Significance
Bacteria regulate the synthesis and degradation of RNA molecules to ensure timely and robust responses to an ever-changing environment. The transcript’s lifetime can be influenced profoundly by a secondary structure that can form in the RNA and that may inhibit or promote its digestion by RNases. The molecular mechanisms by which RNases interact with structured RNAs therefore are of great interest. In this study we used optical tweezers to investigate the mechanistic properties of two such enzymes from Escherichia coli, polynucleotide phosphorylase and RNase R. Our results offer new insights into the functional characteristics of these two enzymes, including the sequence-dependent behavior of RNase R and the presence of discrete steps of six or seven nucleotides taken by polynucleotide phosphorylase.
Keywords: single- molecule biophysics, optical trap, exoribonuclease, processivity, step size
Abstract
Bacterial RNases catalyze the turnover of RNA and are essential for gene expression and quality surveillance of transcripts. In Escherichia coli, the exoribonucleases RNase R and polynucleotide phosphorylase (PNPase) play critical roles in degrading RNA. Here, we developed an optical-trapping assay to monitor the translocation of individual enzymes along RNA-based substrates. Single-molecule records of motion reveal RNase R to be highly processive: one molecule can unwind over 500 bp of a structured substrate. However, enzyme progress is interrupted by pausing and stalling events that can slow degradation in a sequence-dependent fashion. We found that the distance traveled by PNPase through structured RNA is dependent on the A+U content of the substrate and that removal of its KH and S1 RNA-binding domains can reduce enzyme processivity without affecting the velocity. By a periodogram analysis of single-molecule records, we establish that PNPase takes discrete steps of six or seven nucleotides. These findings, in combination with previous structural and biochemical data, support an asymmetric inchworm mechanism for PNPase motion. The assay developed here for RNase R and PNPase is well suited to studies of other exonucleases and helicases.
Both polynucleotide phosphorylase (PNPase) and RNase R are responsible for degrading a variety of transcripts in bacteria, such as mRNAs, defective rRNAs, and antisense regulatory RNAs (1–4). Although strains of Escherichia coli remain viable with a loss of either enzyme, the elimination of both results in an accumulation of rRNA fragments and cell death (3). To degrade RNA, both ribonucleases require an unstructured binding site of at least 7–10 nt at the 3′ end of the substrate (5–7), a sequence that often is supplied by posttranscriptional polyadenylation (8–10). After binding RNA, PNPase and RNase R processively digest the substrate in the 3′-to-5′ direction (11–13).
PNPase consists of a trimer that degrades RNA at catalytic sites within a central channel (9, 14). The enzyme is homologous to eukaryotic and archaeal exosomes and, like these hexameric proteins, possesses a core of six RNase PH domains arranged in a ring-like configuration (two per protomer) (6, 15). Each protomer also carries a KH and an S1 RNA-binding domain at its C terminus, both of which are connected to the body of the enzyme by flexible linkers (6, 16). These domains are thought to bind RNA upstream of the catalytic sites, and recent structural evidence suggests that they may guide the substrate into the central channel through a “hands gripping a rope” mechanism (16, 17). By itself, PNPase may stall on structured RNA, particularly on G:C hairpins comprising more than six base pairs (18). PNPase often associates with the DEAD-box helicase RhlB to move through structured regions and generally does so in the context of the multienzyme degradosome (2, 19). However, there is some evidence that PNPase can digest structured transcripts in the absence of external factors, including tRNA-like molecules with hairpins as long as nine base pairs (1, 20).
In contrast, RNase R is unique among bacterial exoribonucleases, in that it can digest highly structured RNA without additional factors (4), suggesting that the enzyme may have significant helicase activity. In particular, RNase R has been observed to unwind and degrade completely dsRNA substrates of nearly 20 bp (4, 7, 21). Recent single-molecule FRET data indicate that both RNase R and its homolog, the Rrp44 subunit of the eukaryotic exosome (22, 23), advance 3′-to-5′ along dsRNA in steps of approximately four base pairs through internal elastic coupling of their binding and catalytic sites (24). RNase R mutants that lack the nuclease activity required for processive motion nonetheless may separate hybridized strands of RNA (21, 25), presumably through an entirely different, distributive mechanism similar to that described for DEAD-box helicases under certain conditions (26). Although RNase R appears to have a minor preference for adenine (13), no significant sequence-dependent behavior has been reported for this enzyme.
Here, we observed the processive unwinding of structured RNA by PNPase and RNase R directly, at the single-molecule level, through the use of an optical-trapping assay that relies on detecting the intrinsic differences of the contour length between single- and double-stranded RNA (or between a DNA–RNA hybrid and ssDNA). The assay builds on similar arrangements developed to study processive enzymes, such as the T7 DNA polymerase (27, 28), the helicase UrvD (29), λ exonuclease (30), and an RNA-dependent RNA polymerase (31). Here, we extend the assay to study processive exoribonuclease motion with high spatial (near-nucleotide) resolution. We find that RNase R has significantly greater processivity than previously reported, with an individual enzyme capable of unwinding hundreds of base pairs of dsRNA. We find that RNase R exhibits sequence-dependent behavior, with a tendency to pause (or stall) at specific elements on structured RNA. For PNPase, we find a much more limited processivity, with an individual enzyme capable of unwinding around 60 bp of structured RNA, provided that it is rich in adenine and uracil. PNPase mutants lacking the KH and S1 domains exhibited reduced processivity but moved along dsRNA at rates similar to the WT enzyme. Finally, we find that PNPase translocates in discrete steps of six or seven nucleotides, a behavior that suggests it may advance by an asymmetric inchworm mechanism. The single-molecule assay developed for this work should be readily adaptable to the study of additional exonucleases and helicases.
