Significance
Classic physiological studies have documented the endurance-promoting effects of glucocorticoid (GC) hormones on skeletal muscle. Pharmacologic GC therapy also improves muscle function in patients with Duchenne muscular dystrophy (DMD), a genetic muscle-wasting disease. Despite these well-established physiological and clinical observations, the molecular basis underlying the beneficial effects of GCs in skeletal muscle remains obscure. This study shows that physiological effects of GCs on muscle endurance and their therapeutic effect in DMD are mediated, in part, via activation of a potent metabolic gene called Kruppel-like factor 15 (KLF15). Importantly, KLF15 does not drive GC-mediated muscle wasting. These data shed light on the poorly understood ergogenic properties of GCs, findings that may inform steroid-sparing therapies for DMD and other muscle diseases.
Keywords: skeletal muscle metabolism, glucocorticoid, exercise, Duchenne muscular dystrophy, steroid hormone nuclear receptor
Abstract
Classic physiology studies dating to the 1930s demonstrate that moderate or transient glucocorticoid (GC) exposure improves muscle performance. The ergogenic properties of GCs are further evidenced by their surreptitious use as doping agents by endurance athletes and poorly understood efficacy in Duchenne muscular dystrophy (DMD), a genetic muscle-wasting disease. A defined molecular basis underlying these performance-enhancing properties of GCs in skeletal muscle remains obscure. Here, we demonstrate that ergogenic effects of GCs are mediated by direct induction of the metabolic transcription factor KLF15, defining a downstream pathway distinct from that resulting in GC-related muscle atrophy. Furthermore, we establish that KLF15 deficiency exacerbates dystrophic severity and muscle GC–KLF15 signaling mediates salutary therapeutic effects in the mdx mouse model of DMD. Thus, although glucocorticoid receptor (GR)-mediated transactivation is often associated with muscle atrophy and other adverse effects of pharmacologic GC administration, our data define a distinct GR-induced gene regulatory pathway that contributes to therapeutic effects of GCs in DMD through proergogenic metabolic programming.
Synthetic derivatives of the glucocorticoid (GC) class of steroid hormones, which are ligands for the nuclear receptor NR3C1 (also known as the glucocorticoid receptor; GR), are widely used as antiinflammatory drugs (1). The vast majority of literature on muscle GR signaling has focused on muscle wasting, a side effect of excessive or sustained GC exposure that is mediated, in part, by direct GR-dependent transactivation of genes that drive myocyte atrophy (e.g., Foxo3a, Gdf8/Myostatin, Fbxo32/Atrogin1; also known as atrogenes) (1, 2). However, literature dating back to the 1930s, including the classic physiological studies from the laboratory of Dwight Ingle (3, 4), have documented that moderate or transient exposure to GCs can enhance muscle performance and produce ergogenic effects in animals and humans (5–13). Consistent with this known physiological role of GCs in anticipatory metabolic adaptation, surreptitious GC ingestion is a well-known doping strategy used by elite endurance athletes, an act that has prompted disqualifications and has led to the universal banning of these drugs by sports regulatory agencies (14). Although mechanisms governing GC-mediated muscle atrophy have been extensively studied (1, 2), the molecular basis for their ergogenic effects remains poorly understood.
In addition to these ergogenic physiological effects, low-dose GC therapy also improves muscle function, quality of life, and survival in patients with Duchenne muscular dystrophy (DMD) (15–17), a progressive muscle-wasting disease caused by X-linked inheritance of nonsense mutations in the gene encoding Dystrophin (18). Although GCs have been widely used in the treatment of DMD for nearly 25 years (15), the mechanism of action underlying their salutary effects in this condition remains obscure. Several studies in patients and animal models have called into question whether the antiinflammatory properties of GCs adequately explain their therapeutic effect in DMD (15, 19). We hypothesized that the ergogenic properties of GCs and aspects of their therapeutic efficacy in DMD might be mediated, in part, by a defined GR-dependent metabolic transcriptional pathway distinct from that resulting in GC-related muscle atrophy. As GCs cause a myriad of systemic side effects that limit their therapeutic index in DMD, elucidation of such a downstream signaling pathway could inform novel steroid-sparing treatment strategies, a major unmet need for patients suffering from this devastating and currently incurable disease (15, 20).
The transcription factor KLF15, a direct GR-inducible target in multiple cell types (21, 22), is an attractive candidate for mediating important ergogenic effects of GCs in muscle. Studies from our group using systemic KLF15-deficient mice have demonstrated KLF15 to be an important transcriptional regulator of a gene program governing stress-dependent metabolic adaptation in muscle (23, 24). KLF15-deficient mice have impaired ability to catabolize muscle branched-chain amino acids as fuel for gluconeogenic flux during fasting (25, 26). In addition, we have demonstrated that during sustained aerobic exercise, KLF15-deficient mice fail to augment muscle lipid utilization and consequently have impaired endurance exercise capacity (23). Our detailed characterization of KLF15-deficient mice has revealed that these functional defects in muscle substrate flux occur without abnormalities in muscle development, gross histologic structure, fiber type distribution, mitochondrial number, or muscle mass (23).
Although KLF15 is a direct GR target (21, 22, 27, 28), the function of the GC–KLF15 axis in physiology and disease remains largely unknown. The observation that GCs induce KLF15 in rodent myocytes and that adenoviral KLF15 overexpression increases expression of Atrogin-1 has led to the speculation that KLF15 might promote muscle wasting via activation of an atrophy-promoting transcriptional program (22). However, these studies used supraphysiologic levels of KLF15 overexpression and have thus left unanswered whether the physiologic effects of the GC–KLF15 axis are harmful or beneficial in skeletal muscle. Here, we use unbiased transcriptomic profiling and physiologic analysis of mice harboring KLF15 deficiency and skeletal muscle-specific KLF15 overexpression at physiologic levels to gain a deeper understanding of the GC–KLF15 axis in muscle function. We find that the GC–KLF15 axis does not regulate muscle atrophy in vivo but rather activates a metabolic gene program that mediates ergogenic effects in wild-type (WT) mice and ameliorates dystrophic severity in the mdx mouse model of DMD. These studies establish KLF15 as a critical metabolic effector of physiological GC signaling in vivo and suggest that GR-dependent target gene transactivation in skeletal muscle can, in certain contexts, have a therapeutic role.
Results
KLF15 Is Induced by GCs in Live Human Subjects.
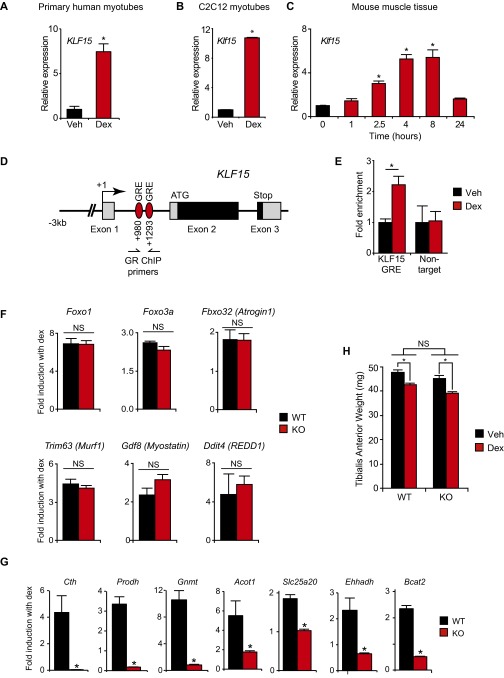
Although GCs can induce KLF15 in rodent muscle tissue (22), it is not known whether GCs regulate KLF15 in human subjects in vivo. Healthy human volunteers underwent skeletal muscle biopsies before and after ingesting the potent GR agonist dexamethasone (dex; 2 mg orally twice daily × 5 d). Quantitative RT-PCR (qRT-PCR) revealed a significant induction of KLF15 expression in human skeletal muscle tissue in vivo (Fig. 1A; FKBP5 is a well-established GR target that serves as a positive control). Similarly, we found that dex rapidly and robustly increased KLF15 expression in primary human myotubes and mouse C2C12 myotubes in vitro (Fig. S1 A and B) and mouse quadriceps in vivo (Fig. S1C). The mammalian KLF15 locus contains two adjacent glucocorticoid response elements (GREs) in the first intron that are highly conserved (schematized in Fig. S1D). We performed ChIP-qPCR in quadriceps tissue of adult mice and treated with or without dex and found dynamic enrichment of endogenous GR at these intronic GREs in the Klf15 locus (Fig. S1E). These data confirm prior observations that KLF15 is a direct GR target in muscle and demonstrate that GC-mediated induction of muscle KLF15 occurs in live human subjects.
Fig. 1.
