Significance
Arsenic holds promise for treating a wide range of tumors. To understand arsenic's antitumor mechanism further, we identified 360 arsenic-binding proteins using a human proteome microarray and found proteins of glycolysis to be highly enriched. In-depth in vitro and in vivo analysis revealed that glycolysis in general and the rate-limiting enzyme hexokinase-2 of the glycolytic pathway in particular play a key role in mediating the anticancer activity of arsenic. These findings shed light on the mode of action of arsenic, and the newly identified arsenic-binding proteins may serve as a rich resource for future studies.
Keywords: arsenic trioxide, human proteome microarray, glycolysis, hexokinase-2I
Abstract
Arsenic is highly effective for treating acute promyelocytic leukemia (APL) and has shown significant promise against many other tumors. However, although its mechanistic effects in APL are established, its broader anticancer mode of action is not understood. In this study, using a human proteome microarray, we identified 360 proteins that specifically bind arsenic. Among the most highly enriched proteins in this set are those in the glycolysis pathway, including the rate-limiting enzyme in glycolysis, hexokinase-1. Detailed biochemical and metabolomics analyses of the highly homologous hexokinase-2 (HK2), which is overexpressed in many cancers, revealed significant inhibition by arsenic. Furthermore, overexpression of HK2 rescued cells from arsenic-induced apoptosis. Our results thus strongly implicate glycolysis, and HK2 in particular, as a key target of arsenic. Moreover, the arsenic-binding proteins identified in this work are expected to serve as a valuable resource for the development of synergistic antitumor therapeutic strategies.
Arsenic and its derivatives have been applied as therapy for a variety of diseases for more than 2,200 y (1). To date, the disease most successfully treated with these types of compounds is acute promyelocytic leukemia (APL). Administration of arsenic trioxide (ATO) combined with all-trans retinoic acid has demonstrated a remarkable 5-y overall survival rate of 85–90% as a consequence of the dramatic down-regulation of the key protein driving APL tumorigenicity, promyelocytic leukemia-retinoic acid receptor α (PML-RARα) (2). However, ATO also has been found to be effective against many other hematologic malignancies and solid tumors. For example, together with imatinib it is a promising treatment for chronic myelocytic leukemia (3), and it has been used alone with some success to treat multiple myeloma (4), myelodysplasia syndrome (5), and non-Hodgkin lymphoma (6). ATO also is under clinical investigation as a possible medication for lung cancer, hepatocellular carcinoma, melanoma, renal cell carcinoma, and colorectal cancer (https://www.clinicaltrials.gov/). At the cellular level, ATO has been shown to inhibit significantly the growth of almost all the cell lines (59 of 60) in the US National Cancer Institute anticancer drug screen that spans nine different tumor types (7). Thus ATO is one of the most promising broadly effective medications against cancer.
Although its mode of action in APL is well established, the underlying mechanisms by which ATO acts in other types of cancers remain poorly understood. A variety of systematic studies, including studies that provided transcriptomic, chemogenomic, or proteomic characterizations (8, 9), have been performed to gain a better understanding of this broader anticancer activity of ATO. However, these studies examined only the cellular consequences of treatment with ATO without identifying the primary proteins directly bound and modulated by ATO. Knowledge of ATO’s binding partners is ultimately crucial for any mechanistic understanding of its effects at the cellular level. To date, about 20 validated ATO-binding human proteins have been reported (10), but inspection of their known functions suggests that they cannot account for the wide and profound effects of ATO on cancer cells. We thus hypothesized that more physiologically relevant ATO-binding proteins remain to be discovered.
In this study, we applied a human proteome microarray containing 16,368 proteins (11) for a systematic identification of arsenic-binding proteins. We identified a total of 360 protein candidates, nearly 20-fold more than previously known. Unexpectedly, we found that proteins involved in glycolysis are significantly enriched within this set, including the key rate-limiting enzyme of glycolysis, hexokinase-1 (HK1). More detailed examination of the homologous enzyme hexokinase-2 (HK2), which is overexpressed in many cancers, confirmed this arsenic binding and demonstrated that its enzymatic activity was inhibited significantly by arsenic, both in vitro and in vivo. Our study thus implicates the glycolytic pathway and HK2 in particular as a general target of ATO in cancer. Because a high level of glycolysis is a common property of many cancers (referred to as the “Warburg effect”) (12), our results provide a possible explanation for the general inhibitory effect of ATO in different types of cancers.
Results
Global Profiling of Arsenic-Binding Proteins Using a Human Proteome Microarray.
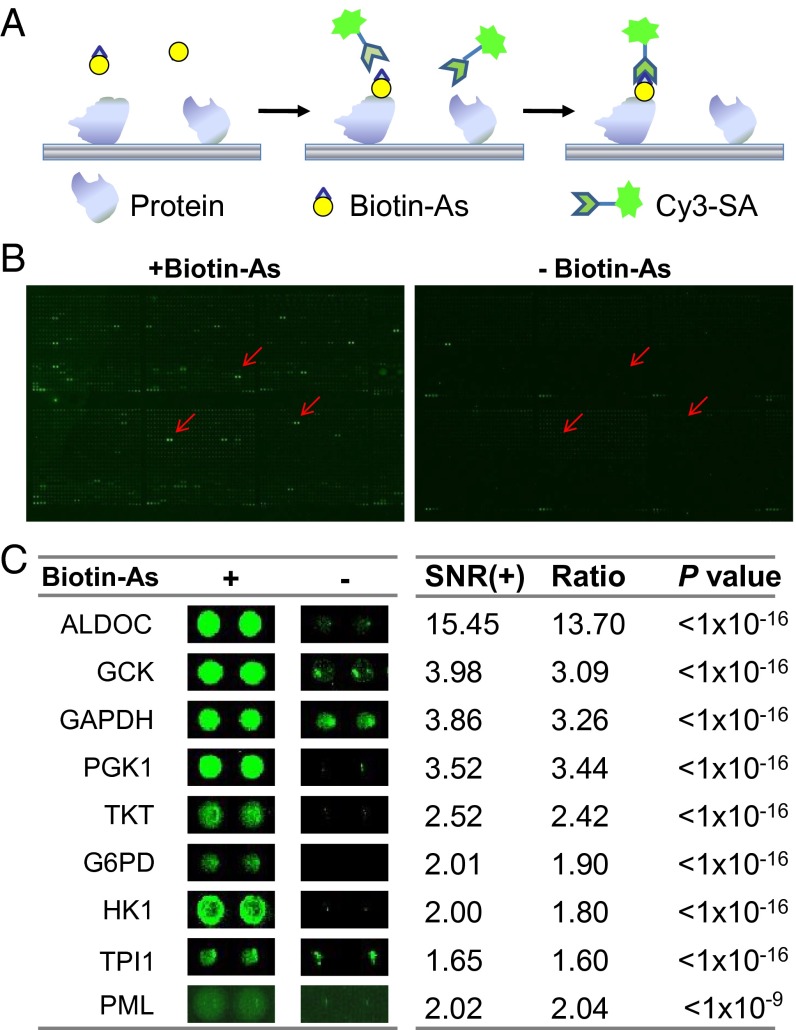
To identify arsenic-binding proteins, we probed a human proteome microarray consisting of 16,368 affinity-purified N-terminally GST-tagged proteins (11) with a biotinylated arsenic molecule (Fig. 1A). Briefly, the arsenic–biotin conjugate, biotinylated p-aminophenylarsenoxide (Biotin–As) (10), was incubated with the human proteome microarray, and proteins with arsenic-binding capacity were identified by adding Cy3-conjugated streptavidin (Cy3–SA). To avoid false-positive detections, we compared this sample with another in which Biotin–As was omitted from the reaction and free biotin was included instead and identified as arsenic-binding candidate proteins those exhibiting markedly greater signals in the former reaction. To assure that this reaction exhibits binding specificity, the human proteome microarray was incubated first with 100 µM ATO, followed by 10 µM Biotin–As with 100 µM ATO. We found that the binding of Biotin–As to the human proteome microarray was abolished completely. Two randomly chosen blocks from the same location in the experimental microarray and the negative control microarray are shown in Fig. 1B. Clearly, many more positive signals are present on the microarray incubated with Biotin than on the negative control microarray. In total, we identified 360 candidate arsenic-binding proteins from these microarrays (Dataset S1); representatives are shown in Fig. 1C. We note that PML was included in this array as a positive control because it is a well-known target of arsenic (13).
