Significance
Using an approach derived from statistical physics, we quantify transcriptome-wide motif usage in human and murine noncoding RNAs (ncRNAs), determining that most have motif usage consistent with the coding genome. However, an outlier subset of tumor-associated ncRNAs comprises repetitive elements whose motif usage patterns are more typically associated with the genomes of inflammatory pathogens. We demonstrate that a key subset of these elements directly activates the cellular innate immune response. We propose that the innate response in tumors partially originates from direct interaction of immunogenic ncRNAs preferentially expressed in cancer cells with innate pattern recognition receptors.
Keywords: noncoding RNA, genome evolution, cancer immunology
Abstract
Recent studies have demonstrated abundant transcription of a set of noncoding RNAs (ncRNAs) preferentially within tumors as opposed to normal tissue. Using an approach from statistical physics, we quantify global transcriptome-wide motif use for the first time, to our knowledge, in human and murine ncRNAs, determining that most have motif use consistent with the coding genome. However, an outlier subset of tumor-associated ncRNAs, typically of recent evolutionary origin, has motif use that is often indicative of pathogen-associated RNA. For instance, we show that the tumor-associated human repeat human satellite repeat II (HSATII) is enriched in motifs containing CpG dinucleotides in AU-rich contexts that most of the human genome and human adapted viruses have evolved to avoid. We demonstrate that a key subset of these ncRNAs functions as immunostimulatory “self-agonists” and directly activates cells of the mononuclear phagocytic system to produce proinflammatory cytokines. These ncRNAs arise from endogenous repetitive elements that are normally silenced, yet are often very highly expressed in cancers. We propose that the innate response in tumors may partially originate from direct interaction of immunogenic ncRNAs expressed in cancer cells with innate pattern recognition receptors, and thereby assign a previously unidentified danger-associated function to a set of dark matter repetitive elements. These findings potentially reconcile several observations concerning the role of ncRNA expression in cancers and their relationship to the tumor microenvironment.
The recent development of total RNA sequencing has allowed a better appreciation of the complexity and breadth of the entire transcriptome (1–4). Analysis by the Encyclopedia of DNA Elements (ENCODE) consortium unexpectedly showed that far more of the mammalian genome than previously appreciated is transcribed into noncoding RNA (ncRNA). Several short ncRNAs have conserved metabolic and regulatory functions, and some antiviral properties have been assigned to novel ncRNA classes, such as eukaryotic siRNA, piRNA (PIWI-interacting) RNA, and prokaryotic CRISPR (clustered regularly interspaced short palindromic repeats) RNA (5). In eukaryotes, long noncoding RNA (lncRNA), such as long-intergenic ncRNA, has been associated with transcriptional, posttranscriptional, and epigenetic regulation (6, 7).
It is now evident that germ-line and cancer cells can have atypical ncRNA transcription, including repetitive elements from regions usually silenced in steady state (8, 9). In eukaryotes, transcription of endogenous retroviruses and mobile elements is mostly repressed epigenetically through processes such as histone modification and DNA methylation, preventing disruptive or deregulatory effects due to integration into coding regions. In mammals, DNA methylation targets the cytosine in CpG motifs to form 5-methylcytosine contributing to down-regulation of transcription for methylated sequences (10). Epigenetic regulation is strongly associated with the developmental process, whereas its deregulation, such as by disruption of DNA methylation, can be associated with dedifferentiation and carcinogenic processes (11, 12).
In cancers, such as those cancers driven by p53 mutations and epigenetic alterations, ncRNA associated with repetitive elements can be induced (8, 9). In a study of mouse and human epithelial malignancies by Ting et al. (9), several repetitive elements emanating from genomic dark matter and often repressed in steady-state conditions, particularly in pericentromeric repeats, such as GSAT (major satellite) in mouse and human satellite repeat II (HSATII) in humans, were only transcribed in cancer cells. Leonova et al. (8) demonstrated a strong induction of repetitive elements from the mouse genome (particularly GSAT, B1, and B2), along with several other ncRNAs, in cells bearing p53 oncogenic mutations and exposed to epigenome-altering demethylating agents. Anomalous expression of the murine repetitive element GSAT was shown to trigger transcription of the repeat-dependent activated IFN response, which can regulate apoptosis-related cell death. Similarly, when expressed, endogenous retroviral RNA can activate the innate immune response via several pathways (13). Altogether, these studies suggest that certain ncRNAs may also have attributes of immunostimulatory nucleic acid sequences.