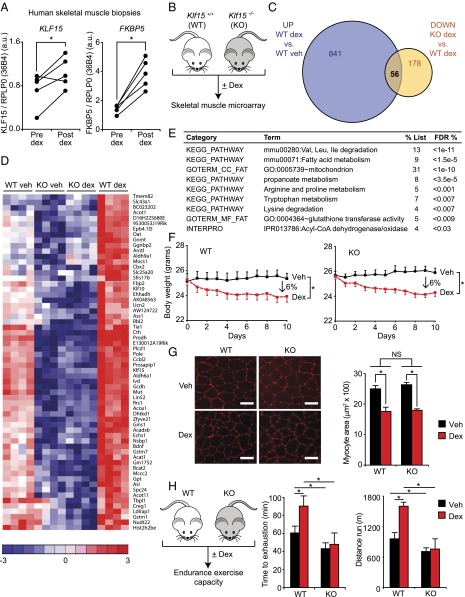
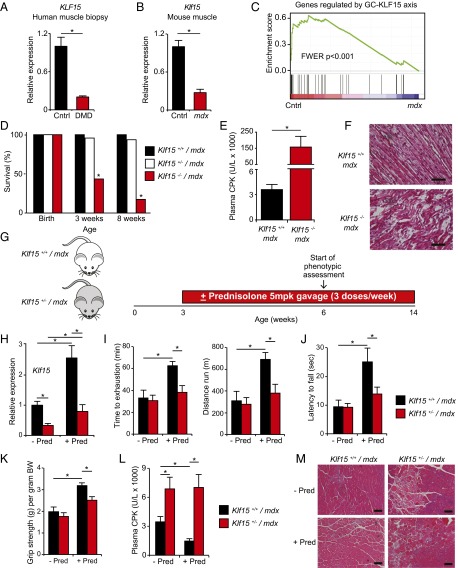
The GC–KLF15 axis dissociates ergogenic physiology from muscle atrophy. (A) KLF15 expression in skeletal muscle of healthy human subjects pre- and postdex administration. FKBP5 is a known direct GR target shown as a positive control (n = 5; *P < 0.05 for indicated comparisons). (B) Schematic for skeletal muscle (quadriceps) microarray study (n = 4 per group). (C) Venn diagram of dex-inducible and KLF15-regulated genes [2 mg/kg i.p. × 1 dose, 8 h time point; fold change > 1.5 and false discovery rate (FDR) < 0.05]. (D) Heat map of genes robustly regulated [family-wise error rate (FWER) < 0.05] by GC–KLF15 axis in mouse skeletal muscle. (E) Functional annotation [Database for Annotation, Visualization, and Integrated Discovery (DAVID)] of genes regulated by GC–KLF15 axis. (F) Body weight and (G) muscle (tibialis anterior) histology [wheat-germ agglutinin (WGA) stain] and cross-sectional area in a mouse model of chronic dex-induced atrophy (1 mg/kg·d s.c. × 10 d; n = 8; *P < 0.05 for indicated comparisons). (Scale bar, 50 µm.) (H) Schematic of dex-induced doping experiment (2 mg/kg i.p. × 1 dose) with time to exhaustion and distance run on a motorized treadmill 18 h post-dex administration (n = 7–9; *P < 0.05 for indicated comparisons). Data are shown as mean ± SEM.
Fig. S1.
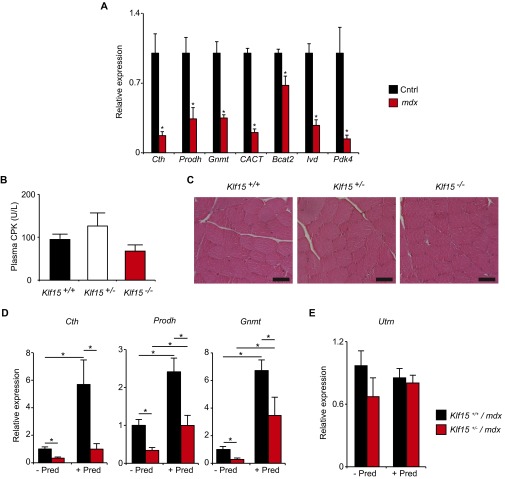
Definition of the GC–KLF15 transcriptional axis. KLF15 expression in (A) primary human myotubes and (B) C2C12 myotubes treated ± dex (100 nM, 6 h; n = 4; *P < 0.05 vs. Veh). (C) Klf15 expression in mouse quadriceps after dex administration (1 mg/kg i.p.; n = 4; *P < 0.05 vs. 0 h). (D) Schematic of the conserved KLF15 locus denoting two highly conserved GREs in intron 1. (E) GR ChIP in adult mouse quadriceps ± dex treatment (1 mg/kg i.p., 6 h) using qPCR primers for the indicated GRE region (n = 4; *P < 0.05 for indicated comparisons). (F) Expression of indicated atrogenes from mouse quadriceps shown as fold induction with dex (2 mg/kg i.p. × 1 dose, 8 h time point; n = 4–5). (G) Expression of indicated genes from mouse quadriceps shown as fold induction with dex (2 mg/kg i.p. × 1 dose, 8 h time point) (*P < 0.05 vs. WT; n = 4–5 per group). (H) Chronic dex-induced muscle atrophy model: tibialis anterior weight of WT and KLF15 KO mice treated ± dex (1 mg·kg·d s.c. × 10 d) (*P < 0.05 for indicated comparisons; n = 8). Data are shown as mean ± SEM.
The GC–KLF15 Axis Regulates an Ergogenic Gene Program That Is Not Involved in Muscle Atrophy.
In an effort to definitively resolve the gene program regulated by the GC–KLF15 axis in vivo, we performed unbiased transcriptomic profiling from skeletal muscle tissue of WT and Klf15−/− (KO) mice treated with or without dex. WT and KO mice were given a single dose of dex (2 mg/kg i.p.) versus vehicle, and quadriceps tissue was isolated for transcript expression profiling by cDNA microarrays (Fig. 1B). Dex induced a total of 897 genes, of which 7% were highly KLF15-dependent, as shown by Venn diagram (Fig. 1C) and heat map of representative genes (Fig. 1D; full list of differential gene expression provided in Excel format in Dataset S1). To our surprise, the microarray study revealed that KLF15 was not required for the robust GC-meditated induction of the canonical atrogene program, findings that were confirmed independently by qRT-PCR (Fig. S1F). Rather, functional annotation of the GC–KLF15-regulated transcriptome revealed highly significant enrichment for genes critical for metabolism of several amino acids, fatty acids, and propanoate (Fig. 1 D and E, Fig. S1G, and Dataset S1).
Based on these unbiased expression profiles, we hypothesized that KLF15 was not involved in muscle wasting but, in contrast, was required for GC-mediated ergogenic effects in vivo. To test this hypothesis, we first subjected mice to a standard model of chronic dex-mediated muscle atrophy (dex 1 mg/kg given daily s.c. × 10 d) (29, 30). We found that both WT mice and KLF15 KO mice had an identical degree of muscle atrophy, as assessed by changes in body weight, muscle weight, and myofiber cross-sectional area (Fig. 1 F and G and Fig. S1H). Thus, KLF15 is not required for GC-mediated muscle atrophy in vivo. To test the requirement of KLF15 in GC-mediated ergogenesis, WT and KO mice were given a single dose of dex and challenged to an endurance exercise trial 16 h after dex treatment (Fig. 1H). Consistent with the gene expression profiling, WT mice treated with dex were able to augment endurance exercise capacity by 50%, whereas KO mice did not demonstrate any significant ergogenic response to dex (Fig. 1H). Hence, these data demonstrate that KLF15 is dispensable for GC-mediated muscle atrophy but required for the ergogenic effects of GCs.
Skeletal Muscle-Specific KLF15 Induction Is Sufficient to Drive an Ergogenic Gene Program in Vivo.
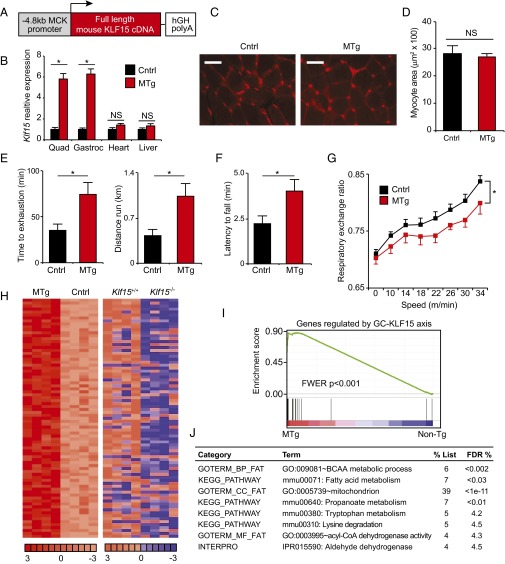
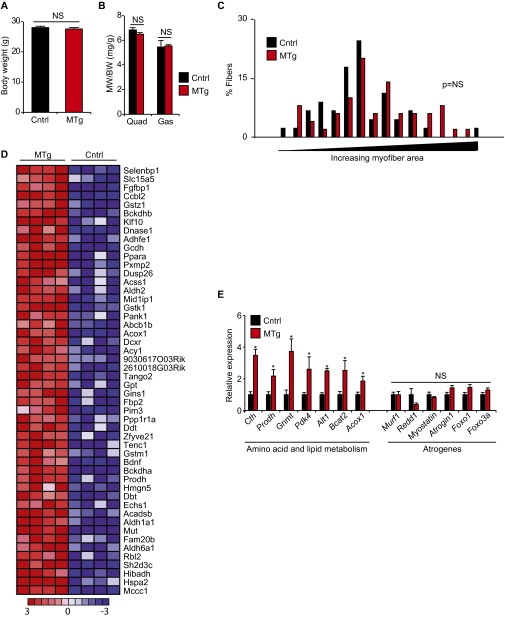
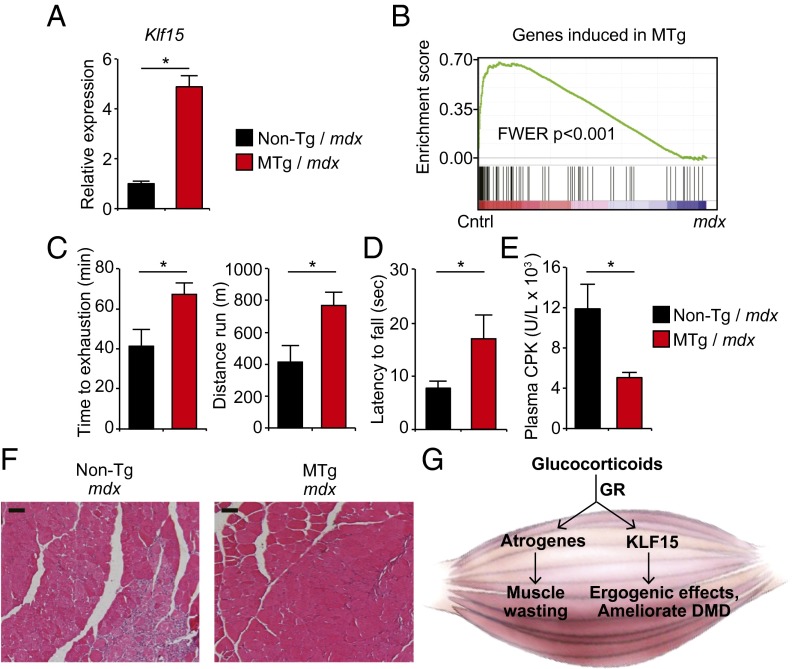
As the data in Fig. 1 were generated using mice harboring systemic KLF15 deficiency, we next asked whether skeletal muscle-specific augmentation of KLF15 expression was sufficient to activate an ergogenic metabolic program in vivo. We used the –4.8 kb MCK promoter/enhancer to generate transgenic mice expressing a full-length mouse KLF15 cDNA in a skeletal muscle-specific manner (Fig. 2A). We studied a transgenic mouse line that expressed KLF15 levels at 5–6 times higher than control exclusively in skeletal muscle with no induction in the heart or other tissues (muscle transgenic, MTg; Fig. 2B). This level of transgenic overexpression of KLF15 is in the physiological range (23) and similar to the increase in KLF15 seen with nanomolar concentrations of dex (Fig. S1 A and B). MTg mice showed no evidence of muscle atrophy as assessed by gravimetry (Fig. S2 A and B) or histology (Fig. 2 C and D and Fig. S2C). In contrast, MTg mice had significantly increased endurance exercise capacity (Fig. 2 E and F). Metabolic exercise testing demonstrated that MTg mice had a decreased respiratory exchange ratio (Fig. 2G), signifying that muscle-specific KLF15 induction leads to a global shift in substrate preference toward amino acids and lipids. To confirm that these physiological observations were associated with KLF15-dependent transcriptional control in skeletal muscle, we performed genome-wide expression profiling in quadriceps of MTg mice using cDNA microarrays. These analyses revealed that skeletal muscle-specific KLF15 overexpression in the physiological range resulted in a signature that was dominated by gene induction (90 induced genes out of 109 total differentially expressed genes; 83%). A representative heat map of the top 50 genes induced in the MTg skeletal muscle is provided in Fig. S2D. As visualized in the global heat maps from MTg mice (Fig. 2H), the transcriptomic profile of MTg mice reflected significant reversal of the metabolic gene expression abnormalities observed in KLF15 KO skeletal muscle. This highly significant overlap between genes induced in the MTg and genes reduced in KLF15 KO muscle was statistically confirmed by gene set enrichment analysis (GSEA; Fig. 2I). Functional annotation of differentially expressed genes in MTg mice (Fig. 2J) revealed robust enrichment for the same pathways regulated by the GC–KLF15 axis (Fig. 1E), with significant induction of genes critical for amino acid, lipid, and propanoate metabolism (Fig. 2J and Fig. S2E; full list of differential gene expression provided in Excel format as Dataset S2). Consistent with our published (23) and current observations in KO mice (Fig. 1), the MTg microarrays did not demonstrate significant regulation of a gene program governing fiber type specification, mitochondrial biogenesis, autophagic flux, or angiogenesis. Again, expression profiles and direct qRT-PCR showed no induction of the canonical atrogene program in MTg mice (Fig. S2E and Dataset S2). Thus, the data in Figs. 1 and 2 demonstrate that KLF15 is not an atrogene but rather functions as a critical downstream transcriptional effector of muscle GR signaling that regulates amino acid and lipid metabolic programs. Furthermore, these data demonstrate that KLF15 is both necessary and sufficient to mediate ergogenic effects of GCs in vivo.
