Significance
The unfolded protein response (UPR) is a cellular mechanism for coping with misfolded proteins in the lumen of the endoplasmic reticulum (ER). UPR pathways are also induced in response to viral and bacterial pathogens, resulting in enhanced proinflammatory cytokine induction. Here we provide mechanistic evidence for how an intracellular pathogen is able to inhibit the IRE1 branch of the UPR by blocking host translation elongation. Given that a broad spectrum of pathogens block protein synthesis specifically at elongation rather than at other steps of translation, this may point to a common mechanism for blocking the UPR and thereby preventing enhanced proinflammatory cytokine signaling.
Keywords: Legionella, translation inhibition, unfolded protein response
Abstract
Cells of the innate immune system recognize bacterial pathogens by detecting common microbial patterns as well as pathogen-specific activities. One system that responds to these stimuli is the IRE1 branch of the unfolded protein response (UPR), a sensor of endoplasmic reticulum (ER) stress. Activation of IRE1, in the context of Toll-like receptor (TLR) signaling, induces strong proinflammatory cytokine induction. We show here that Legionella pneumophila, an intravacuolar pathogen that replicates in an ER-associated compartment, blocks activation of the IRE1 pathway despite presenting pathogen products that stimulate this response. L. pneumophila TLR ligands induced the splicing of mRNA encoding XBP1s, the main target of IRE1 activity. L. pneumophila was able to inhibit both chemical and bacterial induction of XBP1 splicing via bacterial translocated proteins that interfere with host protein translation. A strain lacking five translocated translation elongation inhibitors was unable to block XBP1 splicing, but this could be rescued by expression of a single such inhibitor, consistent with limitation of the response by translation elongation inhibitors. Chemical inhibition of translation elongation blocked pattern recognition receptor-mediated XBP1 splicing, mimicking the effects of the bacterial translation inhibitors. In contrast, host cell-promoted inhibition of translation initiation in response to the pathogen was ineffective in blocking XBP1 splicing, demonstrating the need for the elongation inhibitors for protection from the UPR. The inhibition of host translation elongation may be a common strategy used by pathogens to limit the innate immune response by interfering with signaling via the UPR.
Legionella pneumophila, the causative agent of Legionnaires’ disease, is a Gram-negative intracellular pathogen that replicates within alveolar macrophages during disease. The ability of Legionella to replicate within a host is dependent on its type IVb secretion system (T4SS), termed Icm/Dot (1, 2). Using this apparatus, the bacterium translocates ∼300 Icm/Dot-translocated substrate (IDTS) proteins into the host cell cytosol (3, 4). These proteins play roles in manipulation of host cell membrane trafficking pathways, inhibition of the immune response, and eventual lysis of the host cell (5, 6).
A common strategy used by intracellular bacterial pathogens is to manipulate host membranes to establish a niche within cells (7). L. pneumophila establishes one such niche by replicating within a vacuole that resembles rough endoplasmic reticulum (ER) while avoiding targeting down the endocytic pathway (8, 9). Multiple IDTSs manipulate proteins associated with the ER compartment, resulting in vast reorganization of membranes from this organelle (10, 11). The ability of Legionella to form this vacuolar barrier is necessary for intracellular replication in macrophages, given that an inability to form this compartment results in routing of the bacterium to a degradative pathway, whereas permeabilization of the vacuole results in recognition by cytosolic innate immune receptors (9, 12).
The cell encodes an evolutionarily conserved system to cope with misfolding of proteins within the ER lumen, termed the unfolded protein response (UPR). In mammalian cells, the UPR can be initiated by signaling from three transmembrane proteins: PKR-like endoplasmic reticulum kinase (PERK), inositol-requiring kinase 1 (IRE1), and activating transcription factor 6 (ATF6) (13, 14). The IRE1 pathway is induced by its oligomerization and transautophosphorylation, resulting in activation of an RNase domain within the protein (15). Activation causes cleavage of an intron within the mRNA encoding the transcription factor X-box binding protein 1 (XBP1s), resulting in mRNA that allows translation of the mature transcription factor (16). Recently, this pathway has been observed to induce IRE1-dependent decay (RIDD) of mRNA, limiting the cytosolic pool of mRNA to be translated (17). Induction of UPR pathways limits ER stress through the up-regulation of chaperones, global inhibition of translation, and expansion of the ER (13, 14). An ineffectual response that fails to alleviate ER stress results in the initiation of an apoptotic program (18).
The UPR is linked to signaling pathways of the innate immune response, including activation of NF-κB and the JNK MAPK networks, resulting in production of proinflammatory cytokines (19). Furthermore, pathogen-associated molecular products (PAMPs), particularly those acting through Toll-like receptors (TLRs) (20), induce UPR-dependent cytokine production as a consequence of XBP1s-mediated transcriptional activation (21, 22). This activation results from ubiquitination of IRE1α by tumor necrosis factor receptor-associated factor 6 (TRAF6), preventing dephosphorylation of IRE1α by protein phosphatase 2A, facilitating XBP1 splicing (21).
Pathogen effector proteins also can activate the UPR. Direct binding of unfolded cholera toxin A subunit to IRE1α has been shown to induce RIDD, activating the RIG-I single-stranded innate immune sensing pathway (23). Moreover, Brucella spp. appear to activate the UPR through two different mechanisms. Brucella melitensis activates multiple UPR pathways through the microtubule stabilizing protein, TcpB, and the T4SS substrate, VceC, of Brucella abortus activates UPR, possibly through direct binding to the ER luminal chaperone BiP (24, 25).
Currently little is known about the interaction of Legionella with pathways of the UPR. Given that L. pneumophila intimately interacts with the ER, we hypothesized that this pathogen may use mechanisms to modulate the evolutionarily conserved IRE1α pathway. Here we present evidence that L. pneumophila PAMP activation of the IRE1α pathway is blocked by IDTSs that interfere with the host cell translation elongation machinery, providing a mechanism for blocking one arm of the innate immune response.
Results
L. pneumophila Inhibits Activation of the IRE1α Branch of the UPR.
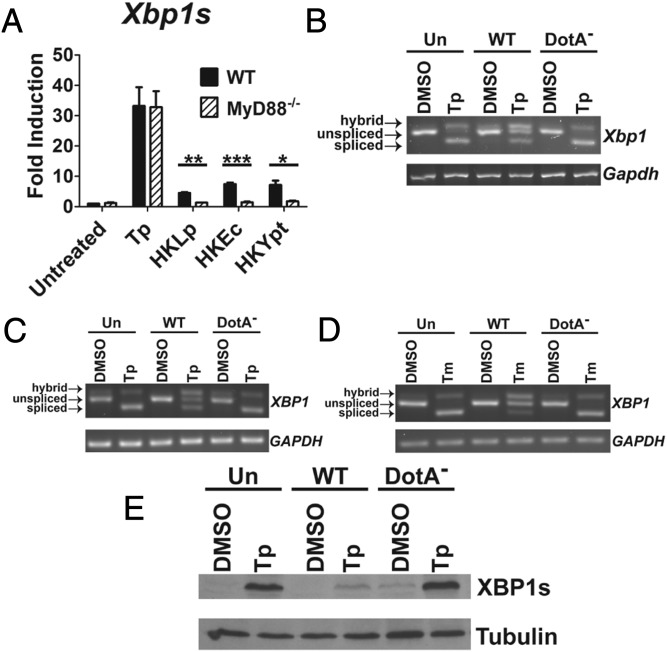
It was previously shown that TLR detection of PAMPs activates IRE1α to induce the cytosolic splicing of mRNA encoding the transcription factor Xbp1s (21, 22). To determine whether L. pneumophila-derived PAMPs induce Xbp1 splicing, bone marrow-derived macrophages (BMDMs) from wild type (WT) and Myd88−/− mice were treated with heat-killed L. pneumophila (HKLp) (Fig. 1A). Cells were also treated with the TLR stimulators heat-killed E. coli (HKEc) and Y. pseudotuberculosis (HKYpt) (20) to induce Xbp1 splicing, and with thapsigargin (Tp) to induce ER stress due to depletion of luminal ER calcium stores. RNA isolated from challenged cells was analyzed by quantitative RT-PCR (qRT-PCR) to detect splicing of Xbp1, and splicing was compared with untreated controls. HKLp induced levels of Xbp1 splicing in WT macrophages similar to those observed in response to the other heat-killed organisms (Fig. 1A). In contrast, no splicing could be observed in the Myd88−/− macrophages, consistent with this response being driven by TLR detection of L. pneumophila (Fig. 1A). Although levels of Xbp1 splicing were low relative to those seen during Tp treatment, they are consistent with those in previous reports showing pattern recognition receptor (PRR)-mediated XBP1 splicing (21, 22).
