Significance
Squashes, pumpkins, and gourds belonging to the genus Cucurbita were domesticated on several occasions throughout the Americas, beginning around 10,000 years ago. The wild forms of these species are unpalatably bitter to humans and other extant mammals, but their seeds are present in mastodon dung deposits, demonstrating that they may have been dispersed by large-bodied herbivores undeterred by their bitterness. However, Cucurbita may have been poorly adapted to a landscape lacking these large dispersal partners. Our study proposes a link between the disappearance of megafaunal mammals from the landscape, the decline of wild Cucurbita populations, and, ultimately, the evolution of domesticated Cucurbita alongside human cultivators.
Keywords: evolutionary ecology, sensory ecology, TAS2R genes, ancient DNA, archaeogenomics
Abstract
The genus Cucurbita (squashes, pumpkins, gourds) contains numerous domesticated lineages with ancient New World origins. It was broadly distributed in the past but has declined to the point that several of the crops’ progenitor species are scarce or unknown in the wild. We hypothesize that Holocene ecological shifts and megafaunal extinctions severely impacted wild Cucurbita, whereas their domestic counterparts adapted to changing conditions via symbiosis with human cultivators. First, we used high-throughput sequencing to analyze complete plastid genomes of 91 total Cucurbita samples, comprising ancient (n = 19), modern wild (n = 30), and modern domestic (n = 42) taxa. This analysis demonstrates independent domestication in eastern North America, evidence of a previously unknown pathway to domestication in northeastern Mexico, and broad archaeological distributions of taxa currently unknown in the wild. Further, sequence similarity between distant wild populations suggests recent fragmentation. Collectively, these results point to wild-type declines coinciding with widespread domestication. Second, we hypothesize that the disappearance of large herbivores struck a critical ecological blow against wild Cucurbita, and we take initial steps to consider this hypothesis through cross-mammal analyses of bitter taste receptor gene repertoires. Directly, megafauna consumed Cucurbita fruits and dispersed their seeds; wild Cucurbita were likely left without mutualistic dispersal partners in the Holocene because they are unpalatable to smaller surviving mammals with more bitter taste receptor genes. Indirectly, megafauna maintained mosaic-like landscapes ideal for Cucurbita, and vegetative changes following the megafaunal extinctions likely crowded out their disturbed-ground niche. Thus, anthropogenic landscapes provided favorable growth habitats and willing dispersal partners in the wake of ecological upheaval.
The wild precursors of domestic squashes (Cucurbita spp.) are adapted for a landscape inhabited by large herbivores. Their robust pepo fruits (1) were dispersed by large mammals, as revealed by intact Cucurbita seeds in mastodon dung deposits (2). Furthermore, Cucurbita is a weedy genus (3) well suited to the mosaic-like landscapes maintained by megafauna, which offer an abundance of disturbed habitat in a niche-diverse ecosystem (4). However, all megafauna >1,000 kg disappeared from the Americas by the early Holocene—through ecological shifts, human predation, or some combination of both (5)—leaving Cucurbita in a turbulent ecosystem lacking its mutualistic partners. Cucurbita is among many such anachronistic New World taxa (6), including avocado (Persea americana), chocolate (Theobroma cacao), Osage orange (Maclura pomifera), honey locust (Gleditsia triacanthos), and tree calabash (Crescentia cujete). Distributions of ancient samples show that some species of Cucurbita were distributed much more broadly in the past (e.g., refs. 7 and 8), suggesting that the Holocene has witnessed extirpation and refugiation of certain wild types. The decline of wild Cucurbita may have stemmed from both its loss of dispersal mutualists and the changing habitat. Other anachronistic plants have partnered with substitute dispersers (9), but most wild Cucurbita have not adapted in this way (10).
Although Cucurbita species declined in the wild, they thrived in domestication. The genus contains at least five domesticated species and has been used extensively throughout the Holocene (11), beginning around 10,000 B.P. in Mexico (12). Cucurbita domestication was a widespread phenomenon involving numerous wild precursor lineages throughout the Americas (11, 13–15), and these taxa now contain hundreds of cultivars and landraces grown worldwide (11, 15). In this study, we use a two-stage approach to study the simultaneous decline of wild Cucurbita and rise of its domestic counterparts. First, to assess aspects of Cucurbita domestication history and phylogeographic patterning, we recover and analyze complete plastid genomes from 91 total archaeological (n = 19) and modern wild (n = 30) and domestic (n = 42) Cucurbita specimens. Second, wild Cucurbita may have adapted poorly to dispersal anachronism partly for physiological reasons. Specifically, it produces a cytotoxic suite of cucurbitacins—harshly bitter triterpenoid compounds (16)—that deter small mammals (17) but appear to be harmless to megafauna. As such, Cucurbita was palatable to some megafauna (2) but possibly not to the majority of smaller New World mammals that survived into the Holocene; thus, Cucurbita survival was threatened by the disruption of its propagation strategy. By screening 46 mammal genomes for bitter taste receptor-encoding taste receptor type 2 (TAS2R) genes, we evaluate the hypothesis that mammal body size may be related directly to the palatability of Cucurbita and other bitter species. If smaller mammals have adapted mechanisms for better detecting and avoiding bitter, potentially toxic compounds in lower doses, Cucurbita may have coadapted specifically for a landscape containing large mammals; such adaptation would increase the potential impacts of a major shift toward an ecosystem lacking them. In sum, we combine archaeogenetic and comparative genomic techniques to probe the natural history of a genus that is ecologically anachronistic in the wild but is prolific in cultivation.
Results
Plastid Genome Analysis.
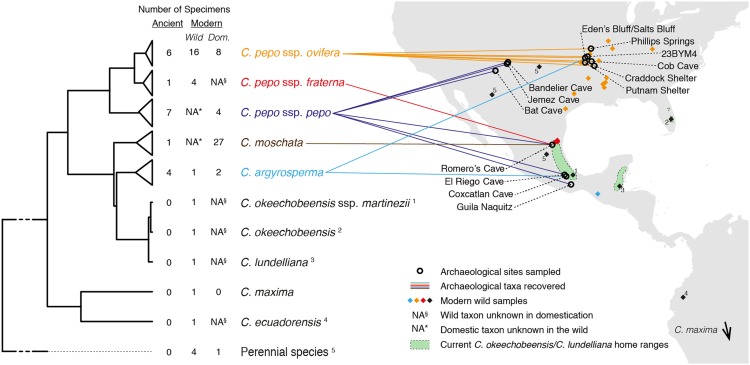
Using Illumina shotgun sequencing, we recovered and assembled complete plastid genomes from 12 of the 14 accepted extant Cucurbita species, representing 18 total species, subspecies, and varietal taxa. Using our completed assemblies for C. pepo ssp. pepo and C. moschata, we then designed and synthesized biotinylated RNA probes (18) to enrich additional modern and ancient DNA isolates for targeted sequencing of complete plastid genomes. Using this enrichment strategy, we tested 32 archaeological Cucurbita samples, yielding 19 ancient plastid genomes sufficiently complete for analysis. We also sequenced an additional 53 modern plastid genomes using a combination of shotgun sequencing and targeted enrichment, bringing the total sample size to 91 (n = 19 ancient; n = 72 modern) (Tables S1–S3), and reconstructed a Cucurbita plastid genome phylogeny from this dataset (Fig. 1).
Table S1.
Sample details for Cucurbita accessions analyzed: archaeological samples
| Laboratory ID | Taxon | Site | Site no. | Accession no. | Tissue |
| 101 | C. pepo ssp. ovifera | Putnam Shelter, AR | 3WA4 | 32-44-291 | Rind |
| 102 | C. pepo ssp. ovifera | Salts Bluff, AR | 3BE18 | 33-12-125 | Rind |
| 103 | C. pepo ssp. ovifera | 23BYM4, MO | 23BYM4 | 32-34-107 | Rind |
| 104 | C. pepo ssp. ovifera | Craddock Shelter, AR | 3CW2 | 34-7-130 | Rind |
| 105 | C. pepo ssp. ovifera | Cob Cave, AR | 3NW6 | 31-72-25 | Rind |
| 106 | C. argyrosperma | Eden's Bluff, AR | 3BE6 | 31-72-25 | Rind |
| 107 | Cucurbita sp. | Salts Cave, KY | 15HT4 | CRF-81 | Rind |
| 108 | Cucurbita sp. | Salts Cave, KY | 15HT4 | CRF-85 | Rind |
| 109 | Cucurbita sp. | Salts Cave, KY | 15HT4 | CRF-95 | Rind |
| 110 | C. pepo ssp. ovifera | Phillips Spring, MO | 23HI216 | 1502 seed 8 | Seed |
| 111 | Cucurbita sp. | Phillips Spring, MO | 23HI216 | 1502 seed 13 | Seed |
| 112 | C. argyrosperma | Coxcatlan Cave, Puebla, Mexico | TC50 | XI 39/5 | Rind |
| 113 | Cucurbita sp. | Coxcatlan Cave, Puebla, Mexico | TC50 | XI 79/6 | Rind |
| 114 | C. argyrosperma | Coxcatlan Cave, Puebla, Mexico | TC50 | VII 70/8 | Rind |
| 115 | C. argyrosperma | Coxcatlan Cave, Puebla, Mexico | TC50 | V 37-4 | Rind |
| 116 | Cucurbita sp. | Romero's Cave, Tamaulipas, Mexico | TMC247 | occ 2 247/14 | Rind |
| 118 | Cucurbita sp. | Romero's Cave, Tamaulipas, Mexico | TMC247 | occ 5 247/72 | Rind |
| 119 | Cucurbita sp. | Romero's Cave, Tamaulipas, Mexico | TMC247 | occ 10 247 32 | Rind |
| 120 | Cucurbita sp. | Romero's Cave, Tamaulipas, Mexico | TMC247 | occ 11 247/93 | Rind |
| 121 | C. pepo ssp. pepo | Romero's Cave, Tamaulipas, Mexico | TMC247 | occ 14 247/243 | Rind |
| 122 | C. moschata | Romero's Cave, Tamaulipas, Mexico | TMC247 | occ 16 51/16 | Rind |
| 123 | C. pepo ssp. fraterna | Romero's Cave, Tamaulipas, Mexico | TMC247 | occ 8 247/89 | Rind |
| 139 | C. pepo ssp. pepo | Guila Naquitz, Oaxaca, Mexico | OC43 | INAH42(6N18) Zona B1 Cuadro C11 | Rind |
| 140 | C. pepo ssp. pepo | Guila Naquitz, Oaxaca, Mexico | OC43 | INAH58(6N20) Zona B(B1) Cuadro B9 | Rind |
| 141 | Cucurbita sp. | Jemez Cave, NM | 759 Lab 16801 | Rind | |
| 142 | C. pepo ssp. pepo | El Riego Cave, Puebla, Mexico | TC50 | Square16(S3E3) Level 3 Peduncle 21 | Peduncle |
| 143 | Cucurbita sp. | Newt Kash Hollow, KY | 15MF1 | 10842 Lab 16390 | Rind |
| 144 | C. pepo ssp. pepo | Bandelier Cave, NM | 2143 B | Rind | |
| 145 | Cucurbita sp. | Cloudsplitter Rockshelter, KY | 15MF36 | FS1580 ES8145 | Rind |
| 146 | C. pepo ssp. pepo | Jemez Cave, NM | 969 Lab 16291 | Seed | |
| 147 | Cucurbita sp. | Mammoth Cave, KY | 15ED1 | 87276 B16 Wartly | Rind |
| 148 | C. pepo ssp. pepo | Bat Cave, NM | LA44182 Bag 2144 SV45A | Rind |
Table S3.