Fig. 2.
Skeletal muscle-specific KLF15 overexpression is sufficient to drive an ergogenic metabolic program in vivo. (A) Schematic of transgenic construct. (B) qRT-PCR from indicated tissues demonstrating skeletal muscle-specific KLF15 overexpression in MTg mice versus control (littermate non-Tg mice; n = 5). (C) Representative histology from quadriceps (WGA staining) and (D) quantification of myocyte cross-sectional area (n = 3 independent mice per group). (Scale bar, 50 µm.) (E) Treadmill exercise (n = 6), (F) wire-hang assay (n = 7–10), and (G) metabolic exercise test (n = 7–10) in MTg and control mice. (H) Heat map of all genes induced in MTg versus non-Tg muscle (Left panel; n = 4). Heat map for the same genes in WT versus KLF15 KO muscle (Right panel, WT levels colored as non-Tg in Left panel) illustrates the reversal in expression versus control in KLF15 KO relative to MTg. (I) GSEA of genes regulated by the GC–KLF15 axis in MTg versus non-Tg muscle. (J) Functional annotation (DAVID) of genes regulated by KLF15 overexpression (*P < 0.05 for indicated comparisons). Data are shown as mean ± SEM.
Fig. S2.
Phenotype of mice with skeletal muscle-specific overexpression of KLF15 in vivo. (A) Body weight, (B) quadriceps (Quad) and gastrocnemius (Gas) muscle weight, and (C) myofiber cross-sectional area distribution in quadriceps from control (Cntrl) and MTg mice (n = 5). (D) Heat map of the top 50 genes induced in MTg versus non-Tg muscle (n = 4). (E) Expression of indicated genes in Cntrl and MTg quads (n = 5; *P < 0.05 vs. Cntrl). Data are shown as mean ± SEM.
The GC–KLF15 Transcriptional Axis Modulates Disease Phenotype in the mdx Mouse Model of DMD.
As the GC–KLF15 axis controls a gene program governing muscle substrate utilization and ergogenesis (Figs. 1 and 2), we asked whether this metabolic signaling pathway might, in part, mediate the salubrious effects of GCs in DMD. Landmark clinical studies have demonstrated that low and intermittent dosing of GCs such as prednisolone can slow disease progression and improve outcomes in DMD patients (15–17). Despite this established efficacy, a defined molecular basis for the salutary effects of GCs in DMD remains poorly understood (15, 20). Although GCs are known to exert antiinflammatory effects via transrepression of NF-κB signaling (1), several clinical and experimental studies cast significant doubt on the contention that GCs ameliorate DMD via a primary antiinflammatory mechanism (15, 19, 31–34). First, non-GC antiinflammatory or immunosuppressant drugs, such as cyclosporine-A or azathioprine, fail to show efficacy in DMD patients (32–34). Second, GC therapy provides benefit in DMD patients even when administered via low or intermittent dosing schemes that do not provide a potent and sustained antiinflammatory effect (15). Third, GC therapy does not dramatically alter the inflammatory cell infiltrate seen in dystrophic muscle (31, 34). Fourth, in the mdx mouse model of DMD (35), the therapeutic effects of GCs are fully preserved in a Rag2−/− background, in which B and T lymphocytes are absent (19).
In contrast, several studies in DMD patients and animal models have documented abnormalities in muscle substrate and energy metabolism (36–47), prompting the term “metabolic crisis” as a hallmark feature of this disease (41). Given the important role for KLF15 in muscle metabolism, we hypothesized that DMD might be characterized by a state of relative KLF15 deficiency and that GCs may mediate important therapeutic effects in DMD, in part, via induction of KLF15. Skeletal muscle biopsies from DMD patients (Fig. 3A) and muscle tissue from male mdx mice (Fig. 3B) had significantly decreased KLF15 expression compared with control tissues. We next used GSEA to statistically test whether the metabolic gene program regulated by the GC–KLF15 axis was deficient in dystrophic muscle. These GSEAs confirmed in an unbiased and quantitative manner that the set of genes decreased in mdx skeletal muscle tissue (curated from ref. 44) was highly enriched for the set of genes regulated by the GC–KLF15 axis (Fig. 3C). These findings were also confirmed by direct qRT-PCR analysis of representative genes (Fig. S3A). Hence, DMD is characterized by decreased expression of KLF15 and its downstream metabolic targets.
Fig. 3.
KLF15 deficiency severely exacerbates DMD, and KLF15 is required for therapeutic efficacy of GCs. qRT-PCR of KLF15 from (A) vastus lateralis biopsies of human subjects with DMD versus healthy controls (n = 10) and (B) quadriceps tissue of mdx versus control mice (n = 5; *P < 0.05 for indicated comparisons). (C) GSEA demonstrating that genes reduced in mdx skeletal muscle tissue (curated from ref. 44) are highly enriched for those regulated by the GC–KLF15 axis. (D) Survival by genotype of male offspring generated from crosses between Klf15+/− mdx males and females. There is significant postnatal death of Klf15−/− mdx males (P < 0.05; n = 157 total mice). (E) Plasma CPK concentration from mice of indicated genotypes (n = 6–7; *P < 0.05 for indicated comparison). (F) Representative H&E-stained section of hindlimb muscle tissue from mice of indicated genotypes before death (age P4–5; representative of n = 3). (Scale bar, 20 µm.) (G) Schematic of experimental design for GC therapeutic trial (± prednisolone 5 mg/kg·dose oral gavage given 3 times weekly). (H) Klf15 expression in quadriceps (n = 4–7; *P < 0.05 for indicated comparisons). (I) Treadmill endurance exercise capacity (n = 6–15), (J) wire-hang endurance assay (n = 6–13), and (K) grip strength (n = 6–11) of indicated genotypes treated ± prednisolone (*P < 0.05 for indicated comparisons). (L) Plasma CPK concentration (n = 6–9; *P < 0.05 for indicated comparisons). (M) Representative H&E-stained sections of quadriceps from mice of indicated genotypes and treatments (representative of n = 3). (Scale bar, 100 µm). Data are shown as mean ± SEM.
Fig. S3.
KLF15 deficiency in mdx mice. (A) Expression of indicated genes in quadriceps of mdx and control mice (*P < 0.05 vs. Cntrl; n = 4–5 per group). (B) Plasma CPK and (C) representative H&E-stained quadriceps sections from male mice of indicated genotypes. Note that the male Klf15-deficient mice shown in B and C harbor a normal Dystrophin allele (age 8 wk; n = 4). (D) Expression of indicated genes in quadriceps of mice of indicated genotypes and treatments (n = 4–7; *P < 0.05 for indicated comparisons). (E) Expression of Utrophin in quadriceps of mice of indicated genotypes and treatments (n = 4–7). Data are shown as mean ± SEM.
