Li3 neurons elaborate dendritic arbors spanning the entire lobula strata 5 and 6 (A). Approximately 30 Li4 neurons, clustered in the posterior lobula cortex have dendritic arbors covering the entire lobula stratum 5.
(A1 and D1) Schematic drawings of Li3 (A1) and Li4 (D1) neurons.
(B-C1, E-F1) Single Li3 (B-C1) or Li4 (E-F1) neurons labeled with mCD8::GFP using the single-cell flip-out technique and visualized in side (B,C,E,F) and plan views (B1,C1,E1,F1) by anti-GFP (green). (B-C1) Each Li3 neuron elaborates a dense dendritic arbor spanning an area corresponding to about 11–20% of the visual field (dotted ellipse, B1 and C1) in lobula strata 5 and 6 (B and C). (E-F1) Each Li4 neuron extends a dendritic arbor that covers a large portion (45–78%) of the visual field (dotted ellipse) in lobula stratum 5 (E1, F1). Occasional distal branches of Li4 invade the neighboring lobula stratum 4 (arrow, E).
(A, B, C, D, E, F) ventral views; (B1, C1, E1, F1) frontal views. A: anterior; L: lateral; D: dorsal. Scale bar: 20 µm in A for A-F1.
Li3, Li4, LT11 and LC14 are potential target neurons for chromatic Tm neurons
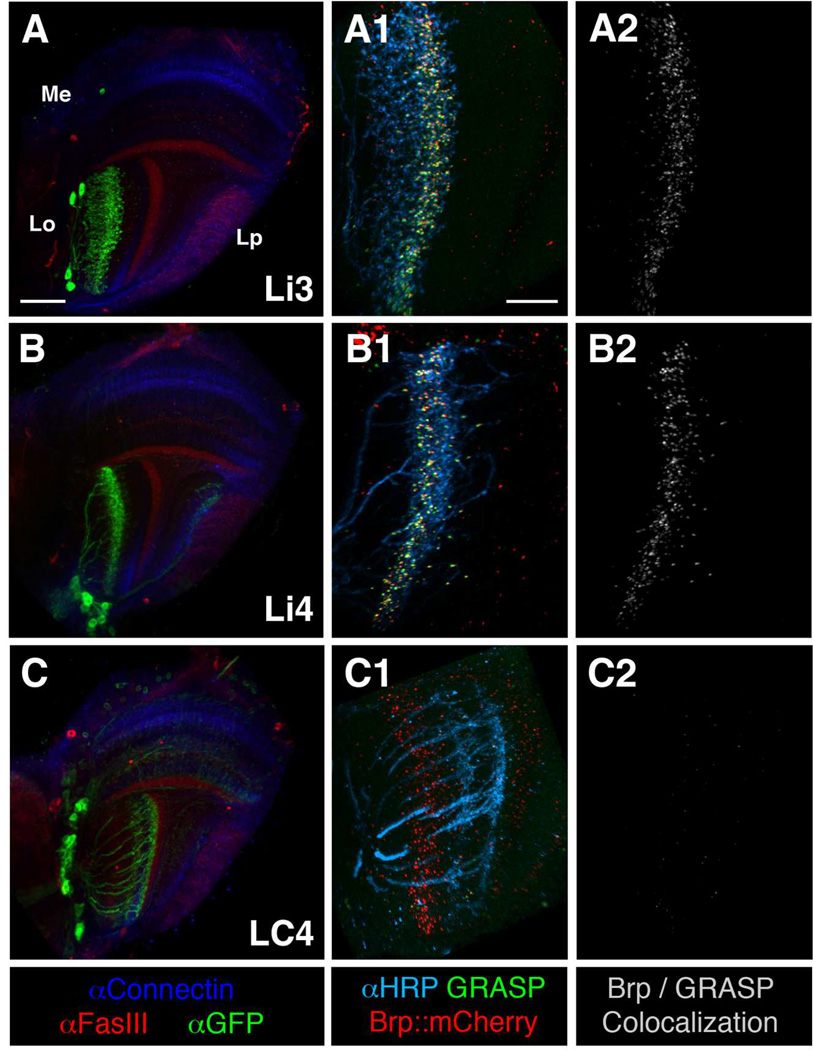
We used the GRASP method to screen for lobula neurons that form membrane contacts with the chromatic Tm neurons. In adult flies we expressed one membrane-tethered split GFP component (spGFP11::CD4) in chromatic Tm neurons and the other (i.e. spGFP1–10::CD4) in different combinations of lobula neuron subsets, and examined reconstituted GFP fluorescence signals in the lobula’s six different strata (see Materials and Methods for details). To differentiate genuine synapses from mere membrane appositions, we additionally marked presynaptic sites of the chromatic Tm neurons using the presynaptic marker BrpD3::mCherry, and examined co-localization of GRASP signals and BrpD3::mCherry. We screened 37 lobula drivers (which marked 24 pathways) from the NP Gal4 enhancer-trap lines and eight additional drivers (marking five types of lobula neurons) from the Janelia FlyLight GAL4 collection (Table 1). We found that four types of lobula neurons (Li3, Li4, LT11, and LC14) consistently showed numerous puncta at which BrpD3::mCherry colocalized with GRASP in deep lobula strata (Lo4–5) across the entire visual field (Figures 3 and 4). These results suggested that these lobula neurons are likely to be postsynaptic partners for chromatic Tm neurons. In contrast, the other lobula neuron subsets, including LC4, showed very little or no BrpD3::mCherry/GRASP co-localization (Figure 3C2), suggesting that they either form limited synaptic contacts with chromatic Tm neurons or lacked these entirely.
Figure 3.