Fig. 1.
L. pneumophila inhibits chemically induced XBP1 splicing. (A) PAMPs derived from L. pneumophila induce Xbp1 splicing in a Myd88-dependent manner. WT and Myd88−/− C57BL/6 BMDMs were incubated with HKLp, HKEc, or HKYpt at an effective MOI of 100, or treated with Tp (500 nM), for 6 h. Total RNA isolated from lysates was used to measure levels of Xbp1s transcript by qRT-PCR (Materials and Methods). (B) L. pneumophila is able to inhibit Tp-induced Xbp1 splicing in an Icm/Dot-dependent manner. A/J BMDMs were challenged with Lp02 (WT) or Lp03 (DotA−) at an MOI of 3 for 7 h, with the addition of Tp (500 nM), as indicated, at 1 h postchallenge. cDNA generated from total RNA lysates was used to analyze Xbp1 splicing by semiquantitative RT-PCR (Materials and Methods). (C) Chemical induction of the IRE1α pathway is also limited in a human macrophage-like cell line. U937 cells, treated with Tp (500 nM) or DMSO as a vehicle control, were challenged with WT or DotA− at an MOI of 5 for 6 h. XBP1 splicing was determined as in B. (D) Similar results were seen in cells treated with Tm (1 µg/mL) to induce ER stress. (E) Induction of XBP1s protein is also limited by WT challenge. Total lysates from challenged U937 cells were probed with antibodies specific to the product of spliced XBP1 transcript (XBP1s) or α-tubulin as a loading control. Data are mean ± SEM of three independent experiments (A) or are representative of three independent experiments (C–E) or a single representative experiment (B). Statistical analyses were performed using the unpaired t test with Welch’s correction where appropriate. *P < 0.05; **P < 0.01; ***P < 0.001.
We next tested whether L. pneumophila-derived PAMPs could induce the IRE1α branch of the UPR during host cell challenge with live bacteria. BMDMs were challenged with the WT strain Lp02 or the Icm/Dot-deficient strain Lp03 (DotA−) to assess for a specific response to IDTS. In addition, Tp was added at 1 h postchallenge to determine how an ER stress-inducing reagent affects UPR in the presence of L. pneumophila infection. Xbp1 splicing was then analyzed by semiquantitative RT-PCR (Materials and Methods). In this system, the spliced product presents as a band that electrophoretically migrates faster than the unspliced product. In addition, a spliced/unspliced hybrid presents as the slowest migrating form, as noted in previous work (26).
In the DMSO-treated cells, there was little evidence for the induction of Xbp1 splicing from the L. pneumophila challenge (Fig. 1B). Tp alone resulted in almost complete loss of the unspliced Xbp1 transcript (Fig. 1B; Un, Tp). In contrast, infection of cells with L. pneumophila WT resulted in clear retention of the unspliced form (WT, Tp). Blockage of Xbp1 splicing required the Icm/Dot system, because the DotA− strain showed no blockage (DotA−, Tp). The blockage of splicing observed in the WT infection was not complete, likely as a consequence of uninfected bystander cells undergoing Tp-induced Xbp1 splicing.
Even at these low levels of infectivity, cytotoxicity of the primary macrophages could be observed. Thus, a macrophage-like cell line that showed lower levels of cytotoxicity was used, to eliminate the possibility that the UPR blockage was caused by cell death. Phorbol ester-differentiated human U937 cells were challenged with the WT or DotA− strain. As seen with BMDMs, WT inhibited Tp-induced XBP1 splicing, but the DotA− strain did not (Fig. 1C; WT, Tp vs. DotA−, Tp). Furthermore, in the absence of Tp, XBP1 splicing was undetectable in cells challenged with WT, with a clear reduction in levels of the hybrid band compared with challenge of macrophages with the DotA− strain or uninfected cells (Fig. 1C; WT, DMSO vs. DotA−, DMSO). Similar results were seen in cells challenged in the presence of tunicamycin (Tm), which induces ER stress by inhibiting N-linked glycosylation, indicating that inhibition of XBP1 splicing was not dependent on the chemical inducing agent being tested (Fig. 1D). Finally, consistent with the ability of WT to inhibit XBP1 splicing, the protein levels of XBP1s were reduced in cells challenged with WT and treated with Tp, relative to the uninfected or DotA−-challenged populations (Fig. 1E). Taken together, these results indicate that L. pneumophila infection suppresses pharmacologically induced XBP1 splicing.
Inhibition of XBP1 Splicing Is Dependent on Translocated Substrates That Limit Host Translation Elongation.
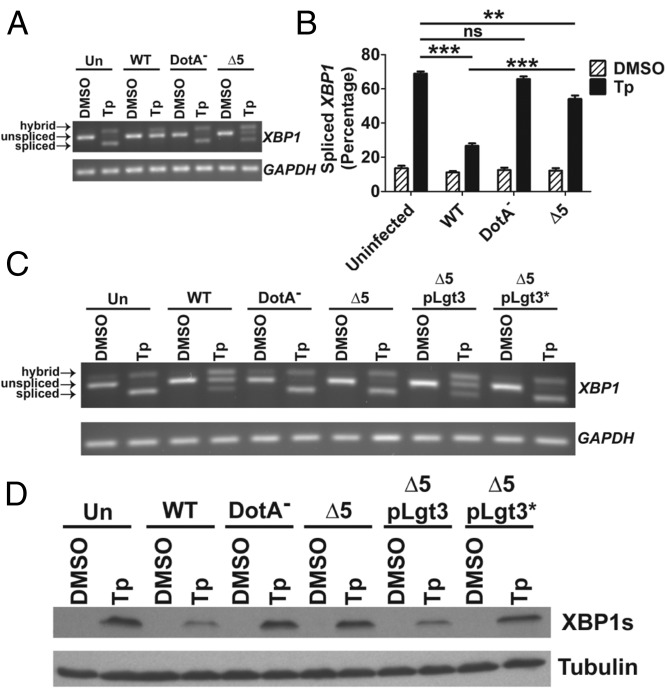
Previous studies have shown that chemical translation elongation inhibitors, such as cycloheximide (CHX), can block pharmacologic induction of the UPR (27). Legionella has been shown to target host translation elongation through IDTSs that inhibit the activity of the eukaryotic elongation factors eEF1A and eEF1Bγ (28, 29). A strain lacking these IDTSs (Lgt1–3 and SidI), as well as an additional IDTS (SidL), named ∆5, exhibits a decreased ability to inhibit host protein translation (30, 31). Thus, we hypothesized that the ability of L. pneumophila to limit the induction of XBP1 splicing is dependent on these elongation inhibitors.