Sample details for Cucurbita accessions analyzed: modern samples
| Laboratory ID | Taxon | Status | Location (wild samples) | US Department of Agriculture accession no. | Sequencing strategy | Percent sites represented |
| 010 | C. pepo ssp. pepo | Cultivar | PI 508469 | Shotgun/LR PCR | 99.85 | |
| 011 | C. pepo ssp. ovifera var. ovifera | Cultivar | PI 518687 | Shotgun/LR PCR | 99.85 | |
| 012 | C. pepo ssp. ovifera var. texana | Wild | Texas | Ames 26891 | Shotgun/LR PCR | 99.80 |
| 013 | C. pepo ssp. ovifera var. ozarkana | Wild | Arkansas | Ames 26607 | Shotgun/LR PCR | 99.80 |
| 014 | C. pepo ssp. fraterna | Wild | Tamaulipas, Mexico | PI 532354 | Shotgun/LR PCR | 99.80 |
| 015 | C. moschata | Domestic | PI 209116 | Shotgun/LR PCR | 99.86 | |
| 016 | C. argyrosperma | Landrace | PI 511929 | Shotgun | 99.86 | |
| 017 | C. argyrosperma var. palmeri | Wild | Sinaloa, Mexico | PI 512211 | Shotgun/LR PCR | 99.11 |
| 018 | C. argyrosperma ssp. sororia | Wild | Chiapas, Mexico | PI 532404 | Shotgun/LR PCR | 99.86 |
| 020 | C. maxima ssp. andreana | Domestic | G 29254 | Shotgun/LR PCR | 99.86 | |
| 021 | C. ecuadorensis | Wild | Casco, Ecuador | PI 432445 | Shotgun/LR PCR | 99.85 |
| 022 | C. okeechobeensis | Wild | Florida | N/A (LAN, sample no. S-600) | Shotgun/LR PCR | 99.86 |
| 023 | C. okeechobeensis ssp. martinezii | Wild | Veracruz, Mexico | PI 540900 | Shotgun/LR PCR | 99.83 |
| 024 | C. lundelliana | Wild | Guatemala | Grif 9452 | Shotgun/LR PCR | 99.85 |
| 025 | C. ficifolia | Domestic | PI 512680 | Shotgun | 99.86 | |
| 026 | C. digitata | Wild | Sonora, Mexico | PI 240879 | Shotgun/LR PCR | 99.84 |
| 027 | C. pedatifolia | Wild | San Luis Potosí, Mexico | PI 442290 | Shotgun/LR PCR | 99.86 |
| 028 | C. cordata | Wild | Mexico | PI 653839 | Shotgun/LR PCR | 99.86 |
| 029 | C. foetidissima | Wild | Oklahoma | PARL 264 | Shotgun | 99.82 |
| 036 | C. pepo ssp. fraterna | Wild | Tamaulipas, Mexico | PI 532356 | Shotgun | 99.77 |
| 037 | C. pepo ssp. ovifera var. ozarkana | Wild | Illinois | Ames 26875 | Shotgun | 99.76 |
| 038 | C. pepo ssp. ovifera var. texana | Wild | Texas | PI 614686 | Shotgun | 99.73 |
| 039 | C. pepo ssp. ovifera var. ovifera | Cultivar | PI 267755 | Shotgun | 99.77 | |
| 040 | C. pepo ssp. ovifera var. ovifera | Cultivar | NSL 180768 | Shotgun | 99.79 | |
| 041 | C. moschata | Domestic | PI 244707 | Shotgun | 99.85 | |
| 042 | C. moschata | Domestic | PI 442263 | Shotgun | 99.85 | |
| 043 | C. moschata | Domestic | PI 634693 | Shotgun | 99.81 | |
| 044 | C. pepo ssp. pepo | Cultivar | PI 615105 | Plastid capture | 99.01 | |
| 045 | C. pepo ssp. pepo | Cultivar | PI 615088 | Plastid capture | 98.34 | |
| 046 | C. pepo ssp. ovifera var. ovifera | Cultivar | PI 595838 | Plastid capture | 98.56 | |
| 047 | C. pepo ssp. ovifera var. ovifera | Cultivar | PI 615111 | Plastid capture | 98.01 | |
| 048 | C. pepo ssp. ovifera var. ovifera | Cultivar | PI 615114 | Plastid capture | 97.31 | |
| 049 | C. pepo ssp. pepo | Cultivar | PI 615108 | Plastid capture | 96.14 | |
| 050 | C. pepo ssp. ovifera var. ovifera | Cultivar | PI 615152 | Plastid capture | 96.87 | |
| 051 | C. pepo ssp. ovifera var. ovifera | Cultivar | PI 615115 | Plastid capture | 97.97 | |
| 052 | C. pepo ssp. ovifera var. texana | Wild | Mississippi | PI 614693 | Plastid capture | 98.01 |
| 053 | C. pepo ssp. ovifera var. texana | Wild | Mississippi | PI 614692 | Plastid capture | 98.44 |
| 054 | C. pepo ssp. ovifera var. texana | Wild | Texas | PI 614689 | Plastid capture | 97.72 |
| 055 | C. pepo ssp. ovifera var. texana | Wild | Texas | PI 614687 | Plastid capture | 92.44 |
| 056 | C. pepo ssp. ovifera var. texana | Wild | Mississippi | PI 614701 | Plastid capture | 93.81 |
| 057 | C. pepo ssp. ovifera var. ozarkana | Wild | Oklahoma | Ames 26610 | Plastid capture | 95.74 |
| 058 | C. pepo ssp. ovifera var. ozarkana | Wild | Illinois | Ames 26616 | Plastid capture | 96.96 |
| 059 | C. pepo ssp. ovifera var. ozarkana | Wild | Louisiana | Ames 26884 | Plastid capture | 93.45 |
| 060 | C. pepo ssp. ovifera var. ozarkana | Wild | Mississippi | Ames 26882 | Plastid capture | 96.12 |
| 061 | C. pepo ssp. ovifera var. ozarkana | Wild | Kentucky | Ames 26617 | Plastid capture | 86.99 |
| 062 | C. pepo ssp. ovifera var. ozarkana | Wild | Missouri | Ames 26612 | Plastid capture | 92.02 |
| 063 | C. pepo ssp. ovifera var. ozarkana | Wild | Arkansas | Ames 26890 | Plastid capture | 91.06 |
| 064 | C. pepo ssp. fraterna | Wild | Tamaulipas, Mexico | PI 532355 | Plastid capture | 92.29 |
| 065 | C. pepo ssp. fraterna | Wild | Tamaulipas, Mexico | PI 614683 | Plastid capture | 88.18 |
| 071 | C. moschata | Domestic | N/A (LAN, “Seminole Pumpkin”) | Plastid capture | 99.83 | |
| 072 | C. moschata | Domestic | PI 211998 | Plastid capture | 99.82 | |
| 073 | C. moschata | Domestic | PI 169409 | Plastid capture | 99.84 | |
| 074 | C. moschata | Domestic | PI 183258 | Plastid capture | 99.80 | |
| 075 | C. moschata | Domestic | PI 483347 | Plastid capture | 99.85 | |
| 076 | C. moschata | Domestic | PI 419083 | Plastid capture | 99.85 | |
| 077 | C. moschata | Domestic | PI 200822 | Plastid capture | 99.78 | |
| 078 | C. moschata | Domestic | PI 249565 | Plastid capture | 99.85 | |
| 079 | C. moschata | Domestic | PI 287532 | Plastid capture | 99.85 | |
| 080 | C. moschata | Domestic | PI 357918 | Plastid capture | 99.85 | |
| 081 | C. moschata | Domestic | PI 357918 | Plastid capture | 99.86 | |
| 082 | C. moschata | Domestic | PI 524427 | Plastid capture | 99.84 | |
| 084 | C. moschata | Domestic | PI 490351 | Plastid capture | 99.85 | |
| 085 | C. moschata | Domestic | PI 200736 | Plastid capture | 99.80 | |
| 086 | C. moschata | Domestic | PI 438548 | Plastid capture | 99.86 | |
| 087 | C. moschata | Domestic | PI 406848 | Plastid capture | 99.85 | |
| 088 | C. moschata | Domestic | PI 369346 | Plastid capture | 99.85 | |
| 089 | C. moschata | Domestic | PI 194570 | Plastid capture | 99.85 | |
| 090 | C. moschata | Domestic | PI 458728 | Plastid capture | 99.86 | |
| 091 | C. moschata | Domestic | PI 441725 | Plastid capture | 99.85 | |
| 092 | C. moschata | Domestic | PI 498429 | Plastid capture | 99.85 | |
| 093 | C. moschata | Domestic | PI 475750 | Plastid capture | 99.84 | |
| 094 | C. moschata | Domestic | PI 458650 | Plastid capture | 99.84 |
Fig. 1.