Results
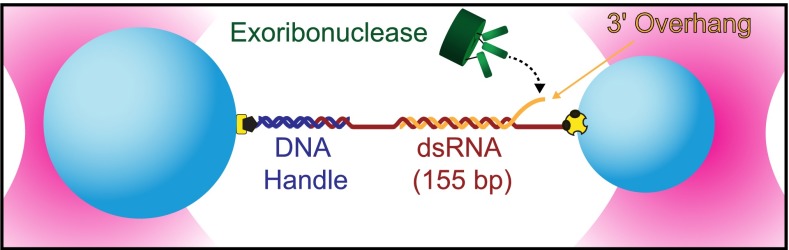
In the exoribonuclease assay, a substrate RNA is hybridized to a complementary strand composed of either DNA or RNA to form a duplex. At its 3ʹ end the duplex carries a short (12- to 14-nt) noncomplementary loading sequence, which overhangs and serves as a target for the initial binding of the enzyme. The hybridized duplex and associated loading sequence are suspended between two optically trapped polystyrene beads using chemical linkages and a DNA “handle” (Fig. 1). Enzyme-driven unwinding of the duplex region, driven by hydrolysis of the bound strand, converts the double-stranded substrate into a single-stranded form that has a longer contour length, resulting in a change in the end-to-end extension by an amount related to the applied load (∼0.2 nm per nucleotide at 20 pN): an RNA duplex has a rise of 0.28 nm per nucleotide (32), whereas ssRNA (or ssDNA) has a force-dependent rise of 0.48 nm per nucleotide at 20 pN (33). Using this approach, we were able to monitor the activity of RNase R and PNPase in real time and at near-nucleotide resolution.
Fig. 1.
Single-molecule exoribonuclease assay (not to scale). A 155-bp segment of dsRNA is placed under tension between polystyrene beads (light blue) held in twin optical traps (pink). One strand of RNA (the substrate, orange) carries a 3′ adenine-rich terminal overhang that serves as a loading site for a processive exoribonuclease (green) that unwinds in the 3′-to-5′ direction. The complementary strand (red) is attached at its ends to the beads directly at its 5′ terminus via a biotin–avidin linkage (yellow and black) and via hybridization to a DNA handle (dark blue) attached with a digoxygenin–antibody linkage (yellow and black). As the exoribonuclease unwinds and digests the RNA substrate, the length of the duplex region is reduced, resulting in an overall increase in the tether extension. In some experiments, the RNA complementary to the enzyme substrate was replaced with DNA.
RNase R: Velocity, Processivity, and Pausing Behavior.
RNase R was assayed on a 155-bp dsRNA construct held at 20 pN with an optical force clamp. The motion was highly processive, with 77% of enzymes (n = 10 of 13) advancing through more than 24 nm and nearly half (46%) unwinding the entire substrate (Fig. 2A). Because the complete unwinding of the RNA substrate corresponded to an extension change of 26.9 ± 0.2 nm (mean ± SE), we calculate that the release of each nucleotide increased the tether length by 0.174 ± 0.001 nm (mean ± SE). This value is close to the predicted value of 0.2 nm per nucleotide (32, 33), and it was used to calculate the number of nucleotides unwound in subsequent experiments with dsRNA. RNase R was similarly processive when degrading RNA hybridized to a DNA complement, with 73% of molecules (n = 11 of 15) advancing over 24 nm, and 45% removing the entire transcript. The conversion factor for unwinding a DNA–RNA hybrid duplex was 0.178 ± 0.004 nm per nucleotide (mean ± SE), the same, within error, as for dsRNA.
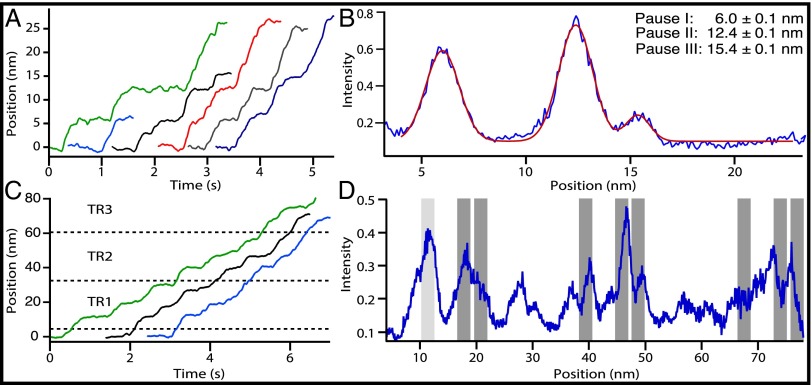
Fig. 2.