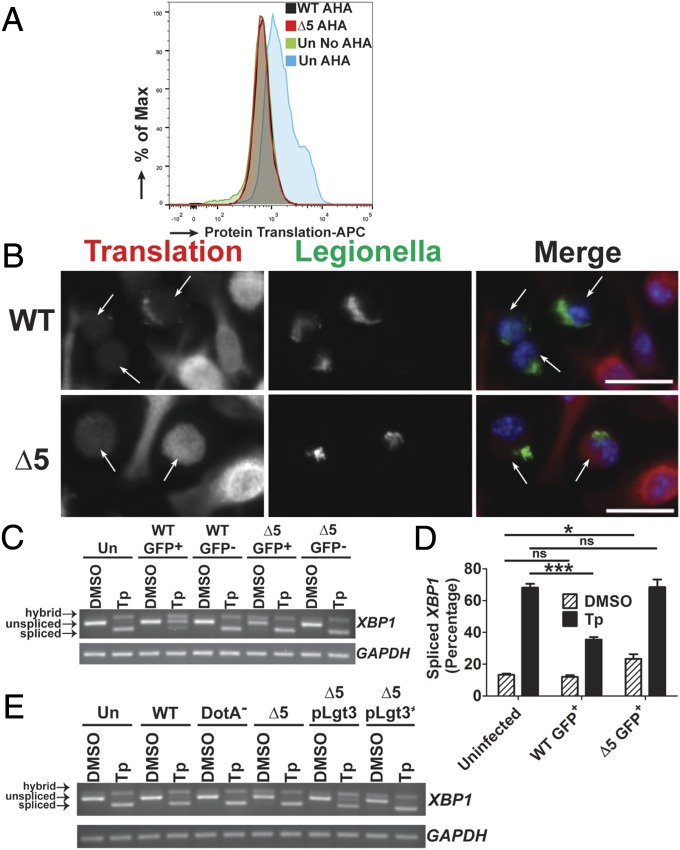
To test this model, we challenged cells with L. pneumophila-GFP, and U937 cells harboring bacteria were sorted from uninfected cells to specifically analyze XBP1 splicing in the population harboring bacteria. In the absence of uninfected bystanders, RNA isolated from cells challenged with WT showed almost complete inhibition of Tp-induced XBP1 splicing, in contrast to cells challenged with ∆5 or DotA− (Fig. 2 A and B). That this action is dependent on the activity of these IDTSs is further supported by the finding that complementation of ∆5 by the IDTS Legionella pneumophila glucosyltransferase 3 (Lgt3), but not a mutant Lgt3 that harbors a catalytically inactive point mutation, limited XBP1 splicing in a population of cells challenged by these strains (Fig. 2C). Western blot analysis of levels of XBP1s protein revealed a similar dependence on the translation elongation inhibitors (Fig. 2D). Moreover, blockage of XBP1s expression was dependent on the biochemical activity of Lgt3 in complementation experiments (Fig. 2D). These results show that bacterial inhibitors of host translation elongation block IRE1 signaling at both XBP1 splicing and downstream XBP1s protein expression.
Fig. 2.
Inhibition of XBP1 splicing is dependent on T4SS substrates that inhibit host translation elongation. (A) Inhibition of XBP1 splicing is largely dependent on host translation elongation inhibitors. U937 cells were challenged with GFP expressing WT, DotA−, or ∆5 at an MOI of 2 for 4 h in the presence or absence of 100 nM Tp. Populations with associated bacteria were obtained through sorting by flow cytometry and lysates were used to determine XBP1 splicing (Materials and Methods). (B) Quantitation of XBP1 splicing from A. (C and D) Expression of Lgt3, but not of a catalytically inactive mutant (Lgt3*), limits Tp-induced XBP1 splicing and XBP1s protein expression. Tp- (100 nM) or DMSO- treated U937 cells were challenged with the indicated strains at an MOI of 2 for 4 h. XBP1 splicing and XBP1s protein levels were determined from total cell lysates (Materials and Methods). Data are representative of at least three independent experiments (A, C, and D) or are the mean ± SEM of three independent experiments (B). Statistical analyses were performed using the unpaired t test. **P < 0.01; ***P < 0.001.
Induction of Transcripts Regulated by XBP1s Is Limited by L. pneumophila Challenge.
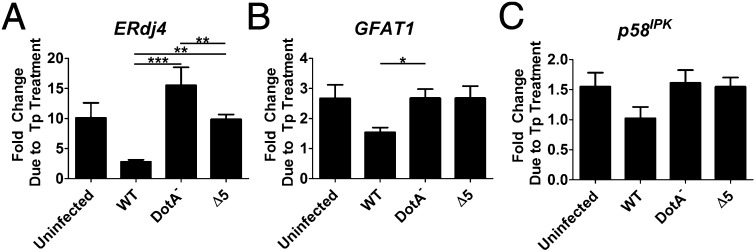
Transcription of a subset of genes has been shown to be specifically up-regulated by XBP1s in response to ER stress (32, 33). To determine whether L. pneumophila challenge attenuates signaling downstream from XBP1s induction, Tp-treated cells were challenged with L. pneumophila-GFP strains, and RNA was isolated from the sorted infected (GFP+) population and analyzed by qRT-PCR. All of the XBP1s-regulated transcripts analyzed—endoplasmic reticulum-localized DnaJ 4 (ERdj4), glutamine:fructose-6-phosphate amidotransferase (GFAT1), and p58IPK (IPK, inhibitor of protein kinase)—exhibited reduced Tp-dependent induction in cells challenged with WT compared with uninfected cells and cells challenged with either DotA− or the ∆5 strain (Fig. 3 A–C). The decreased expression of p58IPK was below the level of significance seen for ERdj4 and GFAT1. On the other hand, the effect was most significant for ERdj4, which showed 80% lower induction in the presence of WT compared with the DotA− strain (Fig. 3A). The extreme attenuation of the response was also dependent on the translocated protein synthesis inhibitors, with greater induction of ERdj4 for the ∆5 strain compared with the WT strain (Fig. 3A). It also should be noted that in the case of ERdj4, the DotA− strain showed an exaggerated response to Tp relative to the uninfected cells. Presumably, in strains with an intact Icm/Dot system, this response was partially attenuated by other effectors among the more than 300 known translocated substrates. These results indicate that inhibition of translation elongation is necessary to demonstrate complete blockage of the response and is consistent with the ineffectual inhibition of Tp-induced XBP1 splicing by the ∆5 strain (Fig. 2 A and B).
Fig. 3.
L. pneumophila blocks transcription of genes controlled by XBP1s. U937 cells were challenged with noted GFP-harboring strains at an MOI of 2 for 4 h, treated with either Tp (100 nM), or DMSO, and sorted into infected and uninfected populations by flow cytometry. RNA isolated from GFP-positive populations was used to measure transcriptional expression of genes regulated by XBP1s by qRT-PCR. Shown is the fold enhancement, in the infected populations, resulting from Tp treatment: (A) ERdj4; (B) GFAT1; (C) p58IPK. Expression is relative to 18S ribosomal RNA. Data are the mean ± SEM of three independent experiments. Statistical analyses were performed using the unpaired t test. *P < 0.05; **P < 0.01; ***P < 0.001.
Induction of UPR Does Not Limit L. pneumophila Intracellular Replication.