Prior studies from our group demonstrate that mice with KLF15 deficiency, which have been extensively characterized in a nondystrophic genetic background, are viable into adulthood, are fertile, have normal muscle development and histology, and show no evidence of muscle damage (23). We confirmed this lack of baseline histopathology in an independent cohort of mice and found that male KLF15-deficient mice in a nondystrophic background showed no evidence of elevated plasma creatine phosphokinase (CPK) concentration or histologic muscle damage (Fig. S3 B and C). Although KLF15 deficiency alone does not cause dystrophic muscle pathology, we hypothesized that KLF15 deficiency might exacerbate dystrophic severity in the mdx mouse model of DMD. To test this hypothesis, we bred KLF15-deficient mice into the mdx background by intercrossing Klf15+/−/Dmdmdx Y males with Klf15+/−/Dmdmdx Dmdmdx females. Although pups of all three expected genotypes were born in predicted Mendelian ratios, we observed significant premature death of male Klf15−/− mdx mice postnatally with only ∼20% surviving to adulthood (Fig. 3D). We did not observe any statistically significant loss of Klf15−/−/Dmdmdx Dmdmdx female mice, a finding that may be related to gender-specific metabolic consequences of KLF15 deficiency or milder aspects of disease severity in Dmdmdx Dmdmdx versus Dmdmdx Y mice (48). Analysis of male Klf15−/− mdx mice before death revealed severe dystrophic pathology in skeletal muscle tissue and massively elevated plasma CPK concentration, an unbiased and quantitative clinical biomarker of global muscle damage in DMD patients (15) (Fig. 3 E and F). Importantly, single-mutant intercrosses of mice harboring either the null Klf15 allele or the mdx allele alone do not produce such mortality or muscle damage (23, 49). Our observations in these compound mutant strains support the contention that there is a genetic interaction between KLF15 deficiency and the dystrophic pathology of the mdx mouse model.
Given the low survival of male Klf15−/− mdx mice, we were unable to perform longitudinal studies with appropriately matched littermate controls. Therefore, we focused our attention on Klf15 haploinsufficient mice, which survive into adulthood (Fig. 3D) and reflect the partial loss of KLF15 expression seen in muscle tissue from DMD patients and mdx mice (Fig. 3 A and B). We subjected male Klf15+/− mdx mice and Klf15+/+ mdx littermate controls to a therapeutic trial of prednisolone using a low and intermittent dosing regimen similar to that used clinically in DMD patients and in prior studies of mdx mice (Fig. 3G) (15, 19, 49). We confirmed that this regimen of chronic prednisolone produced the expected increase in muscle KLF15 and its downstream targets in Klf15+/+ mdx control mice, with blunted expression in Klf15+/− mdx mice (Fig. 3H and Fig. S3D). Klf15+/− mdx mice had minimal baseline perturbations in muscle strength and endurance but had significantly attenuated therapeutic responses to prednisolone therapy, as demonstrated by decreased endurance exercise capacity on treadmill (Fig. 3I) and wire-hang tests (Fig. 3J) and decreased grip strength (Fig. 3K). Klf15+/− mdx mice had histologic evidence of excessive muscle damage and plasma CPK elevation at baseline with no significant therapeutic response to prednisolone therapy (Fig. 3 L and M). Utrophin expression, which is known to compensate for mutant Dystrophin in mice (50), was not affected by prednisolone therapy or KLF15 deficiency (Fig. S3E). The data in Fig. 3 reveal that KLF15 deficiency, which does not cause significant muscle damage in a nondystrophic background (Fig. S3 B and C) (23), exacerbates features of dystrophic severity in the mdx background. Furthermore, the data in the Klf15 haploinsufficient mice demonstrate that partial deficiency of this gene attenuates the therapeutic effect of GCs in the mdx mouse model.
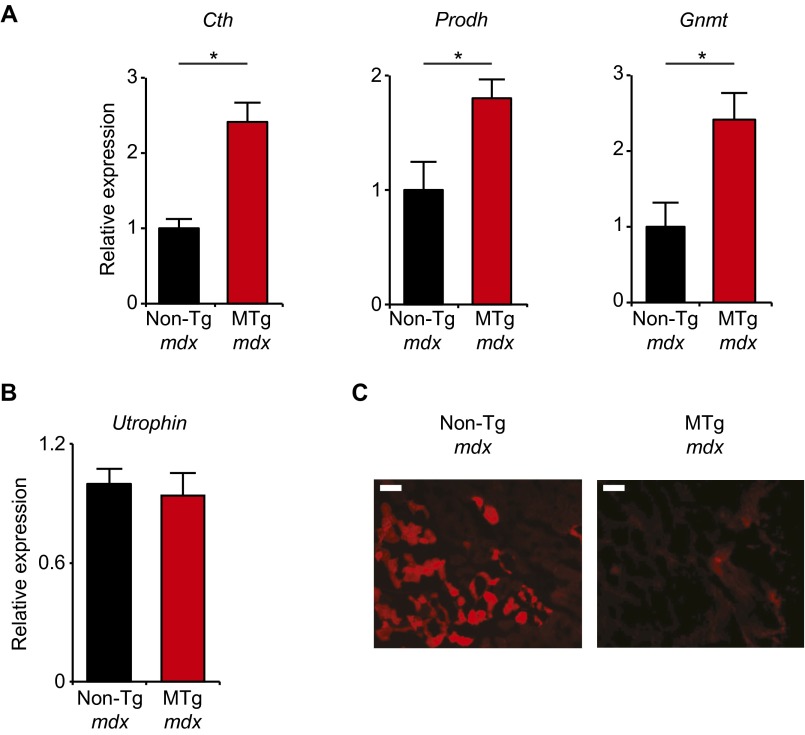
We next asked whether effects of KLF15 in the mdx model were muscle-specific in vivo and whether KLF15 induction was sufficient to exert beneficial effects in the absence of exogenous GCs. To address both of these questions, we crossed KLF15 MTg mice into the mdx background to produce male mice of genotype MTg+/− Dmdmdx and non-Tg Dmdmdx control littermates. The transgene increased total KLF15 expression in mdx skeletal muscle by fivefold, a level of induction that approximates the effect of GCs (Fig. 4A). To test whether the gene expression program induced by the KLF15 transgene was statistically enriched for genes down-regulated in mdx mice in an unbiased manner, we performed GSEA between our transgenic microarray data (Fig. 2) and a curated profile of differentially expressed genes in mdx mouse skeletal muscle (44). GSEA revealed that the metabolic gene program induced in MTg mice was significantly enriched for genes deficient in mdx skeletal muscle (Fig. 4B), findings that were confirmed by qRT-PCR of representative genes (Fig. S4A). There was no differential expression of Utrophin in MTg mdx mice (Fig. S4B). Phenotypic assessment revealed that MTg mdx mice had significantly increased endurance exercise capacity on a treadmill (Fig. 4C) and wire-hang test (Fig. 4D). In addition, MTg mdx mice had lower plasma CPK concentration (Fig. 4E), a reduction in histologic muscle damage (Fig. 4F), and decreased Evans blue dye extravasation (Fig. S4C) compared with non-Tg mdx littermate controls. Taken together, the data in Figs. 3 and 4 demonstrate that DMD tissues have low KLF15 expression and that genetic Klf15 deletion exacerbates dystrophic pathology and attenuates the therapeutic response to GCs in mdx mice. Furthermore, genetic induction of KLF15 in a skeletal muscle-specific and GC-independent manner can ameliorate several pathologic features in mdx mice.
Fig. 4.
Muscle-specific KLF15 overexpression ameliorates DMD phenotype in mdx mice. (A) qRT-PCR of Klf15 in quadriceps from mice of indicated genotypes (n = 5). (B) GSEA demonstrating that genes reduced in mdx skeletal muscle tissue (curated from ref. 44) are highly enriched for those induced by the muscle-specific KLF15 transgene. (C) Treadmill exercise capacity (n = 9 MTg mdx, n = 5 non-Tg mdx) and (D) wire-hang test (n = 9 MTg mdx, n = 15 non-Tg mdx) in mice of indicated genotypes. (E) Plasma CPK concentration (n = 7 MTg mdx, n = 5 non-Tg mdx). (F) Representative H&E-stained quadriceps sections demonstrating histologic improvement of muscle damage in MTg mdx mice (representative of n = 3 per group; *P < 0.05 for indicated comparisons). (Scale bar, 100 µm.) (G) Schematic depicting the role of the GC–KLF15 axis in skeletal muscle. Data are shown as mean ± SEM.
Fig. S4.
Skeletal muscle-specific KLF15 overexpression in mdx mice. (A) Expression of indicated genes in mouse quadriceps (*P < 0.05 vs. non-Tg mdx; n = 5). (B) Expression of Utrophin in quadriceps (n = 5). (C) Representative sections of mouse quadriceps muscle after injection of mice with Evans blue dye. (Scale bar, 100 µm.) Red signal indicates pathological dye extravasation. Data are shown as mean ± SEM.
Discussion
This study defines a molecular mechanism that mediates ergogenic effects of GCs and contributes to their therapeutic efficacy in DMD (schematized in Fig. 4G). The GR directly transactivates KLF15 and regulates a KLF15-dependent metabolic gene program in skeletal muscle that increases endurance exercise capacity in a nondystrophic background and attenuates features of disease severity in the mdx mouse model of DMD. Classic physiological studies from Dwight Ingle and colleagues documented that optimal muscle endurance required an intact adrenal cortex (3, 4). In fact, this ergogenic bioactivity formed the basis of a bioassay that directly facilitated the eventual purification, characterization, and synthesis of cortisone from the adrenal cortex by the teams led by Edward Kendall and Tadeus Reichstein (51). When considered alongside these seminal observations, our current data highlight the evolutionarily conserved role of GC signaling in anticipatory metabolic adaptation to stress and establish the GC–KLF15 axis as a critical transcriptional effector of muscle physiology.
Our in vivo gain- and loss-of-function data also support the contention that KLF15 does not directly participate in skeletal muscle atrophy. Although previous studies have demonstrated that adenoviral overexpression of KLF15 can induce Atrogin-1 and affect cell size in myocytes (22), unbiased gene expression profiling in adult KLF15-deficient mice reveals that KLF15 is not required for GC-mediated induction of canonical atrogenes and GC-mediated muscle wasting in vivo (Fig. 1). Furthermore, skeletal muscle-specific overexpression of KLF15 at levels within the physiological range does not induce atrogenes or produce muscle atrophy (Fig. 2). These data establish the principle that a pathway mediating ergogenic metabolic effects of GCs can be dissociated from the molecular pathways that cause GC-mediated muscle wasting.