RNase R pauses at specific sites as it unwinds dsRNA. (A) Six representative single-molecule records of RNase R activity on the 155-bp dsRNA construct. (B) Positional histogram of aligned RNase R records (blue curve; n = 13). Peaks indicate pauses by RNase R. Data are fit to a sum of three Gaussians (red curve). The legend gives the three main pause locations (mean ± SE). (C) Three representative single-molecule records of RNase R activity on a 1,500-bp dsRNA construct carrying three tandem repeats (TR1–TR3) of the 155-bp sequence (Fig. S2). Approximate beginning and ending locations for each repeat are indicated (dashed lines). (D) Positional histogram of aligned RNase R records (n = 18) for the 1,500-bp construct (blue curve), including data from molecules that did not fully traverse the region (Materials and Methods). The predicted locations of pauses within the repeats are indicated (dark gray bars); these records were left-aligned at the measured location of the first pause (light gray bar at 11.4 ± 0.3 nm), which was found within 1 nm of its predicted location, at 10.5 ± 0.1 nm. The widths of the gray bars (2.0 nm) correspond to the approximate widths of peaks in the pause distributions.
RNase R moved along dsRNA with an average velocity of 88 ± 8 nt/s (mean ± SE), but its motion was not uniform: Most enzymes paused briefly at one or more locations, a behavior reminiscent of elongation by RNA polymerase (34). However, the pauses observed were confined primarily to the first ∼16 nm of the dsRNA substrate, and pauses with durations greater than 0.1 s were not found in the ∼9-nm (50-nt) stretch immediately following this region. Based on the rates of traversal of this comparatively pause-free region, we estimate the pause-free velocity of RNase R to be 151 ± 11 nt/s (SI Materials and Methods). In contrast, the average velocity of RNase R on the DNA–RNA hybrid was significantly faster, 128 ± 14 nt/s (two-tailed t test, P = 0.016), and the corresponding pause-free velocity was 184 ± 20 nt/s (22% faster than on dsRNA). Because RNase R digests an identical RNA sequence in both experiments, the slower velocity over dsRNA must be a consequence of the enzyme’s being differentially sensitive to the structure (35–37) or of the stability (38) of the DNA–RNA hybrid.
RNase R paused with high probability at three specific positions on the dsRNA construct (Fig. 2B and Fig. S1). By aligning single-molecule records, we determined that pauses occurred at extensions of 6.0 ± 0.1 nm (34–36 nt), 12.4 ± 0.1 nm (70–74 nt), and 15.4 ± 0.1 nm (87–91 nt) relative to the starting position. There were no obvious sequence similarities among these three locations, nor did we observe pausing behavior at the equivalent sequence positions on the DNA–RNA hybrid. To confirm that the pauses on dsRNA were sequence dependent, we repeated the experiment on a 1,500-bp dsRNA substrate carrying three tandem repeats of the original 155-bp sequence, each separated by a spacer region of six base pairs (Fig. 2C and Fig. S2). For the enzymes that successfully traversed all three tandem repeats (n = 7 of 18), pauses were observed at each of the expected positions in every segment (Fig. 2D).
The pauses scored in the tandem repeats enabled us to obtain an independent estimate of the conversion factor for relating the change in distance (measured in nanometers) to the number of nucleotides unwound for dsRNA. Our original estimate of 0.174 ± 0.001 nm per nucleotide (mean ± SE) had been calculated based on an assumption that RNase R completely digested the target substrate for the 155-bp construct. By instead measuring the change in distance between the corresponding pauses in tandem repeats, we independently calculated a value of 0.172 ± 0.002 nm per nucleotide (mean ± SE), which is identical within error.
With some probability, the motion of RNase R arrested completely at pause sites; the presence of multiple pause sequences engineered into the 1,500-bp construct likely hindered its full unwinding. Nonetheless, about one third of the enzymes successfully traversed at least 800 bp of dsRNA (n = 5 of 18). We therefore decided to examine RNase R behavior on a second 1,500-bp construct lacking the tandem pause elements to obtain an independent estimate of processivity. On this substrate, RNase R unwound an average of 503 ± 110 bp (mean ± SE; n = 16).
PNPase: Stepping Behavior and Sequence Dependence.
We also studied the behavior of PNPase as it degraded the same (155-bp) dsRNA and DNA–RNA hybrid sequences used for the RNase R study and under an identical 20 pN load. These experiments were performed under buffer conditions that favored phosphorolysis (Materials and Methods). Under these conditions, PNPase removed an average of 23 ± 2 nt (mean ± SE; n = 20) from the dsRNA substrate and the same number of nucleotides (23 ± 1 nt; mean ± SE; n = 14) from the DNA–RNA hybrid (Fig. 3A). The average velocity of PNPase over these two substrates was the same within error: 121 ± 30 nt/s (mean ± SE; n = 15) for dsRNA and 129 ± 40 nt/s (mean ± SE; n = 10) for DNA–RNA.