Sample map and plastid genome phylogeny of Cucurbita. All nodes shown are supported with Bayesian posterior probability of 1 and maximum likelihood support of 100/100 bootstrap replicates, with two exceptions: (i) C. okeechobeensis/C. okeechobeensis ssp. martinezii node: 93 bootstrap replicates; (ii) C. argyrosperma/C. okeechobeensis/C. lundelliana node: 95 bootstrap replicates. Tree tip labels in color represent taxa recovered among our archaeological samples, with color-matching lines indicating sampling sites and diamonds showing sampling locations of modern wild counterparts, if applicable (present for C. pepo ssp. ovifera, C. pepo ssp. fraterna, and C. argyrosperma). Labeled black diamonds show the sampling locations of other wild accessions analyzed as single taxonomic representatives. Among the four wild perennial species analyzed (Tables S1–S3), C. cordata is not shown, because sampling records for this accession are not specific regarding collection locality. C. maxima, a South American species with wild and domestic members, was sampled from a wild accession of C. maxima ssp. andreana from central Argentina and so is not shown.
Table S2.
Read alignment details for Cucurbita accessions analyzed: archaeological samples
| Laboratory ID | Merged read pairs obtained | Percent reads on-target | Percent sites represented | Mean depth of coverage | Nucleotides masked for final consensus |
| 101 | 4,461,973 | 3.49 | 98.30 | 85.21 | 8 |
| 102 | 5,888,667 | 2.50 | 98.04 | 89.39 | 8 |
| 103 | 5,679,328 | 3.80 | 99.15 | 173.55 | 8 |
| 104 | 9,329,829 | 2.16 | 99.13 | 125.02 | 8 |
| 105 | 6,926,007 | 1.27 | 97.01 | 43.79 | 8 |
| 106 | 5,180,946 | 2.15 | 97.69 | 56.13 | 8 |
| 107 | 4,970,331 | 0.03 | 21.40 | 0.75 | N/A |
| 108 | 3,850,699 | 0.06 | 34.08 | 1.10 | N/A |
| 109 | 4,422,071 | 0.04 | 20.50 | 0.89 | N/A |
| 110 | 5,557,862 | 0.76 | 98.48 | 23.28 | 8 |
| 111 | 5,024,510 | 0.08 | 40.41 | 1.72 | N/A |
| 112 | 5,620,282 | 2.96 | 99.23 | 106.34 | 9 |
| 113 | 4,267,521 | 0.02 | 10.88 | 0.46 | N/A |
| 114 | 6,444,043 | 3.01 | 99.06 | 108.83 | 4 |
| 115 | 5,140,918 | 4.01 | 98.78 | 136.50 | 8 |
| 116 | 4,615,057 | 0.05 | 23.18 | 1.12 | N/A |
| 118 | 19,996,947 | 0.02 | 42.40 | 2.04 | N/A |
| 119 | 4,927,607 | 0.03 | 13.75 | 0.69 | N/A |
| 120 | 4,203,398 | 0.03 | 12.00 | 0.63 | N/A |
| 121 | 5,877,218 | 3.57 | 99.50 | 129.44 | 8 |
| 122 | 6,177,243 | 3.78 | 99.77 | 169.71 | 5 |
| 123 | 18,192,689 | 0.14 | 82.22 | 11.18 | 5 |
| 139 | 23,463,065 | 0.04 | 68.10 | 3.96 | 8 |
| 140 | 20,493,606 | 0.09 | 89.37 | 7.43 | 7 |
| 141 | 16,149,579 | 0.02 | 32.47 | 1.39 | N/A |
| 142 | 8,495,477 | 2.73 | 99.76 | 162.22 | 10 |
| 143 | 8,572,258 | 0.03 | 33.88 | 1.51 | N/A |
| 144 | 10,665,311 | 2.13 | 99.79 | 163.77 | 10 |
| 145 | 6,091,069 | 0.05 | 27.64 | 1.50 | N/A |
| 146 | 33,314,376 | 0.02 | 66.54 | 3.95 | 8 |
| 147 | 5,208,273 | 0.05 | 22.79 | 1.25 | N/A |
| 148 | 4,175,335 | 1.33 | 97.53 | 28.47 | 10 |
C. pepo comprises three subspecies: ssp. pepo, ssp. ovifera, and ssp. fraterna. Of these, C. pepo ssp. pepo (zucchini, pumpkins, and summer and winter squashes) appears to have been domesticated in the Oaxaca Valley region around 10 kya (12) and has no known extant wild type. C. pepo ssp. ovifera contains wild varieties found in patches throughout the southeastern United States, including C. pepo ssp. ovifera var. ozarkana, and C. pepo ssp. ovifera var. texana, as well as a wide variety of domestic types (scallop and acorn squashes, ornamental gourds). Archaeologically, it appears that ssp. ovifera domestication took place in eastern North America (ENA) alongside a suite of other crop species (e.g., 19). C. pepo ssp. fraterna is a wild form restricted to a narrow range in northeastern Mexico and unknown as a domesticate (20). However, it is genetically similar to ssp. ovifera and therefore has been hypothesized as an alternate source population of domesticated C. pepo in ENA (14).
We found that members of ssp. ovifera form a well-supported clade that includes modern cultivars and wild gourds from ENA as well as all archaeological ENA C. pepo samples. There was no signal of ssp. ovifera among ancient samples from Mexico, the alternative possible source region (14), nor in modern free-living ssp. fraterna in that region. This absence strongly supports local evolution of domesticated C. pepo ssp. ovifera in ENA, in agreement with archaeological evidence of an increase in seed size over time and molecular data clustering modern ssp. ovifera cultivars with ENA wild populations (21–23). However, although ssp. fraterna is known only in the wild at present and is absent from the ENA archaeological record, we observed a ssp. fraterna plastid sequence in an archaeological rind fragment from Romero’s Cave in northeastern Mexico. The specimen shows a thick (>2 mm) rind and a furrow typical only of domesticated Cucurbita. We obtained a direct date from the rind of 4,614–4,836 calibrated calendar years BP (2σ range). This result might represent cross-pollination of native wild ssp. fraterna by domesticated ssp. pepo leading to the expression of some domestic-type characters or an independent domestication trajectory that did not contribute to modern plastid diversity among cultivars. In either circumstance, this finding demonstrates a previously unknown role of ssp. fraterna in the crop fields of ancient Mexico. Additionally, we observed archaeological ssp. pepo in cave sites ranging from Oaxaca to the southwestern United States, suggesting either prolific human-mediated dispersal following domestication or a geographically widespread evolution of domestic forms.
Finally, we found that three endemic populations spanning the Gulf of Mexico, currently defined as distinct taxa (24), are essentially indistinguishable based on the plastid genome. C. okeechobeensis is restricted to the Florida peninsula, on the shores of Lake Okeechobee and the St. John’s River basin, C. okeechobeensis ssp. martinezii is found on the north coastal plain of Veracruz, and C. lundelliana grows on the limestone plains of the Yucatan Peninsula (24), but the three taxa differ at only six single-nucleotide variants across the 156.6-kb plastid genome (average pairwise nucleotide diversity: π = 0.0043%) despite their geographic separation. Along with the undifferentiated plastid genome structure among patchy C. pepo ssp. ovifera in ENA (Fig. 1), this result may reflect recent fragmentation of a previously contiguous population.
In summary, we observed a pattern that supports hypotheses for widespread Holocene domestication of Cucurbita and suggests that this process coincided with dramatic habitat and range fractionation and wild plant extinction. C. pepo ssp. pepo is thought to be extinct in the wild, but we find seven archaeological examples from this lineage spanning more than 2,000 km and dating back nearly 10,000 y. We observe at least one other independent domestication episode of C. pepo, that of ssp. ovifera in ENA alongside a suite of other native crops, and a possible third, arrested domestication trajectory involving ssp. fraterna in northeastern Mexico. We also observe genetic similarity between disparate endemic taxa (C. okeechobeensis, C. okeechobeensis ssp. martinezii, and C. lundelliana), suggesting recent separation of these populations.
Analysis of Ability to Detect Bitter-Tasting Compounds.
We screened 46 therian mammal genomes to assess general ability to detect bitter-tasting compounds by analyzing functional TAS2R gene number variation. We excluded the poorest quality assemblies [length-weighted midpoint scaffold size of the genomic assembly (scaffold N50) <250,000 nt; n = 8] because of unreliable gene detectability (Materials and Methods). Among the 38 remaining genomes, we detected a combined total of 851 intact TAS2R genes, ranging from a low of 8 in the genome of the West Indian manatee (Trichechus manatus latirostris) to a high of 46 in the genome of the common shrew (Sorex araneus). We tested for relationships among TAS2R count, body mass, and dietary breadth (Materials and Methods) while controlling for phylogenetic nonindependence.