Mapping chromatic lobula neurons by the GRASP method with presynaptic labeling. Lobula intrinsic neurons Li3 and Li4 receive densely packed synaptic contacts from the chromatic Tm neurons across lobula stratum 5. The lobula projection neuron LC4 forms very few or no synaptic contacts with the chromatic Tm neurons. Left panels (A–C): GAL4 lines used to express a membrane-tethered GFP marker (mCD8GFP, green) in three different types of lobula neurons (as indicated) to reveal their morphologies. FasIII (red) and Connectin (blue) antibodies labeling discrete medulla and lobula strata were used as layer-specific landmarks. Middle and right panels (A1-C2): ortC1a-LexA::VP16 drive expression of a split-GFP moiety (spGFP11-HA::CD4), and an active zone marker, BrpD3::mCherry, in four types of chromatic Tm neurons, Tm5a/b/c and Tm20. In the same animals, various lobula GAL4 lines express the other split-GFP (spGFP1-10::CD4) and the membrane marker HRP-CD2 in specific lobula neurons (as indicated). Functional GFP (GRASP) reconstituted at the membrane contacts of these two groups of neurons was examined for native fluorescence (green). Colocalizations (white) of the active zone marker (BrpD3::mCherry) and the GRASP signal indicated potential synaptic contacts (right panels, A2-C2). Lobula neurons are outlined by anti-HRP immunolabeling (cyan, middle panels A1-C1). Scale bar, 30 µm in A for A-C; 15 µm in A1 for A1-C2.
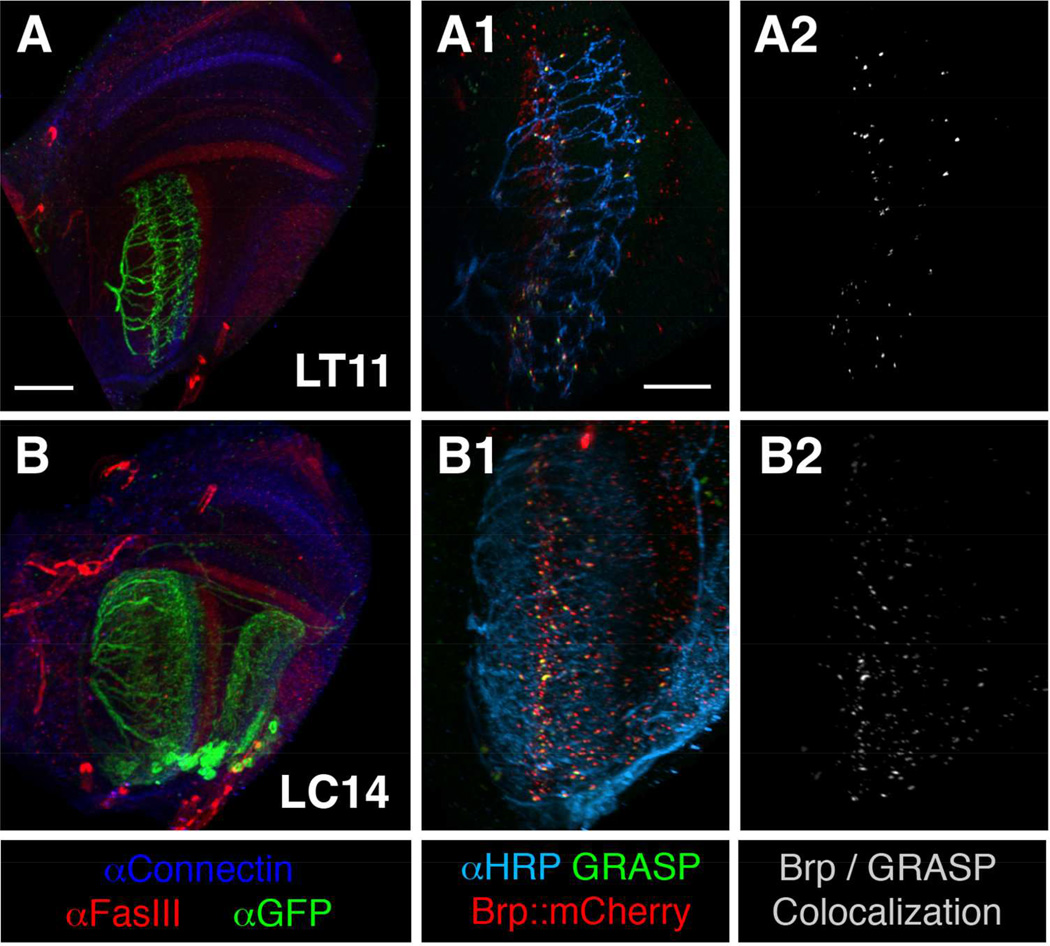
Figure 4.
The lobula projection neurons LT11 and LC14 receive sparse synaptic contacts from the chromatic Tm neurons in lobula strata 3–5. Left panels (A, B): GAL4 expressing lobula neurons were outlined by a membrane GFP (green). FasIII (red) and Connectin (blue) immuno-labeling showed the layer pattern of different neuropils. Middle panels (A1, B1): In GRASP analysis, GRASP signals (green) between presynaptic Tm neurons (Tm5a/b/c and Tm20; αHRP (cyan)) and projection neurons LT11 (A1) and LC14 (B1) were sparsely colocalized with presynaptic active zone marker BrpD3::mCherry (red). Right panels (A2, B2): Colocalizations (white) were defined by using the coloc module of Imaris software. Scale bar, 30 µm in A for A-B; 15 µm in A1 for A1-B2.