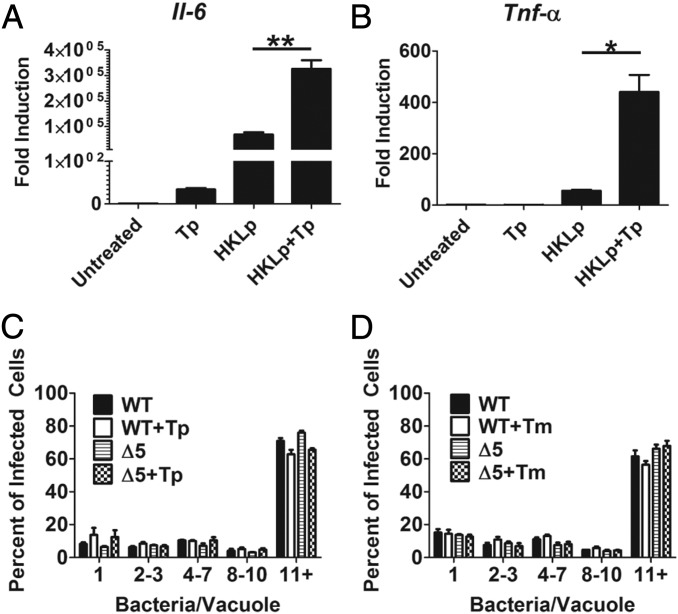
Activation of the UPR through the IRE1α pathway has been implicated in enhanced expression of proinflammatory cytokines (21, 22, 30). This enhanced induction occurs under conditions of activation of PRRs by PAMPs. To determine whether detection of L. pneumophila-derived PAMPs during activation of the UPR enhances the expression of cytokines, BMDMs were treated with HKLp in the presence or absence of Tp. RNA isolated from stimulated macrophages showed markedly increased transcription of both tumor necrosis factor-alpha (Tnf-α) and interleukin 6 (Il-6) in cells treated with both HKLp and Tp compared with cells treated with HKLp alone (Fig. 4 A and B).
Fig. 4.
Legionella replicates in presence of an induced UPR. (A and B) The UPR synergizes with L. pneumophila PAMPs to induce transcription of proinflammatory cytokines. A/J BMDMs were treated with Tp (500 nM), HKLp (effective MOI of 20), or both HKLp and Tp, for 6 h. RNA isolated from total lysates was analyzed for transcription of Il-6 or Tnf-α by qRT-PCR. Expression is plotted relative to Gapdh transcript levels. (C and D) Legionella replicates in presence of an induced UPR. A/J BMDMs, treated with Tp (500 nM), Tm (1 µg/mL), or DMSO were challenged with bacteria for 14 h at an MOI of 0.5. Infected macrophages were fixed and probed with anti-L. pneumophila (Materials and Methods). The number of bacteria/vacuoles was determined for 100 cells in each of three replicates. The percentage of vacuoles with the indicated number of bacteria is plotted. Data are the mean ± SEM of three independent experiments. Statistical analyses were performed using the unpaired t test with Welch’s correction where appropriate. *P < 0.05; **P < 0.01.
The enhanced cytokine transcription under conditions of PRR engagement by L. pneumophila in cells undergoing UPR could create an environment that is limiting for intracellular replication of the bacterium, as been seen for other intracellular pathogens, such as Listeria monocytogenes (34). It is possible that the ability of WT L. pneumophila to inhibit XBP1 splicing may limit the cell’s ability to respond in this manner. To assess this, we challenged BMDMs with either WT or the ∆5 strain, which is defective in inhibition of XBP1 splicing (Fig. 4 C and D). Challenged cells were analyzed by immunofluorescence microscopy to determine bacterial replication. In cells treated with either Tp or Tm, both strains were able to replicate at similar levels, based on counting the number of bacteria/vacuoles. Thus, L. pneumophila was markedly resilient in the presence of UPR inducers, because BMDMs treated with either Tp or Tm caused little or no decrease in intracellular replication (Fig. 4 C and D), consistent with the lack of cell death caused by these inhibitors, as reported previously (35).
The Mechanism of Translation Inhibition Is Critical for Blocking XBP1 Splicing.
It has been shown that levels of protein synthesis in host cells challenged by WT L. pneumophila are ∼95% below that of uninfected cells, whereas translation in cells challenged with the Δ5 strain is still ∼80% below that of uninfected cells at an early time point postchallenge (30). Furthermore, as infection progresses, levels of translation in ∆5-infected cells continue to decrease relative to earlier time points (31). Much of this translation inhibition in the Δ5 strain can be explained by the observation that the host cell shuts down translation initiation in response to the pathogen (36).
Based on the foregoing results, we hypothesized that the ability of ∆5 to replicate during conditions of an induced UPR could be explained by inhibition of XBP1 splicing at late time points during host cell challenge, as a consequence of protein synthesis inhibition by the host. To analyze this issue further, we measured host protein synthesis at late time points. At 10 h postinfection (hpi), host cells were incubated with the amino acid analog l-azidohomoalanine (AHA) for 1 h, and levels of incorporation into nascent polypeptides were determined by flow analysis after fluorescent labeling of the incorporated analog (Materials and Methods). The cellular subpopulations harboring either WT or ∆5 showed label incorporation indistinguishable from background controls, indicating inhibited translation in host cells at late time points after challenge by both strains (Fig. 5A). We verified this using an adaptation of the SUnSET protocol (37), in which translation is analyzed by the incorporation of puromycin into translating ribosomes. In infected cells treated with puromycin at 10 hpi, microscopic examination after an additional 1 h of infection revealed some detectable puromycin incorporation into translating ribosomes in ∆5-infected cells that was distinguishable from the WT infection. The levels of incorporation were much lower than those seen in neighboring uninfected bystander cells, however, again indicating inhibited translation at late time points, even in response to the ∆5 strain (Fig. 5B).
Fig. 5.
The mechanism of translation inhibition is critical for blocking XBP1 splicing. (A) The ∆5 strain inhibits protein synthesis at late time points during infection. U937 cells were challenged with GFP-L. pneumophila strains at an MOI of 1 for 10 h, then incubated with the methionine analog AHA (100 µM) for 1 h. Fixed cells harboring GFP-expressing WT or ∆5 were analyzed for levels of translation by detection of AHA incorporation into nascent peptides using DyLight 650-phosphine and flow cytometry (Materials and Methods). (B) Low levels of translation are observed at late time points during challenge with the ∆5 strain when analyzed by puromycin incorporation into translating ribosomes. A/J BMDMs were challenged with bacteria at an MOI of 0.5 for 10 h then treated with puromycin (1 µg/mL) for another 1 h. Cells were fixed and permeablized, then stained with antibodies specific to L. pneumophila or puromycin, to measure translation (Materials and Methods). (Scale bar: 10 µm.) (C) At late time points, the ∆5 strain induces XBP1 splicing and is unable to inhibit chemically induced XBP1 splicing. U937 cells were challenged with bacteria at an MOI of 1 for 9 h, then treated with either DMSO or Tp (100 nM) for 2 h. Cells were sorted by GFP (for the infected populations), and lysates from each population were used to isolate RNA (Materials and Methods). (D) Quantitation of XBP1 splicing from C. (E) Expression of Lgt3 limits chemically induced XBP1 splicing at late time points. U937 cells were challenged with each strain at an MOI of 1 for 11 h and then treated with DMSO or Tp (100 nM) at 9 h postchallenge. RNA isolated from the total cell population was analyzed for XBP1 splicing (Materials and Methods). Data are representative of three independent experiments (A–C and E) or the mean ± SEM of three independent experiments (D). Statistical analyses were performed using unpaired t test. *P < 0.05; ***P < 0.001.
Given the significant shutdown of host protein synthesis in response to the Δ5 strain at 10–11 hpi, we investigated whether this could interfere with the UPR, by analyzing XBP1 splicing. In U937 cells challenged with GFP-L. pneumophila and treated with Tp for 2 h at 9 hpi, there was no evidence that the ∆5 strain could limit XBP1 splicing in the sorted infected population (Fig. 5 C and D), even though there was little host translation at this time point (Fig. 5 A and B). In fact, at 11 hpi, the ∆5 strain induced XBP1 splicing even in the absence of Tp treatment, as visualized by a hybrid band and faint spliced band (Fig. 5 C and D). Evidence for UPR was not observed in untreated WT challenged cells, consistent with the ability of this strain to limit the response observed in ∆5-challenged cells. As observed at early time points, the ∆5 strain harboring Lgt3 was sufficient to inhibit XBP1 splicing (Fig. 5E). It is notable that, in contrast to earlier time points (Fig. 2C), the strain bearing the plasmid-borne Lgt3 showed enhanced inhibition of Tp- induced XBP1 splicing relative to that observed with the WT strain. Also, at 11 h postchallenge, sorted WT-challenged cells that had been treated with Tp showed clear evidence of a fully spliced XBP1 message (Fig. 5 C and D).