When all species are treated as independent, TAS2R count is significantly negatively correlated with both body mass (Pearson’s product–moment correlation; r2 = 0.236; P = 0.002) (Fig. 2A) and greater dietary specialization (r2 = 0.194; P = 0.0056) (Fig. 2B). When all variables are analyzed simultaneously with a generalized linear model, body mass and dietary breadth are each significant predictors of TAS2R count when controlling for the other variable (P = 0.0043 and P = 0.0122, respectively), but the significance of this relationship is strongest when both variables are considered together, and explains a substantial proportion of the variance in TAS2R count (P = 0.0004; multiple r2 = 0.363). When taxonomic nonindependence among our sample is controlled for by using a phylogenetic generalized least squares test (Materials and Methods), the associations of body mass and dietary breadth with TAS2R count are still significantly stronger than expected from chance (TAS2R–size: P = 0.044; TAS2R–diet: P = 0.015; overall model: P = 0.007, multiple r2 = 0.246). The TAS2R–body size association is most pronounced among smaller-bodied taxa; when only the very largest mammal species are excluded from the analysis, the inverse relationships between the number of intact TAS2R genes and both body size and diet are highly significant even when controlling for phylogeny (among n = 35 mammals <1,000 kg, TAS2R–size: P = 0.0013; TAS2R–diet: P = 0.001; overall model: P = 4.08 × 10−5; multiple r2 = 0.468). To summarize, smaller species, and especially those with more diverse diets, tend to have a larger repertoire of functional bitter taste receptor genes, and thus they likely are better able to detect and avoid potentially toxic bitter compounds while foraging.
Fig. 2.
TAS2R gene count and phenotypic traits in 38 high-quality mammal genomes. (A) Log10-scaled body mass vs. TAS2R count. The regression line assumes taxonomic independence, and point shading corresponds to diet specialization as in B (blue: least specialized; tan: most specialized). Representative species shown from left to right are common shrew, mouse, tarsier, human, giant panda, West Indian manatee, and African elephant. (B) Boxplots of TAS2R count by diet specialization. Category 1 consists of broad dietary generalists (e.g., human, mouse), and category 4 consists of highly specialized feeders (e.g., giant panda).
Discussion
Holocene Anachronism.
The Holocene decline of wild Cucurbita was likely driven, at least in part, by the nearly complete disappearance of herbivores ≥1,000 kg, with whom they appear to have had an important symbiotic relationship. Wild Cucurbita are “too bitter for humans and well-fed livestock” (10), but larger mammals are much more tolerant of bitter plant compounds than smaller ones. Larger species are likely able to metabolize or pass moderate toxins harmlessly as part of a high daily biomass intake. For example, even if we ignore any additional physiological or behavioral detoxification strategies and the large digestive throughput of hindgut fermenters, a fully-grown African elephant [averaging 4,540 kg (22)] would need to ingest an estimated 7.5–23 kg of wild Cucurbita gourds—approximately 75–230 whole fruits—over a short timespan (25, 26) to approach a lethal dose of cucurbitacins (27). Indeed, several bitter species of Cucurbitaceae are eaten and dispersed by African elephants in the present (10). As another illustration of these concepts, the one-horned rhinoceros in lowland Nepal [1,500–2,000 kg (22)] preferentially feeds on the bitter fruits of trewia (Trewia nudiflora, Euphorbiaceae), daily consuming up to hundreds of fruits that are unpalatable to smaller mammals who are concomitantly incapable of dispersing the ∼1-cm-diameter seeds (10, 28). Humans have reported severe symptoms of cucurbitacin toxicity after consuming only “one or two bites” of cucurbitacin-tainted squash (29). The presence of Cucurbita seeds in mastodon dung is clear evidence that, like their extant counterparts, North American megafauna were effective Cucurbitaceae dispersers, physiologically undeterred by cucurbitacin bitterness.
Larger numbers of intact TAS2R genes are associated with greater ability to detect and avoid toxins via their bitterness, and interspecific variation in copy number is hypothesized to reflect ecological and evolutionary diversity (30–32). Previous studies have reported that herbivores have slightly higher TAS2R copies than carnivores, presumably to detect plant-based toxins more acutely (30), and that the common ancestor of modern whales completely lost functional TAS2R copies, probably as a result of feeding behaviors and the switch to a marine environment (33). Similarly, in our analysis, dietary breadth is significantly correlated with TAS2R count, presumably because a wider range of possible food species necessitates sensitivity to a more diverse range of toxic compounds, and because narrow dietary specialists have little need for toxin detection. Because very large-bodied species are more physiologically resilient to moderately toxic compounds, they too require fewer TAS2R gene copies. That is, bitter compounds signal toxicity to foraging animals, but in the absence of serious toxic threat, large mammals do not benefit by acutely recognizing this danger signal. Therefore, selection to maintain a broad TAS2R repertoire in larger species may be relaxed, leading to gene loss. Furthermore, although we cannot test this hypothesis in the present study, TAS2R pseudogenization could even be adaptive for megafauna if the consumption of bitter fruit was a benefit to survival, i.e., if acuity to bitter tastes would be an unnecessary barrier to herbivory.
The now-extinct megafaunal herbivores of the Americas were sufficiently large that they were likely to have been protected against moderate toxins, whereas the smaller surviving mammals are not. Accordingly, smaller mammals, and especially those with broad dietary habits, tend to have larger suites of TAS2R genes to detect and avoid toxic compounds. These smaller species generally would be unable to consume Cucurbita fruits whole and pass the intact seeds in the manner of mastodons; therefore, as seed predators instead of dispersal mutualists, the smaller species pose a threat to Cucurbita. Thus the toxicity of wild Cucurbita and related species may be, in part, an adaptive strategy to facilitate beneficial seed dispersal by very large mammals while warding off smaller seed predators. The resulting unpalatability of Cucurbita to small-bodied mammals, combined with the megafaunal extinctions, would have left the plants lacking plausible dispersal partners and the associated nutrient-rich dung deposits important in seed germination and seedling development (28); thus intergenerational seed propagation would be curtailed and offspring would be more vulnerable to localized stressors.
Moreover, Cucurbita prefers field edges, floodplains, and other disturbed habitats and would have thrived in the niche-diverse, mosaic-like landscapes maintained by large herbivores (4). In the wake of these herbivores’ extinction, large-scale zonal vegetative patterns replaced the fine-grained variation maintained by the megafauna (34), and weedy taxa would have been crowded out as their disturbed-ground niche disappeared. Taken together, these transitions could have been instrumental in driving wild Cucurbita into relict, refugial patches, causing fragmentation of previously contiguous distributions, and ultimately, in some cases, leading to the extinction of wild forms.
Domestication as an Adaptive Strategy.
Against the backdrop of these ecological changes, the anthropogenic landscape provided a disturbed habitat readily colonized by weedy species, and humans fulfilled the dispersal role. Our phylogenetic results support previous hypotheses that the domestication process was widespread, occurring independently many times throughout the Americas, including in ENA (C. pepo ssp. ovifera), Mesoamerica (C. pepo ssp. pepo and C. argyrosperma, possibly C. pepo ssp. fraterna, C. moschata, and C. ficifolia), and South America (C. maxima and possibly C. ecuadorensis and C. moschata) (35). ENA is a long-debated putative source region for Cucurbita pepo ssp. ovifera var. ovifera, a hypothesis that is strongly supported by the clustering of ovifera cultivars with modern and ancient ENA gourds in our analysis. C. pepo ssp. pepo—unknown in the wild—originated in the Valley of Oaxaca around 10,000 y B.P (12) but also was prominent among our samples from northern Mexico and the southwestern United States, several hundred miles to the north. This result suggests either (i) a widespread ancient distribution of this subspecies and the emergence of domestic forms across a broad geographic range, illustrating a range-wide shift to human landscapes as an adaptive strategy; or (ii) prolific human-mediated dispersal of this taxon throughout Mesoamerica, illustrating the massive reproductive and dispersal potential for plants under cultivation. Either possibility underscores the adaptive potential of domestication (36). One ancient C. pepo ssp. fraterna sample from northeastern Mexico also indicates the possible presence of this taxon in ancient crop fields. Presently, C. pepo ssp. fraterna exists only in the wild, suggesting that our ancient fraterna sample may represent a lost domestic lineage or an independent, subsequently arrested, pathway to domestication.
Reproductive Isolation.
The fixation of domesticated phenotypes requires a level of reproductive isolation between cultivated and wild forms to prevent the constant back-crossing of unfavorable traits into populations under human selection (37, 38). For example, emmer wheat required removal from its source region to express the phenotypic effects of domestication robustly (39), and it has been suggested that maize flourished into its dramatic, familiar, hyper-domesticated forms only after being transported out of southern Mesoamerica (40). This requirement may vary by species, with the level of isolation being linked to the genetic configuration and cultural importance of domestication traits. In Cucurbita, the requirement for reproductive isolation is especially pronounced, with cross-fertilization from wild forms producing unpalatable fruits (10), likely through the up-regulation of cucurbitacin production. That is, without isolation from wild stands, it is unlikely that Cucurbita could have evolved into a form suitable for human consumption. During the Holocene, the collapse of wild populations following the onset of a relationship with humans ultimately may have prevented gene flow from wild types constantly recharging crop fields with the allelic predisposition for a tough rind and bitter cucurbitacins. Cucurbita species have thrived in the Holocene largely by partnering with humans through domestication as wild populations dwindled and have been bred to lower their natural defenses for human palatability. At the same time, the fixation of important domestication alleles conferring palatability might have been impossible if wild plants had not largely receded and disappeared.
Materials and Methods
Plastid Genome Analysis.
Sample materials.
Details of all modern and ancient Cucurbita samples analyzed are reported in Tables S1–S3. Modern samples were provided by L.A.N. and the US Department of Agriculture Agricultural Research Service National Plant Germplasm System. Access to ancient samples was granted by B.D.S., the Illinois State Museum, the National Park Service, and the University Museum at the University of Arkansas.