Single-cell GRASP reveals different densities of synaptic contacts between lobula intrinsic and projection neurons
The aforementioned GRASP analyses also revealed that the lobula intrinsic neurons Li3 and Li4 (Figure 3) appear to form more synaptic contacts with chromatic Tm neurons than do the lobula projection neurons, LT11 and LC14 (Figure 4). The apparent differences in contact density could result from selective innervation of different Tm subtypes or different synaptic numbers in lobula neuron subclasses. Furthermore, determining the innervating frequency and selectivity of Tm subtypes to different lobula neurons would be important for understanding the integration mechanisms of chromatic channels in the lobula. To address these issues, we performed a single-cell GRASP assay (Figure 5A; Karuppudurai et al., 2014) to examine the potential contacts between different chromatic Tm neurons and two types of lobula neurons, the intrinsic neuron Li4 and the projection neuron LT11. Single chromatic Tm neurons expressed the presynaptic marker BrpD3:mCherry in addition to an HA-tagged split-GFP, spGFP11::CD4::HA, while the lobula neurons expressed the other split-GFP component (i.e. spGFP1–10::CD4).
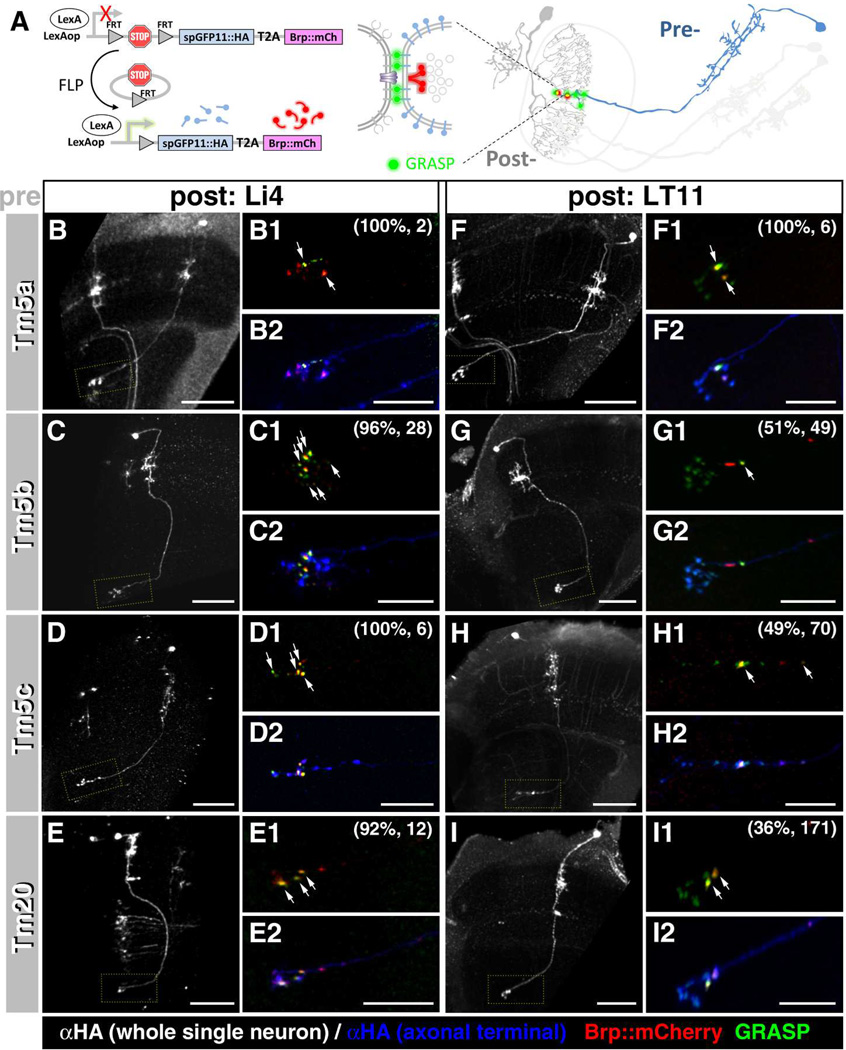
Figure 5.
Single-cell GRASP reveals the number and specificity of synapses between chromatic Tm neurons and their candidate lobula neuron partners.
(A) Schematic illustration of the single-cell GRASP method. After flipase-induced mitotic recombination, a single Tm neuron co-expresses one split-GFP moiety (spGFP11) and the presynaptic active zone marker (Brp-mCherry) while a postsynaptic lobula neuron expresses the other split-GFP (spGFP1-10) in the same brain. Colocalization (yellow, arrows) of the GRASP signal (green) and Brp-mCherry (red) suggests the presence of synaptic contacts between the respective Tm and lobula neurons. (B–I) Single-cell GRASP analyses shows that all four types of Tm neurons form multiple synapses with lobula intrinsic neuron Li4 (B-E2). (B1-I1, B2-I2) high magnification views of (B–I) marked by yellow dotted rectangles. Approximately half of the examined Tm neurons formed one or two synapses with the visual projection neuron LT11 (F-I2) (see main text for details). Specific Tm types were identified based on their morphologies, visualized by anti-HA immunolabeling (white in B-I and blue in B2-I2). The percentage of Tm neurons forming synapses with Li4 or LT11 neurons and the number of neurons examined are shown in parentheses (B1-I1). Scale bar, 30 µm for B-I, 10 µm for B2-I2.
