Significance
Unique among the large number of noncoding RNA species, the pericentromeric human satellite II (HSATII) repeat is massively expressed in a broad set of epithelial cancers but is nearly undetectable in normal tissues. Here, we show that deregulation of HSATII expression is tightly linked to growth under nonadherent conditions, and we uncover an unexpected mechanism by which HSATII RNA-derived DNA (rdDNA) leads to progressive elongation of pericentromeric regions in tumors. The remarkable specificity of HSATII overexpression in cancers, together with the consequences of targeting its RT, points to a potential novel vulnerability of cancer cells.
Keywords: satellites, reverse transcription, repeats, cancer
Abstract
Aberrant transcription of the pericentromeric human satellite II (HSATII) repeat is present in a wide variety of epithelial cancers. In deriving experimental systems to study its deregulation, we observed that HSATII expression is induced in colon cancer cells cultured as xenografts or under nonadherent conditions in vitro, but it is rapidly lost in standard 2D cultures. Unexpectedly, physiological induction of endogenous HSATII RNA, as well as introduction of synthetic HSATII transcripts, generated cDNA intermediates in the form of DNA/RNA hybrids. Single molecule sequencing of tumor xenografts showed that HSATII RNA-derived DNA (rdDNA) molecules are stably incorporated within pericentromeric loci. Suppression of RT activity using small molecule inhibitors reduced HSATII copy gain. Analysis of whole-genome sequencing data revealed that HSATII copy number gain is a common feature in primary human colon tumors and is associated with a lower overall survival. Together, our observations suggest that cancer-associated derepression of specific repetitive sequences can promote their RNA-driven genomic expansion, with potential implications on pericentromeric architecture.
Pericentromeric satellite repeats are essential core centromere-building elements that stabilize interactions with DNA-binding proteins, maintain heterochromatin architecture, sustain kinetochore formation, and drive chromosomal segregation during mitosis, thereby ensuring faithful duplication of the genome (1). Transcription from pericentromeric satellites has been reported in plants and invertebrates, as well as during early stages of vertebrate development, and some types of satellite repeats are induced following environmental stress in cell line models (2). We recently reported the massive overexpression of specific classes of satellite repeats in human epithelial cancers, resulting from aberrant transcription of these pericentromeric domains (3). In almost all cancers analyzed, subsets of pericentromeric satellites are expressed at very high levels (3–5), whereas others show consistently reduced expression compared with normal tissues.
The human satellite II (HSATII) is the most differentially expressed satellite subfamily in epithelial cancers (3). It constitutes the main component of pericentromeric heterochromatin on chromosomes 2, 7, 10, 16, and 22 (UCSC Genome Browser, genome.ucsc.edu), and it is also found at chromosome band 1q12, where it is colocated with satellite III sequences. HSATII is defined by tandemly repeated divergent variants of 23- to 26-bp consensus sequences, organized in long arrays that may span up to thousands of kilobases (6). Although repetitive DNA sequences are frequently hypomethylated in cancer cells (7), the mechanisms underlying their aberrant expression are not well understood. For instance, loss of DNA methylation alone does not result in overexpression of the HSATII satellite (8), suggesting the existence of more complex regulatory networks.
In establishing models to study the molecular basis of HSATII RNA overexpression in cancer, we found that growth of cells under nonadherent conditions is sufficient to trigger induction of this satellite repeat. Unexpectedly, under these and other conditions, we uncovered that these repeated transcripts are reverse-transcribed into DNA/RNA hybrids. The reintegration of HSATII RNA-derived DNA (rdDNA) is correlated with a progressive expansion of host HSATII genomic loci. These results point to an unexpected plasticity of pericentromeric repeat-containing structures during cancer progression.
Results and Discussion
Detection of HSATII Expression in Human Tumors and 3D Cancer Cell Models.
The highly repetitive nature of satellites precludes their precise quantitation and qualitative analysis using PCR-based RNA sequencing approaches. We previously showed that PCR-independent single molecule next-generation sequencing [digital gene expression (DGE) profiling] is uniquely sensitive and quantitative (9), although it is not suited to routine analysis and does not provide qualitative length information. To enable experimental models for the study of HSATII deregulation, we first designed a modified Northern blot HSATII assay (SI Appendix, Fig. S1A). HSATII satellite transcripts encompass arrays of variable lengths derived from multiple different genomic locations (6); thus, Northern blotting generates a pattern of bands ranging from 30 nt to greater than 800 nt in size (SI Appendix, Fig. S1B), consistent with the pattern reported for other repeats, such as murine minor satellites (10) and human satellite III (11). Quantitation of DGE profiles and Northern blot signal intensity for matched primary cancer specimens were highly correlated (SI Appendix, Fig. S1 B and C).