Fig. 1.
Identification of arsenic-binding proteins on the human proteome microarrays. (A) Schematic of the procedure for detecting binding events. (B, Left) Human proteome microarrays were probed with arsenic, followed by Cy3-labeled streptavidin. (Right) A control experiment was carried out without arsenic. By comparing the signals from the two microarrays, 360 candidate arsenic-interacting proteins were identified. (C) Representative arsenic-interacting proteins. Cy3 signals were measured at 532 nm. +, in the presence of Biotin–As; −, in the presence of biotin alone.
Bioinformatics Analysis Reveals a Striking Enrichment of Glycolytic Enzymes.
To gain insight into the functional roles of this set of arsenic-binding proteins, we examined the overrepresented ontological terms and components of molecular pathways of the 360 candidates (Dataset S1) relative to their occurrence in the human proteome using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (14). The candidate proteins then were examined for enrichment in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, biological process, and cellular component.
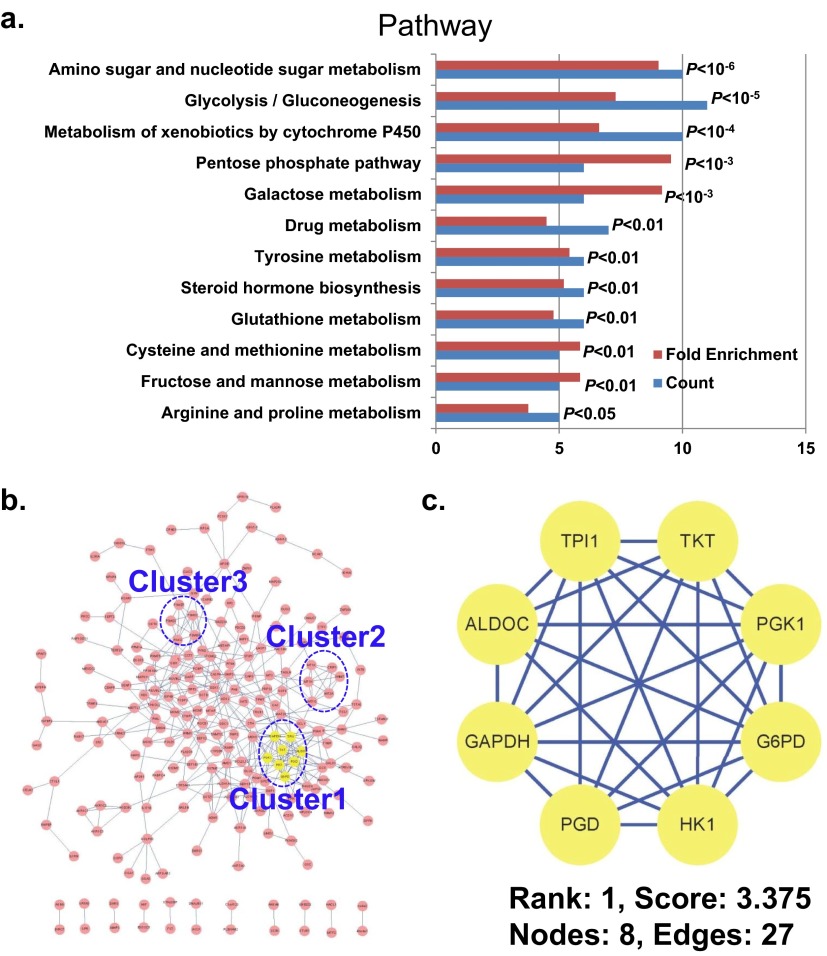
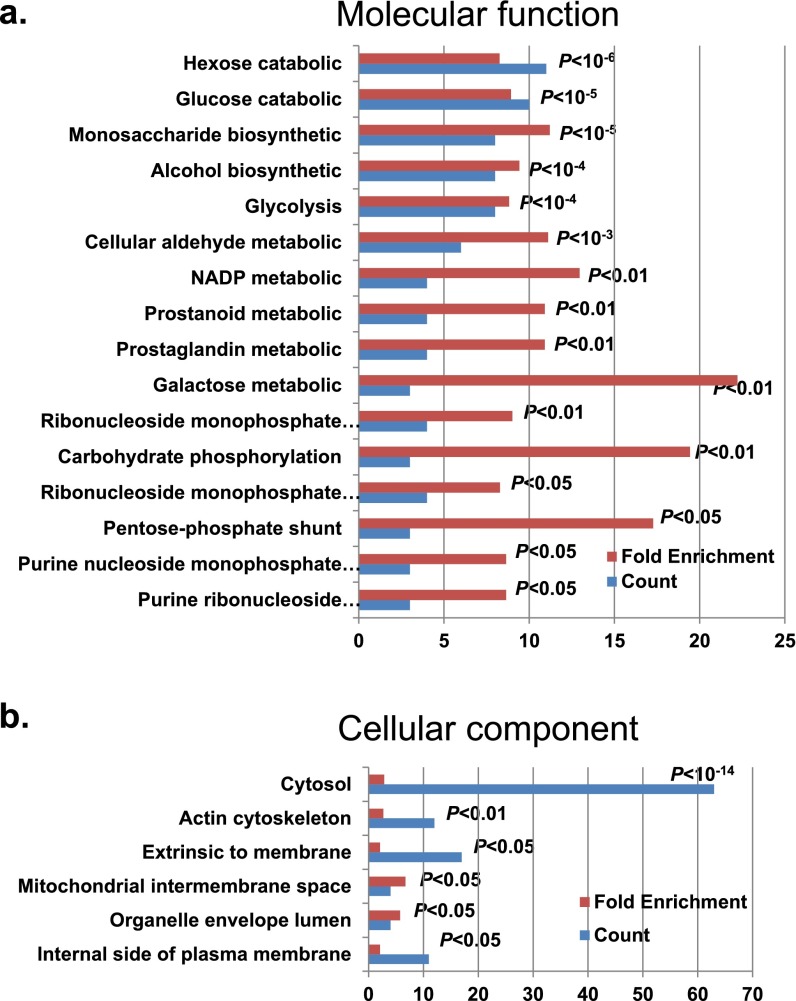
Pathways exhibiting a KEGG term enrichment of P < 0.05 are shown in Fig. S1A. There is a substantial enrichment in glycolysis/gluconeogenesis (P < 10−5). For biological process, the top 16 Gene Ontology (GO) terms with P < 0.01 are shown in Fig. S2A. In this case, the candidate list was substantially enriched in glycolysis (P < 10−4). Hence both biological process and KEGG results indicate many enzymes involved in glycolysis, which may significantly disturb these pathways. The cellular components exhibiting a GO term enrichment of P < 0.05 are shown in Fig. S2B. There is a marked enrichment of candidates localizing in the cytosol (P < 10−14).
Fig. S1.
Bioinformatics analysis of the arsenic-binding proteins. (A) Pathway enrichment analysis by DAVID. (B) The complete interaction network of the arsenic-interacting proteins obtained from STRING with a confidence score >0.7. The three top-ranked and tightly connected network clusters obtained with MCODE are color coded and rendered as separate modules. (C) The top network cluster is glycolysis with a score of 3.375. See Fig. S3 for the other two clusters.
Fig. S2.
GO analysis of the 360 arsenic-binding proteins by DAVID. (A) Cellular component. (B) Molecular function.