We show that KLF15 expression is reduced in dystrophic human and murine muscle and that increasing or decreasing KLF15 levels affects disease phenotype in the mdx mouse model of DMD. Although KLF15-deficient mice have been documented to have abnormalities in adaptive substrate metabolism (23, 52), this mouse strain has no evidence of abnormal myogenesis, myocyte fragility, or histologic muscle damage in a nondystrophic background (23), findings that were confirmed in the present study. However, when introduced into the mdx background, KLF15 deficiency leads to exaggerated muscle damage, suggesting a causal role for decreased muscle KLF15 in DMD disease pathogenesis. One caveat of our murine loss-of-function experiments is that KLF15 is systemically targeted, raising the possibility that KLF15 deficiency in nonmuscle tissues may contribute to the observed phenotypes. However, our transgenic mouse studies support a skeletal muscle-specific role for KLF15 in vivo. Muscle-specific KLF15 overexpression at levels that approximate the GC effect activates a metabolic gene program, drives ergogenic physiology, and ameliorates dystrophic severity in mdx mice. Future studies using conditional deletion of KLF15 in the mdx background will be useful to further annotate the muscle-specific contribution of this metabolic transcription factor in muscular dystrophy.
Our published work detailing the consequences of KLF15 deficiency on muscle physiology (23) and the current study of the GC–KLF15 axis in vivo provide important insight into the putative downstream mechanisms by which activation of this GR target can ameliorate DMD. Collectively, these studies show that KLF15 does not directly regulate a transcriptional program governing myogenesis, myofiber specification, mitochondrial biogenesis, autophagic flux, or muscle mass. Rather, our physiological and transcriptomic studies in mice with KLF15 gain- or loss-of-function reveal a dominant role in regulation of metabolic substrate catabolism, particularly that of critical amino acid and lipid species. Deficiency of KLF15 may lead to accumulation of metabolic intermediates that are toxic to muscle, produce excess reactive oxygen species (ROS), and render myocytes more susceptible to dysfunction and death when combined with the “second hit” of genetic dystrophinopathy. Our finding that KLF15 and a significant number of its downstream targets are deficient in dystrophic muscle is consistent with several studies that have documented abnormalities in lipid and amino acid metabolism in DMD (36–47). The observation that muscle-specific overexpression of KLF15 improves aspects of dystrophic pathology suggests that restoration of the KLF15-dependent metabolic program plays a role in ameliorating DMD. Although the clear common denominator between our gain- and loss-of-function models is regulation of a metabolic gene program, we recognize that KLF15-dependent regulation of other processes such as ROS homeostasis, neuromuscular junction plasticity, or sarcolemmal integrity may also contribute to the phenotypes that are observed in this study.
Previous studies have used locus-specific ChIP to demonstrate the principle that key metabolic genes are direct targets of KLF15 (22, 23, 26, 27). We note that a number of KLF15-regulated targets are also known to be direct targets of the GR (23, 27, 53, 54), suggesting that these two transcription factors can cooperate. Indeed, a recent transcriptomic study from our group in airway cells has established that GR and KLF15 participate in robust and dynamic feed-forward transcriptional signaling at metabolic targets (27), supporting our current finding that KLF15 is a major molecular effector of the GR in vivo. We note that currently available antibodies for KLF15 are not adequate for ChIP-Seq analysis in skeletal muscle (23), precluding the genome-wide definition of the muscle KLF15 cistrome. Based on the importance of the GC–KLF15 axis, we postulate that GR and KLF15 may enrich at common loci genome-wide and functionally interact to fine tune GC-dependent transcriptional responses.
In light of our current observations pertaining to GR, we note that several other members of the nuclear receptor superfamily and associated coregulators have been shown to be potent effectors of adaptive muscle metabolism and ergogenic physiology (55–58). Activation of some of these transcriptional regulators, such as ERRγ (59), the androgen receptor (60), PPARδ (61), and the PGC-1 coactivators (62, 63), have also been shown to improve pathology in animal models of DMD. These observations support the contention that gene regulatory pathways such as the GR–KLF15 axis ameliorate DMD via modulation of muscle metabolic programming and add to an emerging body of evidence that defines intimate functional relationships between KLF family members and nuclear receptors (23, 24, 64–66).
Our data also provide potentially important insights into one of the most commonly prescribed classes of drugs. GCs have been classically associated with muscle wasting and weakness, a side effect of excessive or sustained exposure to pharmacologic GR agonists that is mechanistically linked to direct transactivation of atrogenes (e.g., Myostatin, Foxo3a, Atrogin1, Trim63/Murf1) (2). This has fueled the pervasive view that the beneficial therapeutic effects of GCs largely arise from their transrepressive function (e.g., inhibition of NFκB signaling), whereas GR-dependent gene transactivation is principally responsible for side effects (1, 2), a view that has been the basis for large-scale drug development programs to improve GC-based pharmacotherapy. The current study, however, establishes that transient or moderate exposure to GCs improves muscle performance and ameliorates DMD via direct transactivation of a potent downstream metabolic effector (KLF15), establishing that GR-dependent transactivation can mediate important therapeutic effects in certain settings. Furthermore, our transgenic experiments provide proof-of-principle that muscle-specific activation of KLF15 can ameliorate DMD in a GC-independent manner, suggesting a previously unidentified strategy for development of steroid-sparing therapeutics for DMD and other myopathic diseases.
Materials and Methods
Mouse Models.
All protocols concerning animal use were approved by the Institutional Animal Care and Use Committee at Case Western Reserve University and conducted in strict accordance with the NIH Guide for the Care and Use of Laboratory Animals (67). Mice were housed in a temperature and humidity-controlled barrier facility with a 12-h light/dark cycle and ad libitum access to water and standard laboratory rodent chow. Dex studies in congenic WT mice were performed with age- and sex-matched littermate controls (12 wk old, male, pure C57BL/6 background). Klf15−/− mice in the C57BL/6 background have been previously described (25). KLF15 MTg mice were generated by cloning a full-length mouse KLF15 cDNA downstream of the MCK (–4.8 kb) promoter/enhancer with a 3′ hGH-polyA sequence (68). The 3.2 kb backbone (pBS II SK+) was released via digestion with XhoI and NotI. The linearized transgene was injected into pure C57BL/6 ES cells via the Case Western Reserve University Transgenic and Targeting Core. F0 offspring were screened for transgene expression, and germ-line transmission was established in several lines. After screening several lines, we focused studies on a line with fivefold KLF15 overexpression that was highly skeletal muscle-specific and lacking any elevation of total KLF15 in the heart. The mdx mice in the C57BL/10-ScSn background and C57BL/10-ScSn controls were purchased from Jackson Laboratory (cat. no. 001801 and 000476). To generate KLF15-deficient mice harboring the mutant mdx allele, we first crossed Klf15−/− male mice with homozygous mdx female mice to generate the F1 generation (i.e., male Klf15+/− mice homozygous for the mdx allele and female Klf15+/− mice heterozygous for the mdx allele). F1 intercrosses produced both male and female mice that were Klf15+/− and homozygous for the mdx allele (F2 generation). All subsequent study mice were generated via intercrosses of littermate males and females that were Klf15+/− and homozygous for the mdx allele. All data were generated using F3–F6 generations and strict littermate controls to control for the mixed background of the parental strains. To generate KLF15 MTg mice harboring the mdx allele, male MTg mice harboring a single copy of the KLF15 transgene were mated to female mice homozygous for the mdx allele. We studied male offspring of this cross, which were homozygous for the mdx allele and either carried the muscle transgene or were nontransgenic littermate controls. Each cohort of study mice were bred from the original parental strains, and all studies were performed using strict littermate controls. Genotyping for the mdx allele was performed using endpoint PCR as detailed on the Jackson Laboratories website (jaxmice.jax.org/strain/001801.html).
Statistical Analysis.
All pooled results are expressed as means, and error bars depict SEM. Statistical analysis to detect for genotype effect on energy flux during exercise was performed using ANOVA (69). Statistical analyses to probe for effects of GC administration or genotype were performed using two-way ANOVA followed by a Bonferroni posttest. Survival analysis of KLF15-deficient mice in the mdx background was performed on a total of 157 mice, and statistical analysis was performed using a χ2 test with the expected number of mice determined from predicted Mendelian inheritance ratios of the targeted Klf15 allele and the X-linked mutant mdx allele. For experiments comparing means of two independent and normally distributed datasets, two-tailed Student’s t tests for unpaired data were used. Statistical significance was defined as P < 0.05. Statistical analysis of the human dex study, microarrays, and GSEA are described in SI Materials and Methods.
Human Skeletal Muscle Biopsies.
Skeletal muscle samples were obtained from five healthy male subjects as previously described (70) in accordance with the Declaration of Helsinki and was approved by the Ethics Committee and Institutional Review Board of Copenhagen and Frederiksberg communities. For further experimental details of human tissue studies, see SI Materials and Methods.
SI Materials and Methods
Human Skeletal Muscle Samples Pre- and Post-Dex.
Skeletal muscle samples were obtained from five healthy male subjects as previously described (70) in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of Copenhagen and Frederiksberg communities. The subjects ingested 2 mg of dex by mouth at 9:00 AM and at 8:00 PM for 5 d. Before the treatment and ∼11 h after the last ingestion of dex, a muscle biopsy was obtained from vastus lateralis. On the day before the biopsies, the subjects consumed a standardized meal in the evening and refrained from physical activity. On the following day, the subjects consumed a standardized breakfast and reported to the laboratory between 8:00 AM and 10:00 AM. The two biopsy days were separated by exactly 2 wk. Muscle biopsies were obtained under local anesthesia using a Bergström needle with suction. Visible blood, fat, and connective tissue were immediately removed, and the biopsy was frozen in liquid N2 no more than 30 s after sampling. RNA and cDNA preparation was as described in subsequent sections. Validated TaqMan qRT-PCR assays for human KLF15, FKBP5, and RPLP0 (36B4) were purchased from Applied Biosystems. Relative expression was calculated using the ΔΔCt method. Statistical analysis for dex-dependent effects was determined using a two-tailed, paired, Student’s t test, with statistical significance defined as P < 0.05.