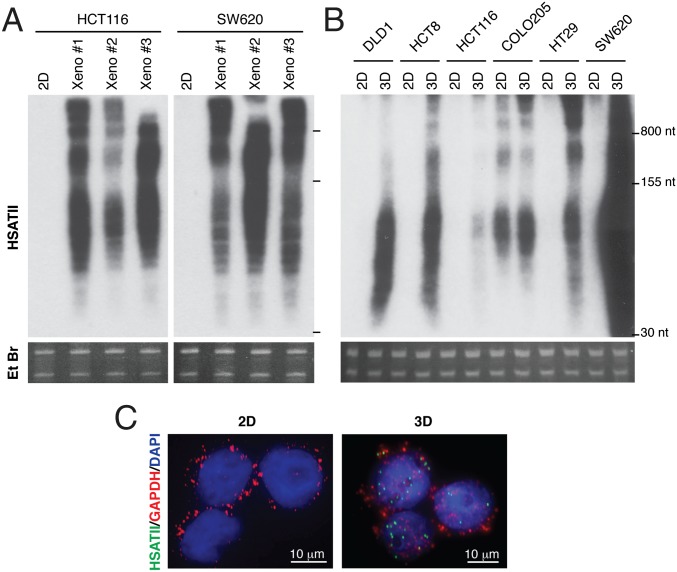
We observed that human colorectal cancer cell lines do not express HSATII under standard in vitro adherent (2D) culture conditions, but strongly up-regulate its expression when grown as tumor xenografts (Fig. 1A). To define specific experimental conditions that modulate HSATII expression within tumors, we tested multiple stimuli associated with cellular stress and tumorigenesis, including hypoxia, UV irradiation, heat shock, oxidative stress, overconfluence, treatment with demethylating agents, coculturing with stromal-derived feeder layers, and culture under anchorage-free conditions (SI Appendix, Fig. S2 A–D). Remarkably, only culture under nonadherent conditions, as 3D tumor spheres in solution or in soft agar, led to robust induction of HSATII in five colorectal cancer cell lines, as detected by Northern blotting (Fig. 1B). The specific induction of HSATII RNA by anchorage-independent growth was also demonstrated using RNA in situ hybridization, an imaging assay that does not involve nucleic acid extraction or denaturation, confirming the RNA specificity of the hybridization signal (Fig. 1C). A sixth colorectal cancer line, COLO205, noteworthy for its semiadherent growth pattern, was unique in expressing HSATII RNA at baseline under standard culture conditions (Fig. 1B). Notably, the elevated RNA levels detected in tumor spheres and xenografts from two independent cell lines were rapidly lost upon reestablishing the growth of these cells under adherent 2D culture (SI Appendix, Fig. S2E). Consistent with our previous findings in primary tumors (3), HSATII transcripts were present in both sense (S) and antisense (AS) orientations (SI Appendix, Fig. S2F), and they were primarily localized to the nuclear compartment in a panel of colon cancer cell lines (SI Appendix, Fig. S2G), similar to other repetitive noncoding RNAs such as telomeric repeat-containing RNA (TERRA) (12).
Fig. 1.
HSATII is expressed in human tumors and 3D cancer cell models. (A) Northern blot analysis of HSATII expression in HCT116 and SW620 cells grown as 2D cultures or xenografts (Xeno). (B) Northern blot analysis of HSATII expression in colon cancer cell lines grown as 2D cultures or tumor spheres (3D). Ethidium bromide (Et Br) stainings of gels are shown for each Northern blot analysis as loading controls. (C) RNA in situ hybridization (with the indicated fluorescent probes) of SW620 cells cultured under 2D conditions or as tumor spheres (3D). HSATII/DAPI colocalization coefficient measured by confocal imaging: R = 0.6 ± 0.04.
Previous studies have shown that satellite expression in both human and mouse cells may be due to a combination of environmental stimuli and genetic factors. For instance, human satellite III is expressed in response to cellular stress, including UV-C, oxidative, heat shock, and hyperosmotic stress (13), whereas mouse pericentromeric major satellites can be induced by growth to confluence or treatment with hypomethylating, apoptotic, or differentiation-inducing agents (10). Genetic lesions in the tumor suppressor BRCA1 lead to Alpha-satellite sequence derepression in human breast epithelial cells (5), and deletion of Trp53 in mouse cells results in murine major satellite expression (14). Remarkably, the HSATII pericentromeric repeat is resistant to general environmental stressors and differences in oncogene or tumor suppressor genotypes under standard adherent culture. Only growth under nonadherent conditions is sufficient to induce robust HSATII expression across different cancer cell lines, an interesting phenomenon that merits further investigation.
Identification of Medium/Small-Molecular-Weight HSATII DNA.
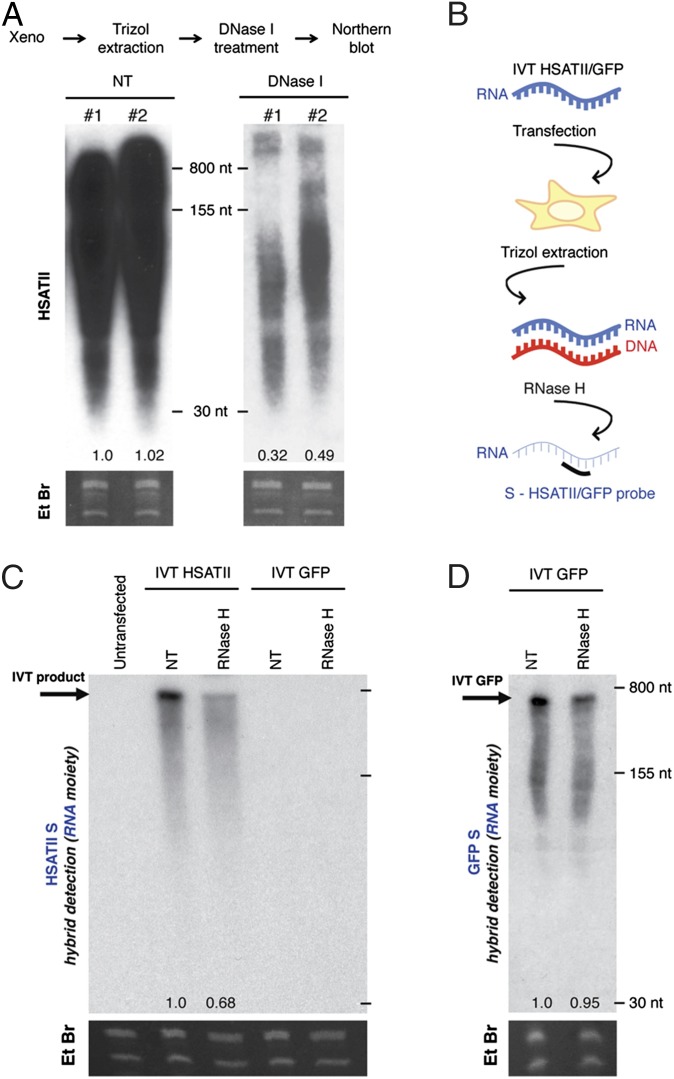
Unexpectedly, we observed that a fraction of the xenograft-induced HSATII sequences present within medium/small-molecular-weight nucleic acids (extraction with Trizol; ThermoFisher) was sensitive to DNase I (Fig. 2A). This event may result from genomic DNA (gDNA) contamination or from the existence of small dsDNA, dsRNA partially susceptible to DNase I, and/or DNA/RNA hybrids. To exclude the possibility of gDNA contamination, we performed multiple controls. First, during nucleic acid extraction, we applied a solid barrier to prevent any cross-contamination between the aqueous phase and the organic, gDNA-containing phase (phase lock gel tubes) (SI Appendix, Fig. S3A). Second, we showed that high-molecular-weight HSATII gDNA from a colon cancer xenograft is readily distinguished by Northern blotting from separately processed small-molecular-weight RNA/DNA from the same tissue, without evidence of cross-contamination (SI Appendix, Fig. S3B). Third, using multiple cell lines, we