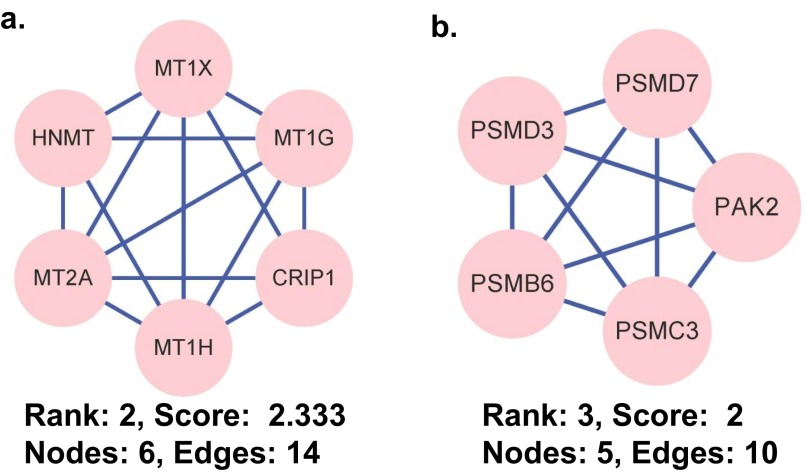
We constructed biological interaction networks for the arsenic-binding proteins to identify meaningful connections between these proteins using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) system (15) with a high-confidence setting and then analyzed the networks for highly connected regions using the Cytoscape plugin, MCODE (“molecular complex detection”). As shown in Fig. S1 B and C, several complexes and cellular functions formed prominent, highly connected clusters. For example, the densest cluster (cluster 1) is formed by many enzymes involved in or highly related to the glycolysis pathway, including triosephosphate isomerase 1 (TPI1), transketolase (TKT), phosphoglycerate kinase 1 (PGK1), glucose-6-phosphate dehydrogenase (G6PD), HK1, phosphogluconate dehydrogenase (PGD), GAPDH, and aldolase C (ALDOC). The majority of the proteins in the second densest cluster (cluster 2) are metallothioneins (Fig. S3A), which are rich in cysteine residues. Because arsenic binds strongly to cysteine (16), the enrichment of the metallothioneins in this list is not unexpected and in fact provides further evidence of the reliability of the data presented here. The third densest cluster (cluster 3) is composed of several key components of the proteasome (Fig. S3B), suggesting that arsenic may disturb the proper functioning of the proteasome, as previously proposed (9).
Fig. S3.
The top-ranked and tightly connected network clusters obtained with MCODE are color coded and rendered as separate modules. (A) The rank 2 cluster is composed of metallothioneins. (B) The rank 3 cluster is rich in proteasome components. CRIP1, cysteine-rich protein 1; HNMT, histamine N-methyltransferase; MT1G, metallothionein 1G; MT1H, metallothionein 1H; MT1X, metallothionein 1X; MT2A, metallothionein 2A; PAK2, p21 protein (Cdc42/Rac)-activated kinase 2; PSMB6, proteasome subunit beta type-6; PSMC3, 26S protease regulatory subunit 6A; PSMD3, 26S proteasome non-ATPase regulatory subunit 3; PSMD7, 26S proteasome non-ATPase regulatory subunit 7.
To assess whether consensus sequences were present within the newly identified arsenic-binding proteins (Dataset S1), we performed a MEME motif search based on the primary amino acid sequences (17). A consensus motif was identified that includes many cysteines (E-value = 1.4e + 47) (Fig. S4). This result also is consistent with the known high affinity of arsenic for cysteine, especially for many adjacent cysteines (16).
Fig. S4.
Motif analysis of the arsenic-binding proteins. The top 100 arsenic-targeting proteins identified in this study (Dataset S1) were analyzed by MEME (version 4.8.1). Here x represents any amino acid, K is lysine, C is cysteine, and A is alanine.
Arsenic Binds to HK2 and Inhibits Its Activity in Vitro.
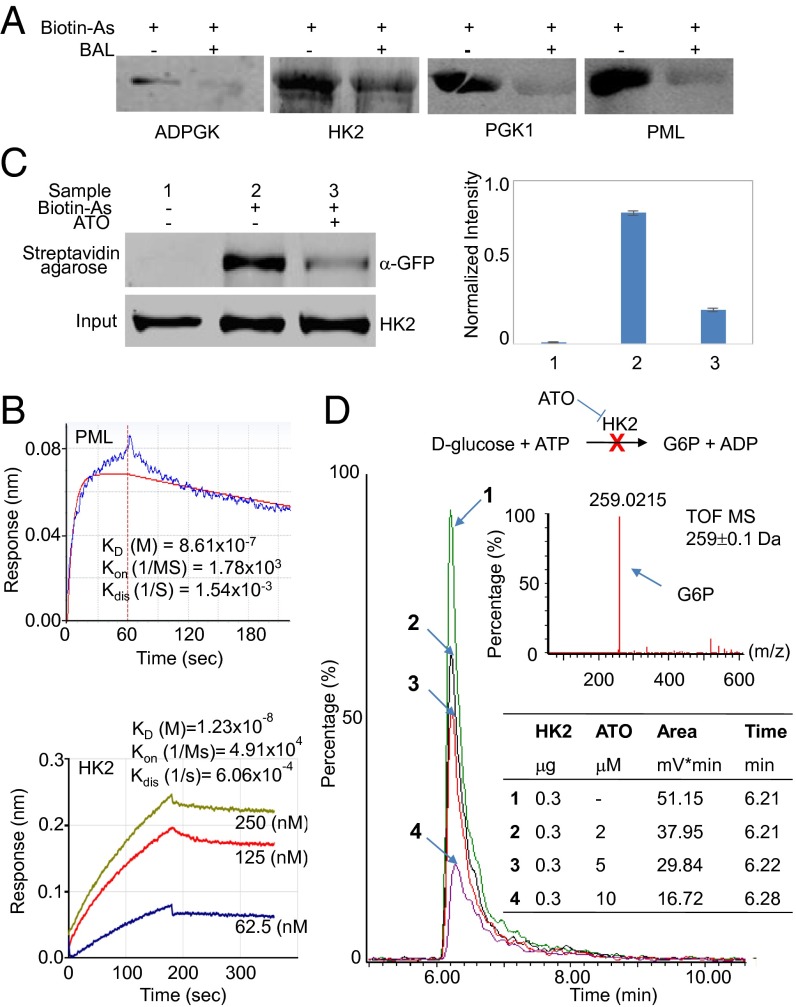
With the preponderance of arsenic-binding proteins identified as glycolytic enzymes, we undertook more detailed in vitro characterizations of arsenic binding and its functional consequences on key enzymes of glycolysis. Because HK1 was identified as an arsenic-binding protein, we also investigated HK2. Although HK2 was not on our human proteome microarray, this protein is known to play a key role in maintaining the high glucose catabolic rates of rapidly growing tumors and is highly homologous to HK1 (2), especially in their N and C termini. We therefore reasoned that arsenic also could bind HK2. In particular, using Biotin–As and Cy3–SA) we examined arsenic binding to PGK1, ADP-dependent glucokinase (ADPGK), and HK2, the first two identified by the aforementioned microarray experiments. PML also was studied as a positive control. As shown in Fig. 2A, each of these proteins readily bound to arsenic. To rule out possible nonspecific interactions, a control reaction with the same concentration of Biotin–As together with an excess of 2,3-dimercapto-1-propanol (BAL), which can effectively neutralize the toxicity of arsenic (18), was performed also. In each case, the interaction of the proteins with arsenic was significantly reduced in the presence of BAL (Fig. 2A).
Fig. 2.
Arsenic binds to HK2 and inhibits its activity in vitro. (A) Streptavidin blotting of selected arsenic-binding proteins. Purified proteins were incubated with 100 µM Biotin–As with and without 100 mM BAL. After 10% SDS/PAGE electrophoresis, proteins were transferred to a PVDF membrane and were incubated with IRDye 800CW streptavidin. PML was included as positive control. (B) BLI analysis of the binding of arsenic to HK2. PML was included as a positive control. Proteins were immobilized on streptavidin-coated biosensors and exposed to Biotin–As in buffer. Binding was measured by coincident change in the interference pattern. Results presented are representative of at least three experiments. (C) A streptavidin agarose affinity assay in which Biotin–As was used as the probe indicates that arsenic binds with HK2. Error bars represent the standard deviation of three replicates. (D) An HK2 activity assay was performed using glucose and ATP as substrates. The major product of this reaction, G6P, was measured by UPLC-MS. The enzymatic activity assays were performed with various concentrations of ATO, and a control reaction without ATO was carried out in parallel.
As an independent approach, we used bio-layer interferometry (BLI) to determine the binding affinity of PML and HK2 for arsenic and found that both interacted reasonably strongly (Kd of 860 nM and 12.3 nM, respectively) with arsenic (Fig. 2B).
We further investigated whether arsenic binds directly to HK2 within cells using a streptavidin agarose affinity assay. In this experiment, after HEK293T cells were incubated with Biotin–As, the cell lysate was incubated with streptavidin-agarose to isolate arsenic-binding proteins, which then were dissociated from the agarose by denaturation and were probed with anti-HK2. As shown in Fig. 2C, arsenic indeed binds to HK2 within cells, and this binding could be attenuated by the prior addition of ATO (Fig. 2C).