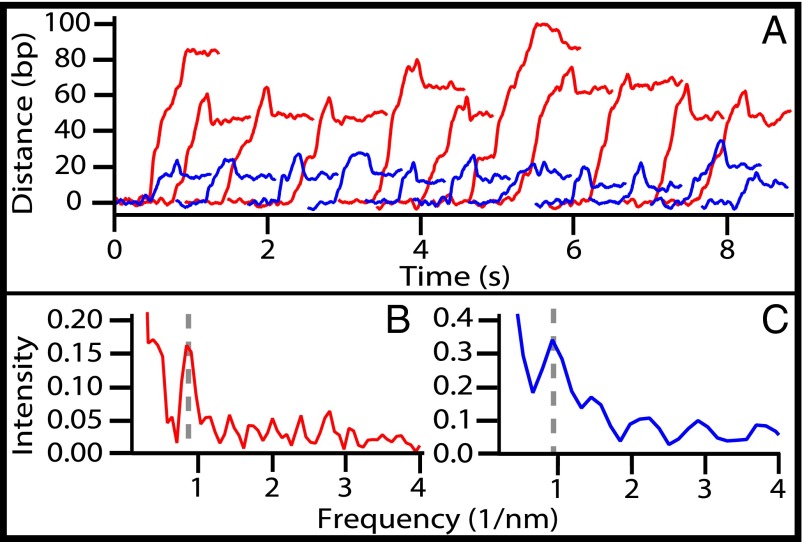
Fig. 3.
PNPase motion along RNA. (A) Twenty-two representative single-molecule records of PNPase advancing on 155-bp DNA–RNA hybrid constructs. Enzymes traversing RNA with 70% A+U content over the first 26 nt (blue traces, n = 11) moved an average distance of 23 ± 1 nt (mean ± SE). Enzymes traversing RNA with 70% A+U content over the first 50 nt (AU50; red traces, n = 11) moved a longer average distance of 62 ± 3 nt (mean ± SE). Records were smoothed with a 100-point boxcar window (t = 50 ms). (B) Periodogram analysis of records from the AU50 construct (n = 27) yielded a peak centered at 0.87 ± 0.14 nm−1 (mean ± SD), corresponding to a step size of 6.6 ± 1.1 nt. (C) Periodogram analysis of records of the unmodified 155-bp construct (n = 25; data from the dsRNA and DNA–RNA hybrid are combined) yielded a peak centered at 0.94 ± 0.22 nm−1 (6.1 ± 1.4 nt; mean ± SD).
The 155-nt substrate had an A+U content of 46%, overall. However, this content was not uniformly distributed but was enriched to a concentration of 69% in the 26-bp region initially digested by PNPase. To investigate whether the processivity of PNPase depended on the base composition, we studied its motion on a DNA–RNA hybrid enriched for A+U in which the first 48 bp were replaced with a randomized sequence so that the first 50 bp carried an A+U concentration of 70% (hereafter referred to as “AU50”). On AU50, PNPase moved an average of 62 ± 3 nt (mean ± SE; n = 27), unwinding more than twice as much RNA as before while moving at the same rate, within error (137 ± 8 nt/s; mean ± SE).
In single-molecule records of PNPase motion on AU50, we occasionally observed brief pauses that were highly suggestive of enzyme stepping. Although the signal-to-noise ratio of individual records was not sufficiently good for the direct detection of steps, we were able to perform a periodogram analysis of the combined datasets (i.e., a Fourier analysis of the distribution of extension changes) to investigate the frequency of the stepwise events (39). The analysis yielded a strong peak at a spatial frequency of 0.87 ± 0.14 nm−1 (mean ± SD), corresponding to an underlying step size of 6.6 ± 1.1 nt (Fig. 3B). We separately analyzed PNPase records collected on the original 155-bp construct (Fig. 3C) and obtained a peak at 0.94 ± 0.22 nm−1 (mean ± SD) corresponding to a step size of 6.1 ± 1.4 nt, which was the same, within error. We conclude that the observed periodicity is unlikely to be a consequence of any sequence-dependent pausing but instead reflects a series of discrete steps of six to seven nucleotides taken by PNPase molecules. As an additional control, we carried out an analogous analysis of records obtained with RNase R (Fig. S3). Although those data were collected and analyzed using the identical apparatus, software, and sampling rate (2 kHz) and with enzymes that moved at similar velocities, we saw no evidence for such steps of six or seven nucleotides in the records of RNase R.
PNPase mutants in which the KH and S1 RNA-binding domains were deleted have been reported to display reduced nucleolytic activity (17, 40). We considered whether removal of these domains had any effect on velocity or processivity. Using a PNPase ΔKH–ΔS1 mutant, we observed a twofold reduction in processivity on dsRNA (Fig. S4) relative to WT (14 ± 1 bp; mean ± SE; n = 10), but the velocity was the same within error (137 ± 43 nt/s; mean ± SE). Our findings suggest that the ∼100-fold loss of nuclease activity reported for truncated mutants (40) may be a consequence primarily of reduced binding affinity, because a twofold drop in processivity alone, with no concomitant change in velocity, is insufficient to explain the pronounced loss of activity displayed by the mutants. This explanation is consistent with the comparatively infrequent observation of records of unwinding, which require productive binding of the enzyme, in assays using these mutants, in which higher concentrations of the enzyme (roughly eightfold greater than wild-type) were routinely required. The reduced processivity of the PNPase mutant led to short records of motion that precluded a periodogram analysis.