showed that the presence of medium/small-molecular-weight HSATII sequences is only evident following culture under 3D or xenograft conditions. Identical and simultaneous analysis of these cells cultured under 2D conditions showed no evidence of these small HSATII sequences (Fig. 1 A and B and SI Appendix, Fig. S2). Finally, under identical processing conditions, the rapid loss of HSATII signal following replating of 3D cultures into 2D cultures further excluded gDNA contamination as a source for the HSATII DNA signal on Northern blotting (SI Appendix, Fig. S2E). We therefore concluded that deregulated HSATII satellite transcripts coexist with matched DNA fragments within the medium/small-molecular-weight nucleic acid fraction of cells that overexpress this satellite repeat. Given the presence of small RNA and DNA species, we hypothesized that HSATII RNA may be reverse-transcribed into DNA/RNA hybrids and, ultimately, dsDNA (SI Appendix, Fig. S3C), a phenomenon known to occur with other repetitive elements, such as long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), and long terminal repeat (LTR) retrotransposons (15).
Fig. 2.
Ectopic HSATII RNA gives rise to DNA/RNA intermediates. (A) Northern blot analysis of HSATII in untreated (NT) or DNase I-treated extracts obtained from SW620 xenografts. Numbers below indicate relative signal quantitation. (B) TRIzol extracts obtained from 293T cells 24 h after transfection with HSATII/GFP IVT products were subjected to RNase H treatment, followed by Northern blotting and hybridization to detect the RNA strand (S-HSATII) of the hybrid. (C) Northern blot analysis of extracts from 293T cells either untransfected or transfected with IVT HSATII or GFP, subjected to the indicated nuclease treatment and probed for HSATII S. (D) Northern blot analysis of extracts from 293T cells after transfection with IVT GFP, treated with RNase H and probed for GFP S. Numbers below indicate relative signal quantitation.
Generation of DNA/RNA Hybrids upon Ectopic Introduction of HSATII RNA.
To capture the HSATII RNA-to-DNA conversion, we first developed an assay to introduce synthetically produced HSATII RNA generated by in vitro transcription (IVT) directly into 2D-cultured 293T cells that lack endogenous HSATII expression (SI Appendix, Fig. S3D). To assess the formation of DNA/RNA hybrids in cells transfected with single-stranded IVT HSATII RNA (HSATII-chr10), we subjected nucleic acid extracts to treatment with RNase H, which specifically digests the RNA moiety of DNA/RNA hybrids but does not affect either ssRNA or the DNA component of DNA/RNA hybrids (Fig. 2B). Indeed, RNase H treatment caused a strong reduction in the Northern blot signal identified for the HSATII S (but not AS) sequence (Fig. 2C and SI Appendix, Fig. S3E). Thus, consistent with the generation of rdDNA, a fraction of the IVT-produced RNA is within a complex with a cDNA strand. Transfection of comparable amounts of IVT GFP RNA produced the expected RNA signal but showed no significant sensitivity to RNase H (Fig. 2D), and introduction of GFP RNA did not lead to the induction of HSATII satellite RNA (Fig. 2C, last two lanes). These findings exclude the occurrence of nonspecific responses to RNA transfection. We conclude that introduction of purified S HSATII RNA generates a DNA/RNA hybrid in IVT RNA-transfected cells. Because IVT using T7 polymerase relies on a PCR-generated DNA template as starting material, we included multiple controls to ensure the absence of DNA template contamination within the IVT product itself, as well as any genomic HSATII sequences in the cellular extracts (SI Appendix, Fig. S3 F–L). These results indicate that ectopically introduced single-stranded HSATII RNA is capable of generating cDNA within transfected cells.
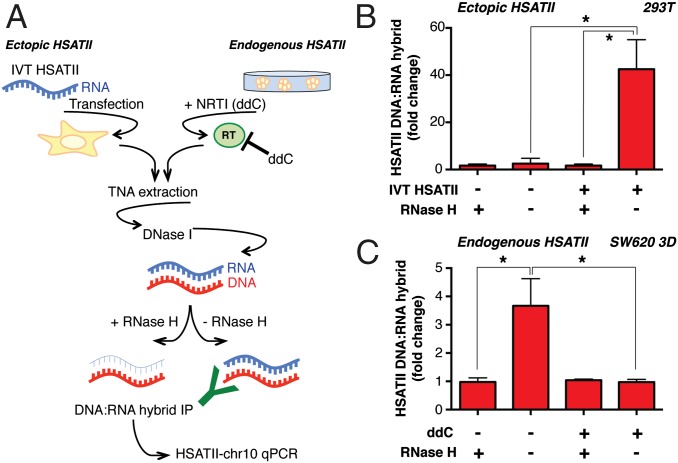
To validate these results further, we used the S9.6 monoclonal antibody, which is highly specific in its recognition of DNA/RNA hybrids (16–19). We established a DNA/RNA hybrid immunoprecipitation (DRIP) assay using quantitative PCR (qPCR) of S9.6 immunoprecipitates (HSATII-chr10 qPCR), which was applied to nucleic acids from untransfected or IVT HSATII RNA-transfected cells. Samples were first subjected to complete DNase I digestion (which removes all dsDNA but does not affect DNA/RNA hybrids), followed by DRIP analysis (Fig. 3A, Left). HSATII DNA/RNA duplexes were present only in 293T cells transfected with HSATII RNA, and treatment of extracts with RNase H effectively abolished immunoprecipitation of the HSATII DNA/RNA hybrids (Fig. 3B). Taken together, the formation of HSATII RNA/DNA hybrids in a controlled IVT model and the detection of these species by DRIP are suggestive of RT.