We next measured the activity of HK2 in a well-established in vitro assay including glucose and ATP to test directly whether the interaction of HK2 with ATO alters its activity (Fig. 2D). Indeed, we found that 2 µM ATO sharply inhibits the activity of HK2 in vitro, substantially decreasing the glucose-6-phosphate (G6P) reaction product, as is consistent with the idea that HK2 activity is inhibited by the binding of ATO.
Overexpression of HK2 Reverses ATO-Induced Apoptosis in SGC7901 Cells.
When cancer cells are treated with ATO, most cells exhibit pronounced growth inhibition and apoptosis (7). As described above, arsenic binds tightly to HK2 and significantly inhibits its activity in vitro. Therefore we hypothesized that one of the possible mechanisms of these deleterious effects of arsenic on cancer cells could be mediated through the inhibitive binding of arsenic to HK2.
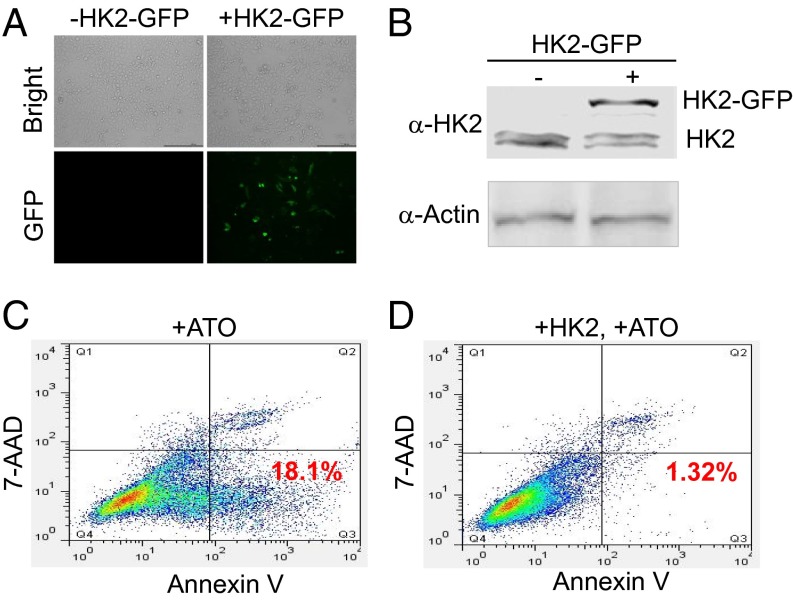
To test this hypothesis, we first confirmed the apoptotic response of the gastric cancer cell line, SGC7901, to arsenic. Consistent with previous studies (19), significant arsenic-induced growth inhibition was observed in these cells 12 and 24 h after ATO treatment. HK2 then was cloned and overexpressed in SGC7901 cells (Fig. 3 A and B). These cells were treated with ATO, and apoptosis was monitored by flow cytometry. A control experiment in which HK2 was not overexpressed was carried out also. We found that, upon treatment with ATO for 12 h, significant apoptosis (18.1%) was induced in the control cells (Fig. 3C), but the apoptosis rate dropped to 1.32% in the cells in which HK2 was overexpressed (Fig. 3D).
Fig. 3.
In the SGC7901 cell line overexpression of HK2 could reverse the apoptosis induced by ATO treatment. (A and B) The overexpression of HK2 in the SGC7901 cell line. SGC7901 cells were transfected with the plasmid carrier HK2-GFP, and the overexpression of HK2 was monitored by fluorescence microscopy (A) and Western blotting (B) using an anti-HK2 antibody. (C and D) SGC7901 cells were transfected with the plasmid carrier HK2-GFP. An apoptosis detection kit was applied. Annexin V- and 7-amino-actionomycin (7-AAD)–stained cells then were analyzed by flow cytometry.
Metabolomics Analysis Revealed That HK2 Is Targeted by Arsenic in Vivo.
Because arsenic binds to a significant portion (5/10) of the enzymes that are directly involved in the glycolysis pathway and inhibits HK2 activity, we hypothesized that there would be significant changes in many of the metabolites of this pathway in cancer cells treated with arsenic. To test this possibility, a systems-wide analysis of the changes in these metabolites was carried out in the gastric cancer cell line SGC7901 studied above.
Cells were harvested after treatment with 2 µM arsenic for 6, 12, or 24 h, as were vehicle controls without arsenic treatment. To ensure the reliability of this analysis, five biological replicates were carried out for both the treated and control group.
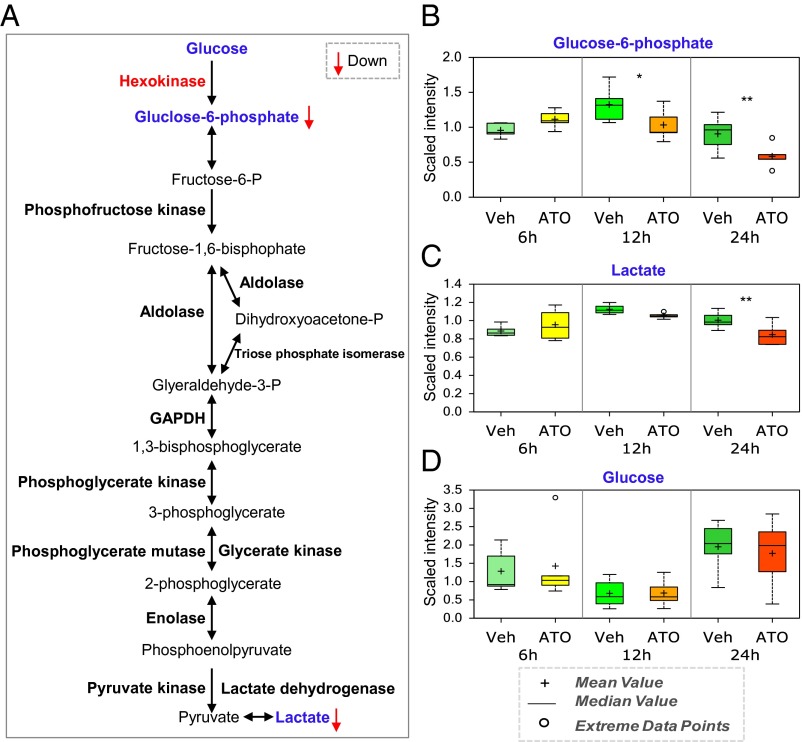
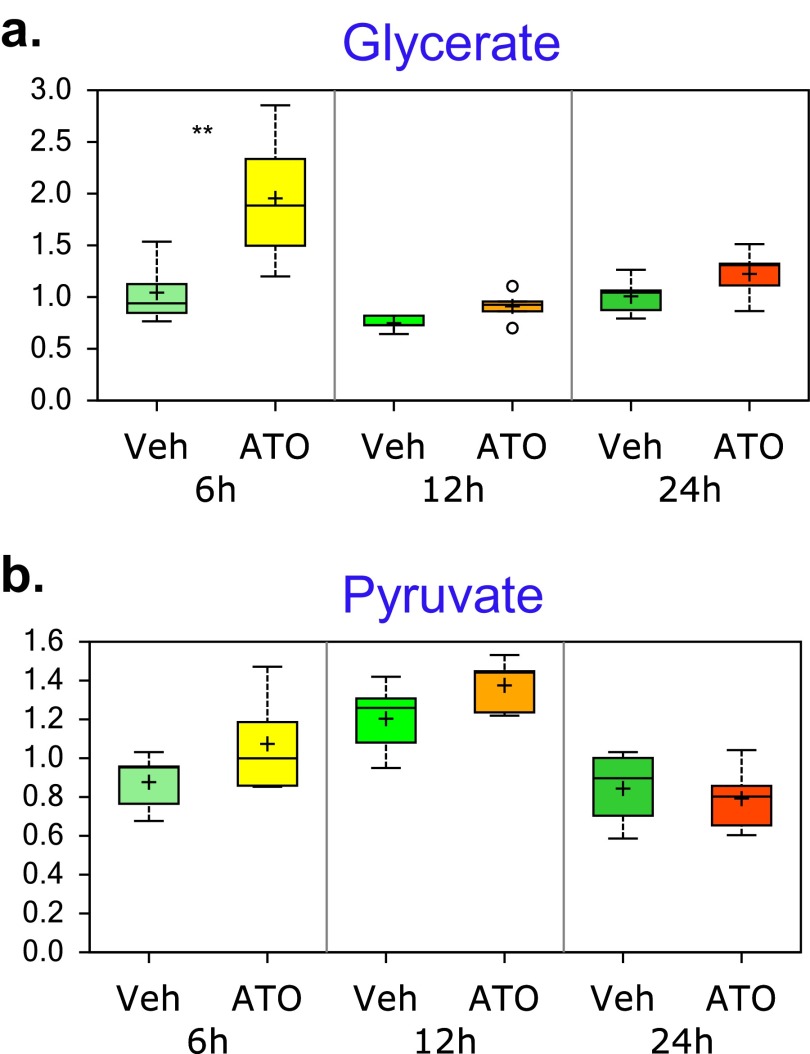
The first, and rate-limiting, step of glycolysis is catalyzed by hexokinase (HK2 in cancer cells) (Fig. 4A). Our metabolomics analysis revealed that the concentration of the direct product of hexokinase, G6P, was lower in the cells treated with arsenic than in the control cells at the 12-h time point and was even more significantly lower at the 24-h time point (Fig. 4B). In addition, the concentration of lactate, an index of the extent of glycolysis, was lower in the cells treated with arsenic for 24 h than in the control cells (Fig. 4C). To rule out the possibility that treatment with arsenic could change the cellular concentration of glucose and lead to the lower concentrations of G6P and lactate, we also investigated the concentration of glucose in SGC7901 cells. Although the glucose concentration changed over time, the changes in the arsenic-treated cells were similar to those in the control cells at all time points (Fig. 4D). In addition, the concentration of glycerate, a metabolite derived from glycine metabolism that feeds into glycolysis, was elevated significantly in the arsenic-treated cells as compared with the control cells at the earliest time point (Fig. S5). These results thus indicate that arsenic significantly inhibits glycolysis and suggest that the inhibition of hexokinase activity is a major factor underlying this effect within cells.