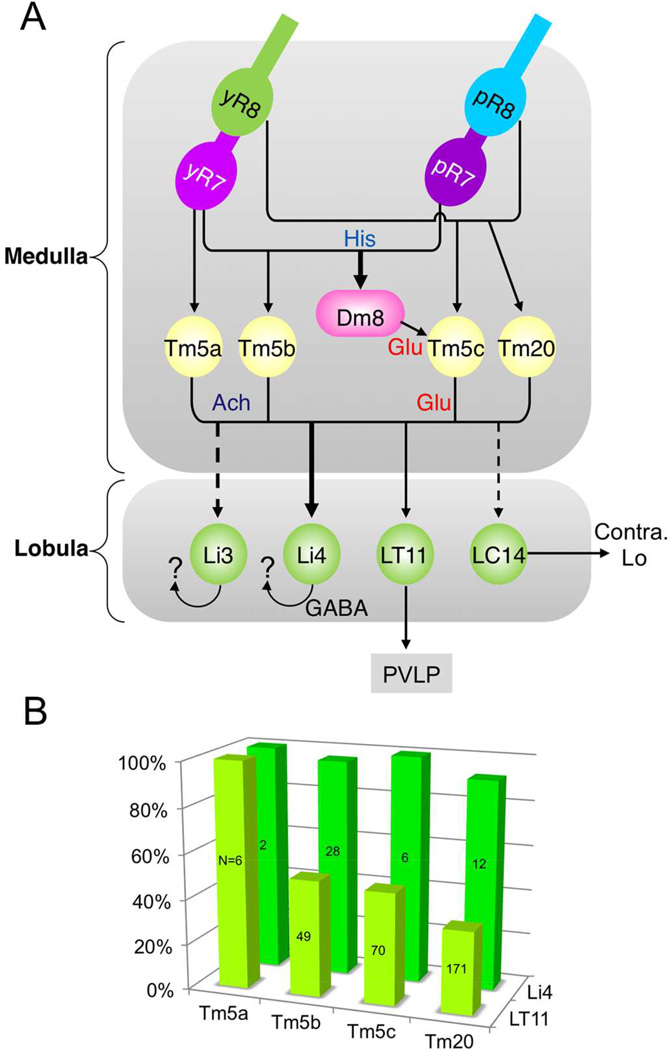
We found that Li4 neurons form many synaptic contacts with all four types of chromatic Tm neurons (Tm5a: 100% [n=2/2]; Tm5b: 96.4% [n=27/28]; Tm5c: 100% [n=6/6]; Tm20: 91.7% [n=11/12]; Figures 5B–E, 7B). Multiple GRASP/Brp puncta were observed at the axon terminals of Tm neurons in lobula stratum 5 (Figure 5), suggesting that each Tm neuron forms multiple synapses with one or more Li4 neurons. In contrast, only one or two GRASP/Brp double-labeled puncta were observed between the lobula projection neuron LT11 and individual Tm neurons (Figures 5F–I, 7B) even though, as indicated by the GRASP puncta lacking BrpD3::mCherry, they apparently form multiple membrane contacts. The number of synapses is also lower for LT11, there being fewer Tm neuron synapses with LT11 (for Tm5a: 100% [n=6/6]; Tm5b: 51% [n=25/49]; Tm5c: 48.5% [n=34/70]; and Tm20: 36.3% [n=62/171]). In summary, the lobula intrinsic neuron Li4 receives synaptic input from all four types of chromatic Tm neuron both in large numbers and at high densities while the projection neuron LT11 is postsynaptic to the chromatic Tm neurons with greater selectivity and at lower frequency and density.
Figure 7.
(A) Connectivity graph of Drosophila chromatic visual circuit. Arrows represent directionality and weighted by synapse numbers. (B) Bar graph of the synaptic frequency of Tm5a/b/c/20 neurons to Li4 (green) and LT11 (chartreuse yellow), calculated from Figure 5.
“Two-tag” double labeling EM
LT11 is a single centripetal lobula tangential (LT) neuron with dendrites arborizing throughout the entire lobula depth encompassed by strata 3–5 and is involved in spectrum selective phototaxis behavior (Otsuna and Ito, 2006; Otsuna et al., 2014). Our GRASP results suggest that LT11 receives synaptic inputs from all four classes of chromatic Tm neurons but that the number of these contacts is much lower than the inputs these form to Li4. To examine the synaptic contacts between Tm and LT neurons at the electron microscopic (EM) level, we have developed a “two-tag” double labeling system that can highlight both pre- and postsynaptic neurons in the same preparation, by combining two orthogonal expression systems and two different peroxidases, HRP (horseradish peroxidase; Larsen et al., 2003; Edwards and Meinertzhagen, 2009) and APX (ascorbate peroxidase; Martell et al., 2012). In this system, Gal4-driven HRP::DsRed::GPI (Han et al., 2012) labels the cell membrane of postsynaptic dendrites (LT11), whereas LexA-driven mito::APX::HA, APX41F fused to a mitochondrion matrix targeting peptide highlights mitochondria and identifies presynaptic terminals (Tm5c). We took advantage of mitochondria because they are present abundantly in presynaptic terminals but their membrane is separate from the cell’s plasma membrane (Figures 6B–C and 6D). We validated cell-specific mitochondrial labeling by anti-HA immunohistochemistry at the light microscopic level (Figures 6A and A1). The double-labeled optic lobes were reacted in a single reaction with 3,3-diaminobenzidine (DAB) to visualize both peroxidases simultaneously and sections of the lobula were subjected to EM analyses to seek appositions between synaptic profiles (Figures 6B–C and E).