The presence of a spliced message after WT challenge in the presence of Tp was surprising, given the lack of evidence for this level of XBP1 splicing in WT-challenged cells and the scant evidence of host translation occurring in the presence of the WT strain at earlier time points (Fig. 5 A and B). The ability to block XBP1 splicing appears to decay over time despite the lack of host protein synthesis. In contrast, interference of XBP1 splicing can be maintained at late time points by expression of an unregulated translation elongation inhibitor (Lgt3) harbored on a plasmid. Therefore, blocking of elongation may be specifically required for limiting the XBP1 arm of the UPR.
Pharmacologic Inhibition of Translation Elongation Limits Both Chemically and Bacterially Induced XBP1 Splicing.
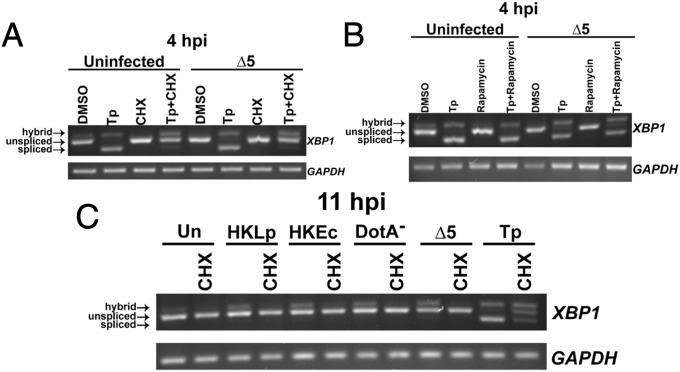
The finding that a bacterial effector specifically targets translation elongation, and not initiation, to limit XBP1 splicing indicates that elongation blockage may be required to disrupt this arm of the UPR. To determine whether we could rescue inhibition of XBP1 splicing in U937 cells challenged with the ∆5 strain, we treated cells with the elongation inhibitor CHX throughout a 4-h challenge with this strain. Incubation of the ∆5 strain simultaneously with CHX resulted in the inhibition of Tp-induced XBP1 splicing, with greatly reduced amounts of the fully spliced form (Fig. 6A), consistent with the requirement of an elongation block for blockage of XBP1 splicing. In contrast, treatment with rapamycin, which interferes with translation initiation and results in reduced, but not totally eliminated, protein synthesis (36), did not result in the inhibition of Tp-induced XBP1 splicing in either in the presence or absence of the ∆5 strain (Fig. 6B). Therefore, either the mechanism of translation inhibition plays a critical role in blocking XBP1 splicing or the high efficiency of CHX translation inhibition relative to rapamycin is responsible for the block.
Fig. 6.
Inhibition of translation elongation inhibits chemical and PRR mediated XBP1 splicing. (A) U937 cells were challenged with ∆5 at an MOI of 2 for 4 h in the presence of Tp (100 nM), CHX (2 µg/mL), or Tp+CHX. CHX inhibits Tp-induced XBP1 splicing in uninfected cells and those challenged with the ∆5 strain. (B) Challenge of U937 cells by ∆5 at an MOI of 2 for 4 h with the addition of Tp (100 nM), rapamycin (100 nM), or rapamycin+Tp. Rapamycin treatment does not inhibit Tp-mediated XBP1 splicing. (C) CHX limits XBP1 splicing induced by L. pneumophila. U937 cells treated with HKLp or HKEc at an effective MOI of 100, with DotA− at an MOI of 20, or with ∆5 at an MOI of 2, all for 11 h, were treated with CHX (2 µg/mL) throughout the experiment. Data are representative of two (B) or three (A and C) independent experiments.
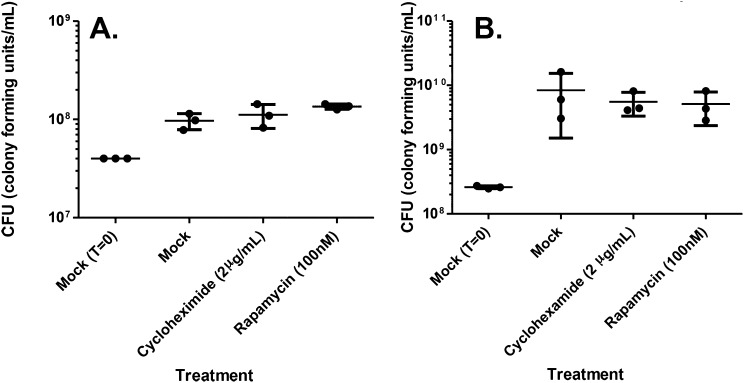
Whereas CHX had been previously shown to inhibit chemically induced XBP1 splicing (27) (Fig. 6A), the effect of inhibiting host translation elongation on PAMP-induced XBP1 splicing remained unknown. To determine this, we challenged U937 cells with HKLp, HKEc, DotA−, or the ∆5 strain for 11 h, a time point at which the ∆5 strain induces XBP1 splicing (Fig. 5 C and D), and evaluated the effect of CHX treatment. In the absence of CHX, treatment of cells with heat-killed bacteria or challenge with DotA- or ∆5 induced XBP1 splicing, as demonstrated by the hybrid band in the figure. In contrast, CHX completely inhibited XBP1 splicing in all cases (Fig. 6C). This result is consistent with the lack of induction of XBP1 splicing by WT at 11 h postchallenge relative to ∆5-challenged cells (Fig. 5 C and D), and shows that inhibition of translation elongation blocks XBP1 splicing induced by varying signals. As was true for rapamycin, CHX had no effect on growth of L. pneumophila in culture, arguing that any effects observed are predicted to be directed toward the host cell (Fig. S1).
Fig. S1.
CHX and rapamycin have no effect on L. pneumophila yields in broth. To determine whether drugs used to manipulate the UPR alter growth of L pneumophila, strain Lp02 (Materials and Methods) was grown to A600 = 0.2, at which time either CHX or rapamycin in DMSO (1 μL/mL culture) was added at the concentrations noted, with Mock (1 μL/mL DMSO) serving as a control. The Mock control was allowed to approximately double (A) or was grown to postexponential phase (B), and yields of bacteria were determined for each culture by measuring CFU on CYE agar medium (Materials and Methods).
Discussion
In this study, we have shown that Legionella is able to inhibit the activation of the IRE1α branch of the UPR. Inhibition is dependent on the Icm/Dot T4SS, specifically five IDTSs that interfere with host translation elongation. In the absence of these translocated proteins, Legionella induces XBP1 splicing at late time points, even in the absence of a chemically induced UPR. This is consistent with bacterial-mediated inhibition of host translation elongation blocking XBP1 splicing that results from pathogen detection.
Interference with the host cell UPR during the intracellular replication of bacterial pathogens is an emerging theme in bacterial pathogenesis, as documented previously with viral pathogens (38, 39). The ability of Legionella to inhibit UPR pathways, through bacterial proteins that inhibit protein synthesis, has been documented recently (40). Although that study showed that L. pneumophila could block chemically induced UPR pathways, our work points to the role of this inhibition in blocking bacterially induced UPR activation observed at late time points in cells challenged by the ∆5 strain. Another recent report has demonstrated that, similar to L. pneumophila, the chlamydial organism Simkania negevensis replicates in a vacuole closely associated with the ER and mitochondria (41). Interestingly, this pathogen does not induce an ER stress response and is able to inhibit chemically induced activation of UPR pathways, including that of IRE1α (41). Although the mechanism by which S. negevensis limits UPR pathways is unknown, an analogous mechanism for limiting UPR through bacterially mediated inhibition of host translation is possible.