Fig. 4.
Glycolysis and HK2 are targeted by arsenic in vivo. (A) Metabolomic analysis showed that glycolysis is heavily targeted by arsenic. SGC7901 cells were treated with 2 μM ATO and were harvested at various time points until 24 h after treatment. Then metabolomics analysis was performed on the resuspended and washed cells. Vehicle controls were carried out without ATO treatment. Statistical analysis of the data was performed using JMP and R. Welch’s two-sample t-tests were performed on the log-transformed data to compare the vehicle- and drug-treated groups. (B–D) The levels of two key metabolites of glycolysis, i.e., G6P (B) and lactate (C), and of glucose (D) during ATO treatment. *, 0.05 < P < 0.1; **P ≤ 0.05.
Fig. S5.
Metabolomics analysis showed that glycolysis is heavily targeted by arsenic. The box-and-whisker plots show the levels of glycerate (A) and pyruvate (B) during ATO treatment.
Arsenic Binds Directly to Cys256 and Cys704 in HK2.
As a first step in understanding the mechanism by which arsenic inhibits the activity of HK2, we sought to identify the amino acid residues that could be involved directly in the arsenic–protein interaction. To this end, potential arsenic-binding sites of HK2 were evaluated with MALDI-TOF MS. Within the spectra, we observed a mass peak of 2,336.13, consistent with a calculated value of 2,236.1 for the C-terminal fragment MC(256)INMEWGAFGDDGSLNDIR plus AsO [that is, As(OH)2 with a loss of a water] and one H (Fig. 5A). In addition, there was a mass peak of 2,372.12 that is consistent with an expected mass of the peptide MC(704)VNMEWGAFGDNGC(717)LDDFR plus two AsO (Fig. S6A). Because arsenic is known to bind preferentially to cysteine residues, these results suggest that there are three binding sites of arsenic on HK2, namely, Cys256, Cys704, and Cys717.
Fig. 5.
Arsenic inhibits HK2 activity through the binding of C256 and C704. (A) The arsenic-binding sites of HK2 were identified by MALDI-TOF MS. Affinity-purified HK2 was incubated with ATO, and after tryptic digestion the sample was analyzed by MALDI-TOF MS. Tentative arsenic-binding peptides were identified on the basis of their mass fingerprinting. The arsenic-binding site Cys256 was identified by the predicted peptide mass plus AsO [As(OH)2 with a loss of a water] and minus one H (x + 74.9 + 16.0–1.0). (B and C) Close-up views of the active site of the N-terminal (B) and C-terminal (C) domain. Shown are all residues within 3 Å of the glucose ligand; negatively charged residues are red, positively charged are blue, and polar residues are green. Cys256 (B) and Cys704 (C) are within 10 Å of the respective active sites.
Fig. S6.
Arsenic inhibits HK2’s activity by binding of C256 and C704. (A) Identifying the arsenic-binding sites of HK2 by MALDI-TOF MS. Affinity-purified HK2 was incubated with ATO. After tryptic digestion the sample was analyzed with MALDI-TOF MS. Tentative arsenic-binding peptides were identified on the basis of their mass fingerprinting. The arsenic-binding site Cys704 was identified by the predicted peptide mass plus AsO [As(OH)2 with a loss of a water] and minus one H (x + 74.9 + 16.0 − 1.0). (B) HK2 is composed of two similar major domains (one N-terminal, one C-terminal) connected by a single helix (gray). Within each domain, there is a large subdomain (pink) and a small subdomain (cyan). Also shown is the glucose ligand (red and cyan stick representation) and Cys256 and Cys704 (yellow) that interact with arsenic directly.
Inspection of the crystal structure of the human HK2 (Fig. S6B) reveals that Cys256 and Cys704 are each in close proximity (10 Å) to one of the two active sites of this protein (Fig. 5 B and C) (20). This enzyme consists of two functionally active (and nearly identical) domains connected by a long alpha helix. Each domain is composed of a small and a large subdomain, and glucose binding occurs within the interface between these domains, concurrent with a closing of the cleft between these subdomains (20). Cys256 and Cys704 are located at identical relative positions in the two domains, close to the interface between the two subdomains. In contrast, Cys717 is more than 15 Å from the active site, and thus it is not immediately clear how arsenic binding to this site might affect the enzymatic activity. With the close proximity of Cys256 and Cys704 to the functionally important regions of this enzyme, it is highly likely that the activity of HK2 could be affected by the binding of arsenic to these residues, as observed in both the in vitro and in vivo data presented in this work.
Discussion
In this study, we developed a strategy to identify arsenic-binding proteins globally by combining biotinylated arsenic with a human proteome microarray (11). In this way we identified 360 arsenic-binding proteins, among which proteins involved in glycolysis are significantly enriched. In particular, these results led to the discovery that HK2 is a major target of arsenic-induced inhibition.
One of the most effective ways of elucidating the mechanism of action of a drug compound is to identify the proteins to which it binds. Traditionally, this identification can be done by affinity pull-down assays using a biotinylated compound followed by MS, as was performed in the identification of eIF4A as a target for pateamine A (21). However, this strategy is labor intensive and time-consuming, and, because of the intrinsic limitations of MS, it is challenging for both low- and high-abundant proteins. Furthermore, the proteins identified by this strategy might not interact with the drug directly but might act through an intermediary protein instead.
In comparison, proteomic microarray-based strategies identify the proteins directly targeted by a drug. As demonstrated in this study, a global drug–protein interaction discovery experiment consuming a limited number of microarrays can be performed readily in less than 4 h. Furthermore, all of the 16,368 human proteins on the proteome microarray are expressed and affinity purified in a high-throughput manner following the same procedure. As a result, distinct regions on the proteome microarray can be made highly homogeneous in terms of surface protein densities, with a dynamic range of protein amount per spot of less than 102 (11). In contrast, at a given time point, at least 5,000 proteins are detectable by MS with a dynamic range of 107 (22). Thus, by using the human proteome microarray, three times more proteins can be screened at high confidence in a single experiment.
The arsenic-binding proteins identified in this study represent a broad range of biological functions and are involved in a variety of cellular signaling pathways, such as apoptosis (caspase CASP14L), protein acetylation/deacetylation (SIRT5), and protein kinase signaling (MAPK11). Most of these proteins play key roles in biological processes (23–25). The existence of a multitude of arsenic-binding proteins suggests that there is a dynamic equilibrium for arsenic binding to different proteins upon the exposure of cancer cells to the drug. It is worth noting that many of the arsenic-binding proteins contain a zinc finger or ring finger domain rich in cysteines (Dataset S2). Because zinc binds to these proteins through multiple cysteine residues (26), arsenic may compete with zinc in binding to these residues and thereby may interfere with the transcription, translation, or protein degradation functions of these proteins.