DNA isolation, sequencing, and assembly.
In modern samples, we used sterile razor blades to excise embryonic tissue from seeds and extracted whole genomic DNA from the embryos using Qiagen DNeasy Plant Mini Kits, following the manufacturer’s protocol after grinding tissue in the kit lysis buffer with sterile pellet pestles. In modern-sample representatives from each separate taxon, we first PCR-amplified long, overlapping fragments of the plastid genome with primers in conserved regions (see Table S4 for primer sequences) designed using an alignment of cucumber (Cucumis sativus, Cucurbitaceae) and karaka (Corynocarpus laevigatus, Corynocarpaceae) plastid genomes, both order Cucurbitales. We were unable to amplify complete plastid genomes in any taxon using long-range PCR (LR-PCR), but we recovered several long fragments to assist with plastid genome assembly. We separately sheared LR-PCR product pools and whole genomic DNA to ∼350 bp using a Covaris model S2, size-selected the sheared DNA at ∼350 bp on a 1.6% low-melt agarose gel, and prepared barcoded Illumina libraries (41). We pooled all barcoded libraries equally among samples, with 20% of estimated pool molarity comprising LR-PCR products and the remainder whole genomic DNA. We sequenced the pool on one lane of an Illumina HiSeq 2000 flow cell with a 101-nt paired-end configuration. To acquire additional C. pepo and C. moschata plastid genome sequences to compare with the initial assemblies, we shotgun-sequenced eight additional plastid genomes on one lane of an Illumina HiSeq 2500 flow cell with a 151-nt paired-end configuration in Rapid Run mode.
Table S4.
LR-PCR primer sequences
| Primer ID | Primer sequence (5′ – 3′) |
| CuLR1F_14 | ACGGGAATTGAACCCGCGCATGGTG |
| CuLR1R_10234 | GCGGGAATCGAACCCGCATCGTTAGC |
| CuLR2F_7978 | GCCAAATTGCCCGAGGCCTACGCTTT |
| CuLR2R_19445 | GCCGAGTAGGCGGATTGGTCCGAGT |
| CuLR3F_17745 | AGATCGCTTCTTCCAAAGCACGCCC |
| CuLR3R_28990 | ATTTGCAGTCCCCCGCCTTACCGCT |
| CuLR4F_27271 | CCTACTCACACGAGCCCATATCCTTGCT |
| CuLR4R_36971 | GGAGCACGCAGATCCCAAAAACGCA |
| CuLR5F_35924 | TGGGCCGGGAATGCCCGACTTATCA |
| CuLR5R_44861 | TTCCGACCGGGGAGAACAGGCCATT |
| CuLR6F_43879 | CTCGAGAAATGACCCGGTCTGGCCC |
| CuLR6R_53080 | CGGGAATAGAACCAATGGGCGATGCT |
| CuLR7F_51582 | GGGCCATCCTACCCAACTTTCAGGCA |
| CuLR7R_61940 | GCCGGAAATACTAGGCCCACTAAAGGCA |
| CuLR8F_60937 | GGAGGAGCACGCATGCAAGAAGGAAG |
| CuLR8R_70507 | TCGGGGGCAAACGCCTACGAAAAGA |
| CuLR9F_70068 | TGGCCAAGGGTAAAGATGCCCGAGT |
| CuLR9R_79605 | CATTGGGCCATGCGGGTTCTCCGTA |
| CuLR10F_79451 | TCATGTCCGGTTCCTTCGGGGGATGG |
| CuLR10R_88670 | ACTGGATGCACGCCAATGGGACCCT |
| CuLR11F_IR_88137 | CTACGGCTCCATTGCGTGTGCTCGG |
| CuLR11R_IR_97912 | TGGGTTCAAGCTTTCCCCAGCCCCT |
| CuLR12F_IR_97208 | CGTGCATGAGACTTTCATCTCGCACGGC |
| CuLR12R_IR_107874 | TTCCTTGACCTTCCGGCACTGGGCA |
| CuLR13F_107734 | AAGGGGTGCCTCCTCACAAAGGGGG |
| CuLR13R_117642 | TCATGGATTTATTGGCGCTGCGCTT |
| CuLR14F_115832 | ATCTCGGTTCGAGTCCGAGTGGCGG |
| CuLR14R_124372 | GCTTCTCATCTGTTATGGCTTGGCCCT |
| CuLR15F_124203 | AGCCGCGACTCCCCCGATACGAAAA |
| CuLR15R_134752 | CGCCGAAGATGAACGGGGCTAAGCG |
| CuLR16F_end_146561 | TGAATAGGACGAACCGCCCCGTGGT |
| CuLR16R_end_668 | TAGCTGGTGTATTCGGCGGCTCCCT |
| CuLR_alt_8F_61002 | TGGAAAAGCAAGTTGATCGGTTAAT |
| CuLR_alt_8R_70245 | CTTCTAGAATCCTTTGTTCCCTTCA |
| CuLR_alt_7F_52080 | CTAAGCGGGCTCACATAACAGAAAT |
| CuLR_alt_7R_61259 | TAGAGTTCGGGTTCGAATTCCATAG |
| CuLR_alt_5F_36058 | GTGGTAGAGTAACGCCATGGTAAGG |
| CuLR_alt_5R_44044 | AATCTTTATGCTCAAAACCCCGATT |
| CuLR_alt_2F_10163 | GTCAGTACCTAGCCGGGCTTTTT |
| CuLR_alt_2R_18853 | GAGATGATGGAAGCAGGAGTTCATT |
| CuLR_alt_13BF_111751 | CTCCCAATTGGTTGGACCGTAG |
| CuLR_alt_13BR_116450 | AGAAAAGTGGGAGAGAAGGGGTTT |
We divided LR-PCR–derived sequence data into groups of 100,000 read pairs and shotgun sequence data into 1,000,000 pair subsets to normalize coverage for de novo assembly. We used Velvet 1.2.08 to iterate over LR-PCR subsets to produce long, high-quality contigs from the high-coverage enriched plastid regions. We combined all LR-PCR contigs per sample and then used Velvet (42) to iterate over shotgun data subsets using stringent parameters (long k-mer value and high coverage cutoff) and included LR-PCR contigs as long reads to anchor the assembly. We combined all resulting contigs and reiterated over shotgun subsets using new contigs as long reads until no substantial increase in contig length and quality was observed (two to five iterations). We used Burrows–Wheeler Aligner (BWA) (43) to align resulting contigs from one taxon, Cucurbita pepo ssp. fraterna, to the plastid genome of cantaloupe (Cucumis melo, Cucurbitaceae), a relative with an estimated 30-Mya divergence date from Cucurbita (44). We then used the resulting alignment to identify contigs covering the majority of the plastid genome and used pairwise alignment to assemble them to the cantaloupe reference sequence. We used bash utilities to close the remaining short gaps by reiteratively querying contig terminal sequences from the short-read data and realigning matching short reads to the working assembly until reaching the next adjacent contig. We used YASRA (45) with LASTZ (46) to map shotgun reads from other taxa to the circularized C. pepo ssp. fraterna plastid reference assembly reiteratively and closed gaps using de novo contigs and manual gap-filling as described above. Two short stretches (estimated <200 nt based on comparison with the cantaloupe reference sequence) in the long single-copy region containing tandem repeats were unresolvable using short-read data, one in the rps4-ndhJ intergenic spacer region and the other apparently within the coding region of accD. These regions were hard-masked for analysis. After the initial assembly of plastid genomes across all taxa, we used BWA (43) and SAMtools (47) to align short reads from subsequent samples to the appropriate reference and call consensus sequences.
Following plastid genome assembly for all available taxa, we used the C. pepo ssp. pepo and C. moschata plastid consensus sequences to design target capture probes for plastid DNA enrichment by RNA hybridization (18). We extracted DNA and prepared sequencing libraries from 22 additional modern specimens of C. pepo and 23 additional modern C. moschata. In addition, we extracted DNA from 33 archaeological Cucurbita rind, seed, and peduncle specimens using an N-phenacylthilazolium bromide (PTB)-based extraction protocol (48) and prepared sequencing libraries (41). Because of the fragmentary, low-copy, and contamination-prone nature of ancient DNA, stringent protocols were observed during ancient DNA extraction and library preparation. All pre-PCR handling of ancient tissue and DNA was carried out in a dedicated sterile laboratory physically isolated from any molecular biology building, with HEPA filtration, positive pressure, frequent decontamination using a strong bleach solution, and workflow protocols to prevent modern contamination of the clean laboratory. We enriched modern and ancient DNA sequencing libraries for plastid DNA using the MYbaits kit (Mycroarray), according to the manufacturer’s protocol but with two extra wash repetitions and an 18-cycle PCR reamplification using primers IS5 and IS6 (41) after hybridization. For the modern samples, we combined the barcoded libraries into pools of 10 samples before the capture step and carried each pool through a single capture reaction. The enriched libraries were sequenced in parallel on an Illumina HiSeq 2500.