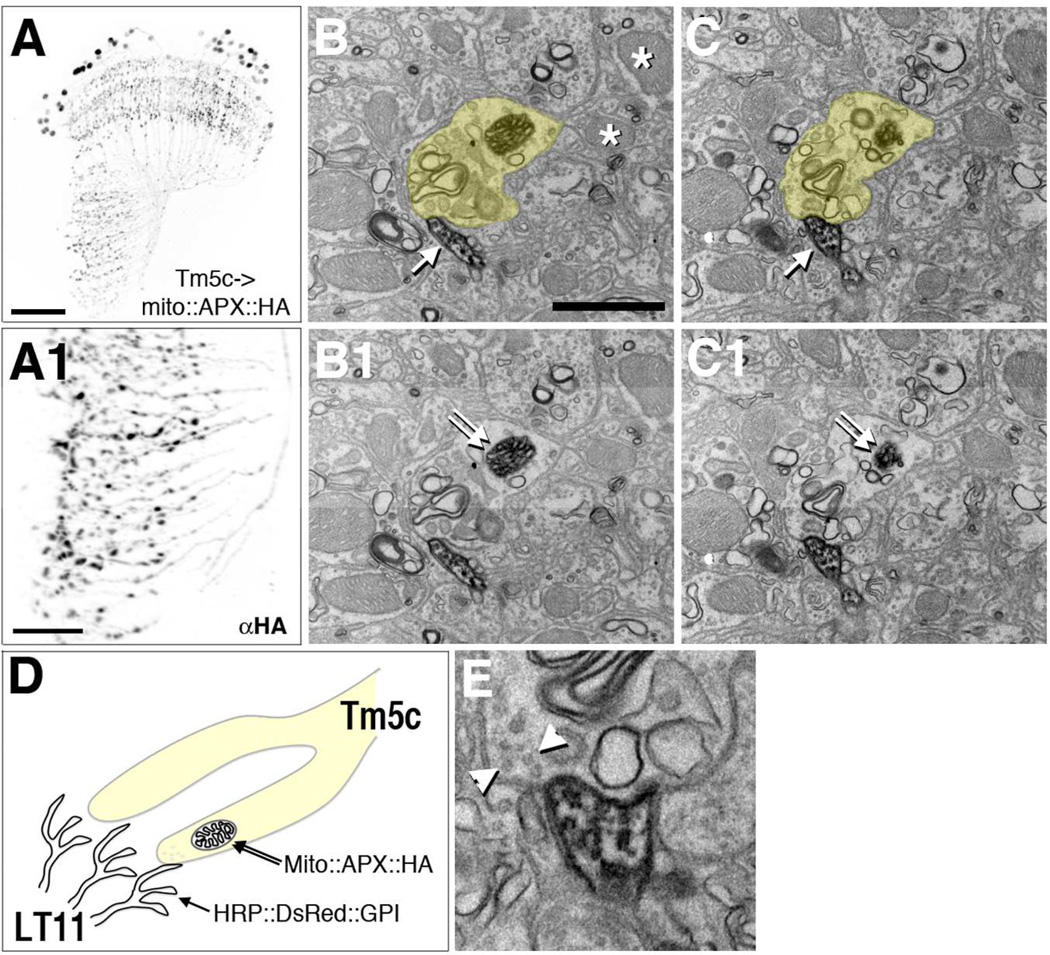
Figure 6.
“Two-tag” EM double labeling reveals Tm5c to LT11 contacts.
Tm5c neurons express HA-tagged APX targeted to the mitochondrion matrix (mito::APX::HA). Confocal microscopic analyses reveal Tm5c mitochondria labeled by anti-HA (αHA) in axon terminals in the lobula (A1) as well as dendrites and cell bodies (A). (A1) high magnification of the lobula of (A). Scale bar, 30 µm for A, 10 µm for A1.
Tm5c axon terminals identified by the presence of APX-labeled mitochondria and LT11 dendritic membrane labeled by membrane-tethered HRP (HRP::DsRed::GPI). (B-C1, E) EM sections of double-labeled lobula neuropil stained with DAB. A Tm5c mitochondrion identified by its dense DAB staining and mitochondrial morphology (double arrows, B1 and C1). Unlabeled mitochondria are indicated with asterisks (B). A Tm5c terminal (with a labeled mitochondria) is pseudocolored yellow in B and C, and raw images of the same area are presented in B1 and C1, respectively. LT11 dendrites identified by the DAB staining of the cytoplasmic membrane (single arrows in consecutive sections, B and C).
(D) A schematic illustration of “two-tag” double labeling method.
(E) High magnification view of the contact between Tm5c terminal and LT11 dendrites. Arrowheads indicate synaptic vesicles in Tm5c, compatible with the terminal being presynaptic to LT11 dendrites.
Consecutive 50-nm EM sections of double-labeled preparations reveal numerous scattered dendritic profiles in lobula stratum Lo4 (Figures 6B–C). These reveal a rather dense pattern of innervation. A single Tm5c terminal (Figures 6B and 6C) with a labeled mitochondrion (Figures 6B1 and 6C1), clearly darker than two unlabeled mitochondria in other nearby cell profiles (Figure 6B) contacts an LT11 dendrite through consecutive sections. In a following section, evidence for a presynaptic role in Tm5c is revealed by the presence of clear ~30-nm synaptic vesicles (Figure 6E). At many synapses in Drosophila, a presynaptic site is revealed by the presence of a bipartite T-bar ribbon, but these are seen neither in all brain regions (for example not in all synapses of the mushroom body calyx, Butcher et al., 2012, nor at all neuromuscular synapses, Atwood et al., 1993), and were also not seen at this depth in the lobula, despite their presence at more distal lobula strata 1 and 2 (Shinomiya et al., 2014). It is possible that during DAB incubation they become lost or less visible, and would be seen in unlabeled tissue. It is also theoretically possible they would be seen at other synapses than the contacts we illustrate. The probability to capture in a single image the tiny profile of a T-bar ribbon along with the mitochondrial profile used to identify the presynaptic neuron at sites of contact with HRP labeled postsynaptic membrane, would in any case depend on the number and distribution of synaptic contacts, and the low probability to capture all three organelles in a single image, for which a cumulus of presynaptic vesicles is a reliable proxy. What is clear is that membrane contact between Tm5c and LT10 is very often seen between labeled cell partners.
DISCUSSION
Primary chromatic pathway to lobula intrinsic and output neurons
The lobula is the gateway of the visual system to the central brain. Its deep strata receive retinotopic inputs from medulla Tm and TmY neurons and outputs to various central brain regions. By screening the NP enhancer trap collections, a previous study identified two classes of visual projection neurons, the LC and LT types neurons, which communicate between the lobula and central brain (Otsuna and Ito, 2006). Subsequent studies using the NP collection further identified two complex neuronal types, CT and CC, which connect the lobula with multiple brain regions (Shinomiya et al., 2011). In this study, we extended these efforts by examining the recently available Janelia collection and identified three classes of lobula intrinsic neurons and two likely new LC-type neurons (Table 1). Using the flip-out mosaic method, we further examined single-cell morphologies of the lobula intrinsic neurons, Li3 and Li4. The Li3 and Li4 neurons extend medium and large dendritic trees in distinct lobula strata, respectively. The difference in their dendritic tree sizes and stratum distribution reflects their receptive field sizes and perhaps, their distinct roles in spatial integration of retinotopic inputs.