Fig. 3.
DRIP reveals the presence of ectopic as well as endogenous HSATII hybrids whose production is affected by RT inhibition. (A) Outline of the experimental layout. Total nucleic acids (TNAs) were isolated from IVT HSATII-transfected 293T cells or SW620 tumor spheres cultured in the presence of ddC or DMSO (ddC−). TNAs were treated with DNase I digestion to remove all potential gDNA contamination. DNA/RNA hybrids were then purified by immunomagnetic pull-down using a hybrid-specific antibody, and their relative quantities were measured by HSATII-chr10 qPCR. Pretreatment of TNA samples with RNase H as indicated demonstrates abrogation of DNA/RNA hybrid detection. (B) Fold change in the enrichment of DNA/RNA hybrids in HSATII-transfected 293T cells measured by qPCR after DRIP. (C) Fold enrichment of endogenous HSATII DNA/RNA hybrids in SW620 tumor spheres analyzed by HSATII-chr10 qPCR after DRIP. Fold changes were calculated based on percent input values, and the RNase H-treated samples were set at 1. For all charts, values represent the average of three independent experiments ± SEM. *P < 0.05 (t test).
RT of Endogenous HSATII RNA into DNA/RNA Hybrids.
To extend these analyses to a more physiological context, we assessed the presence of endogenous HSATII DNA/RNA hybrids by applying the DRIP assay to SW620 colon cancer cells grown as 3D tumor spheres (Fig. 3A, Right). RNase H-sensitive DNA/RNA HSATII hybrids were immunoprecipitated using the DRIP assay in SW620 spheroids (Fig. 3C). DNA/RNA hybrids were also identified in COLO205 semiattached cell cultures (SI Appendix, Fig. S3M), which are characterized by baseline expression of HSATII transcripts (Fig. 1B).
To evaluate the consequences of RT inhibition on the formation of these hybrids, we tested the effect of the nucleoside analog reverse transcriptase inhibitor (NRTI) 2′,3′-dideoxycytidine (ddC) in HSATII-expressing cells (Fig. 3A, Right). Notably, ddC is very poorly incorporated by replicative polymerases (20, 21), although it displays high specificity for multiple classes of RT, including LINE-1 (22). Treatment of SW620 spheroids and COLO205 cells with ddC significantly reduced the levels of endogenous HSATII DNA/RNA hybrids, as measured by the DRIP assay (Fig. 3C and SI Appendix, Fig. S3M). These observations are consistent with RT activity in HSATII-expressing cells, contributing to the generation of DNA/RNA structures derived from satellite transcripts.
RT activity in mammalian cells is derived primarily from retrotransposons, including LINE-1, the human endogenous retrovirus (HERV) family members, and human telomerase reverse transcriptase (hTERT). Definitive identification of the cellular RTs responsible for HSATII RT is complicated by the diversity of the retrovirally encoded enzyme families and the limited reagents available for their analysis. Attempts to reduce HERV and LINE-1 levels in our cell line models with previously published siRNAs (23) were unsuccessful. Because tools to evaluate hTERT were readily available, we undertook RNA immunoprecipitation analysis of endogenous hTERT from nuclear extracts of SW620 and HCT116 cell lines, followed by RT-qPCR for HSATII species. Significant enrichment of HSATII RNA with hTERT immunoprecipitates was evident, relative to control IgG (SI Appendix, Fig. S4 A and B). Importantly, this enrichment was only observed when the cancer cells were grown as xenografts, but not when they were cultured under standard 2D in vitro conditions. As a control, the coprecipitation with hTERT of telomerase RNA component (TERC), its primary RNA template for telomere elongation, was unaffected by growth conditions. Negligible coprecipitation with hTERT was observed for an unrelated noncoding RNA target. Furthermore, TERT knockdown using three independent siRNAs demonstrated significant reduction, although not complete depletion, of HSATII rdDNA species (SI Appendix, Fig. S4 C and D). Thus, hTERT may mediate HSATII RT, although the contribution of other known cellular RTs cannot be excluded.
Progressive Expansion of Pericentromeric Loci Through Stable Reintegration of HSATII DNA Sequences.