Human Skeletal Muscle Samples from DMD Patients.
Human muscle specimens were sourced from the Telethon Network of Genetic Biobanks (Neuromuscular Tissue Bank) as previously described (71). Biopsies were taken from the vastus lateralis muscles of patients with DMD (aged 1–9 y) or healthy control subjects (aged 18–25 y) using either a Bergström or open biopsy technique. For the healthy control subjects, informed consent was obtained, and the procedures were approved by the Deakin University Human Ethics Committee. All biopsies were frozen immediately in liquid nitrogen and stored at −80 °C until analysis. RNA extraction, cDNA synthesis, and qRT-PCR for human KLF15 and RPLP0 are as above.
Cell Culture.
C2C12 myoblasts were purchased from ATCC and grown in DMEM supplemented with 10% (vol/vol) FBS. C2C12 myoblasts were passaged before reaching 60% confluence to maintain cells in the undifferentiated state. All studies used low-passage C2C12 (passages 3–6). C2C12 differentiation was induced at 80% confluence by switching media to DMEM supplemented with 2% (vol/vol) horse serum. Differentiation medium was changed every 2 d. Studies were performed on mature myotubes at differentiation day 6 in fresh differentiation medium containing 2% (vol/vol) horse serum. Primary normal human skeletal myoblasts were purchased from Invitrogen, Life Technologies (cat. no. A12555) and were differentiated per the manufacturer’s instructions. Stimulation with 100 nM dex was performed after 48 h of incubation in differentiation medium.
ChIP.
ChIP was performed from mouse quadriceps as previously described (53). For each experiment, bilateral quadriceps muscles from a single mouse (∼300 mg tissue) were minced, crushed, and fixed with 1.1% formaldehyde for 10 min, followed by chromatin preparation and sonication using a BioRuptor (Diagnode). The sonicated chromatin was immunoprecipitated using anti-GR polyclonal antibodies (Santa Cruz, cat. no. sc-1002) bound to Dynabeads (Invitrogen) followed by extensive washing and elution. Chromatin was then reverse cross-linked followed by purification of genomic DNA. Target and nontarget regions of genomic DNA were amplified by qPCR in both the immunoprecipitates and input samples. Relative abundance in immunoprecipitates was expressed as percentage of abundance in input samples as previously described (72). Mouse 28S locus was used as a nontarget control (53). ChIP primer sequences were as follows: mouse Klf15 GRE target region, forward 5′-cgtgatttgcacgctgac-3′ and reverse 5′-caggcctgctgtttatcctc-3′; mouse nontarget region, forward 5′-ctgggtataggggcgaaagac-3′ and reverse 5′-ggccccaagacctctaatcat-3′.
RNA Extraction and RT-qPCR.
Tissue samples were disrupted/homogenized in Qiazol (Qiagen) in a Tissue-lyzer (Qiagen) using stainless steel beads (30 Hz for a total of 4 min). An initial extraction of the aqueous phase was performed with chloroform. Subsequently, RNA was purified from the aqueous phase using the BioRAD Aurum kit with on-column DNase treatment (cat. no. 732–6820). Reverse transcription and synthesis of cDNA was performed using the iScript kit (Bio-Rad, cat. no. 170–8841) following the manufacturer’s instructions. qRT-PCR was performed with the TaqMan method (using the Roche Universal Probe Library System) on an Roche Light Cycler Real-Time PCR System. Relative expression was calculated using the ΔΔCt method with normalization to constitutive genes as follows: All human samples were normalized to RPLP0 (36B4), C2C12 samples and WT mouse samples were normalized to Cyclophilin-B (Ppib), and mdx samples were normalized to Actb. Specific primer/probe sequences are available on request.
Microarray Studies in Klf15−/− Mice.
Quadriceps RNA was isolated from WT and KO mice 8 h after treatment with dex versus vehicle (n = 4 per group). Sample preparation, labeling, and array hybridizations were performed according to standard protocols from the University of California San Francisco Shared Microarray Core Facilities and Agilent Technologies (www.arrays.ucsf.edu and www.agilent.com). Samples were collected directly into lysis buffer, and amplified cDNA was made using the WT-Ovation one-direct amplification system kits following the manufacturer’s protocol (NuGen Technologies, Inc.). Cy3-CTP labeling was performed using NimbleGen one-color labeling kits (Roche-NimbleGen Inc.). The size distribution and quantity of amplified and labeled products were assessed using the Agilent 2100 Bioanalyzer and the Nanodrop ND-8000 (Nanodrop Technologies, Inc.), respectively. Equal amounts (2 µg) of the Cy3-labeled target were hybridized to Agilent whole mouse genome 4 × 44K ink-jet arrays for 14 h, according to the manufacturer’s protocol. Hybridizations were performed for 14 h, according to the manufacturer’s protocol. Arrays were scanned using the Agilent microarray scanner, and raw signal intensities were extracted with Feature Extraction v10.6 software. The dataset was normalized using the quantile normalization method (73). No background subtraction was performed, and the median feature pixel intensity was used as the raw signal before normalization. A one-way ANOVA linear model was used to fit to the comparisons and to estimate the FDR and adjusted P value (FWER) for each gene. All procedures were carried out using functions in the R package limma in Bioconductor (74, 75). Pathway-based categorization of transcripts was performed using DAVID as previously described (76).
Microarray Studies in KLF15 MTg Mice.
Quadriceps RNA from adult MTg and non-Tg mice (n = 4) was prepared as described above. Sample preparation, labeling, and array hybridizations were performed according to standard protocols from the Dana Farber Cancer Institute Gene Expression Core. Briefly, cDNA was generated using the Ambion WT Expression Kit and labeled using the Affymetrix GeneChip WT Terminal Labeling and Hybridization protocol. The labeled, fragmented DNA was hybridized to Mouse 430A 2.0 arrays (Affymetrix) for 16–18 h in a GeneChip Hybridization oven 645 (Affymetrix, Inc.) at 45 °C with rotation (60 rpm). The hybridized samples were washed and stained using an Affymetrix fluidics station 450. After staining, the genechip arrays were immediately scanned using an Affymetrix GeneArray Scanner 3000 7G Plus. Bioconductor (bioconductor.org) packages affyQCReport and affy were used to perform quality control and normalization of arrays. Custom BrainArray Entrez Gene probe set annotations (brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/19.0.0/entrezg.asp) were used to summarize expression by Robust Multichip Average (77). Probe sets called “present” in at least one sample by affy mas5call and having an interquartile range > 10% were examined for differential expression. Differential expression was assessed as for the KO arrays. Genes with a fold change > 1.2 and FDR < 0.05 were retained for further analysis. Comparisons between KO, MTg, and published microarray studies (GSE52766) (44) were done on the basis of Entrez Gene IDs after collapsing multiple probe sets for a given gene, if necessary, based on maximal interquartile range. To that end, all annotations were updated to common gene IDs using the National Center for Biotechnology Information’s gene2accession database (retrieved January 11, 2015).
GSEA.
Gene sets of differentially expressed genes were compiled, again using gene IDs as identifiers. GSEA software (version 2.07, www.broad.mit.edu/GSEA) was used to determine the significance of correlation between gene sets and expression. The signal-to-noise ratio of linear (not log2) data were used as the metric with gene set permutation. Microarray data complying with minimum information about a microarray experiment (MIAME) annotation standards have been deposited in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) under accession no. GSE74625.
Mouse Treadmill Exercise.
Mice were run on a motorized, speed-controlled, modular treadmill system (Columbus Instruments) equipped with an electric shock stimulus (set at 1 Hz, 20% output) and adjustable inclination angle. To assess the ergogenic response to dex administration in WT mice, males (12 wk age) were pretreated with a single dose of dex (2 mg/kg) or vehicle 18 h before exercise. This GC dose has been previously used to augment aerobic exercise endurance in rodents (9). Mice were acclimatized to the treadmill for 10 min at 8 m/min for 3 consecutive days before an experiment. After a warm-up period (8 m/min × 10 min, 0% incline), mice were run at a fixed speed/incline (16 m/min, 0%) until exhaustion. To assess the ergogenic effect of KLF15 overexpression in non-mdx mice, 10-wk-old MTg and non-Tg control mice were acclimatized to the treadmill for 10 min at 8 m/min for 3 consecutive days before an experiment. After a warm-up period of 10 m/min × 10 min, the belt was increased to 12 m/min × 2 min, 14 m/min × 48 min, 15 m/min × 30 min, and finally 16 m/min (all 0% incline) until exhaustion was reached. When assessing endurance exercise capacity in mdx mice harboring KLF15 gain- or loss-of-function, 9-wk-old males were slowly acclimatized to the treadmill at a 20° downhill angle for 10 min 3 consecutive days before the experiment at 3 m/min × 5 min and increasing to 5 m/min × 2 min, 7 m/min × 2 min, and 8 m/min × 1 min. When assessing time to exhaustion, mdx mice were run at a 20° downhill angle at 3 m/min × 5 min, 5 m/min × 2 min, 7 m/min × 2 min, 8 m/min × 1 min, 9 m/min × 2 min, 10 m/min × 10 min, 12 m/min × 20 min, 13 m/min × 1 min, 14 m/min × 30 min, and finally 16 m/min until exhaustion was reached. Exhaustion was defined as the time point when mice were unable to avoid repetitive electrical shocks for 5 continuous seconds as previously described (78).
Passive Wire Hang and Grip Strength.
For passive wire-hang studies, mice were placed on the cage top, which was then inverted and suspended above the home cage. The latency to when the animal falls was recorded. Latency to fall was calculated as an average of three consecutive trials per mouse, as previously described (23). For grip-strength studies, the mouse’s forelimbs were placed on a tension bar while the mouse was restrained by the base of the tail. The mouse was gently pulled back until it lost its grip from the bar. The maximal force generated was measured in grams of resistance by a grip strength meter (Columbus Instruments) and was repeated five consecutive times per mouse. Grip strength was calculated by taking the average of the three highest measurements and was normalized to body weight, as previously described (79). For studies in the non-mdx background, these assays were performed at 10 wk of age. For studies of mice harboring KLF15 gain- or loss-of-function in the mdx background, these assays were performed at 6–8 wk of age.