Using this composite collection of lobula Gal4 drivers, which label 28 classes of lobula neurons (Table 1), we screened for lobula neurons that receive direct synaptic inputs from chromatic Tm neurons. Using a combination of the GRASP method and presynaptic labeling, we found four types of lobula neurons that form potential synaptic contacts with chromatic Tm neurons. These include two visual projection neurons (VPN), LT11 and LC14, and two lobula intrinsic neurons, Li3 and Li4. Interestingly, the intrinsic neurons appeared to form more synaptic contacts with chromatic Tm neurons than did the VPNs, as judged by their more numerous Brp/GRASP puncta. It may be that the number of these puncta reveals the respective strengths of the pathways, but the further implications of synapse number must in fact await physiological studies. To provide a better estimate of the specificity, spatial distributions and numbers of synaptic contacts, we carried out single-cell GRASP analyses for Li4 and LT11. We found that LT11 receives mostly single synaptic contacts from approximately half of the Tm5b, Tm5c and Tm20 neurons but does so with 100% of Tm5a. Tm5c and Tm20 are present in essentially every medulla column and receive retinotopic R8 inputs, regardless of whether these are of the yellow or pale types. In contrast, Tm5a receives inputs from the yellow type only and not the pale type of R8 photoreceptors and therefore likely responds maximally to light in the green region of the spectrum (Karuppudurai et al., 2014). LT11 has been implicated in spectrum-selective phototatic behavior (Otsuna et al., 2014). Whether this modest connection specificity between LT11 and different Tm neurons reflects the spectrum-selective function of LT11 remains to be determined.
In contrast to LT11, Li4 receives multiple synaptic contacts from nearly 100% of all four types of chromatic Tm neuron classes. Our single-cell morphological analyses show that each Li4 neuron extends a large dendritic tree that covers some 60% of the lobula neuropil. Given the high areal density of its synapses and the large size of its dendritic tree, Li4 likely integrates inputs from all four types of chromatic Tm neurons over a large receptive field and provides inputs in turn to other lobula neurons. Based on its immunolabeling with anti-GABA, Li4 appears to be GABAergic (Lin and Lee, unpublished), suggesting that its output is inhibitory. It is tempting to speculate that Li4 might serve to “normalize” chromatic inputs over a large receptive field. The functions of Li4 in processing color information remain to be determined from behavioral and activity studies. It is interesting to note that the chromatic Tm neurons have mixed neurotransmitter phenotypes: Tm5c is glutamatergic while Tm5a and Tm5b have a cholinergic phenotype (Karuppudurai et al., 2014). Thus, both LT11 and Li4 must express receptors for both glutamate and acetylcholine to receive all four chromatic Tm inputs.
Determining the connectivity of large neurons in complex neuropils
Identifying connections between specific neurons in the neuropil requires electron microscopy and this can in principle be achieved using powerful connectomic approaches (Takemura et al., 2013). However, the approach requires reconstructing most neurites and identifying cell types solely based on the morphologies of reconstructed profiles. It remains very challenging by this method to assess the connectivity of large and complex neurons, such as LT11, which elaborate large dendritic trees and receive inputs from distant neurons. Indeed, in a recent connectome study for the medulla neuropil, most tangential neurons have not been reconstructed (Takemura et al., 2013). To overcome this problem, we combined light and electron microscopic approaches. We took advantage of the Gal4 drivers that label specific presynaptic neurons in the medulla and postsynaptic neurons in the lobula, and identified candidate connections based on a modified GRASP method (Karuppudurai et al., 2014). To identify both pre- and postsynaptic neurons at the ultrastructural level, we additionally developed a “two-tag” double-labeling method for electron microscopic analyses. We expressed a membrane-tethered HRP in LT11 to identify its dendrites (Han et al., 2014). To identify presynaptic neurons, we complementarily attempted to label the ubiquitous profiles of mitochondria, which are abundant in the presynaptic terminals but which have a membrane that is separated from the cytoplasmic membrane. In various preliminary experiments, we confirmed that HRP fails to label mitochondria. HRP is in fact inactive in most cellular compartments, probably because its four structurally essential disulfide bonds and two Ca2+ binding sites do not form in reducing and Ca2+-scavenged environments (Hopkins et al., 2000). We therefore used an alternative peroxidase, an engineered ascorbate peroxidase (APXW41F), that withstands strong fixation for EM, gives excellent EM imaging of mammalian organelles, and is active in the mitochondrial matrix (Martell et al., 2012). We further optimized the incubation parameters to co-express both peroxidase signals in the same brain preparation. With this method, we successfully labeled both pre- (Tm5c) and postsynaptic (LT11) neurons in single preparations and confirmed their synaptic connections at the EM level. We envision this method would be generally applicable to map the connectivity of other complex or amorphous neuropils in Drosophila as well as in the brains of other genetically tractable animals.
ACKNOWLEDGMENTS
Support or grant information:
This work was supported by the Intramural Research Programs of the National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development (grant Z01-HD008776 to C.-H.L) and the Natural Sciences and Engineering Research Council, Ottawa, Canada (Discovery grant A-0000065 to I.A.M.), and Ministry of Science and Technology, Taiwan (MOST103-2311-B-016 −001 -MY2 to T.-Y.L).