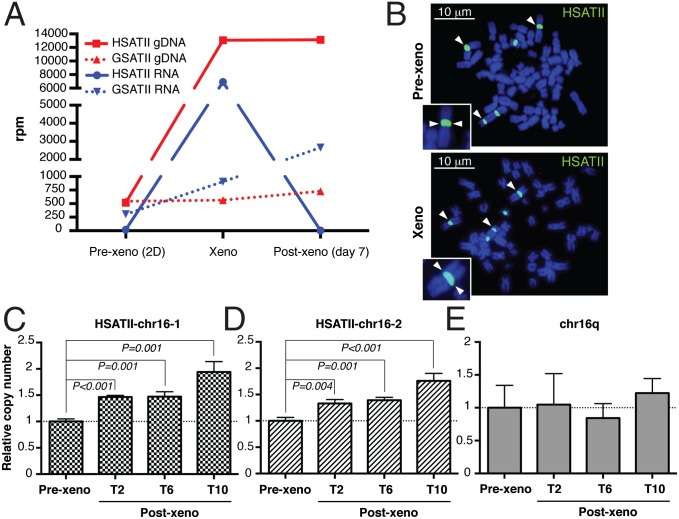
HSATII-derived rdDNA fragments within the cell nuclear fraction (SI Appendix, Fig. S2G) may either give rise to extrachromosomal elements or be integrated at chromosomal loci, leading to stable expansion of HSATII genomic sequences. By analogy, RT of LINE-1 transcripts, followed by retrotransposition at chromosomal loci, has been described in epithelial cancers, including colon carcinoma (24). To address this possibility, we first analyzed the dynamics of global HSATII RNA- and DNA-level changes using single molecule sequencing in SW620 colon cancer cells transitioned from 2D in vitro culture conditions to growth as mouse xenografts, and vice versa. As expected, the number of HSATII RNA reads was minimal when cells were cultured in 2D conditions, induced 360-fold as the cells gave rise to xenografts in mice, and then promptly down-regulated as xenograft-derived tumor cells were returned to in vitro 2D cultures (Fig. 4A, blue solid line). Remarkably, total cellular HSATII DNA copy number, which was already abundant at baseline, increased by 25-fold as 2D-cultured cells were transitioned to xenografts (taking into account the multiple-length variants of the HSATII tandem repeat unit). The amplified HSATII DNA sequences remained stably expanded despite the down-regulation of HSATII RNA transcripts when cells were reestablished under 2D culture conditions in vitro (Fig. 4A, red solid line). As a control, we analyzed gamma satellite II (GSATII), which is structurally similar to HSATII but whose expression is not deregulated in cancer (3). SW620 cells showed negligible GSATII changes, in either RNA or DNA content, as cells transitioned between 2D in vitro and xenograft culture conditions (Fig. 4A). Notably, the cancer-enriched human alpha satellite (ALR/Alpha) and simple satellite repeat (CATTC)n RNAs were also induced at high levels in xenografts with conjugate DNA copy number gains, suggesting a common mechanism of RT-mediated genomic expansion of particular repeat classes (SI Appendix, Table S1).
Fig. 4.
HSATII rdDNA is reintegrated at the same original loci in the genome, leading to pericentromere elongation in colon cancer xenografts. (A) DGE (RNA) and copy number (gDNA) analysis of satellite repeats (HSATII and GSATII) in the indicated samples (SW620) quantitated by single molecule sequencing. (B) Representative HSATII DNA FISH (white arrowheads) on metaphase spreads of prexenograft (Pre-xeno) 2D cultures and xenografts obtained from SW620 cells. (Insets) Enlarged (1,000×) HSATII-positive chromosomes. CNV in SW620 cells was assessed by qPCR on the HSATII–chr16-1 locus (C), HSATII–chr16-2 locus (D), and chromosome 16q arm (E). Cycle threshold values for all samples were normalized against β-actin, and DNA CNV is expressed relative to SW620 cells before xenograft implants, which was set at 1 (T2, T6, T10 = 1 wk of culture after the second, sixth, and 10th serial transplants, respectively). Error bars represent SD (n = 3).
To address the localization of the amplified HSATII DNA sequences, we performed HSATII DNA FISH analysis of 40 xenograft-derived metaphase spreads that did not reveal detectable extrachromosomal elements, and no hybridization signal was visible outside the five chromosomal loci known to harbor long arrays of pericentromeric HSATII (Fig. 4B). We could not discern increased fluorescence intensity or size of hybridization signal to demonstrate local expansion due to the limitations of this assay. However, consistent with the FISH data, alignment of HSATII gDNA reads obtained by single molecule sequencing from SW620 xenografts showed that the additional HSATII sequences were distributed among the various endogenous preexisting HSATII pericentromeric loci (SI Appendix, Fig. S5 A and B).
To model HSATII DNA copy gain over time and as a function of tumor progression, we serially transplanted SW620 cells as xenografts over multiple generations of mice. Progressive amplification of HSATII gDNA was evident over 10 successive rounds of in vivo tumor initiation, as measured in cultures derived from xenografted cells. This amplification was assessed using an ∼170-bp qPCR-based copy number variation (CNV) assay at the two highest density HSATII pericentromeric regions on chromosome 16q (HSATII–chr16-1 and HSATII–chr16-2; Fig. 4 C and D and SI Appendix, Fig. S5 C and D). An adjacent chromosomal region showed no xenograft transplantation-associated copy number changes, ruling out nonspecific gains in the 16q chromosomal arm or in ploidy (Fig. 4E and SI Appendix, Fig. S5E). We did not observe a single chromosome locus duplication event during tumor progression leading to a discrete increase in HSATII gDNA. Instead, the pericentromeric genomic loci demonstrated a gradual increase in HSATII gene copy number over time, all within preexisting satellite domains. Such a time line would be consistent with multiple rdDNA-mediated reintegration events.
Common Pericentromeric Expansion of HSATII Repeats in Human Colorectal Cancers.