Discussion
We developed a high-resolution single-molecule assay for studies of helicases and exonucleases and used it to examine the RNA-unwinding activities of RNase R and PNPase. The experimental approach relies on the increase in the length of the associated tether produced by the conversion of a duplex region to a single-stranded region. Because this increase is proportional to the applied stretching force (33), experiments performed under comparatively high external loads (20 pN) are able to achieve near-nucleotide resolution.
The load applied to RNA was well below that required for spontaneous unwinding as the result of force-induced shearing (>35 pN) (41). Therefore, given the absence of a separate source of energy, the processive unwinding of RNA must be driven by the degradation of the RNA substrate. Nevertheless, the application of an external load is expected to perturb the energy landscape for the unwinding reaction, favoring the melting of any double-stranded regions and thereby influencing the velocity and processivity of PNPase and RNase R. This effect may explain why we observed greater processivity for PNPase than suggested by a previous estimate (18). Because the external load primarily affects the melting of the duplex, RNase R and PNPase are expected to move at least as fast and as far on ssRNA where no additional energy is required for unwinding. Therefore, the velocity and processivity values returned by our assay likely represent lower bounds for single-stranded substrates and upper bounds for double-stranded substrates.
The results reveal RNase R to be highly processive. The enzyme moves fairly rapidly and at a rate sensitive to whether it is acting on dsRNA or a DNA–RNA hybrid. We found that the propensity to pause (or arrest) is sensitive to sequence, and we identified three pause sites within the coding region of the rpoB gene from E. coli. The existence of pauses suggests that some RNAs may carry sequence elements that render them comparatively less sensitive to degradation by RNase R and therefore may indicate a role for this enzyme in the maturation of transcripts.
One recent study using single-molecule FRET (24) reported that both RNase R and its eukaryotic homolog, Rrp44, take steps of approximately four nucleotides along RNA. However, a periodogram analysis of our RNase R records did not yield a step of this size (Fig. S3), although the same approach successfully identified steps of six or seven nucleotides for PNPase. The discrepancy in the behavior of RNase R might be attributable to the different reaction conditions; we studied RNase R at 26 °C, but the FRET study was performed at a comparatively low temperature (12 °C) at which the enzyme velocity was reported to be approximately 50-fold slower than in our assay.
For PNPase, we found that the velocity and processivity were independent of whether it unwound dsRNA or a DNA–RNA hybrid. However, processivity was strongly influenced by the A+U content of the substrate, a result consistent with previous reports that the susceptibility of RNA hairpins to PNPase digestion depends on their thermodynamic stability (20).
Immediately following the initial phase of unwinding by PNPase, we typically observed an ∼2 nm decrease in records of the tether extension, which took place on a timescale of 0.1–0.2 s, resulting in an apparent backtracking motion (Fig. 3A). Because the PNPase catalytic site is buried deep inside the enzyme core and therefore is spatially distinct from the point where the protein initially binds RNA, tether shortening may occur when PNPase releases a short fragment of unwound RNA that subsequently rehybridizes to its complement. In support of this explanation, we note that the average backtrack distance corresponded to the release of 12 ± 1 nt (mean ± SE; n = 44), as is consistent with current models for the PNPase–RNA interaction ahead of the catalytic site (16).
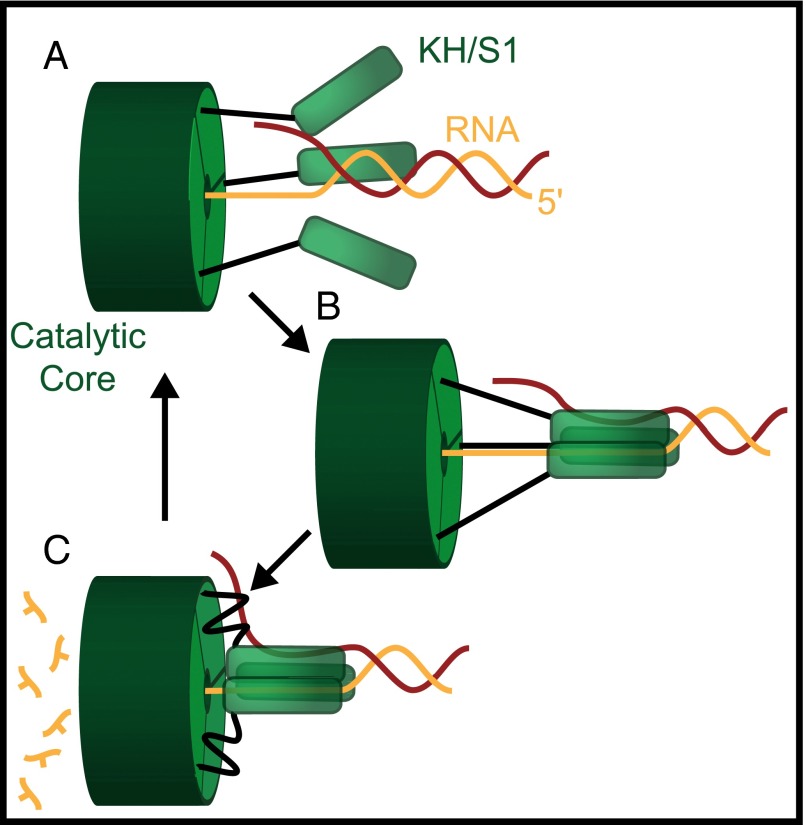
The observation of steps of six or seven nucleotides, combined with structural and biochemical data for PNPase, enables us to postulate a mechanism by which this protein may advance along RNA (Fig. 4). In this asymmetric inchworm model, the KH and/or S1 domains bind to RNA on the 5′ side of the catalytic core and (for structured substrates) melt six or seven base pairs. The catalytic domains in the enzyme core then degrade RNA on the 3′ side, one nucleotide at a time, advancing until they encounter the bound KH/S1 domains. The latter domains then dissociate from the transcript and rebind downstream in the 5′ direction, repeating the cycle. In this model, the increases in tether extension observed in our single-molecule records correspond to the abrupt melting steps of approximately six or seven nucleotides, and the pauses between steps correspond to the degradation of the RNA by the PNPase core domains. The model relies on the presence of flexible linkers connecting the KH and S1 domains to the remainder of the protein; structural data suggest that these linkers could provide the comparatively large conformational changes required to execute a step of six or seven nucleotides (16). The precise roles of the individual RNA-binding domains and whether they bind via a concerted (as depicted in Fig. 4) or a hand-over-hand mechanism (16) remain to be determined.