We thank Dr. Chun Han for providing UAS-HRP::DsRed::GPI flies, Dr. Greg Macleod (Florida Atlantic University) for suggesting the APX mitochondrial targeting sequences, and Dr. Julie Simpson (Janelia Research Campus) for valuable discussions and reagents for alternative approaches to two-color electron microscopy.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ROLE OF AUTHORS
All authors had access to all data presented in this study. The authors accept responsibility for the integrity and accuracy of the data and data analysis. Study concept and design: TYL, CHL, IAM. Preparation and acquisition of data: TYL, JL, KS, CYT, ZL. Analysis and interpretation of data: TYL, CHL. Drafting of the manuscript: TYL, IAM, CHL.
LITERATURE CITED
- Atwood HL, Govind CK, Wu C-F. Differential ultrastructure of synaptic terminals on ventral longitudinal abdominal muscles in Drosophila larvae. J Neurobiol. 1993;24:1008–1024. doi: 10.1002/neu.480240803. [DOI] [PubMed] [Google Scholar]
- Bell ML, Earl JB, Britt SG. Two types of Drosophila R7 photoreceptor cells are arranged randomly: a model for stochastic cell-fate determination. J Comp Neurol. 2007;502:75–85. doi: 10.1002/cne.21298. [DOI] [PubMed] [Google Scholar]
- Butcher NJ, Friedrich AB, Lu Z, Tanimoto H, Meinertzhagen IA. Different classes of input and output neurons reveal new features in microglomeruli of the adult Drosophila mushroom body calyx. J Comp Neurol. 2012;520:2185–2201. doi: 10.1002/cne.23037. [DOI] [PubMed] [Google Scholar]
- Cajal SR, Sanchez D. Contibucion al conocimiento de los centros nerviosos de los insectos. Trab Lab Invest Biol. 1915;13:1–167. [Google Scholar]
- Cardona A, Saalfeld S, Schindelin J, Arganda-Carreras I, Preibisch S, Longair M, Tomancak P, Hartenstein V, Douglas RJ. TrakEM2 software for neural circuit reconstruction. PLoS One. 2012;7:e38011. doi: 10.1371/journal.pone.0038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Akin O, Nern A, Tsui CY, Pecot MY, Zipursky SL. Cell-type-specific labeling of synapses in vivo through synaptic tagging with recombination. Neuron. 2014;81:280–293. doi: 10.1016/j.neuron.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen F, Zube C, Andlauer TF, Wichmann C, Fouquet W, Owald D, Mertel S, Leiss F, Tavosanis G, Luna AJ, Fiala A, Sigrist SJ. Presynapses in Kenyon cell dendrites in the mushroom body calyx of Drosophila. J Neurosci. 2011;31:9696–9707. doi: 10.1523/JNEUROSCI.6542-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TN, Meinertzhagen IA. Photoreceptor neurons find new synaptic targets when misdirected by overexpressing runt in Drosophila. J Neurosci. 2009;29:828–841. doi: 10.1523/JNEUROSCI.1022-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, Bargmann CI. GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- Fischbach K-F, Dittrich APM. The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res. 1989;258:441–475. [Google Scholar]
- Franceschini N, Kirschfeld K, Minke B. Fluorescence of photoreceptor cells observed in vivo. Science. 1981;213:1264–1267. doi: 10.1126/science.7268434. [DOI] [PubMed] [Google Scholar]
- Gao S, Takemura SY, Ting CY, Huang S, Lu Z, Luan H, Rister J, Thum AS, Yang M, Hong ST, Wang JW, Odenwald WF, White BH, Meinertzhagen IA, Lee CH. The neural substrate of spectral preference in Drosophila. Neuron. 2008;60:328–342. doi: 10.1016/j.neuron.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MD, Scott K. Motor control in a Drosophila taste circuit. Neuron. 2009;61:373–384. doi: 10.1016/j.neuron.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Song Y, Xiao H, Wang D, Franc NC, Jan LY, Jan YN. Epidermal cells are the primary phagocytes in the fragmentation and clearance of degenerating dendrites in Drosophila. Neuron. 2014;81:544–560. doi: 10.1016/j.neuron.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie RC, Franceschini N, McIntyre PD. Electrophysiological analysis of fly retina II. Spectral and polarisation sensitivity in R7 and R8. J Comp Physiol A. 1979;133:23–39. [Google Scholar]
- Hardie RC, Kirschfeld K. Ultraviolet sensitivity of fly photoreceptor-R7 and photoreceptor-R8—evidence for a sensitizing function. Biophys Struct Mech. 1983;9:171–180. [Google Scholar]
- Hertel H. Chromatic properties of identified interneurons in the optic lobes of the bee. J Comp Physiol A. 1980;137:215–231. [Google Scholar]
- Hopkins C, Gibson A, Stinchcombe J, Futter C. Chimeric molecules employing horseradish peroxidase as reporter enzyme for protein localization in the electron microscope. Methods Enzymol. 2000;327:35–45. doi: 10.1016/s0076-6879(00)27265-0. [DOI] [PubMed] [Google Scholar]
- Jagadish S, Barnea G, Clandinin TR, Axel R. Identifying functional connections of the inner photoreceptors in Drosophila using Tango-Trace. Neuron. 2014;83:630–644. doi: 10.1016/j.neuron.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, Iyer N, Fetter D, Hausenfluck JH, Peng H, Trautman ET, Svirskas RR, Myers EW, Iwinski ZR, Aso Y, DePasquale GM, Enos A, Hulamm P, Lam SC, Li HH, Laverty TR, Long F, Qu L, Murphy SD, Rokicki K, Safford T, Shaw K, Simpson JH, Sowell A, Tae S, Yu Y, Zugates CT. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karuppudurai T, Lin TY, Ting CY, Pursley R, Melnattur KV, Diao F, White BH, Macpherson LJ, Gallio M, Pohida T, Lee CH. A hard-wired glutamatergic circuit pools and relays UV signals to mediate spectral preference in Drosophila. Neuron. 2014;81:603–615. doi: 10.1016/j.neuron.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen CW, Hirst E, Alexandre C, Vincent JP. Segment boundary formation in Drosophila embryos. Development. 2003;130:5625–5635. doi: 10.1242/dev.00867. [DOI] [PubMed] [Google Scholar]
- Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ellisman MH, Ting AY. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni EO, Celik A, Wernet MF, Vasiliauskas D, Johnston RJ, Cook TA, Pichaud F, Desplan C. Iroquois complex genes induce co-expression of rhodopsins in Drosophila. PLoS Biol. 2008;6(4):e97. doi: 10.1371/journal.pbio.0060097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows LA, Gell D, Broadie K, Gould AP, White RA. The cell adhesion molecule, connectin, and the development of the Drosophila neuromuscular system. J Cell Sci. 1994;107(Pt 1):321–328. doi: 10.1242/jcs.107.1.321. [DOI] [PubMed] [Google Scholar]
- Melnattur KV, Pursley R, Lin TY, Ting CY, Smith PD, Pohida T, Lee CH. Multiple redundant medulla projection neurons mediate color vision in Drosophila. J Neurogenet. 2014;28:374–388. doi: 10.3109/01677063.2014.891590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikeladze-Dvali T, Desplan C, Pistillo D. Flipping coins in the fly retina. Curr Top Dev Biol. 2005;69:1–15. doi: 10.1016/S0070-2153(05)69001-1. [DOI] [PubMed] [Google Scholar]
- Morante J, Desplan C. The color-vision circuit in the medulla of Drosophila. Curr Biol. 2008;18:553–565. doi: 10.1016/j.cub.2008.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuna H, Ito K. Systematic analysis of the visual projection neurons of Drosophila melanogaster. I. Lobula-specific pathways. J Comp Neurol. 2006;497:928–958. doi: 10.1002/cne.21015. [DOI] [PubMed] [Google Scholar]
- Otsuna H, Shinomiya K, Ito K. Parallel neural pathways in higher visual centers of the Drosophila brain that mediate wavelength-specific behavior. Front Neural Circuits. 2014;8:8. doi: 10.3389/fncir.2014.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulk AC, Phillips-Portillo J, Dacks AM, Fellous JM, Gronenberg W. The processing of color, motion, and stimulus timing are anatomically segregated in the bumblebee brain. J Neurosci. 2008;28:6319–6332. doi: 10.1523/JNEUROSCI.1196-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Zipursky SL. Design principles of insect and vertebrate visual systems. Neuron. 2010;66:15–36. doi: 10.1016/j.neuron.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaitmann C, Garbers C, Wachtler T, Tanimoto H. Color discrimination with broadband photoreceptors. Curr Biol. 2013;23:2375–2382. doi: 10.1016/j.cub.2013.10.037. [DOI] [PubMed] [Google Scholar]
- Schnaitmann C, Vogt K, Triphan T, Tanimoto H. Appetitive and aversive visual learning in freely moving Drosophila. Front Behav Neurosci. 2010;4:10. doi: 10.3389/fnbeh.2010.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomiya K, Karuppudurai T, Lin TY, Lu Z, Lee CH, Meinertzhagen IA. Candidate neural substrates for off-edge motion detection in Drosophila. Curr Biol. 2014;24:1062–1070. doi: 10.1016/j.cub.2014.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomiya K, Matsuda K, Oishi T, Otsuna H, Ito K. Flybrain neuron database: a comprehensive database system of the Drosophila brain neurons. J Comp Neurol. 2011;519:807–833. doi: 10.1002/cne.22540. [DOI] [PubMed] [Google Scholar]
- Snow PM, Bieber AJ, Goodman CS. Fasciclin III: a novel homophilic adhesion molecule in Drosophila. Cell. 1989;59:313–323. doi: 10.1016/0092-8674(89)90293-6. [DOI] [PubMed] [Google Scholar]
- Strausfeld NJ, Okamura JY. Visual system of calliphorid flies: organization of optic glomeruli and their lobula complex efferents. J Comp Neurol. 2007;500:166–188. doi: 10.1002/cne.21196. [DOI] [PubMed] [Google Scholar]
- Takemura SY, Bharioke A, Lu Z, Nern A, Vitaladevuni S, Rivlin PK, Katz WT, Olbris DJ, Plaza SM, Winston P, Zhao T, Horne JA, Fetter RD, Takemura S, Blazek K, Chang LA, Ogundeyi O, Saunders MA, Shapiro V, Sigmund C, Rubin GM, Scheffer LK, Meinertzhagen IA, Chklovskii DB. A visual motion detection circuit suggested by Drosophila connectomics. Nature. 2013;500:175–181. doi: 10.1038/nature12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CY, Herman T, Yonekura S, Gao S, Wang J, Serpe M, O'Connor MB, Zipursky SL, Lee CH. Tiling of R7 axons in the Drosophila visual system is mediated both by transduction of an activin signal to the nucleus and by mutual repulsion. Neuron. 2007;56:793–806. doi: 10.1016/j.neuron.2007.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CY, Gu S, Guttikonda S, Lin TY, White BH, Lee CH. Focusing transgene expression in Drosophila by coupling Gal4 with a novel split-LexA expression system. Genetics. 2011;188:229–233. doi: 10.1534/genetics.110.126193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CY, McQueen PG, Pandya N, Lin TY, Yang M, Reddy OV, O'Connor MB, McAuliffe M, Lee CH. Photoreceptor-derived activin promotes dendritic termination and restricts the receptive fields of first-order interneurons in Drosophila. Neuron. 2014;81:830–846. doi: 10.1016/j.neuron.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AM, Wang JW, Axel R. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell. 2002;109:229–241. doi: 10.1016/s0092-8674(02)00707-9. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Desplan C, Heisenberg M. Contribution of photoreceptor subtypes to spectral wavelength preference in Drosophila. Proc Natl Acad Sci U S A. 2010;107:5634–5639. doi: 10.1073/pnas.0809398107. [DOI] [PMC free article] [PubMed] [Google Scholar]