Of note, our findings are in good agreement with the results of other studies of the biology of arsenic-related systems. Ge et al. (9) monitored global changes in protein expression in a multiple myeloma cell line treated with arsenic. The most significant changes were observed for carbohydrate metabolism and nucleotide metabolism. Pan et al. (27), using a chemogenomic approach in Saccharomyces cerevisiae, found that the folding of actin and tubulin was disrupted upon the addition of arsenic as a result of its binding to the chaperonin complex TRiC (also called “chaperonin containing TCP-1,” CCT) (27). Interestingly, several members of human CCT complex are in our list of arsenic-binding proteins (Dataset S1), including CCT7 and CCT8. Finally, Zheng et al. (28) treated a leukemia cell line, NB4, with arsenic and found that the transcription of some of the apoptosis regulators and some of the stress response-related genes were modulated by the compound. On the proteome level, a strong reduction of cytoskeleton proteins was observed also.
Deregulating cellular energetics is an emerging hallmark of cancer (12). Most cancer cells rely on aerobic glycolysis (the Warburg effect) for cellular processes. Here, we found that the glycolysis pathway was significantly targeted by arsenic. Arsenic bound to five of the key enzymes involved in glycolysis, i.e., hexokinase, aldolase, triose phosphate isomerase, GAPDH, and phosphoglycerate kinase. A previous study (10) identified another set of four enzymes involved in glycolysis as targets of arsenic, i.e., PKM2, enolase, pyruvate carboxylase, and l-lactate dehydrogenase B chain. Thus it is clear that the majority of the enzymes in the glycolysis pathway can be targeted directly or indirectly by arsenic. The targeting of these enzymes may explain why arsenic is generally effective on most of the solid tumor cell lines so far investigated (7).
HK2 has been widely characterized as a facilitator of glycolysis and as a repressor of apoptosis in several types of cancer. Our biochemical assays showed that the activity of HK2 can be inhibited by arsenic; furthermore, detailed metabolomics analysis revealed that G6P, the direct product of hexokinase, is down-regulated significantly in the gastric cancer cell line SGC7901 when treated with arsenic. Taken together, these results strongly suggest that HK2 and glycolysis are inhibited by arsenic both in vitro and in vivo.
Consistent with previous studies, arsenic treatment could cause significant apoptosis of SGC7901 cells (19). Interestingly, the apoptosis could be rescued effectively by the overexpression of HK2. One plausible explanation is that apoptosis may be promoted by direct inhibition of the activity of HK2 upon binding to arsenic. HK2 is localized mainly at the outer membrane of mitochondria where it interacts with some key proteins, such as voltage-dependent anion channel (VDAC) (29). It is possible that arsenic may inhibit the physical interactions of HK2 with other proteins on mitochondria outer membranes. Together with the inhibition of the enzymatic activity, this inhibition may be the cause, at lease in part, of arsenic-induced apoptosis.
Finally, MS analysis showed that Cys256 and Cys704 are the arsenic-binding sites; these sites lie within 10 Å of the active sites of HK2, suggesting that the binding of arsenic to these two sites may interfere with the active sites of the enzyme and thus affect the enzyme’s activity. The two glucokinase-equivalent domains of HK2 are symmetric, and both exhibit catalytic activity (2). It is worth noting that Cys256 and Cys704 are equivalent sites on the two domains; thus these cysteine residues may represent key regulatory points of the biological function of HK2
In summary, we have performed a comprehensive survey of the direct binding proteins of arsenic, and the arsenic-binding proteins that we identified in this study provide a valuable resource. By correlating the proteomic microarray data with functional studies and metabolomics analysis, we have verified glycolysis as a key pathway that is significantly targeted by arsenic; this targeting could represent a general mechanism of action of the suppressive effects of arsenic on cancer cells.
Materials and Methods
Human Proteome Microarray Fabrication.
Human proteome microarray fabrication was carried out as previously described (11).
Probe of Biotin–As on the Human Proteome Microarray.
Biotin–As was synthesized as described (10). Proteome microarrays were blocked with blocking buffer (1% BSA in 0.1% Tween 20; TBST) for 1 h at room temperature with gentle agitation. Biotin–As was diluted to 10 μM in blocking buffer and incubated on the proteome microarray at room temperature for 1 h. The microarrays were washed with TBST three times for 5 min each washing and were incubated with Cy3-Streptavidin at 1:1,000 dilution (Sigma) for 1 h at room temperature, followed by three 5-min washes in TBST. The microarrays were spun dry at 250 × g for 3 min and were scanned with a GenePix 4200A microarray scanner (Axon Instruments) to visualize and record the results. GenePix Pro-6.0 softwarewas used for data analysis.
Protein Microarray Data Processing.
An R script was applied to process the protein microarray data (SI Materials and Methods). Briefly, the signal-to-noise ratio (SNR) was defined as the ratio of the median of foreground signal to the median of background signal and was calculated for each protein. The SNRs for microarrays incubated with and without Biotin–As were set as SNR(+) and SNR(−), respectively. Another index, “Ratio,” was defined as SNR (+)/SNR(−). The mean SNR was used to represent the signal of the protein. To call the candidates, the cutoff was set as a P value ≤2.2 E-10, SNR(+) ≥1.4, and ratio ≥1.4.
Validating Arsenic–Protein Interaction by Streptavidin Blotting.
HK2 (25 μg/mL) and the positive control PML (600 μg/mL) were incubated with 100 μM Biotin–As in PBS buffer at room temperature for 1 h. Control experiments were carried out with the same amount of protein, 100 μM Biotin–As, and 100 mM BAL (Tokyo Chemical Industry). The proteins were electrophoresed and transferred to a nitrocellulose membrane. The membrane was probed with 1:1,000 IRDye 800CW streptavidin (LI-COR Bioscience) at room temperature for 30 min and was washed three times with TBST for 5 min each and once with double-distilled water for 5 min. The results were recorded by a LI-COR Odyssey scanner (LI-COR Bioscience) and quantified by the application software v3.0.21 of the Odyssey infrared imaging system.
HK2 Activity Assay Using Ultra HPLC.
The HK2 activity assay was performed in a 100-mL reaction composed of 0.1675 M glucose, 0.825 mM ATP, 2.5 mL HK2 (0.125 mg/mL) in 0.05 M Tris⋅HCl with 1.33 mM MgCl2 at pH 8.0. Parallel reactions were carried out except for the addition of ATO at several different concentrations (i.e., 2, 5, or 10 µM) for 1 h before reaction. The product of this reaction, G6P, was monitored by LC-coupled high-resolution MS (LC-HRMS). LC-HRMS was performed on a Waters ACQUITY ultra HPLC (UPLC) system equipped with a binary solvent delivery manager and a sample manager, coupled with a Waters Micromass Q-TOF Premier Mass Spectrometer equipped with an electrospray interface (Waters Corporation).
Metabolomics Analysis.
For metabolic analysis, 1 × 107 SGC7901 cells were seeded in 150-mm plates and were cultured overnight for attachment. Then the supernatant was replaced by fresh medium additionally supplemented with 2 μM of ATO for the cells in the treatment group. Cells were harvested after 6, 12, or 24 h of culture. The cells were trypsinized with 0.25% trypsin for 5 min. The cell suspensions were spun, and the supernatant was removed completely.
The cell pellets were flash-frozen and stored at −80 °C before metabolomics analysis. The metabolomic analysis was performed by the Shanghai Jiao Tong University-Metabolon Joint Metabolomics Laboratory (Shanghai, China).
3D Visualization of Arsenic-Binding Sites in HK2.
The images of the crystal structure of HK2 (Protein Data Bank ID 2NZT) were prepared using VMD (30).
SI Materials and Methods
Human Proteome Microarray Fabrication.
Human proteome microarray fabrication was carried out as previously described (11). Briefly, yeast transformants were grown at 30 °C and inoculated into SC-Ura/raffinose medium. The cultures were grown overnight, induced at an OD600 of 0.6–0.8 to a final concentration of 2% (wt/wt) galactose, and harvested by centrifugation after 6 h. The cells were lysed by shaking with glass beads, and the supernatant was incubated with Glutathione-Sepharose (GE Healthcare Biosciences). The beads were washed, and the proteins were eluted with 40 mM glutathione. The purified proteins were aliquoted to 384-well plates and were printed onto Full Moon slides (Full Moon BioSystems), using a 48-pin contact microarrayer (Bio-Rad ChipWriter Pro). Each protein was spotted in duplicate. Control proteins, including histone H3, histone H4, BSA, and biotinylated BSA, also were spotted in duplicate.