Recent studies of the inhibition of host protein translation during Legionella challenge have identified at least two mechanisms in play, one mediated by the bacterium and the other mediated by the host cell (28, 30, 36). The bacterial-mediated inhibition of translation most notably involves inactivation of elongation factors eEF1A and eEF1Bγ (28, 30). The host response to pathogenic Legionella results in the ubiquitination of positive regulators of mTOR, limiting its activity and leading to the inhibition of translation initiation (36). In a previous study, we observed that as infection progresses, cells challenged by the ∆5 strain show low levels of protein translation, despite the lack of translation elongation inhibitors in this strain (31). Consistent with that observation, here we report low levels of protein translation at 11 hpi after challenge with either the WT or ∆5 strain. Based on our data, we propose that in the first few hours after infection, there is limited global translation in the host cell owing to bacterial inhibition of host translation elongation. As the infection proceeds, a second layer of inhibition of protein synthesis occurs as a consequence of host-promoted self-inhibition of translation initiation. We think it likely that the bacterial inhibition of elongation is necessary to inhibit XBP1 splicing, considering that at 9–11 hpi, treatment with Tp strongly induced XBP1 splicing in cells challenged with the ∆5 strain, even though there little host protein synthesis occurred. Furthermore, the ability of WT L. pneumophila to inhibit Tp-induced XBP1 splicing at these later time points appears impaired relative to the more robust inhibition observed just a few hours earlier. Expression of Lgt3 harbored on a plasmid reverses this effect at late time points, allowing more robust blockage of Tp-induced XBP1 splicing than that observed in WT. This finding supports a model in which the activity of the bacterially derived elongation inhibitors translocated shortly after formation of the L. pneumophila replication vacuole are the primary down-modulators of UPR, and host-mediated translation initiation inhibition does not effectively interfere with this response.
The ability of CHX to inhibit PAMP-induced XBP1 splicing was surprising, given that the mechanism of XBP1 splicing in response to microbial ligands is not supposed to occur via induction of luminal ER protein misfolding (21). We propose that microbial ligands, through TLR signaling, cause TRAF6-dependent ubiquitination of IRE1α, allowing for the maintenance of low levels of IRE1α phosphorylation resulting from physiological, or NOX2-mediated, protein misfolding in the ER (21, 22). When translation elongation is blocked due to CHX treatment, microbial ligands cannot induce XBP1 splicing owing to the lack of misfolding-driven IRE1α phosphorylation in the cell. The ability of WT Legionella to inhibit host protein translation elongation may limit luminal ER protein folding to levels insufficient to induce IRE1α phosphorylation.
We observed that Legionella was able to replicate to high levels in cells pharmacologically induced to undergo UPR, despite the strong transcriptional up-regulation of proinflammatory cytokines under conditions of L. pneumophila-derived PAMPs and chemically induced ER stress. Other studies have found that the effects of UPR on intracellular replication is dependent on the pathogen, because chemical induction of the UPR inhibits replication of Listeria, whereas induction of the IRE1 pathway supports intracellular replication of Brucella (25, 34, 42). It is possible that UPR pathways may play a role in limiting the replication of L. pneumophila in its environmental host, amoeba, given that the IRE1 branch has been shown to play important roles in the innate immune response in other lower eukaryotes (43). Consistent with this hypothesis, the UPR-inducing ∆5 strain is defective for intracellular replication in Dictyostelium discoideum (30). L. pneumophila antagonism of the UPR likely provides a selective advantage for the bacterium during growth in environmental hosts to counteract this evolutionarily ancient antimicrobial response (43, 44).
This study illuminates a mechanism by which a bacterial pathogen inhibits the induction of the IRE1α branch of the UPR. Given that this response to PRR engagement induces downstream innate immune signaling, and that other pathogens similarly interfere with host translation elongation, the mechanism provided here may be shared by a number of other pathogens.
Materials and Methods
Bacterial Culture and Media.
The L. pneumophila strains used in this study are described in Table 1. Strains were propagated in liquid culture in ACES-buffered yeast extract broth and on solid medium containing buffered charcoal yeast extract (BCYE). Strains harboring the pGFP CmR plasmid, encoding an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible GFPmut3 (45), were cultured on BCYE containing 5 µg/mL Cm and 0.1 mg/mL thymidine, with the addition of 1 mM IPTG during growth in broth.
Table 1.
Bacterial strains and plasmids
| Strain | Genotype | Description | Reference |
| Lp02 | WT: Philadelphia 1, thyA rpsL hsdR | Wild type strain | (1) |
| Lp03 | DotA−: Lp02 dotA03 | Translocation deficient | (1) |
| ∆5 | Lp02, lgt1-3, sidI, sidL | Translation inhibition deficient | (22) |
| Lp02 pGFP | Lp02, pEC101 | This study | |
| Lp03 pGFP | Lp03, pEC101 | This study | |
| ∆5 pGFP | ∆5, pEC101 | This study | |
| Lp02 pJB908 | Lp02, pJB908 | Thy+, empty vector | (45) |
| Lp03 pJB908 | Lp03, pJB908 | Thy+, empty vector | (45) |
| ∆5 pLgt3 | ∆5, pLgt3 | FLAG-Lgt3; sidF promoter | (22) |
| ∆5 pLgt3* | ∆5, pLgt3* | FLAG-Lgt3*; sidF promoter | (22) |
Eukaryotic Cell Culture.
BMDMs were isolated from femurs of female mice (8, 31). The animal studies were approved by the Institutional Animal Care and Use Committee of Tufts University.
U937 cells (American Type Culture Collection) were cultured in RPMI supplemented with 10 mM glutamine and 10% (vol/vol) FBS. Cells were differentiated by treatment with 10 ng/mL 12-tetradecanoyl phorbol 13-acetate (TPA) for 24–48 h. Differentiated U937 cells were plated overnight in the absence of TPA before challenge.
Intracellular Replication.
BMDMs isolated from A/J mice were plated on glass coverslips at a density of 2 × 105/well in 24-well plates. Before challenge, medium was replaced with RPMI, 200 µg/mL thymidine, and either DMSO, 500 nM Tp (Sigma-Aldrich), or 1 µg/mL Tm (Sigma-Aldrich). Cells were challenged at a multiplicity of infection (MOI) of 0.5 with postexponential bacteria, and plates were centrifuged at 400 × g for 5 min. The incubation was allowed to proceed for 1 h at 37 °C, followed by three washings with warm medium and then an additional 13 h of incubation. Coverslips were washed three times with PBS, fixed with 4% (wt/vol) paraformaldehyde for 20 min at room temperature, and washed again three times with PBS. After blocking with 4% (vol/vol) goat serum, extracellular Legionella was detected using anti-L. pneumophila rat serum (1:5,000) and goat anti-rat IgG Alexa Fluor 594 (1:500). Cells were permeabilized with 0.1% Triton X-100 for 10 min and probed with anti-L. pneumophila rabbit serum (1:5,000) for 1 h, followed by detection with goat anti-rabbit IgG Alexa Flour 488 (1:500). The number of bacteria per cell was determined for 100 cells per coverslip by immunofluorescence microscopy.
Analysis of XBP1 Splicing by qRT-PCR.