GC-Induced Muscle Atrophy.
WT and Klf15−/− mice were treated with dex (1 mg·kg·d s.c. × 10 d) versus an equivalent volume of vehicle (PBS) at 4:00 PM each day as previously described (29, 30). All mice were provided access to standard chow and water ad libitum. Measurement of body weights and harvesting of all tissue were performed 24 h after drug/vehicle administration.
Histological Analysis.
Mouse skeletal muscles were either fixed in 10% formalin and embedded in paraffin for H&E staining or frozen in optimal cutting tissue compound (OCT). Myocyte cross-sectional area was determined by staining sections with rhodamine-conjugated WGA (Vector Laboratories RL-1022). To evaluate muscle fiber cross-sectional area, 50 fibers were randomly chosen per muscle section, and cross-sectional area was measured using NIH ImageJ.
Measurement of Exercise Respiratory Exchange Ratio in Mice.
For exercise energy-expenditure experiments (performed at NIH MMPC at Vanderbilt University), MTg and non-Tg (n = 7–10) mice were studied in an enclosed treadmill attached to the Comprehensive Laboratory Animal Monitoring System (Columbus Instruments), as previously described (80). After an acclimatization period, the running protocol began at 10 m/min at 0% grade and increased by 4 m/min increments every 3 min to a maximum of 34 m/min. Data analysis was performed using Oxymax software (Columbus Instruments).
Mouse Prednisolone Studies.
Prednisolone was administered to male mice as previously described (49) with a dosing regimen similar to that used in DMD patients. Briefly, prednisolone syrup was administered via oral gavage at 5 mg/kg per dose, given three times per week. Treatment was initiated at the time of weaning (age 3 wk) and continued until the end of the experimental protocol (age 14 wk).
Plasma CPK Activity in Mice.
Blood was harvested from the inferior vena cava, placed into heparin-coated collection tubes, and plasma was prepared by centrifugation. Plasma CPK activity was detected by the University Hospitals Case Medical Center Clinical Laboratory Core.
Evans Blue Dye Analysis.
Evans blue dye was administered to male mdx mice as previously described (63). Briefly, mice were injected with a 1% solution of Evans blue dye (in PBS) at a dose of 1% volume to gram body weight 16 h before harvesting. Muscles were isolated and frozen in OCT. Muscles were analyzed for dye extravasation by fluorescence microscopy.
Supplementary Material
Acknowledgments
We thank Dr. Louise Lantier and the Vanderbilt Mouse Metabolic Phenotyping Center for assistance with mouse metabolic exercise studies and Sarah McMahon for assistance with Gene Expression Omnibus datasets. This work was supported by The Hartwell Foundation and NIH Grants R01 DK093821 and R01 HL127240 (to S.M.H.), T32 HL105338 (to A.M.-N.), R01 HL119195 (to M.K.J.), and R01 HL109557 (to A.N.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE74625).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512968112/-/DCSupplemental.
References
- 1.Patel R, Williams-Dautovich J, Cummins CL. Minireview: New molecular mediators of glucocorticoid receptor activity in metabolic tissues. Mol Endocrinol. 2014;28(7):999–1011. doi: 10.1210/me.2014-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schakman O, Gilson H, Kalista S, Thissen JP. Mechanisms of muscle atrophy induced by glucocorticoids. Horm Res. 2009;72(Suppl 1):36–41. doi: 10.1159/000229762. [DOI] [PubMed] [Google Scholar]
- 3.Heron WT, Hales WM, Ingle DJ. Capacity of skeletal muscle in rats to maintain work output. Am J Physiol. 1934;110(2):357–361. [Google Scholar]
- 4.Visscher MB. Dwight Joyce Ingle: September 4, 1907-July 28, 1978. Biogr Mem Natl Acad Sci. 1992;61:247–268. [PubMed] [Google Scholar]
- 5.Arlettaz A, et al. Effects of acute prednisolone intake on substrate utilization during submaximal exercise. Int J Sports Med. 2008;29(1):21–26. doi: 10.1055/s-2007-964994. [DOI] [PubMed] [Google Scholar]
- 6.Arlettaz A, et al. Effects of short-term prednisolone intake during submaximal exercise. Med Sci Sports Exerc. 2007;39(9):1672–1678. doi: 10.1249/mss.0b013e3180dc992c. [DOI] [PubMed] [Google Scholar]
- 7.Casuso RA, Melskens L, Bruhn T, Secher NH, Nordsborg NB. Glucocorticoids improve high-intensity exercise performance in humans. Eur J Appl Physiol. 2014;114(2):419–424. doi: 10.1007/s00421-013-2784-7. [DOI] [PubMed] [Google Scholar]
- 8.Eagle E, Britton S, Kline R. The influence of cortico-adrenal extract on energy output. Am J Physiol. 1932;102(3):707–713. doi: 10.1126/science.75.1938.221. [DOI] [PubMed] [Google Scholar]
- 9.Gorostiaga EM, Czerwinski SM, Hickson RC. Acute glucocorticoid effects on glycogen utilization, O2 uptake, and endurance. J Appl Physiol (1985) 1988;64(3):1098–1106. doi: 10.1152/jappl.1988.64.3.1098. [DOI] [PubMed] [Google Scholar]
- 10.Ingle D. The time for the work capacity of the adrenalectomized rats treated with cortin. Am J Physiol. 1934;116:622–625. [Google Scholar]
- 11.Ingle DJ, Morley EH, Nezamis JE. The work performance of normal rats given continuous intravenous injections of cortisone and of corticotropin. Endocrinology. 1952;51(6):487–491. doi: 10.1210/endo-51-6-487. [DOI] [PubMed] [Google Scholar]
- 12.Jakobi JM, Killinger DW, Wolfe BM, Mahon JL, Rice CL. Quadriceps muscle function and fatigue in women with Addison’s disease. Muscle Nerve. 2001;24(8):1040–1049. doi: 10.1002/mus.1108. [DOI] [PubMed] [Google Scholar]
- 13.Tharp GD. The role of glucocorticoids in exercise. Med Sci Sports. 1975;7(1):6–11. [PubMed] [Google Scholar]
- 14.Duclos M. Evidence on ergogenic action of glucocorticoids as a doping agent risk. Phys Sportsmed. 2010;38(3):121–127. doi: 10.3810/psm.2010.10.1817. [DOI] [PubMed] [Google Scholar]
- 15.Angelini C. The role of corticosteroids in muscular dystrophy: A critical appraisal. Muscle Nerve. 2007;36(4):424–435. doi: 10.1002/mus.20812. [DOI] [PubMed] [Google Scholar]
- 16.Drachman DB, Toyka KV, Myer E. Prednisone in Duchenne muscular dystrophy. Lancet. 1974;2(7894):1409–1412. doi: 10.1016/s0140-6736(74)90071-3. [DOI] [PubMed] [Google Scholar]
- 17.Mendell JR, et al. Randomized, double-blind six-month trial of prednisone in Duchenne’s muscular dystrophy. N Engl J Med. 1989;320(24):1592–1597. doi: 10.1056/NEJM198906153202405. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51(6):919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 19.Golumbek PT, Keeling RM, Connolly AM. Strength and corticosteroid responsiveness of mdx mice is unchanged by RAG2 gene knockout. Neuromuscul Disord. 2007;17(5):376–384. doi: 10.1016/j.nmd.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Malik V, Rodino-Klapac LR, Mendell JR. Emerging drugs for Duchenne muscular dystrophy. Expert Opin Emerg Drugs. 2012;17(2):261–277. doi: 10.1517/14728214.2012.691965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuno K, et al. Expression profiling identifies Klf15 as a glucocorticoid target that regulates airway hyperresponsiveness. Am J Respir Cell Mol Biol. 2011;45(3):642–649. doi: 10.1165/rcmb.2010-0369OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimizu N, et al. Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell Metab. 2011;13(2):170–182. doi: 10.1016/j.cmet.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Haldar SM, et al. Kruppel-like factor 15 regulates skeletal muscle lipid flux and exercise adaptation. Proc Natl Acad Sci USA. 2012;109(17):6739–6744. doi: 10.1073/pnas.1121060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prosdocimo DA, et al. Kruppel-like factor 15 is a critical regulator of cardiac lipid metabolism. J Biol Chem. 2014;289(9):5914–5924. doi: 10.1074/jbc.M113.531384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gray S, et al. The Krüppel-like factor KLF15 regulates the insulin-sensitive glucose transporter GLUT4. J Biol Chem. 2002;277(37):34322–34328. doi: 10.1074/jbc.M201304200. [DOI] [PubMed] [Google Scholar]
- 26.Jeyaraj D, et al. Klf15 orchestrates circadian nitrogen homeostasis. Cell Metab. 2012;15(3):311–323. doi: 10.1016/j.cmet.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sasse SK, et al. The glucocorticoid receptor and KLF15 regulate gene expression dynamics and integrate signals through feed-forward circuitry. Mol Cell Biol. 2013;33(11):2104–2115. doi: 10.1128/MCB.01474-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asada M, et al. DNA binding-dependent glucocorticoid receptor activity promotes adipogenesis via Krüppel-like factor 15 gene expression. Lab Invest. 2011;91(2):203–215. doi: 10.1038/labinvest.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clarke BA, et al. The E3 Ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 2007;6(5):376–385. doi: 10.1016/j.cmet.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 30.Gilson H, et al. Myostatin gene deletion prevents glucocorticoid-induced muscle atrophy. Endocrinology. 2007;148(1):452–460. doi: 10.1210/en.2006-0539. [DOI] [PubMed] [Google Scholar]
- 31.Fisher I, et al. Prednisolone-induced changes in dystrophic skeletal muscle. FASEB J. 2005;19(7):834–836. doi: 10.1096/fj.04-2511fje. [DOI] [PubMed] [Google Scholar]
- 32.Griggs RC, et al. Duchenne dystrophy: Randomized, controlled trial of prednisone (18 months) and azathioprine (12 months) Neurology. 1993;43(3 Pt 1):520–527. doi: 10.1212/wnl.43.3_part_1.520. [DOI] [PubMed] [Google Scholar]
- 33.Kirschner J, et al. Treatment of Duchenne muscular dystrophy with ciclosporin A: A randomised, double-blind, placebo-controlled multicentre trial. Lancet Neurol. 2010;9(11):1053–1059. doi: 10.1016/S1474-4422(10)70196-4. [DOI] [PubMed] [Google Scholar]