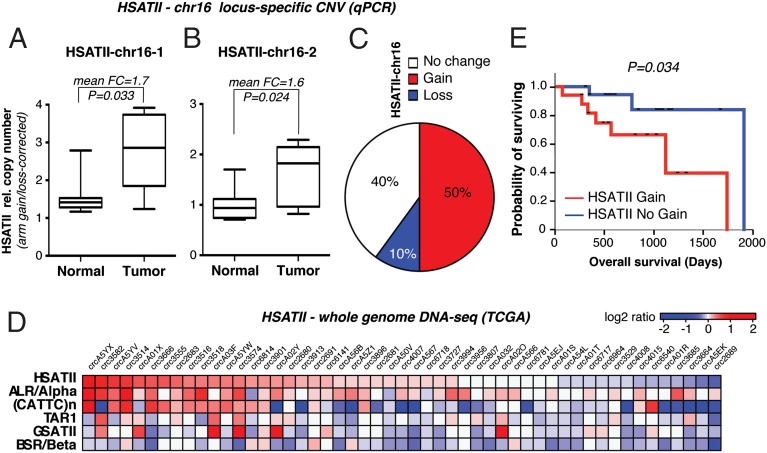
To determine whether HSATII copy number gains occur in primary human colon cancer, we analyzed CNV in 10 pairs of primary tumors and their matched adjacent normal tissue, focusing again on the chromosome 16q (HSATII–chr16-1 and HSATII–chr16-2) loci. After correcting for chr16q arm loss or gain, significantly increased HSATII copy number was evident at either or both of the two independent HSATII loci tested in five of 10 (50%) colon cancers (Fig. 5 A–C and SI Appendix, Fig. S6A). Among other cancers similarly analyzed, HSATII gene copy gain was evident in five of 13 (38%) kidney cancers (SI Appendix, Fig. S6B).
Fig. 5.
Pericentromeric HSATII repeats expand both locally and genome-wide in primary human colon cancer samples. CNV analysis of HSATII–chr16-1 (A) and HSATII–chr16-2 (B) loci on the indicated paired colon specimens (n = 10) is shown. For each sample, values were normalized for β-actin DNA and corrected for chr16q arm changes. Probability was measured by the paired t test. FC, mean fold change. (C) Relative percentage of HSATII copy number changes in colon tumor/normal pairs according to combined HSATII–chr16-1 and HSATII–chr16-2 CNV analysis, including correction for chr16 arm gains/losses. (D) Heat map of whole-genome sequencing data on the indicated primary colon cancer specimens based on a log2 ratio cutoff of 0.1. (E) Kaplan–Meier curve of overall survival (days) of patients with primary colon cancers with HSATII CNV gain or no gain. P = 0.034 (log-rank P value).
To extend our study of HSATII gene copy changes at specific loci, we performed a genome-wide survey of all such satellite repeats using a novel satellite CNV algorithm to undertake computational analyses of whole-genome sequencing from The Cancer Genome Atlas Project (TCGA). After correction of these data for large genomic alterations, comparable in size to HSATII stretches, we found that in fully annotated genomic sequences of 51 colorectal cancers (Dataset S1), 23 (45%) had statistically significant genomic gain of HSATII compared with their matched normal germ line (Fig. 5D). We extended this analysis to include additional satellite repeats, which revealed higher copy number gains in particular in the satellites whose expression was enriched in cancers [ALR/Alpha, HSATII, and (CATTC)n], but not in GSATII and other repeats whose expression is not deregulated in cancer cells (Fig. 5D and SI Appendix, Fig. S6C). HSATII copy gain co-occurred more frequently with ALR/Alpha and (CATTC)n, indicating a common mechanism for repeat expansion in some groups of repeats that is not shared with others (SI Appendix, Fig. S6C). In addition, alignments indicate that repeat expansions occur in the same locations consistent with our xenograft models. Of the 51 TCGA samples, 46 had annotated overall survival data that we analyzed to compare tumors with HSATII gain vs. no gain. Notably, Kaplan–Meier analysis demonstrated a significant reduction in overall survival in the HSATII gain vs. no-gain tumors (Fig. 5E; median overall survival: 1,096 vs. 1,881 d; log-rank P value = 0.034). Taken together, our data show that gene copy gains at HSATII-encoding pericentromeric repeats and other cancer-enriched satellites occur at preexisting repeat arrays and are a common and negative prognostic feature of colorectal cancers.
The mechanism underlying HSATII satellite repeat expansion in cancer remains to be defined, but it may involve an RT/reintegration phenomenon analogous to the phenomenon described for other major repetitive elements, such as LINE-1 (24) and telomeres (25, 26). Although the RT-directed expansion of pericentromeric sequences has not been described in human cells, there is ample precedent for retroelement-mediated integration of such repeats in plants (27, 28). In mammalian cells, integration of nonretroelement sequences within centromeres has not been reported, with the exception of the marsupial tammar wallaby, whose exceptionally short centromeres harbor signatures of retroviral insertions alongside domains of satellite-rich sequences (29). Thus, by analogy with retroviral elements, LINE-1, and telomeres, pericentromeric repeats may expand through the activity of endogenous RT enzymes. However, we cannot unequivocally discriminate between candidate RTs or exclude other mechanisms, including replication-induced errors or epigenetic modifier-induced site-specific copy gain (30–32).
Suppression of HSATII RT Inhibits Tumor Growth and Impairs Copy Number Gains.