U937 cells (8 × 105 to 1 × 106) were challenged at the indicated MOI. Plates were centrifuged for 5 min at 400 × g, then incubated at 37 °C for 2 h. Cells were washed three times with warm media, then replaced with RPMI containing the initial chemicals and allowed to incubate further. BMDMs isolated from A/J mice (4 × 105) were challenged at an MOI of 3, centrifuged for 5 min at 400 × g, and then incubated at 37 °C for 1 h. The medium was replaced, followed by incubation for another 6 h. At the time of analysis, cells were washed three times with HBSS and then lysed in Buffer RLT (Qiagen).
For sorted cell experiments, six wells of U937 cells plated at 2 × 106 were challenged with L. pneumophila at the indicated MOI for 2 h at 37 °C, then treated as above for unsorted samples for the remainder of the challenge. Before sorting, cells were washed with HBSS, then lifted with trypsin, washed with HBSS, and resuspended in PBS plus 1 mM EDTA. A total of 1.5 × 106 cells were collected on the BD Influx sorter at the Tufts University Flow Cytometry Core. Sorted cells were pelleted and resuspended in buffer RLT.
RNA isolation from buffer RLT lysates was performed using the RNeasy Kit (Qiagen), followed by treatment with TURBO DNA-free (Life Technologies). cDNA was generated with SuperScript III (Invitrogen) using oligo dT and 100–750 ng RNA as the template. XBP1 was amplified using the human- specific primer set (5ʹ-TTACGAGAGAAAACTATGGCC and 3ʹ-GGGTCCAAGTTGTCCAGAATGC) and GAPDH (5ʹ-TTGCCATCAATGACCCCTTCA and 3ʹ-CGCCCCACTTGATTTTGGA). Mouse transcripts were amplified with Xbp1 (5ʹ-GAACCAGGAGTTAAGAACACG and 3ʹ-AGGCAACAGTGTCAGAGTCC) and Gapdh (5ʹ-AGGCCGGTGCTGAGTATGTC and 3ʹ-TGCCTGCTTCACCACCTTCT). Products were analyzed by separation on 2.5% (wt/vol) agarose gels and imaged with the Gel Logic 100 Imaging System (Kodak). Quantification of XBP1 splicing was performed using Fiji software to determine the mean pixel intensity for spliced and unspliced products, as well as the hybrid product, which was accounted for as one-half spliced products and one-half unspliced products.
Immunoblotting.
U937 cells were challenged for 4 h, as done for XBP1 splicing analysis. After the challenge, cells were washed three times with HBSS, then lysed with 2× Laemmli sample buffer, followed by boiling for 10 min. SDS/PAGE was performed, followed by transfer to PVDF. Protein detection was performed with antibodies to XBP1s (BioLegend; 1:1,000) or α-tubulin (Sigma-Aldrich; 1:10,000).
qRT-PCR.
For qRT-PCR analysis of Xbp1 splicing, WT and Myd88−/− C57BL/6 BMDMs, plated at 4 × 105 to 8 × 105, were treated with Tp, HKLp, HKEc, or HKYpt (heat-killed by heating at 60 °C for 1 h) at an effective MOI of 100. Plates were centrifuged at 400 × g for 5 min, then incubated at 37 °C for 6 h. Cells were washed three times with PBS, then lysed with buffer RLT. RNA was isolated as for qRT-PCR analysis. Xbp1 splicing was then detected as described previously (46) with the following modifications. Transcripts were measured with the RNA-to-Ct 1-Step Kit (Applied Biosystems) using the mouse Xbp1s primer pair (5ʹ-TGCTGAGTCCGCAGCAGGTG and 3ʹ-ACTAGCAGACTTGGGGAAG) and normalized to 18S ribosomal RNA using 5ʹ-CGCCGCTAGAGGTGAAATTCT and 3ʹ-GCTTTCGTAAACGGTTCTTCA.
For detection of transcripts regulated by XBP1s, RNA isolated from sorted experiments as described above was analyzed by qRT-PCR using the RNA-to-Ct 1-Step Kit. The following primer pairs were used: human ERdj4, 5ʹ-AAAATAAGAGCCCGGATGCT and 3ʹ-CGCTTCTTGGATCCAGTGTT; human GFAT1, 5ʹ-GGACAGCACAACCTGCCTTT and 3ʹ-CAGCACTTGCATCAGAAGCAA; human p58IPK, 5ʹ-CTCAGTTTCATGCTGCCGTA and 3ʹ-TTGCTGCAGTGAAGTCCATC; and human 18S ribosomal RNA, 5ʹ-CGCCGCTAGAGGTGAAATTCT and 3ʹ-CATTCTTGGCAAATGCTTTCG.
For detection of cytokine transcripts during conditions of UPR induction in the presence of L. pneumophila-derived PAMPs, A/J BMDMs were plated at 8 × 105 cells/well. Wells were either uninfected or treated with HKLp at an effective MOI of 30. Plates were spun down at 400 × g, then incubated at 37 °C for 6 h. Wells were washed three times in PBS and then lysed in buffer RLT, and RNA preparation and qRT-PCR were performed as described above. The following primer pairs were used for detection of transcripts: mouse Il6, 5ʹ-GAGGATACCACTCCCAACAGACC and 3ʹ AAGTGCATCATCGTTGTTCATACA; mouse Tnfα, 5ʹ-GCACCACCATCAAGGACTCAA and 3ʹ- GCTTAAGTGACCTCGGAGCT; and mouse Gapdh, 5ʹ-TGTGTCCGTCGTGGATCTGA and 3ʹ- CCTGCTTCACCACCTTCTTGAT.
Translation, Labeling, and Quantification.
To measure host cell translation, U937 cells were challenged with WT-GFP and the ∆5-GFP strain at an MOI of 1 for 9 h. The medium was replaced with RPMI lacking methionine (Invitrogen), and cells were incubated for 1 h at 37 °C. The medium was then replaced with fresh methionine-free medium containing 50 µM AHA (Invitrogen), and cells were incubated for an additional 1 h at 37 °C. Cells were washed with HBSS, lifted with trypsin, washed with PBS, fixed with 4% (wt/vol) paraformaldehyde for 20 min at room temperature, and then washed three times with PBS and stored at 4 °C. Cells were blocked with 1% BSA for 30 min at room temperature, then incubated for 1–3 h at 37 °C in 1% BSA with 100 µM APC-phosphine. Washing was performed with 0.5% Tween-20, followed by two washes with PBS. Flow cytometry analysis was performed on 20,000 cells in a live cell gate using a BD FACSCalibur system.
An adaptation of the SUnSET immunofluorescence microscopy protocol was used to determine levels of translation (37). A/J BMDMs plated at 2 × 105 on coverslips were challenged at an MOI of 0.5, centrifuged at 400 × g for 5 min, and then incubated at 37 °C for 1 h. Cells were washed three times with warm medium, then incubated at 37 °C for another 9 h. The medium was replaced with RPMI containing 1 µg/mL puromycin (Sigma-Aldrich) for another 1 h. Cells were fixed and stained, as for intracellular replication, with the addition of anti-puromycin (12D10; Millipore) at 1:200 to detect incorporation of puromycin into ribosomes.
Acknowledgments
We thank Connor Murphy for technical assistance and Seble Asrat, Kim Davis, Dervla Isaac, and Vinay Ramabhadran for a critical reading of the manuscript. The strain ∆5 and plasmids allowing for bacterial expression of Lgt3 or Lgt3* were graciously donated by Zhao-Qing Luo, PhD. Femurs used to isolate MyD88−/− macrophages were provided by Tanja Petnicki Ocwieja, PhD. Sorting experiments were performed at the Tufts University Flow Cytometry Core, with helpful assistance from Stephen Kwok. R.R.I. is an investigator of the Howard Hughes Medical Institute (HHMI), and this work was supported by HHMI. A.D.H. was supported by National Institutes of Health Training Grants T32AI007422 and T32GM007310.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508716112/-/DCSupplemental.