GO Analysis.
To identify the enrichment of the candidates, the potential candidates (Dataset S1) were analyzed using DAVID 6.7 (14). The protein symbol of each candidate was used as input data. Significantly enriched categories in the subontology of KEGG pathway, biological process, and cellular component with a P value <0.01 or <0.05 were chosen.
Network Analysis.
Protein interaction networks of the candidate proteins (Dataset S1) were built automatically by the STRING system (string-db.org/) (15). The system was operated with the default setting except that organism, confidence (score), and interactors shown were set to “Homo sapiens,” “0.70,” and “no more than 20 interactors,” respectively. A network of protein–protein interactions was generated in this manner and then was visualized by Cytoscape v2.8.1 (www.cytoscape.org) software. This interaction network was analyzed further for densely connected regions using a graph theoretic clustering algorithm MCODE (31). MCODE is part of the plug-in toolkit in Cytoscape (32). The top three high-ranking modules were selected for further analysis.
Motif Enrichment by MEME.
The top 100 arsenic-targeting proteins (Dataset S1) were analyzed by MEME (version 4.8.1) (17) using the following parameters: (i) occurrences of a single motif were set to zero or one per sequence; (ii); the minimum number of sites was 50; (iii) the minimum width of each motif was 10 amino acids, and the maximum width was 20 amino acids.
The Algorithm for Processing Protein Microarray Data.
Normalization was performed for the experimental microarray and the vehicle control assuming the median SNR of each array is equal. The positive signals were identified based on the assumption that the background values across the array follow a normal distribution. The median SNR of all proteins was used to estimate the mean of the normal distribution, and the median absolute deviation (MAD) was used to estimate the SD of the normal distribution. Proteins with P values <0.01 were considered positive. To identify the proteins whose SNRs were significantly higher on the experimental microarray than on those of the vehicle control, the ratio of experiment to vehicle was defined as r and calculated. The r values were assumed not to be changed between experiment and vehicle and to follow a normal distribution. The median and MAD of r of all proteins were used to estimate the mean and the SD of the normal distribution, respectively. Proteins with P values <0.01 were considered significant. We defined the SNR for each spot as the ratio of median foreground signal to median background signal. The SNR of a protein was averaged from the two duplicated spots on each microarray; the SNRs for microarrays incubated with and without Biotin–As were set as SNR(+) and SNR(−), respectively. Another index, “Ratio,” was defined as SNR (+)/SNR(−), and both the Ratio index and P value were calculated for all proteins.
Monitoring the Kinetics of Arsenic and Protein Interaction by BLI.
Super streptavidin tips (ForteBio) were prewet with 0.01 M PBS buffer supplemented with 5‰ DMSO (pH 7.4), which served as the background buffer for the immobilization. The Octet QKe system from ForteBio was used to monitor the binding kinetics. A stable baseline (60 s) was established by coupling 20–50 μg/mL biotinylated protein, and the nonspecific binding was washed off with PBS buffer (60 s). Final immobilization levels were 0.8 ± 0.1 nm. A new baseline was established in 0.01 M PBS buffer supplemented with 5‰ DMSO (pH 7.4). Biotin–As was prepared as a serial dilution (2, 10, and 100 μM) and was allowed to bind the target protein-saturated tips for 60 s and then was dissociated in 0.01 M PBS buffer (pH 7.4) supplemented with 1 mM BAL and 5 ‰ DMSO for 300 s. The results were recorded and processed by Octet Software v7.x from ForteBio.
Streptavidin Agarose Affinity Assay.
The streptavidin agarose affinity assay was performed according to Zhang et al. (13). Briefly, HEK293T cells were grown in DMEM with 10% (vol/vol) FBS, were transfected with HK2-GFP, and then were treated with 10 μM Biotin–As for 4 h. For ATO blocking, cells were pretreated with 80 μM ATO for 2 h, followed by treatment with 10 μM Biotin–As for 4 h in fresh cell medium. The cells then were lysed in Nonidet P-40 lysis buffer (Beyotime Institute of Biotechnology). The cell lysates were incubated with streptavidin agarose overnight at 4 °C. After washing with Nonidet P-40 lysis buffer, the streptavidin agarose beads were denatured and resuspended in SDS/PAGE loading buffer. The final results were revealed by an anti-HK2 antibody.
HK2 Activity Assay Using UPLC-MS.
Briefly, LC was performed on a BEH Amide column (50 × 2.1 mm, 1.7 µm) (Waters). The column was eluted with 5 mM ammonium acetate (pH 9) and 95% (vol/vol) acetonitrile in gradient mode at a flow rate of 0.30 mL/min at 30 °C. MS was performed using negative polarity, 2.4 KV capillary voltage, 30 V sampling cone, 4 eV collision energy, a source temperature of 110 °C, and a desolvation temperature of 350 °C. The flow rate for the desolvation gas was set at 600 L/h. The scan range was set to m/z 50∼1,000, scan time to 0.3 s, and interscan time to 0.02 s.
Treatment of SGC-7901 with ATO.
The SGC7901 cell line was cultured in RPMI-1640 medium (Invitrogen), supplemented with 10% (vol/vol) FBS (Invitrogen), 100 IU/mL penicillin, and 100 μg/mL streptomycin in humidified air at 37 °C with 5% (vol/vol) CO2. Cells (2 × 104) were seeded in 96-well plates and were cultured overnight; then 10 μL of 40 μM ATO was added in the treatment group. After incubation, the viable cells were measured by the CCK-8 assay (Dojindo Molecular Technologies) according to the manufacturer’s instructions. The absorbance at 450 nm was measured using a Synergy 2 microplate reader (BioTek Instruments, Inc.). The apoptosis of cells was determined by evaluating nuclear condensation after cell nuclei were stained with DAPI. Cells were washed two times with PBS containing 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.4 mM KH2PO4 (pH 7.2), were fixed with 4% paraformaldehyde for 20 min, and then were incubated with 300 nM DAPI (Sigma Chemical Co.). After labeling, cells were visualized using an Olympus microscope (Olympus) under light or a filter designed for DAPI fluorescence. Cells were considered apoptotic when they showed either fragmented or condensed nuclei.
Apoptosis Assay by Flow Cytometry.
HK2-GFP plasmid was obtained from GeneCopoeia, and the sequence accuracy was verified by DNA sequencing. The SGC 7901 cell line was transfected with the plasmid carrier HK2-GFP. The cells were grown in RPMI-1640/10% (vol/vol) FBS/1% penicillin-streptomycin medium at 37 °C in a 5% (vol/vol) CO2 incubator. Cultured cells were harvested and dissociated into a single-cell suspension using Accutase (Millipore). The PE Annexin V Apoptosis Detection Kit I was purchased from BD Biosciences. Cells were resuspended in 1× binding buffer [10 mM Hepes/NaOH (pH 7.4), 140 mM NaCl, 2.5 mM CaCl2] and then were incubated with phycoerythrin-conjugated Annexin V (1:100 dilution) and 7-AAD) (1:100 dilution) (BD Biosciences) for 15 min at 4 °C in the dark. Cells were resuspended in 1× binding buffer and were analyzed on a BD Biosciences FACScan flow cytometer using CellQuest Pro software, both from BD Biosciences.
In-Gel Trypsin Digestion.
Proteins were resolved on an 8% SDS/PAGE gel and stained with Coomassie blue G250, and the gel was cut slices. Each slice was diced into ∼1-mm3 cubes for in-gel digestion. Each section was washed in water and completely destained using 50 mM ammonium bicarbonate in 50% (vol/vol) acetonitrile at room temperature. Digestion was carried out using 20 μg/mL sequencing-grade modified trypsin in 50 mM NH4HCO3. Sufficient trypsin solution was added to swell the gel pieces, which were kept at 4 °C for 45 min and then were incubated at 37 °C overnight. The supernatants were transferred into a new microcentrifuge tube, and the gels were sonicated twice with extraction buffer [67% acetonitrile containing 5% (vol/vol) trifluoroacetic acid]. The peptide extract and the supernatant of the gel slice were combined and then were dried completely in a vacuum centrifuge.