Fig. 4.
Asymmetric inchworm model for PNPase. (A) PNPase has three KH/S1 RNA-binding domains (green rods), each of which is loosely coupled to its catalytic core via flexible linkers (black). (B) These domains bind to the RNA substrate downstream of the catalytic site, where they melt six or seven base pairs of dsRNA (digested strand in orange, complementary strand in red). (C) The catalytic core digests and releases individual nucleoside monophosphates as it advances 3′ to 5′ until it reaches the RNA-binding domains. When the core catches up, the KH/S1 domains are released and become available to rebind further downstream along the RNA to complete additional cycles of degradation.
In summary, the single-molecule assay presented here is well suited for studying the nanomechanical behavior of helicases and exonucleases, including the measurement of such properties as velocity, processivity, step size, and sequence-dependent effects. The assay is simple and convenient, and it has the advantage that WT motors can be studied without any of the chemical modifications required for fluorescent labeling or direct surface attachments. We anticipate that this approach should be applicable to the study of other processive exoribonucleases at the single-molecule level, including the human and archaeal exosomes, RNase II and RNase J.
Materials and Methods
RNases.
PNPase and the truncated core of PNPase lacking the S1 and KH domains were purified as described (42). Ribonuclease R (RNR07250; Epicentre) was stored at −20 °C.
DNA/RNA Constructs.
Sequences and oligomers are described in SI Materials and Methods. Except where otherwise described, RNA was obtained by in vitro transcription (MEGAscript T7 kit, AM1333M; Life Technologies) of a DNA template, derived by PCR, from a 155-bp segment of the pALB3 plasmid (43). For RNase R assays with the 1,500-bp dsRNA substrate, RNA was transcribed from a pUC19 vector (44) carrying three tandem repeats of the 155-bp pALB3 sequence inserted between the EcoRI and HindIII restriction sites. Processivity data for RNase R were collected on a similar 1,500-bp construct that had a single modified copy of the pALB3 sequence.
For DNA–RNA hybrid experiments, substrate RNA was hybridized to a chemically synthesized, cDNA strand carrying a 5′ biotin tag (Integrated DNA Technologies). For corresponding dsRNA assays, biotin-labeled RNA was generated by ligating a 20-nt chemically synthesized RNA oligomer with a 5′ biotin tag (Integrated DNA Technologies) to a transcript synthesized in vitro. The ligation was performed using T4 RNA ligase 2 (M0239S; New England BioLabs) in the presence of a cDNA splint that subsequently was digested by DNase I. The full-length RNA was purified by PAGE.
Substrate-complement strands were hybridized at their ends to one or more DNA handles generated by autosticky PCR (45). Experiments used either a 3,057-bp or a 2,250-bp digoxygenin-labeled handle derived from the M13mp18 plasmid (44) and carrying a 25-nt 5′ overhang attached to the 3′ end of the substrate complement via a 46-nt DNA splint. For 1,500-bp dsRNA constructs, a second biotin-labeled 1,044-bp handle was generated from the pRL732 plasmid (34) and was annealed to the 5′ end of the substrate complement.
The substrate RNA, its complement, and the DNA handles for each construct were hybridized and annealed in 80% formamide (SI Materials and Methods), ethanol precipitated, resuspended in TE buffer, and stored at −80 °C.
Assay.