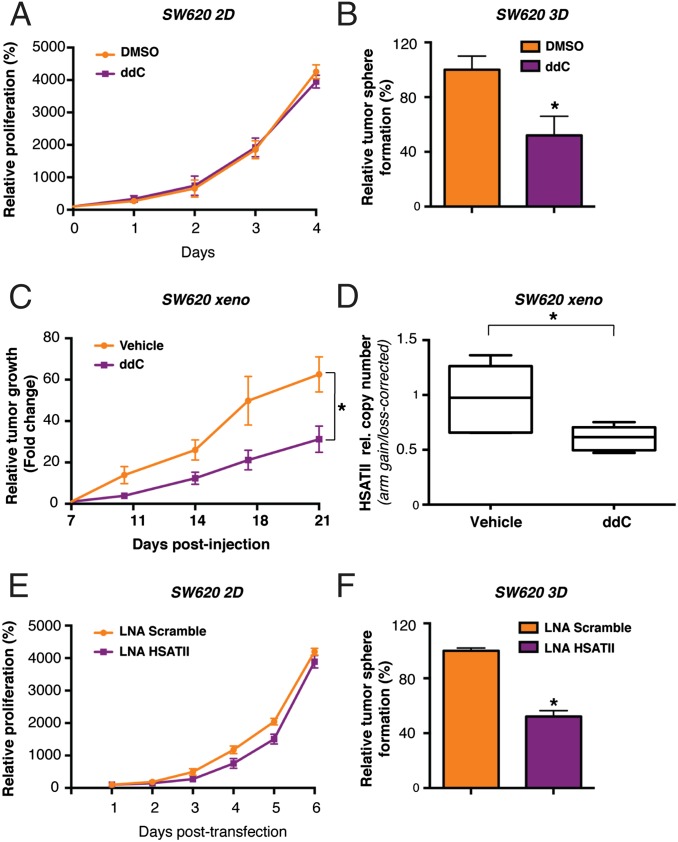
The critical role of pericentromeric repeats in preserving chromosomal integrity is well established (1). However, the unexpected RT-associated mechanism by which pericentromeres are expanded in cancer cells raises the possibility that its disruption may affect tumor growth. Given the inhibition of HSATII rdDNA formation by treatment of cells with the nucleoside analog ddC, we first tested the effect of this NRTI on cell proliferation. Treatment of SW620 cells with ddC inhibited their proliferation under 3D conditions but had minimal effect in 2D culture (Fig. 6 A and B). Mouse tumor xenografts generated from SW620 cells also showed sensitivity to ddC, with a 50% reduction in tumor diameter at 21 d (P = 0.03; Fig. 6C). The antiproliferative effect of ddC was accompanied by a reduction of HSATII copy gain in tumor xenografts (Fig. 6D). In a second colon cancer cell line, HCT116, the growth of tumor xenografts was suppressed using a combination of two RT inhibitors, ddC and 2′,3′-didehydro-2′,3′-dideoxythymidine (d4T), but not using ddC alone, and this effect was again associated with a reduced HSATII copy number gain (SI Appendix, Fig. S7 A–C). To test an alternative strategy to target HSATII, we synthesized a locked nucleic acid (LNA) oligonucleotide complementary to the HSATII sequence. Treatment of SW620 cells with this HSATII-directed LNA had no effect when the cells were grown in two dimensions, but it had a strong inhibitory effect on tumor sphere formation (Fig. 6 E and F). This effect was associated with accumulation of cells in the G0/G1 phase of the cell cycle (SI Appendix, Fig. S7D). Taken together, these results raise the intriguing possibility that targeting HSATII transcripts or suppressing cellular reverse transcriptase activity may selectively suppress proliferation of cancer cells under anchorage-independent conditions (SI Appendix, Fig. S7E). Beyond their potential contribution to the viability of proliferating cancer cells, HSATII transcripts may also play a role in shaping tumor–host interactions, as shown in the accompanying paper (33). Thus, the disruption of HSATII RT may have direct effects on cancer cells, as well as modulating the immune response against tumor cells. Given the very high frequency of HSATII deregulation in epithelial cancers (3), such a therapeutic vulnerability might have broad significance.
Fig. 6.
RT blockage, as well as LNA-mediated inhibition of HSATII transcripts, affects tumor sphere growth, impairs tumorigenesis, and prevents pericentromeric copy number gains in vivo. (A) Proliferation assay on DMSO- and ddC-treated SW620 cells. Values (%) were normalized against the signal derived from viable cells on the day of seeding, which was set at 100%. (B) Tumor sphere-forming ability of SW620 cells was tested upon culture in the presence of ddC for 15 d. Values (%) were normalized against the amount of spheres in the DMSO control, which was set at 100%. *P < 0.05 (t test). (C) In vivo tumor growth of SW620 cell xenografts. Mice were treated daily by i.p. injection of 25 mg/kg ddC or vehicle alone, starting 1 wk after tumor cell injection. Tumor size at this stage was set at 1 to calculate relative size fold change over time. Error bars represent SEM (n = 6). *P < 0.05 (t test). (D) CNV analysis of HSATII by qPCR on the chr16-1 locus in tumors recovered from untreated (Vehicle) or ddC-treated mice (n = 6). Values were normalized for β-actin and expressed as HSATII/chr16q arm ratios. *P < 0.05 (t test). (E) Proliferation assay on SW620 cells upon transfection with an HSATII-specific or Scramble LNA. Values (%) were normalized against the signal derived from viable cells 1 d after transfection, which was set at 100%. (F) Tumor sphere-forming ability of SW620 cells was tested upon transfection with an HSATII-specific or Scramble LNA. Values (%) were normalized against the amount of spheres in the control, which was set at 100%. *P < 0.05 (t test). In A, B, E, and F, error bars represent SD (n = 3).
Materials and Methods
Human normal and tumor tissues were deidentified and discarded excess tissue obtained from the Massachusetts General Hospital (MGH) according to an MGH Institutional Review Board (IRB)-approved protocol (2013P001854). The IRB determined consent was not needed for this study. Total RNA from normal human pancreas was purchased from Clontech. The results here are based, in part, upon data generated by the TCGA Research Network (cancergenome.nih.gov/). TCGA data are available from the TCGA portal. Helicos sequencing data were only analyzed for satellite expression, which is shown in SI Appendix. Given that it is not standard expression, we did not deposit the sequence into National Center for Biotechnology Information Gene Expression Omnibus. Further experimental details are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank M. Miri from the Massachusetts General Hospital (MGH) Tissue Repository for providing pathological specimens. We also thank N. J. Dyson and M. R. Motamedi for critically revising the manuscript, as well as R. Taulli and all laboratory members for helpful discussions. This work was supported by the Howard Hughes Medical Institute (D.A.H.), the National Foundation for Cancer Research (D.A.H.), National Cancer Institute (NCI) Grant R01CA129933 (to D.A.H. and F.B.), the Burroughs Wellcome Trust (D.T.T.), NIH Grant K12CA087723-11A1 (to D.T.T.), Department of Defense Grant W81XWH-13-1-0237 (to D.T.T.), the Warshaw Institute for Pancreatic Cancer Research (D.T.T.), the Verville Family Pancreatic Cancer Research Fund (D.T.T.), the Eleanor and Miles Shore Fellowship (to E.L.), the William Randolph Hearst Fund (E.L.), Susan G. Komen for the Cure Grant KG09042 (to S.M.), and the NCI Federal Share Program and Income (S.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 15008.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518008112/-/DCSupplemental.