References
- 1.Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7(1):7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 2.Marra A, Blander SJ, Horwitz MA, Shuman HA. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc Natl Acad Sci USA. 1992;89(20):9607–9611. doi: 10.1073/pnas.89.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang L, et al. The E Block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol. 2011;13(2):227–245. doi: 10.1111/j.1462-5822.2010.01531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu W, et al. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS One. 2011;6(3):e17638. doi: 10.1371/journal.pone.0017638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isaac DT, Isberg R. Master manipulators: An update on Legionella pneumophila Icm/Dot translocated substrates and their host targets. Future Microbiol. 2014;9(3):343–359. doi: 10.2217/fmb.13.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 7.Asrat S, de Jesús DA, Hempstead AD, Ramabhadran V, Isberg RR. Bacterial pathogen manipulation of host membrane trafficking. Annu Rev Cell Dev Biol. 2014;30:79–109. doi: 10.1146/annurev-cellbio-100913-013439. [DOI] [PubMed] [Google Scholar]
- 8.Swanson MS, Isberg RR. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun. 1995;63(9):3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swanson MS, Isberg RR. Identification of Legionella pneumophila mutants that have aberrant intracellular fates. Infect Immun. 1996;64(7):2585–2594. doi: 10.1128/iai.64.7.2585-2594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kagan JC, Roy CR. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat Cell Biol. 2002;4(12):945–954. doi: 10.1038/ncb883. [DOI] [PubMed] [Google Scholar]
- 11.Neunuebel MR, Machner MP. The taming of a Rab GTPase by Legionella pneumophila. Small GTPases. 2012;3(1):28–33. doi: 10.4161/sgtp.18704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Creasey EA, Isberg RR. The protein SdhA maintains the integrity of the Legionella-containing vacuole. Proc Natl Acad Sci USA. 2012;109(9):3481–3486. doi: 10.1073/pnas.1121286109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 14.Schröder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 15.Liu CY, Xu Z, Kaufman RJ. Structure and intermolecular interactions of the luminal dimerization domain of human IRE1alpha. J Biol Chem. 2003;278(20):17680–17687. doi: 10.1074/jbc.M300418200. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 17.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313(5783):104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 18.Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006;7(9):880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasnain SZ, Lourie R, Das I, Chen AC, McGuckin MA. The interplay between endoplasmic reticulum stress and inflammation. Immunol Cell Biol. 2012;90(3):260–270. doi: 10.1038/icb.2011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4(7):499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 21.Qiu Q, et al. Toll-like receptor-mediated IRE1α activation as a therapeutic target for inflammatory arthritis. EMBO J. 2013;32(18):2477–2490. doi: 10.1038/emboj.2013.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11(5):411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho JA, et al. The unfolded protein response element IRE1α senses bacterial proteins invading the ER to activate RIG-I and innate immune signaling. Cell Host Microbe. 2013;13(5):558–569. doi: 10.1016/j.chom.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.de Jong MF, et al. Sensing of bacterial type IV secretion via the unfolded protein response. MBio. 2013;4(1):e00418–e12. doi: 10.1128/mBio.00418-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith JA, et al. Brucella induces an unfolded protein response via TcpB that supports intracellular replication in macrophages. PLoS Pathog. 2013;9(12):e1003785. doi: 10.1371/journal.ppat.1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shang J, Lehrman MA. Discordance of UPR signaling by ATF6 and Ire1p-XBP1 with levels of target transcripts. Biochem Biophys Res Commun. 2004;317(2):390–396. doi: 10.1016/j.bbrc.2004.03.058. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto K, Tashiro E, Imoto M. Quinotrierixin-inhibited ER stress-induced XBP1 mRNA splicing through inhibition of protein synthesis. Biosci Biotechnol Biochem. 2011;75(2):284–288. doi: 10.1271/bbb.100622. [DOI] [PubMed] [Google Scholar]
- 28.Belyi Y, et al. Legionella pneumophila glucosyltransferase inhibits host elongation factor 1A. Proc Natl Acad Sci USA. 2006;103(45):16953–16958. doi: 10.1073/pnas.0601562103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen X, et al. Targeting eEF1A by a Legionella pneumophila effector leads to inhibition of protein synthesis and induction of host stress response. Cell Microbiol. 2009;11(6):911–926. doi: 10.1111/j.1462-5822.2009.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fontana MF, et al. Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog. 2011;7(2):e1001289. doi: 10.1371/journal.ppat.1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asrat S, Dugan AS, Isberg RR. The frustrated host response to Legionella pneumophila is bypassed by MyD88-dependent translation of pro-inflammatory cytokines. PLoS Pathog. 2014;10(7):e1004229. doi: 10.1371/journal.ppat.1004229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23(21):7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang ZV, et al. Spliced X-box binding protein 1 couples the unfolded protein response to hexosamine biosynthetic pathway. Cell. 2014;156(6):1179–1192. doi: 10.1016/j.cell.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pillich H, Loose M, Zimmer KP, Chakraborty T. Activation of the unfolded protein response by Listeria monocytogenes. Cell Microbiol. 2012;14(6):949–964. doi: 10.1111/j.1462-5822.2012.01769.x. [DOI] [PubMed] [Google Scholar]
- 35.Raciti M, Lotti LV, Valia S, Pulcinelli FM, Di Renzo L. JNK2 is activated during ER stress and promotes cell survival. Cell Death Dis. 2012;3:e429. doi: 10.1038/cddis.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanov SS, Roy CR. Pathogen signatures activate a ubiquitination pathway that modulates the function of the metabolic checkpoint kinase mTOR. Nat Immunol. 2013;14(12):1219–1228. doi: 10.1038/ni.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6(4):275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 38.Pavio N, Romano PR, Graczyk TM, Feinstone SM, Taylor DR. Protein synthesis and endoplasmic reticulum stress can be modulated by the hepatitis C virus envelope protein E2 through the eukaryotic initiation factor 2alpha kinase PERK. J Virol. 2003;77(6):3578–3585. doi: 10.1128/JVI.77.6.3578-3585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tardif KD, Mori K, Kaufman RJ, Siddiqui A. Hepatitis C virus suppresses the IRE1-XBP1 pathway of the unfolded protein response. J Biol Chem. 2004;279(17):17158–17164. doi: 10.1074/jbc.M312144200. [DOI] [PubMed] [Google Scholar]
- 40.Treacy-Abarca S, Mukherjee S. Legionella suppresses the host unfolded protein response via multiple mechanisms. Nat Commun. 2015;6:7887. doi: 10.1038/ncomms8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mehlitz A, et al. The chlamydial organism Simkania negevensis forms ER vacuole contact sites and inhibits ER-stress. Cell Microbiol. 2014;16(8):1224–1243. doi: 10.1111/cmi.12278. [DOI] [PubMed] [Google Scholar]
- 42.Qin QM, et al. RNAi screen of endoplasmic reticulum-associated host factors reveals a role for IRE1alpha in supporting Brucella replication. PLoS Pathog. 2008;4(7):e1000110. doi: 10.1371/journal.ppat.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richardson CE, Kooistra T, Kim DH. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature. 2010;463(7284):1092–1095. doi: 10.1038/nature08762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreno AA, et al. IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS One. 2012;7(2):e31944. doi: 10.1371/journal.pone.0031944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Connor TJ, Boyd D, Dorer MS, Isberg RR. Aggravating genetic interactions allow a solution to redundancy in a bacterial pathogen. Science. 2012;338(6113):1440–1444. doi: 10.1126/science.1229556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Schadewijk A, van’t Wout EF, Stolk J, Hiemstra PS. A quantitative method for detection of spliced X-box binding protein-1 (XBP1) mRNA as a measure of endoplasmic reticulum (ER) stress. Cell Stress Chaperones. 2012;17(2):275–279. doi: 10.1007/s12192-011-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]