To produce a dumbbell assay (Fig. 1), DNA/RNA tethers were incubated at ∼1:1:1 stoichiometry with avidin-coated (600-nm diameter) and digoxygenin antibody-coated (915-nm diameter) beads for 1 h at room temperature (21.5 ± 0.5 °C). These complexes then were diluted in the appropriate assay buffer (below), placed in an ∼5 µL flow chamber, and rinsed with 16 µL additional buffer to remove excess beads. Single tethers were identified as described (46) and were held by optical tweezers at constant force for ∼45 s, after which an assay buffer containing either PNPase or RNase R was introduced into the flow cell. In the dumbbell assay, tethers were maintained under a tension of 20 pN, which is sufficiently high to prevent the formation of secondary structure within the growing single-stranded component (45) but is sufficiently low to minimize tether breakage during the experiments. Changes in tether length extension typically were observed within 1 min of enzyme flow. PNPase experiments were performed in buffer [50 mM Hepes (pH 8.0), 5 mM MgCl2, 10 mM K2HPO4, 0.1 mM Na2EDTA, and 0.1 mM DTT] plus an oxygen-scavenging system [8.3 mg/mL glucose (Sigma), 46 U/mL glucose oxidase (Calbiochem), 94 U/mL catalase (Sigma)] at 26 ± 1 °C, with the exception of five records in which the phosphate concentration was reduced to 1 mM, with no apparent effect on unwinding. These assays were carried out at a variety of protein concentrations, with WT data collected primarily at 100 nM and mutant data collected primarily at 820 nM. RNase R experiments were performed in RNase R Buffer [20 mM Tris⋅HCl (pH 8.0), 0.1 mM MgCl2, and 100 mM KCl] at a concentration of 200 nM in the presence of the oxygen-scavenging system (see above).
Data Collection and Analysis.
Data collection and analysis are discussed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank I. Fischer-Hwang and A. Chakraborty for careful reading of the manuscript. F.M.F. was supported by a National Science Foundation Graduate Research Fellowship. B.F.L. is supported by the Wellcome Trust. This work was supported by National Institute of General Medical Sciences Grant R37GM057035 (to S.M.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.C.N. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1514028112/-/DCSupplemental.
References
- 1.Xu F, Cohen SN. RNA degradation in Escherichia coli regulated by 3′ adenylation and 5′ phosphorylation. Nature. 1995;374(6518):180–183. doi: 10.1038/374180a0. [DOI] [PubMed] [Google Scholar]
- 2.Py B, Higgins CF, Krisch HM, Carpousis AJ. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381(6578):169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 3.Cheng ZF, Deutscher MP. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc Natl Acad Sci USA. 2003;100(11):6388–6393. doi: 10.1073/pnas.1231041100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng ZF, Deutscher MP. An important role for RNase R in mRNA decay. Mol Cell. 2005;17(2):313–318. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 5.Godefroy T. Kinetics of polymerization and phosphorolysis reactions of Escherichia coli polynucleotide phosphorylase. Evidence for multiple binding of polynucleotide in phosphorolysis. Eur J Biochem. 1970;14(2):222–231. doi: 10.1111/j.1432-1033.1970.tb00281.x. [DOI] [PubMed] [Google Scholar]
- 6.Symmons MF, Jones GH, Luisi BF. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure. 2000;8(11):1215–1226. doi: 10.1016/s0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- 7.Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem. 2006;281(40):29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- 8.O’Hara EB, et al. Polyadenylylation helps regulate mRNA decay in Escherichia coli. Proc Natl Acad Sci USA. 1995;92(6):1807–1811. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrade JM, Pobre V, Silva IJ, Domingues S, Arraiano CM. The role of 3′-5′ exoribonucleases in RNA degradation. Prog Mol Biol Transl Sci. 2009;85:187–229. doi: 10.1016/S0079-6603(08)00805-2. [DOI] [PubMed] [Google Scholar]
- 10.Carpousis AJ, Vanzo NF, Raynal LC. mRNA degradation. A tale of poly(A) and multiprotein machines. Trends Genet. 1999;15(1):24–28. doi: 10.1016/s0168-9525(98)01627-8. [DOI] [PubMed] [Google Scholar]
- 11.Thang MN, Guschlbauer W, Zachau HG, Grunberg-Manago M. Degradation of Transfer Ribonucleic Acid by Polynucleotide Phosphorylase. I. Mechanism of Phosphorolysis and Structure of tRNA. J Mol Biol. 1967;26(3):403–421. [Google Scholar]
- 12.Kasai T, Gupta RS, Schlessinger D. Exoribonucleases in wild type Escherichia coli and RNase II-deficient mutants. J Biol Chem. 1977;252(24):8950–8956. [PubMed] [Google Scholar]
- 13.Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J Biol Chem. 2002;277(24):21624–21629. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- 14.Lin-Chao S, Chiou NT, Schuster G. The PNPase, exosome and RNA helicases as the building components of evolutionarily-conserved RNA degradation machines. J Biomed Sci. 2007;14(4):523–532. doi: 10.1007/s11373-007-9178-y. [DOI] [PubMed] [Google Scholar]