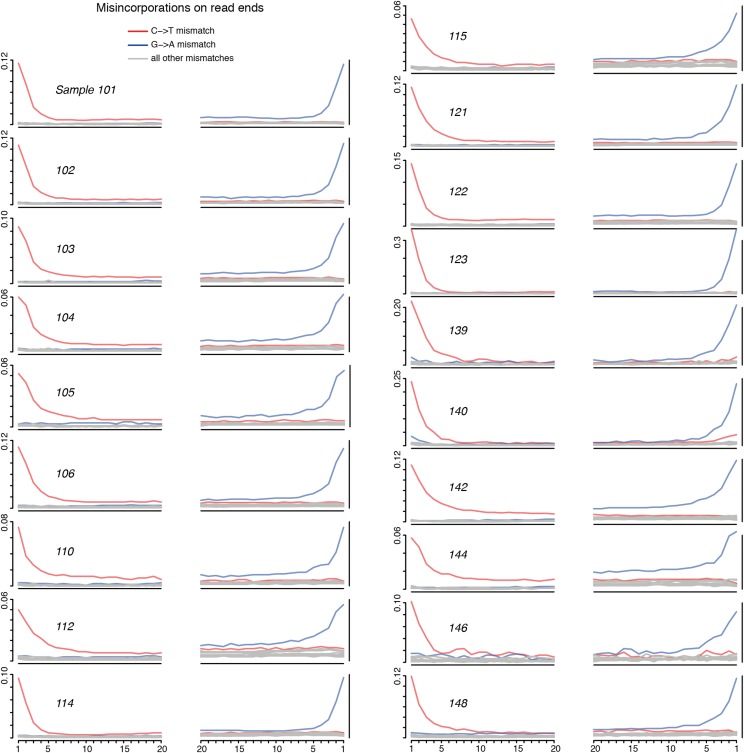
For modern libraries, we used BWA (43) to map reads to the appropriate reference plastid assembly and then used the SAMtools (47) mpileup function and a custom Perl script to call majority-rule haplotype consensus sequences. For ancient samples, we first merged overlapping paired-end reads after Kircher (49) and then BWA-mapped (43) merged reads to the C. pepo ssp. fraterna plastid reference sequence. We estimated the level of cytosine deamination at fragment ends—characteristic ancient DNA damage most prevalent in single-stranded overhangs (50)—by analyzing base mismatches to the reference sequence relative to base position on the fragment, revealing an average 8-nt interval of elevated 5′ C->T bias and 3′ G->A bias (Fig. S1). This observation helped authenticate our ancient DNA data and indicated that the region within 8 nt of read ends, on average, might be prone to damage-derived sequence error. To mitigate any effects of these biases in our analysis, we hard-masked all potentially damaged positions (5′ thymines and 3′ adenines) within appropriate distance of fragment ends (Fig. S1 and Tables S1–S3). We then called haplotype consensus sequences as above, enforcing a minimum 2× nonredundant coverage and 80% site identity for a consensus base call. Under these requirements, we recovered plastid assemblies between 10.9% and 99.8% complete for all ancient samples (Tables S1–S3). We discarded samples with <65% coverage of the single-copy regions and one of the two inverted repeat regions, leaving a total of 19 ancient samples for analysis, 15 of which had ≥97% coverage.
Fig. S1.
DNA misincorporation patterns resulting from ancient DNA deamination. X-axis: distance on mapped read from 5′ (Left) and 3′ (Right) termini; Y-axis: proportion of reference C matched with read T (Left, red), and reference G match with read A (Right, blue).
We aligned all plastid genome sequences (n = 91) using MAFFT (51), manually checked the alignment, and manually masked any short regions of poor mapping quality caused by long homopolymer repeats prone to polymerase and sequencing error (aligned and curated sequences provided as Dataset S1). We used a log-likelihood ratio test to select the Hasegawa, Kishino, and Yano (HKY) substitution model with four gamma-distributed rate categories, and used PhyML (52) with 100 bootstrap replicates to reconstruct a phylogenetic tree (Fig. 1). We also confirmed the tree topology by running a Bayesian phylogenetic analysis to convergence using BEAST 2 (53), enforcing a strict molecular clock and implementing the same evolutionary model described for the maximum likelihood analysis.
Mammalian TAS2R Analysis.
TAS2R gene identification.
We retrieved 46 mammal genome assemblies from public repositories (Tables S5 and S6), representing all scaffold- or chromosome-level therian mammal genome assemblies available at the time of data acquisition (April, 2014), excluding the bottlenose dolphin. Extant whales possess only pseudogenized TAS2R homologs because of the complete loss of functional TAS2R genes in a common ancestor living 36–53 Mya (33). Therefore, dolphins have never possessed a suite of functional bitter taste receptors (33), rendering analysis of subsequent evolution of functional TAS2R suites alongside phenotypic and behavioral variables moot. To identify intact, putatively functional TAS2R gene copies in the mammalian genomes, we first adapted the strategy of Li and Zhang (30) by generating BLAST databases (54) from each genome assembly and queried them for the annotated human (n = 25) and mouse (n = 35) TAS2R protein sequences (30) using TBlastN, enforcing an e-value threshold of ≤10−10. This approach previously has proven robust for localizing TAS2R variants across diverse species, with conserved transmembrane domains in TAS2R genes providing reliable amino acid targets for the TBlastN queries (30). From the TBlastN output, we generated a bed file for each species containing genomic coordinates of candidate TAS2R regions, merged overlapping regions, added 100 nucleotides to each flank, and used BEDtools (55) to generate nucleotide FASTA files for the corresponding regions from the genome assemblies. We then used a Perl script to translate these nucleotide data into amino acid sequences in all six reading frames and return the longest putative intact gene from each region, defined previously (30) as having at least 270 amino acid residues, starting and ending with start and stop codons, and being uninterrupted by premature stop codons. We used MAFFT (51) to align all resulting protein sequences and visually inspected the result to ensure plausible multiple alignment among all copies, validating these genes as TAS2R homologs. This strategy resulted in the recovery of 933 intact TAS2R copies among the 46 species analyzed. To minimize any remaining taxonomic or dietary acquisition biases associated with using human and mouse query sequences, we repeated the entire screening process a second time, using all TAS2R genes recovered in the first pass as input queries. In the second pass, we recovered 20 additional genes across 14 different species, bringing the total TAS2R count to 953 to carry into our analyses (provided as Datasets S2 and S3).
Table S5.
Mammal genome assemblies analyzed, TAS2R count, and assembly quality information
| Species | Common name | Assembly | TAS2R copies, first pass | TAS2R copies, second pass | Body mass estimate, kg | Diet specialization | Scaffold N50 | Included |
| Ailuropoda melanoleuca | Giant panda | ailMel1 | 16 | 16 | 102.5 | 4 | 1281781 | Yes |
| Bos taurus | Cow | bosTau7 | 21 | 21 | 755 | 3 | 2599288 | Yes |
| Callithrix jacchus | Common marmoset | calJac1 | 21 | 23 | 0.33 | 3 | 5167444 | Yes |
| Canis familiaris | Dog | canFam3 | 15 | 16 | 51.5 | 3 | 45876610 | Yes |
| Cavia porcellus | Guinea pig | cavPor3 | 31 | 31 | 0.9 | 3 | 27942054 | Yes |
| Ceratotherium simum | White rhinoceros | cerSim1 | 27 | 27 | 2,520 | 3 | 26277727 | Yes |
| Choloepus hoffmanni | Two-toed sloth | choHof1 | 4 | 4 | 6 | 4 | 9667 | No |
| Dasypus novemcinctus | Nine-banded armadillo | dasNov3 | 10 | 10 | 5.65 | 2 | 1687935 | Yes |
| Daubentonia madagascariensis | Aye-aye | dauMad | 10 | 11 | 2.5925 | 4 | 13597 | No |
| Dipodomys ordii | Ord's kangaroo rat | dipOrd1 | 9 | 10 | 0.0755 | 2 | 36427 | No |
| Echinops telfairi | Lesser hedgehog tenrec | echTel2 | 14 | 17 | 0.2025 | 2 | 45764842 | Yes |
| Equus caballus | Horse | equCab2 | 21 | 22 | 1,150 | 3 | 46749900 | Yes |
| Erinaceus europaeus | Western European hedgehog | eriEur2 | 20 | 20 | 1 | 2 | 3264618 | Yes |
| Felis catus | Cat | felCat5 | 14 | 14 | 4.75 | 3 | 4658941 | Yes |
| Gorilla gorilla gorilla | Western lowland gorilla | gorGor3 | 24 | 24 | 180 | 3 | 913458 | Yes |
| Heterocephalus glaber | Naked mole rat | hetGla2 | 21 | 21 | 0.055 | 4 | 20532749 | Yes |
| Homo sapiens | Human | hg19 | 24 | 24 | 70 | 1 | 46395641 | Yes |
| Loxodonta africana | African bush elephant | loxAfr3 | 20 | 20 | 4,540 | 2 | 46401353 | Yes |
| Macaca mulatta | Rhesus macaque | rheMac3 | 25 | 25 | 8 | 2 | 1660975 | Yes |
| Macropus eugenii | Tammar wallaby | macEug2 | 19 | 20 | 6.55 | 3 | 36602 | No |
| Microcebus murinus | Gray mouse lemur | micMur1 | 10 | 10 | 0.06 | 2 | 140884 | No |
| Monodelphis domestica | Gray short-tailed possum | monDom5 | 27 | 27 | 0.1225 | 2 | 59809810 | Yes |
| Mus musculus | House mouse | mm10 | 36 | 36 | 0.021 | 1 | 52589046 | Yes |
| Mustela putorius furo | Ferret | musFur1 | 15 | 15 | 1.5 | 3 | 9335154 | Yes |
| Myotis lucifugus | Little brown bat | myoLuc2 | 28 | 29 | 0.0095 | 3 | 4293315 | Yes |
| Nomascus leucogenys | Northern white-cheeked gibbon | nomLeu3 | 17 | 17 | 5.7 | 3 | 52956880 | Yes |
| Ochotona princeps | American pika | ochPri2 | 15 | 15 | 0.1485 | 2 | 88760 | No |
| Oryctolagus cuniculus | European rabbit | oryCun2 | 28 | 29 | 2 | 2 | 35972871 | Yes |
| Otolemur garnettii | Northern greater galago | otoGar3 | 22 | 22 | 0.7715 | 3 | 13852661 | Yes |
| Ovis aries | Sheep | oviAri3 | 15 | 15 | 110 | 3 | 100079507 | Yes |
| Pan troglodytes | Chimpanzee | panTro4 | 26 | 26 | 48 | 1 | 8925874 | Yes |
| Papio anubis | Olive baboon | papAnu2 | 29 | 29 | 19.5 | 1 | 528927 | Yes |
| Pongo abelii | Sumatran orangutan | ponAbe2 | 23 | 24 | 60 | 3 | 747460 | Yes |
| Procavia capensis | Rock hyrax | proCap1 | 15 | 15 | 4 | 2 | 24297 | No |
| Pteropus vampyrus | Large flying fox | pteVam1 | 16 | 16 | 0.85 | 4 | 124060 | No |
| Rattus norvegicus | Brown rat | rn5 | 36 | 36 | 0.32 | 1 | 2178346 | Yes |
| Saimiri boliviensis | Black-capped squirrel monkey | saiBol1 | 22 | 22 | 0.75 | 2 | 18744880 | Yes |
| Sarcophilus harrisii | Tasmanian devil | sarHar1 | 19 | 19 | 8 | 3 | 1847106 | Yes |
| Sorex araneus | Common shrew | sorAra2 | 44 | 46 | 0.0095 | 2 | 22794405 | Yes |
| Spermophilus tridecemlineatus | Thirteen-lined ground squirrel | speTri2 | 19 | 19 | 0.125 | 2 | 8192786 | Yes |
| Sus scrofa | Pig | susScr3 | 15 | 16 | 169 | 1 | 576008 | Yes |
| Tarsius syrichta | Philippine tarsier | tarSyr2 | 23 | 25 | 0.125 | 3 | 401181 | Yes |
| Trichechus manatus latirostris | West Indian manatee | triMan1 | 6 | 8 | 400 | 3 | 14442683 | Yes |
| Tupaia chinensis | Chinese tree shrew | tupChi_1.0 | 35 | 35 | 0.385 | 2 | 3670124 | Yes |
| Ursus maritimus | Polar bear | ursMar | 14 | 14 | 475 | 4 | 16000000 | Yes |
| Vicugna pacos | Alpaca | vicPac2 | 12 | 12 | 60 | 4 | 7263804 | Yes |
Table S6.