Metabolomics Analysis.
In brief, the cells were extracted and split into three equal aliquots for analysis by a combination of three independent methods: UHPLC/MS/MS optimized for basic species, UHPLC/MS/MS optimized for acidic species, and GC/MS. After chromatographic separation and full-scan mass spectra were obtained, the ion features captured from the samples were compared with a reference library of chemical standards that included retention time, molecular weight (m/z), preferred adducts, and in-source fragments as well as their associated MS/MS2 spectra. The missing values for a given metabolite were imputed to be the observed minimum detection value on the assumption that they were below the limits of detection. Statistical analysis of the data was performed using JMP (SAS; www.jmp.com/en_us/home.html) and R (https://cran.r-project.org/) statistical software. Welch’s two-sample t-tests were performed on the log-transformed data to compare the vehicle- and drug-treated groups.
MALDI-TOF MS Data Processing.
Arsenic binding to protein was analyzed by MALDI-TOF MS on an ultrafleXtreme MALDI-TOF/TOF mass spectrometer (Bruker Daltonics) in the linear positive ion mode. All peptides were premixed with MALDI matrix solution [30% (vol/vol) acetonitrile containing 0.1% (vol/vol) TFA] and were saturated with α-Cyano-4-hydroxycinnamic acid, spotted onto a gold-surfaced MALDI target plate, and allowed to dry. Each spectrum was produced by accumulating data from 500 consecutive laser shots. Molecular mass calibration was carried out using #222570 Peptide Calibration Standard II that is composed of angiotensin II, angiotensin I, substance P, bombesin, and an ACTH clip (Bruker Daltonics). The spectra were analyzed using MASCOT 2.3 Peptide Mass Fingerprint (Matrix Science) with the human protein database. Search parameters were MS tolerance, 150.00 ppm; enzyme, trypsin. In the probability-based molecular weight search score, the protein score was −10*log (P), where P is the probability that the observed match is a random event. Although a score higher than 58 indicates identity or extensive homology (P < 0.05), the higher the score and the more peptides that matched, the more likely it was that real binding took place.
Supplementary Material
Acknowledgments
We thank S. Quan and Y. M. Shen for expert research assistance, J. Fleming for editing the manuscript, and Zhi Xie and Yongjun Dang for critical reading. This study was supported in part by National High Technology Research and Development Program of China Grants 2012AA020103 and 2012AA020203, National Natural Science Foundation of China Grants 31370813 and 31370750, and Key Project Specialized for Infectious Diseases of the Chinese Ministry of Health Grant 2013ZX10003006.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521316112/-/DCSupplemental.
References
- 1.Chen S-J, et al. From an old remedy to a magic bullet: Molecular mechanisms underlying the therapeutic effects of arsenic in fighting leukemia. Blood. 2011;117(24):6425–6437. doi: 10.1182/blood-2010-11-283598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen Y, et al. Mutations of epigenetic modifier genes as a poor prognostic factor in acute promyelocytic leukemia under treatment with all-trans retinoic acid and arsenic trioxide. EBioMedicine. 2015;2(6):563–571. doi: 10.1016/j.ebiom.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mao JH, et al. As4S4 targets RING-type E3 ligase c-CBL to induce degradation of BCR-ABL in chronic myelogenous leukemia. Proc Natl Acad Sci USA. 2010;107(50):21683–21688. doi: 10.1073/pnas.1016311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berenson JR, et al. Efficacy and safety of melphalan, arsenic trioxide and ascorbic acid combination therapy in patients with relapsed or refractory multiple myeloma: A prospective, multicentre, phase II, single-arm study. Br J Haematol. 2006;135(2):174–183. doi: 10.1111/j.1365-2141.2006.06280.x. [DOI] [PubMed] [Google Scholar]
- 5.Zheng WL, et al. Arsenic trioxide, thalidomide and retinoid acid combination therapy in higher risk myelodysplastic syndrome patients. Leuk Res. 2008;32(2):251–254. doi: 10.1016/j.leukres.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 6.Chang JE, et al. Phase II study of arsenic trioxide and ascorbic acid for relapsed or refractory lymphoid malignancies: A Wisconsin Oncology Network study. Hematol Oncol. 2009;27(1):11–16. doi: 10.1002/hon.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Q, et al. The NRF2-mediated oxidative stress response pathway is associated with tumor cell resistance to arsenic trioxide across the NCI-60 panel. BMC Med Genomics. 2010;3:37. doi: 10.1186/1755-8794-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ong PS, Chan SY, Ho PC. 2012. Microarray analysis revealed dysregulation of multiple genes associated with chemoresistance to As(2)O(3) and increased tumor aggressiveness in a newly established arsenic-resistant ovarian cancer cell line, OVCAR-3/AsR. Eur J Pharm Sci 45(3):367–378.
- 9.Ge F, et al. Proteomic and functional analyses reveal a dual molecular mechanism underlying arsenic-induced apoptosis in human multiple myeloma cells. J Proteome Res. 2009;8(6):3006–3019. doi: 10.1021/pr9001004. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, et al. Identification of arsenic-binding proteins in human breast cancer cells. Cancer Lett. 2007;255(1):95–106. doi: 10.1016/j.canlet.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeong JS, et al. 2012. Rapid identification of monospecific monoclonal antibodies using a human proteome microarray. Mol Cell Proteomics 11(6):016253.
- 12.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Zhang XW, et al. Arsenic trioxide controls the fate of the PML-RARalpha oncoprotein by directly binding PML. Science. 2010;328(5975):240–243. doi: 10.1126/science.1183424. [DOI] [PubMed] [Google Scholar]
- 14.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 15.Szklarczyk D, et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(Database issue):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhattacharjee H, Rosen BP. Spatial proximity of Cys113, Cys172, and Cys422 in the metalloactivation domain of the ArsA ATPase. J Biol Chem. 1996;271(40):24465–24470. doi: 10.1074/jbc.271.40.24465. [DOI] [PubMed] [Google Scholar]
- 17.Bailey TL, et al. 2009. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res 37(Web Server issue):W202–208.
- 18.Vilensky JA, Redman K. British anti-Lewisite (dimercaprol): An amazing history. Ann Emerg Med. 2003;41(3):378–383. doi: 10.1067/mem.2003.72. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, et al. Arsenic trioxide induces apoptosis and G2/M phase arrest by inducing Cbl to inhibit PI3K/Akt signaling and thereby regulate p53 activation. Cancer Lett. 2009;284(2):208–215. doi: 10.1016/j.canlet.2009.04.035. [DOI] [PubMed] [Google Scholar]
- 20.Berg JM. Zinc-finger proteins. Curr Opin Struct Biol. 1993;3(1):11–16. [Google Scholar]
- 21.Low WK, et al. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol Cell. 2005;20(5):709–722. doi: 10.1016/j.molcel.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 22.Pirmoradian M, et al. Rapid and deep human proteome analysis by single-dimension shotgun proteomics. Mol Cell Proteomics. 2013;12(11):3330–3338. doi: 10.1074/mcp.O113.028787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beardmore VA, et al. Generation and characterization of p38beta (MAPK11) gene-targeted mice. Mol Cell Biol. 2005;25(23):10454–10464. doi: 10.1128/MCB.25.23.10454-10464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137(3):560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du J, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334(6057):806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou X, et al. Arsenite interacts selectively with zinc finger proteins containing C3H1 or C4 motifs. J Biol Chem. 2011;286(26):22855–22863. doi: 10.1074/jbc.M111.232926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan X, et al. Trivalent arsenic inhibits the functions of chaperonin complex. Genetics. 2010;186(2):725–734. doi: 10.1534/genetics.110.117655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng PZ, et al. Systems analysis of transcriptome and proteome in retinoic acid/arsenic trioxide-induced cell differentiation/apoptosis of promyelocytic leukemia. Proc Natl Acad Sci USA. 2005;102(21):7653–7658. doi: 10.1073/pnas.0502825102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krasnov GS, Dmitriev AA, Lakunina VA, Kirpiy AA, Kudryavtseva AV. Targeting VDAC-bound hexokinase II: A promising approach for concomitant anti-cancer therapy. Expert Opin Ther Targets. 2013;17(10):1221–1233. doi: 10.1517/14728222.2013.833607. [DOI] [PubMed] [Google Scholar]
- 30.Laity JH, Lee BM, Wright PE. Zinc finger proteins: New insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11(1):39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 31.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.