- 15.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127(6):1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 16.Hardwick SW, Gubbey T, Hug I, Jenal U, Luisi BF. Crystal structure of Caulobacter crescentus polynucleotide phosphorylase reveals a mechanism of RNA substrate channelling and RNA degradosome assembly. Open Biol. 2012;2(4):120028. doi: 10.1098/rsob.120028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong AG, McBurney KL, Thompson KJ, Stickney LM, Mackie GA. S1 and KH domains of polynucleotide phosphorylase determine the efficiency of RNA binding and autoregulation. J Bacteriol. 2013;195(9):2021–2031. doi: 10.1128/JB.00062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spickler C, Mackie GA. Action of RNase II and polynucleotide phosphorylase against RNAs containing stem-loops of defined structure. J Bacteriol. 2000;182(9):2422–2427. doi: 10.1128/jb.182.9.2422-2427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carpousis AJ. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- 20.Coburn GA, Mackie GA. Reconstitution of the degradation of the mRNA for ribosomal protein S20 with purified enzymes. J Mol Biol. 1998;279(5):1061–1074. doi: 10.1006/jmbi.1998.1842. [DOI] [PubMed] [Google Scholar]
- 21.Awano N, et al. Escherichia coli RNase R has dual activities, helicase and RNase. J Bacteriol. 2010;192(5):1344–1352. doi: 10.1128/JB.01368-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell. 1997;91(4):457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 23.Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell. 2008;29(6):717–728. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 24.Lee G, Bratkowski MA, Ding F, Ke A, Ha T. Elastic coupling between RNA degradation and unwinding by an exoribonuclease. Science. 2012;336(6089):1726–1729. doi: 10.1126/science.1216848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hossain ST, Malhotra A, Deutscher MP. The Helicase Activity of Ribonuclease R Is Essential for Efficient Nuclease Activity. J Biol Chem. 2015;290(25):15697–15706. doi: 10.1074/jbc.M115.650176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci USA. 2008;105(51):20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wuite GJ, Smith SB, Young M, Keller D, Bustamante C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000;404(6773):103–106. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
- 28.Maier B, Bensimon D, Croquette V. Replication by a single DNA polymerase of a stretched single-stranded DNA. Proc Natl Acad Sci USA. 2000;97(22):12002–12007. doi: 10.1073/pnas.97.22.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dessinges MN, Lionnet T, Xi XG, Bensimon D, Croquette V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc Natl Acad Sci USA. 2004;101(17):6439–6444. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Oijen AM, et al. Single-molecule kinetics of lambda exonuclease reveal base dependence and dynamic disorder. Science. 2003;301(5637):1235–1238. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- 31.Dulin D, et al. Elongation-competent pauses govern the fidelity of a viral RNA-dependent RNA polymerase. Cell Reports. 2015;10(6):983–992. doi: 10.1016/j.celrep.2015.01.031. [DOI] [PubMed] [Google Scholar]
- 32.Horton NC, Finzel BC. The structure of an RNA/DNA hybrid: a substrate of the ribonuclease activity of HIV-1 reverse transcriptase. J Mol Biol. 1996;264(3):521–533. doi: 10.1006/jmbi.1996.0658. [DOI] [PubMed] [Google Scholar]
- 33.Seol Y, Skinner GM, Visscher K. Elastic properties of a single-stranded charged homopolymeric ribonucleotide. Phys Rev Lett. 2004;93(11):118102. doi: 10.1103/PhysRevLett.93.118102. [DOI] [PubMed] [Google Scholar]
- 34.Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115(4):437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 35.Fedoroff OYu, Salazar M, Reid BR. Structure of a DNA:RNA hybrid duplex. Why RNase H does not cleave pure RNA. J Mol Biol. 1993;233(3):509–523. doi: 10.1006/jmbi.1993.1528. [DOI] [PubMed] [Google Scholar]
- 36.Shen LX, Cai Z, Tinoco I., Jr RNA structure at high resolution. FASEB J. 1995;9(11):1023–1033. doi: 10.1096/fasebj.9.11.7544309. [DOI] [PubMed] [Google Scholar]
- 37.Saenger W. Principles of Nucleic Acid Structure. Springer; New York: 1984. [Google Scholar]
- 38.Lesnik EA, Freier SM. Relative thermodynamic stability of DNA, RNA, and DNA:RNA hybrid duplexes: Relationship with base composition and structure. Biochemistry. 1995;34(34):10807–10815. doi: 10.1021/bi00034a013. [DOI] [PubMed] [Google Scholar]
- 39.Block SM, Svoboda K. Analysis of high resolution recordings of motor movement. Biophys J. 1995;68(4) Suppl:230S–239S, discussion 239S–241S. [PMC free article] [PubMed] [Google Scholar]
- 40.Stickney LM, Hankins JS, Miao X, Mackie GA. Function of the conserved S1 and KH domains in polynucleotide phosphorylase. J Bacteriol. 2005;187(21):7214–7221. doi: 10.1128/JB.187.21.7214-7221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lang MJ, Fordyce PM, Engh AM, Neuman KC, Block SM. Simultaneous, coincident optical trapping and single-molecule fluorescence. Nat Methods. 2004;1(2):133–139. doi: 10.1038/nmeth714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nurmohamed S, Vaidialingam B, Callaghan AJ, Luisi BF. Crystal structure of Escherichia coli polynucleotide phosphorylase core bound to RNase E, RNA and manganese: Implications for catalytic mechanism and RNA degradosome assembly. J Mol Biol. 2009;389(1):17–33. doi: 10.1016/j.jmb.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319(5863):630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 45.Dalal RV, et al. Pulling on the nascent RNA during transcription does not alter kinetics of elongation or ubiquitous pausing. Mol Cell. 2006;23(2):231–239. doi: 10.1016/j.molcel.2006.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koslover DJ, Fazal FM, Mooney RA, Landick R, Block SM. Binding and translocation of termination factor rho studied at the single-molecule level. J Mol Biol. 2012;423(5):664–676. doi: 10.1016/j.jmb.2012.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.