Species and dietary notes
| Species | Dietary notes |
| Ailuropoda melanoleuca | >99% bamboo, rarely some fruits and small animals |
| Bos taurus | Grass specialist, some tougher vegetation |
| Callithrix jacchus | Mostly plant exudates, various other miscellany |
| Canis familiaris | Terrestrial vertebrates |
| Cavia porcellus | Grass specialist, but some other plants |
| Ceratotherium simum | Grazer |
| Choloepus hoffmanni | Strict folivore |
| Dasypus novemcinctus | Generalist carnivore |
| Daubentonia madagascariensis | Canarium and larvae only |
| Dipodomys ordii | Primarily seeds, grasshoppers and moths seasonally |
| Echinops telfairi | Invertebrates, molluscs, fruit |
| Equus caballus | Grazer |
| Erinaceus europaeus | Omnivorous, primarily insects |
| Felis catus | Terrestrial vertebrates |
| Gorilla gorilla gorilla | Primarily folivore, some other plants and occasional insects |
| Heterocephalus glaber | Tuberous structures |
| Homo sapiens | Broad-spectrum omnivore |
| Loxodonta africana | Generalist herbivore |
| Macaca mulatta | Generalist herbivore |
| Macropus eugenii | Grass specialist |
| Microcebus murinus | Primarily insects, small reptiles and amphibians, some plants |
| Monodelphis domestica | Generalist omnivore |
| Mus musculus | Broad-spectrum omnivore |
| Mustela putorius furo | Terrestrial vertebrates |
| Myotis lucifugus | Insects only |
| Nomascus leucogenys | Fruit, some other plant matter, and insects |
| Ochotona princeps | Generalist herbivore |
| Oryctolagus cuniculus | Generalist herbivore |
| Otolemur garnettii | Fruit and insects |
| Ovis aries | Grass specialist |
| Pan troglodytes | Broad-spectrum omnivore |
| Papio anubis | Broad-spectrum omnivore |
| Pongo abelii | Primarily fruit, other plant matter, and insects/eggs to 5% |
| Procavia capensis | Generalist herbivore |
| Pteropus vampyrus | Flowers, nectar, fruit |
| Rattus norvegicus | Broad-spectrum omnivore |
| Saimiri boliviensis | Primarily fruit and insects, but with other contributers |
| Sarcophilus harrisii | Scavenger carnivore |
| Sorex araneus | Invertebrates only |
| Spermophilus tridecemlineatus | Generalist omnivore |
| Sus scrofa | Broad-spectrum omnivore |
| Tarsius syrichta | Obligate carnivore, birds, reptiles and amphibians, insects, arthropods |
| Trichechus manatus latirostris | Sea grasses, occasional fish and invertebrates |
| Tupaia chinensis | General omnivore, but some uncertainty and interspecies variation |
| Ursus maritimus | Terrestrial vertebrates, prefers sea mammals, lipivore |
| Vicugna pacos | Grazer with strict physiological requirements |
Phenotypic character assignments.
Body mass was assigned based on typical values for adult males of each species from the University of Michigan Animal Diversity Web (ADW) (56) or from adults in general when sexual dimorphism data were not available. For dietary breadth, we assigned each mammal species a value from 1 to 4, where 1 signifies a broad-spectrum dietary generalist (omnivorous species with a wide range of foraging behaviors, e.g., mouse, pig, human) and 4 indicates a narrow dietary specialist (species with extremely specialized dietary behaviors, especially species for which the diet may consist almost entirely of a single species or a few similar species, e.g., bamboo for the giant panda or highland grasses for the alpaca), using ADW descriptions of diet as guidelines (Tables S5 and S6). Diet breadth assignations were made without prior knowledge of TAS2R count to avoid undue influence on category choice.
Analysis of TAS2R copy number variation.
To test for the effects of genome assembly quality on the sensitivity of our technique to detect TAS2R copies in poorer assemblies, we first tested for a relationship between log10 scaffold N50 and TAS2R count using a linear model alongside log10 body mass and diet breadth, using the R statistical package (57). We identified a highly significant correlation between scaffold N50 and TAS2R count (P = 0.003). This effect apparently is caused by difficulty in identifying intact TAS2R variants in more fragmentary, lower-quality genome assemblies. When assemblies with N50 <50 kb (n = 5) were excluded, significance decreased dramatically (P = 0.103). Conservatively, we chose to exclude assemblies with N50 <250 kb (n = 8) from analysis, eliminating the significant relationship between assembly quality and intact TAS2R count (P = 0.863). To analyze associations between phenotypic characters and TAS2R gene count while correcting for phylogenetic influence, we implemented a phylogenetic generalized least squares (pGLS) approach within the “caper” R package (58). We constructed an ultrametric phylogenetic mammal supertree manually from the literature (Fig. S2 and Dataset S4) (59–61). We used maximum likelihood optimizations of the delta, kappa, and lambda branch length transformations implemented in the pGLS (detailed in ref. 58).
Fig. S2.
Ultrametric supertree used for pGLS analysis of 38 mammal genomes.
Supplementary Material
Acknowledgments
We thank Craig Praul and Candace Price from the Pennsylvania State University Huck Institutes DNA Core Laboratory for assistance with sequence data collection and the US Department of Agriculture Agricultural Research Service National Plant Germplasm System, the Illinois State Museum, the National Park Service, and the University Museum at the University of Arkansas for access to modern and ancient sample materials. Research was supported by The Pennsylvania State University Huck Institutes of the Life Sciences and College of the Liberal Arts (G.H.P.), Wenner–Gren post-PhD Research Grant 8770 and Natural Environment Research Council Independent Research Fellowship NE/L012030/1 (to L.K.), and the Smithsonian Institution (B.D.S.). Instrumentation was funded by the National Science Foundation through Grant OCI–0821527.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.M. is a guest editor invited by the Editorial Board.
Data deposition: Illumina short read sequence data reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive (study accession no. SRP064244). Annotated plastid genome sequences for taxonomic representatives have been deposited in GenBank database (accession nos. KT898803–KT898821).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516109112/-/DCSupplemental.
References
- 1.Simpson MG. Plant Systematics. Elsevier/Academic; Boston: 2006. [Google Scholar]
- 2.Newsom LA, Mihlbachler MC. Mastodons (Mammut americanum) diet foraging patterns based on analysis of dung deposits. In: Webb SD, editor. First Floridians and Last Mastodons: The Page-Ladson Site in the Aucilla River. Springer; Dordrecht, The Netherlands: 2006. pp. 263–331. [Google Scholar]
- 3.Cowan CW, Smith BD. New perspectives on a wild gourd in Eastern North America. J Ethnobiol. 1993;13(1):17–54. [Google Scholar]
- 4.Johnson CN. Ecological consequences of Late Quaternary extinctions of megafauna. Proc Biol Sci. 2009;276(1667):2509–2519. doi: 10.1098/rspb.2008.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnosky AD, Koch PL, Feranec RS, Wing SL, Shabel AB. Assessing the causes of late Pleistocene extinctions on the continents. Science. 2004;306(5693):70–75. doi: 10.1126/science.1101476. [DOI] [PubMed] [Google Scholar]
- 6.Janzen DH, Martin PS. Neotropical anachronisms: The fruits the gomphotheres ate. Science. 1982;215(4528):19–27. doi: 10.1126/science.215.4528.19. [DOI] [PubMed] [Google Scholar]
- 7.Fritz GJ. Gender and the early cultivation of gourds in Eastern North America. Am Antiq. 1999;64(3):417–429. [Google Scholar]
- 8.Crook MR. 2008. Bilbo (9Ch4) and Delta (38Ja23): Late Archaic and Early Woodland shell mounds at the mouth of the Savannah River. Occasional Papers in Cultural Resource Management no. 17.