References
- 1.Bierhoff H, Postepska-Igielska A, Grummt I. 2013. Noisy silence: Non-coding RNA and heterochromatin formation at repetitive elements. Epigenetics 9(1):53–61. [DOI] [PMC free article] [PubMed]
- 2.Eymery A, Callanan M, Vourc’h C. The secret message of heterochromatin: New insights into the mechanisms and function of centromeric and pericentric repeat sequence transcription. Int J Dev Biol. 2009;53(2-3):259–268. doi: 10.1387/ijdb.082673ae. [DOI] [PubMed] [Google Scholar]
- 3.Ting DT, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331(6017):593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eymery A, et al. A transcriptomic analysis of human centromeric and pericentric sequences in normal and tumor cells. Nucleic Acids Res. 2009;37(19):6340–6354. doi: 10.1093/nar/gkp639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Q, et al. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477(7363):179–184. doi: 10.1038/nature10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeanpierre M. Human satellites 2 and 3. Ann Genet. 1994;37(4):163–171. [PubMed] [Google Scholar]
- 7.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1(2):239–259. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tilman G, et al. 2012. Cancer-linked satellite 2 DNA hypomethylation does not regulate Sat2 non-coding RNA expression and is initiated by heat shock pathway activation. Epigenetics 7(8):903–913.
- 9.Lipson D, et al. Quantification of the yeast transcriptome by single-molecule sequencing. Nat Biotechnol. 2009;27(7):652–658. doi: 10.1038/nbt.1551. [DOI] [PubMed] [Google Scholar]
- 10.Bouzinba-Segard H, Guais A, Francastel C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci USA. 2006;103(23):8709–8714. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizzi N, et al. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell. 2004;15(2):543–551. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318(5851):798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 13.Valgardsdottir R, et al. Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res. 2008;36(2):423–434. doi: 10.1093/nar/gkm1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leonova KI, et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci USA. 2013;110(1):E89–E98. doi: 10.1073/pnas.1216922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 2011;12(9):615–627. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boguslawski SJ, et al. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J Immunol Methods. 1986;89(1):123–130. doi: 10.1016/0022-1759(86)90040-2. [DOI] [PubMed] [Google Scholar]
- 17.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12(3):711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Hu Z, Zhang A, Storz G, Gottesman S, Leppla SH. An antibody-based microarray assay for small RNA detection. Nucleic Acids Res. 2006;34(7):e52. doi: 10.1093/nar/gkl142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rigby RE, et al. RNA:DNA hybrids are a novel molecular pattern sensed by TLR9. EMBO J. 2014;33(6):542–558. doi: 10.1002/embj.201386117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kukhanova M, et al. L- and D-enantiomers of 2′,3′-dideoxycytidine 5′-triphosphate analogs as substrates for human DNA polymerases. Implications for the mechanism of toxicity. J Biol Chem. 1995;270(39):23055–23059. doi: 10.1074/jbc.270.39.23055. [DOI] [PubMed] [Google Scholar]
- 21.Louat T, et al. Antitumor activity of 2′,3′-dideoxycytidine nucleotide analog against tumors up-regulating DNA polymerase beta. Mol Pharmacol. 2001;60(3):553–558. [PubMed] [Google Scholar]
- 22.Dai L, Huang Q, Boeke JD. Effect of reverse transcriptase inhibitors on LINE-1 and Ty1 reverse transcriptase activities and on LINE-1 retrotransposition. BMC Biochem. 2011;12:18. doi: 10.1186/1471-2091-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oricchio E, et al. Distinct roles for LINE-1 and HERV-K retroelements in cell proliferation, differentiation and tumor progression. Oncogene. 2007;26(29):4226–4233. doi: 10.1038/sj.onc.1210214. [DOI] [PubMed] [Google Scholar]
- 24.Lee E, et al. Cancer Genome Atlas Research Network Landscape of somatic retrotransposition in human cancers. Science. 2012;337(6097):967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hastie ND, et al. Telomere reduction in human colorectal carcinoma and with ageing. Nature. 1990;346(6287):866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 26.Counter CM, et al. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11(5):1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang J, Birchler JA, Parrott WA, Dawe RK. A molecular view of plant centromeres. Trends Plant Sci. 2003;8(12):570–575. doi: 10.1016/j.tplants.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 28.Neumann P, et al. Plant centromeric retrotransposons: A structural and cytogenetic perspective. Mob DNA. 2011;2(1):4. doi: 10.1186/1759-8753-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carone DM, et al. A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma. 2009;118(1):113–125. doi: 10.1007/s00412-008-0181-5. [DOI] [PubMed] [Google Scholar]
- 30.Smith GP. Evolution of repeated DNA sequences by unequal crossover. Science. 1976;191(4227):528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- 31.Cohen Z, Bacharach E, Lavi S. Mouse major satellite DNA is prone to eccDNA formation via DNA Ligase IV-dependent pathway. Oncogene. 2006;25(33):4515–4524. doi: 10.1038/sj.onc.1209485. [DOI] [PubMed] [Google Scholar]
- 32.Black JC, et al. KDM4A lysine demethylase induces site-specific copy gain and rereplication of regions amplified in tumors. Cell. 2013;154(3):541–555. doi: 10.1016/j.cell.2013.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanne A, et al. Distinguishing the immunostimulatory properties of noncoding RNAs expressed in cancer cells. Proc Natl Acad Sci USA. 2015;112:15154–15159. doi: 10.1073/pnas.1517584112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.