- 34.Kissel JT, et al. Mononuclear cell analysis of muscle biopsies in prednisone- and azathioprine-treated Duchenne muscular dystrophy. Neurology. 1993;43(3 Pt 1):532–536. doi: 10.1212/wnl.43.3_part_1.532. [DOI] [PubMed] [Google Scholar]
- 35.Sicinski P, et al. The molecular basis of muscular dystrophy in the mdx mouse: A point mutation. Science. 1989;244(4912):1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- 36.Haslett JN, et al. Gene expression profiling of Duchenne muscular dystrophy skeletal muscle. Neurogenetics. 2003;4(4):163–171. doi: 10.1007/s10048-003-0148-x. [DOI] [PubMed] [Google Scholar]
- 37.Sharma U, Atri S, Sharma MC, Sarkar C, Jagannathan NR. Skeletal muscle metabolism in Duchenne muscular dystrophy (DMD): An in-vitro proton NMR spectroscopy study. Magn Reson Imaging. 2003;21(2):145–153. doi: 10.1016/s0730-725x(02)00646-x. [DOI] [PubMed] [Google Scholar]
- 38.Barbiroli B, Funicello R, Ferlini A, Montagna P, Zaniol P. Muscle energy metabolism in female DMD/BMD carriers: A 31P-MR spectroscopy study. Muscle Nerve. 1992;15(3):344–348. doi: 10.1002/mus.880150313. [DOI] [PubMed] [Google Scholar]
- 39.Lott DJ, et al. Assessment of intramuscular lipid and metabolites of the lower leg using magnetic resonance spectroscopy in boys with Duchenne muscular dystrophy. Neuromuscul Disord. 2014;24(7):574–582. doi: 10.1016/j.nmd.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishio H, et al. Glucose, free fatty acid and ketone body metabolism in Duchenne muscular dystrophy. Brain Dev. 1990;12(4):390–402. doi: 10.1016/s0387-7604(12)80071-4. [DOI] [PubMed] [Google Scholar]
- 41.Chen YW, Zhao P, Borup R, Hoffman EP. Expression profiling in the muscular dystrophies: Identification of novel aspects of molecular pathophysiology. J Cell Biol. 2000;151(6):1321–1336. doi: 10.1083/jcb.151.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunn JF, Tracey I, Radda GK. Exercise metabolism in Duchenne muscular dystrophy: A biochemical and [31P]-nuclear magnetic resonance study of mdx mice. Proc Biol Sci. 1993;251(1332):201–206. doi: 10.1098/rspb.1993.0030. [DOI] [PubMed] [Google Scholar]
- 43.Even PC, Decrouy A, Chinet A. Defective regulation of energy metabolism in mdx-mouse skeletal muscles. Biochem J. 1994;304(Pt 2):649–654. doi: 10.1042/bj3040649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kainulainen H, et al. Myostatin/activin blocking combined with exercise reconditions skeletal muscle expression profile of mdx mice. Mol Cell Endocrinol. 2015;399:131–142. doi: 10.1016/j.mce.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 45.Kuznetsov AV, et al. Impaired mitochondrial oxidative phosphorylation in skeletal muscle of the dystrophin-deficient mdx mouse. Mol Cell Biochem. 1998;183(1-2):87–96. doi: 10.1023/a:1006868130002. [DOI] [PubMed] [Google Scholar]
- 46.Porter JD, et al. A chronic inflammatory response dominates the skeletal muscle molecular signature in dystrophin-deficient mdx mice. Hum Mol Genet. 2002;11(3):263–272. doi: 10.1093/hmg/11.3.263. [DOI] [PubMed] [Google Scholar]
- 47.Rayavarapu S, et al. Identification of disease specific pathways using in vivo SILAC proteomics in dystrophin deficient mdx mouse. Mol Cell Proteomics. 2013;12(5):1061–1073. doi: 10.1074/mcp.M112.023127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grounds MD, Radley HG, Lynch GS, Nagaraju K, De Luca A. Towards developing standard operating procedures for pre-clinical testing in the mdx mouse model of Duchenne muscular dystrophy. Neurobiol Dis. 2008;31(1):1–19. doi: 10.1016/j.nbd.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keeling RM, Golumbek PT, Streif EM, Connolly AM. Weekly oral prednisolone improves survival and strength in male mdx mice. Muscle Nerve. 2007;35(1):43–48. doi: 10.1002/mus.20646. [DOI] [PubMed] [Google Scholar]
- 50.Grady RM, et al. Skeletal and cardiac myopathies in mice lacking utrophin and dystrophin: A model for Duchenne muscular dystrophy. Cell. 1997;90(4):729–738. doi: 10.1016/s0092-8674(00)80533-4. [DOI] [PubMed] [Google Scholar]
- 51.Glyn J. The discovery and early use of cortisone. J R Soc Med. 1998;91(10):513–517. doi: 10.1177/014107689809101004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gray S, et al. Regulation of gluconeogenesis by Krüppel-like factor 15. Cell Metab. 2007;5(4):305–312. doi: 10.1016/j.cmet.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phuc Le P, et al. Glucocorticoid receptor-dependent gene regulatory networks. PLoS Genet. 2005;1(2):e16. doi: 10.1371/journal.pgen.0010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuo T, et al. Genome-wide analysis of glucocorticoid receptor-binding sites in myotubes identifies gene networks modulating insulin signaling. Proc Natl Acad Sci USA. 2012;109(28):11160–11165. doi: 10.1073/pnas.1111334109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Narkar VA, et al. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134(3):405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Narkar VA, et al. Exercise and PGC-1α-independent synchronization of type I muscle metabolism and vasculature by ERRγ. Cell Metab. 2011;13(3):283–293. doi: 10.1016/j.cmet.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Handschin C, Spiegelman BM. The role of exercise and PGC1alpha in inflammation and chronic disease. Nature. 2008;454(7203):463–469. doi: 10.1038/nature07206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamamoto H, et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell. 2011;147(4):827–839. doi: 10.1016/j.cell.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matsakas A, Yadav V, Lorca S, Narkar V. Muscle ERRγ mitigates Duchenne muscular dystrophy via metabolic and angiogenic reprogramming. FASEB J. 2013;27(10):4004–4016. doi: 10.1096/fj.13-228296. [DOI] [PubMed] [Google Scholar]
- 60.Cozzoli A, et al. GLPG0492, a novel selective androgen receptor modulator, improves muscle performance in the exercised-mdx mouse model of muscular dystrophy. Pharmacol Res. 2013;72:9–24. doi: 10.1016/j.phrs.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 61.Miura P, et al. Pharmacological activation of PPARbeta/delta stimulates utrophin A expression in skeletal muscle fibers and restores sarcolemmal integrity in mature mdx mice. Hum Mol Genet. 2009;18(23):4640–4649. doi: 10.1093/hmg/ddp431. [DOI] [PubMed] [Google Scholar]
- 62.Handschin C, et al. PGC-1alpha regulates the neuromuscular junction program and ameliorates Duchenne muscular dystrophy. Genes Dev. 2007;21(7):770–783. doi: 10.1101/gad.1525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chan MC, et al. Post-natal induction of PGC-1α protects against severe muscle dystrophy independently of utrophin. Skelet Muscle. 2014;4(1):2. doi: 10.1186/2044-5040-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prosdocimo DA, et al. KLF15 and PPARα cooperate to regulate cardiomyocyte lipid gene expression and oxidation. PPAR Res. 2015;2015:201625. doi: 10.1155/2015/201625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liao X, et al. Kruppel-like factor 4 is critical for transcriptional control of cardiac mitochondrial homeostasis. J Clin Invest. 2015;125(9):3461–3476. doi: 10.1172/JCI79964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oishi Y, et al. SUMOylation of Krüppel-like transcription factor 5 acts as a molecular switch in transcriptional programs of lipid metabolism involving PPAR-delta. Nat Med. 2008;14(6):656–666. doi: 10.1038/nm1756. [DOI] [PubMed] [Google Scholar]
- 67.National Research Council (2011) Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC) 8th Ed. [Google Scholar]
- 68.Akimoto T, et al. Exercise stimulates Pgc-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. J Biol Chem. 2005;280(20):19587–19593. doi: 10.1074/jbc.M408862200. [DOI] [PubMed] [Google Scholar]
- 69.Laird NM, Ware JH. Random-effects models for longitudinal data. Biometrics. 1982;38(4):963–974. [PubMed] [Google Scholar]
- 70.Nordsborg N, Goodmann C, McKenna MJ, Bangsbo J. Dexamethasone up-regulates skeletal muscle maximal Na+,K+ pump activity by muscle group specific mechanisms in humans. J Physiol. 2005;567(Pt 2):583–589. doi: 10.1113/jphysiol.2005.087031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Church JE, et al. Alterations in Notch signalling in skeletal muscles from mdx and dko dystrophic mice and patients with Duchenne muscular dystrophy. Exp Physiol. 2014;99(4):675–687. doi: 10.1113/expphysiol.2013.077255. [DOI] [PubMed] [Google Scholar]
- 72.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38(3):369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 73.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 74.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 76.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 77.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 78.Wang YX, et al. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2004;2(10):e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Connolly AM, Keeling RM, Mehta S, Pestronk A, Sanes JR. Three mouse models of muscular dystrophy: The natural history of strength and fatigue in dystrophin-, dystrophin/utrophin-, and laminin alpha2-deficient mice. Neuromuscul Disord. 2001;11(8):703–712. doi: 10.1016/s0960-8966(01)00232-2. [DOI] [PubMed] [Google Scholar]
- 80.Calvo JA, et al. Muscle-specific expression of PPARgamma coactivator-1alpha improves exercise performance and increases peak oxygen uptake. J Appl Physiol (1985) 2008;104(5):1304–1312. doi: 10.1152/japplphysiol.01231.2007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.