- 9.Guimarães PR, Galetti M, Jordano P. Seed dispersal anachronisms: Rethinking the fruits extinct megafauna ate. PLoS One. 2008;3(3):e1745. doi: 10.1371/journal.pone.0001745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barlow C. The Ghosts of Evolution. Basic Books; New York: 2000. [Google Scholar]
- 11.Nee M. The domestication of Cucurbita (Cucurbitaceae) Econ Bot. 1990;44(3):56–68. [Google Scholar]
- 12.Smith BD. The initial domestication of Cucurbita pepo in the Americas 10,000 years ago. Science. 1997;276(5314):932–934. [Google Scholar]
- 13.Decker-Walters DS. In: Evidence for multiple domestications of Cucurbita pepo. Biology and Utilization of the Cucurbitaceae. Bates DM, Robinson RW, Jeffrey C, editors. Cornell Univ Press; Ithaca, NY: 1990. pp. 96–101. [Google Scholar]
- 14.Sanjur OI, Piperno DR, Andres TC, Wessel-Beaver L. Phylogenetic relationships among domesticated and wild species of Cucurbita (Cucurbitaceae) inferred from a mitochondrial gene: Implications for crop plant evolution and areas of origin. Proc Natl Acad Sci USA. 2002;99(1):535–540. doi: 10.1073/pnas.012577299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferriol M, Picó B, Belen P. 2008. Pumpkin and winter squash. Handbook of Plant Breeding. Vol 1. Vegetables I., eds Prohens J, Nuez F (Springer, Heidelberg), pp. 317–349.
- 16.Tallamy DW, Krischik VA. Variation and function of cucurbitacins in Cucurbita: An examination of current hypotheses. Am Nat. 1989;133(6):766–786. [Google Scholar]
- 17.David A, Vallance DK. Bitter principles of Cucurbitaceae. J Pharm Pharmacol. 1955;7(1):295–296. [Google Scholar]
- 18.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27(2):182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith BD. Eastern North America as an independent center of plant domestication. Proc Natl Acad Sci USA. 2006;103(33):12223–12228. doi: 10.1073/pnas.0604335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andres TC. 1987. Cucurbita fraterna, the closest wild relative and progenitor of C. pepo. Cucurbit Genet Coop Rep (10):69–71.
- 21.Decker-Walters DS, Staub JE, Chung SM, Nakata E, Quemada HD. Diversity in free-living populations of Cucurbita pepo (Cucurbitaceae) as assessed by random amplified polymorphic DNA. Syst Bot. 2002;27(1):19–28. [Google Scholar]
- 22.Decker-Walters DS, Walters T, Cowan CW, Smith BD. Isozymic characterization of wild populations of Cucurbita pepo. J Ethnobiol. 1993;13(1):55–72. [Google Scholar]
- 23.Smith BD. Seed size increase as a marker of domestication in squash (Cucurbita pepo) In: Zeder MA, , Bradley D, Emshwiller ESmith BD, , editors. Documenting Domestication: New Genetic and Archaeological Paradigms. Univ of California Press; Berkeley, CA: 2006. pp. 25–31. [Google Scholar]
- 24.Walters TW, Decker-Walters DS. Systematics of the endangered okeechobee gourd (Cucurbita okeechobeensis: Cucurbitaceae) Syst Bot. 1993;18(2):175–187. [Google Scholar]
- 25.Ferguson JE, Metcalf RL. Cucurbitacins: Plant-derived defense compounds for diabroticites (Coleoptera: Chrysomelidae) J Chem Ecol. 1985;11(3):311–318. doi: 10.1007/BF01411417. [DOI] [PubMed] [Google Scholar]
- 26.Bemis WP, Curtis LD, Weber CW, Berry J. The feral buffalo gourd, Cucurbita foetidissima. Econ Bot. 1978;32(1):87–95. [Google Scholar]
- 27.Gry J, Søborg I, Andersson H. Cucurbitacins in Plant Food. TemaNord; Copenhagen: 2006. [Google Scholar]
- 28.Dinerstein E, Wemmer C. Fruits rhinoceros eat: Dispersal of Trewia nudiflora (Euphorbiaceae) in lowland Nepal. Ecology. 1988;69(6):1768–1774. [Google Scholar]
- 29.Kusin S, Angert T, Von Derau K, Horowitz BZ, Giffin S. 2012. Toxic squash syndrome: A case series of diarrheal illness following ingestion of bitter squash. Poster presentation at the annual meeting of the North American Congress of Clinical Toxicology.
- 30.Li D, Zhang J. Diet shapes the evolution of the vertebrate bitter taste receptor gene repertoire. Mol Biol Evol. 2014;31(2):303–309. doi: 10.1093/molbev/mst219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong D, Jones G, Zhang S. Dynamic evolution of bitter taste receptor genes in vertebrates. BMC Evol Biol. 2009;9:12–20. doi: 10.1186/1471-2148-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Thomas SD, Zhang J. Relaxation of selective constraint and loss of function in the evolution of human bitter taste receptor genes. Hum Mol Genet. 2004;13(21):2671–2678. doi: 10.1093/hmg/ddh289. [DOI] [PubMed] [Google Scholar]
- 33.Feng P, Zheng J, Rossiter SJ, Wang D, Zhao H. Massive losses of taste receptor genes in toothed and baleen whales. Genome Biol Evol. 2014;6(6):1254–1265. doi: 10.1093/gbe/evu095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams J, Shuman BN, Webb T, Bartlein PJ, Leduc PL. Late-Quaternary vegetation dynamics in North America: Scaling from taxa to biomes. Ecol Monogr. 2004;74(2):309–334. [Google Scholar]
- 35.Piperno DR, Stothert KE. Phytolith evidence for early Holocene Cucurbita domestication in southwest Ecuador. Science. 2003;299(5609):1054–1057. doi: 10.1126/science.1080365. [DOI] [PubMed] [Google Scholar]
- 36.Allaby RG, et al. Archaeogenomic insights into the adaptation of plants to the human environment: Pushing plant–hominin co-evolution back to the Pliocene. J Hum Evol. 2015;79:150–157. doi: 10.1016/j.jhevol.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 37.Jones M, Brown T. Selection, cultivation, and reproductive isolation: A reconsideration of morphological and molecular signals of domestication. In: Denham T, Iriarte J, Vrydaghs L, editors. Rethinking Agrictulture: Archaeological and Ethnoarchaeological Perspectives. Left Coast; Walnut Creek, California: 2009. pp. 36–49. [Google Scholar]
- 38.Dempewolf H, Hodgins KA, Rummell SE, Ellstrand NC, Rieseberg LH. Reproductive isolation during domestication. Plant Cell. 2012;24(7):2710–2717. doi: 10.1105/tpc.112.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Civáň P, Ivaničová Z, Brown TA. Reticulated origin of domesticated emmer wheat supports a dynamic model for the emergence of agriculture in the fertile crescent. PLoS One. 2013;8(11):e81955. doi: 10.1371/journal.pone.0081955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webster DL. Backward Bottlenecks. Curr Anthropol. 2011;52(1):77–104. [Google Scholar]
- 41.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010;2010(6):t5448. doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 42.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaefer H, Heibl C, Renner SS. 2009. Gourds afloat: A dated phylogeny reveals an Asian origin of the gourd family (Cucurbitaceae) and numerous oversea dispersal events. Proc R Soc B 276(1658):843–851.
- 45.Ratan A. 2009. Assembly Algorithms for Next-Generation Sequence Data. PhD dissertation (The Pennsylvania State University, State College, PA)
- 46.Harris RS. 2007. Improved Pairwise Alignment of Genomic DNA. PhD dissertation (The Pennsylvania State University, State College, PA)
- 47.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kistler L. Ancient DNA extraction from plants. In: Shapiro B, Hofreiter M, editors. Ancient DNA: Methods and Protocols, Methods in Molecular Biology. Vol 840. Humana; New York: 2012. pp. 71–79. [DOI] [PubMed] [Google Scholar]
- 49.Kircher M. Analysis of high-throughput ancient DNA sequencing data. In: Shapiro B, Hofreiter M, editors. Ancient DNA: Methods and Protocols, Methods in Molecular Biology. Vol 840. Humana; New York: 2012. pp. 197–228. [DOI] [PubMed] [Google Scholar]
- 50.Gilbert MTP, et al. Recharacterization of ancient DNA miscoding lesions: Insights in the era of sequencing-by-synthesis. Nucleic Acids Res. 2007;35(1):1–10. doi: 10.1093/nar/gkl483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 53.Bouckaert R, et al. BEAST 2: A software platform for Bayesian evolutionary analysis. PLOS Comput Biol. 2014;10(4):e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 55.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Myers P, et al. 2015 The Animal Diversity Web. Available at animaldiversity.org. Accessed January 1, 2015.
- 57.R Development Core Team 2013 R: A language and environment for statistical computing. (R Foundation for Statistical Computing, Vienna) Available at www.R-project.org/. Accessed April 1, 2015.
- 58.Orme D, et al. 2013. caper: Comparative Analyses of Phylogenetics and Evolution in R. R Package version 052. Available at CRAN.R-project.org/package=caper. Accessed April 1, 2015.
- 59.Pozzi L, et al. Primate phylogenetic relationships and divergence dates inferred from complete mitochondrial genomes. Mol Phylogenet Evol. 2014;75(1):165–183. doi: 10.1016/j.ympev.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huchon D, et al. Multiple molecular evidences for a living mammalian fossil. Proc Natl Acad Sci USA. 2007;104(18):7495–7499. doi: 10.1073/pnas.0701289104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meredith RW, et al. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science. 2011;334(6055):521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.