We use a set of mathematical tools originally developed to analyze potentially immunostimulatory motif use in viral and host genome coding sequences. These methods were recently recast in the language of statistical physics and are extended here to analyze ncRNA motif use (14, 15). We analyze for the first time, to our knowledge, large-scale patterns of motif use in human and murine transcriptomes, which we use to find anomalies in ncRNA expressed in cancer transcriptomes (5, 16). As a result, we are able to characterize features of ncRNA overexpressed in cancerous cells relative to normal cells (8, 9, 17). Our analysis includes several large datasets of functionally characterized ncRNA, in addition to pseudogenes and repetitive elements, such as satellite DNA, endogenous retroviruses, and long and short interspersed elements. We demonstrate many ncRNAs preferentially expressed in cancerous cells display anomalous motif use patterns compared with the vast majority of ncRNAs whose patterns of motif use we show to be consistent with those patterns of motif use in coding regions. Based on their unusual pattern of motif use and differential expression in cancerous vs. normal cells, we predicted that HSATII and GSAT incorporate immunostimulatory motifs in humans and mice, respectively. Remarkably, we validate our prediction demonstrating that both directly stimulate antigen-presenting cells and accordingly label them immunostimulatory ncRNAs (i-ncRNAs).
Results
General Motif Use Patterns in lncRNAs.
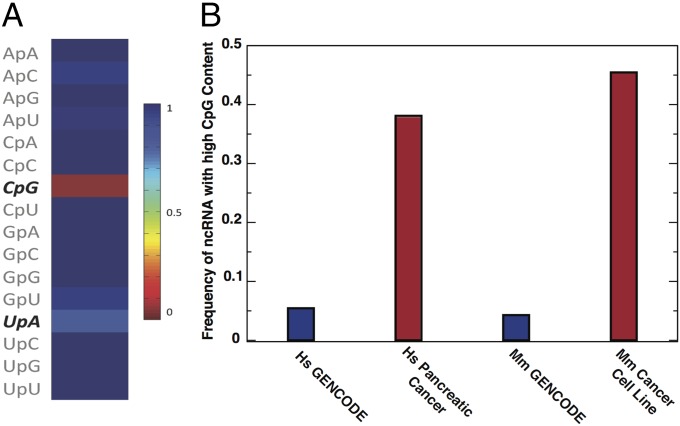
Using the GENCODE database of lncRNA transcripts from humans and mice (versions 19 and 2 for humans and mice, respectively) we calculated the strength of statistical bias (referred to as a force) on sequence motif use for all contained lncRNAs as described in Materials and Methods. GENCODE lncRNA established a baseline of sequence motif use expressed in a broad array of cells and tissues so that we could compare these patterns of motif use with those patterns of motif use of ncRNAs expressed in certain cancers. For each sequence, we calculate the force on all two- and three-nucleotide motifs and use Eq. 5 in Materials and Methods to calculate the probability of observing a sequence with that number of motifs. The number of sequences in GENCODE for which a given dinucleotide is aberrantly expressed is illustrated in Fig. 1A. CpG dinucleotides are vastly underrepresented, as indicated by their negative forces in SI Appendix, Table S1. UpA dinucleotides are often underrepresented, although to a lesser extent. As in our previous work, these patterns cannot be explained by nucleotide frequencies, such as guanine-cytosine (GC) content, which are accounted and normalized for in our method.
Fig. 1.
ncRNAs expressed in cancer differ from general lncRNA motif use patterns. (A) Fraction of GENCODE human lncRNA sequences where a motif occurs the expected number of times as defined by corresponding to a probability greater than 0.05 (Eq. 5). (B) Fraction of GENCODE lncRNA sequences in humans (Hs) and mice (Mm) where CpG motifs occur the expected number of times compared with the CpG motifs expressed in human cancerous cells and mouse cancer cell lines.
These dinucleotide motif use patterns are similar in human and mouse genomes across the wide array of cells and cell lines contained in GENCODE (2, 3). Strikingly, avoidance of the CpG and UpA dinucleotide motifs in this dataset is stronger than in coding regions (SI Appendix, Fig. S1). One can conclude that the patterns previously observed in virus and host coding genes are not due to effects from coding regions, such as codon use patterns (18–20). Rather, such constraints in coding regions likely weaken the strength of a statistical bias that comes from the same underlying mechanisms. This pattern suggests selective restrictions on dinucleotide frequencies observed in ncRNAs preserving a function or avoiding a detrimental consequence, such as a chronic autoinflammatory response that could result from presenting danger-associated molecular patterns (DAMPs). Adaptation of dinucleotide motif use in these elements over time is analogous to the viral mimicry of host patterns of sequence motif use (14, 21). When an avian influenza virus enters the human population, one can observe adaptation to analogous patterns emerging over time (14, 15, 22, 23). In that case, mutation rates in influenza are very high, so one can follow these evolutionary adaptations over far shorter time periods.
Trinucleotide motifs with significant forces are listed in the SI Appendix, Table S1, along with dinucleotide motifs. Trinucleotide motifs with significant forces acting on them are conserved between humans and mice, as was the case for dinucleotides, with the exception of UAC and UAG (which are significant in humans but less so in mice). Except for UAG (chain termination codons used in coding RNAs), whenever a trinucleotide motif is significantly enhanced or avoided in humans, its reverse complement is also significantly enhanced or avoided, suggesting avoidance of complementary motifs. The strongest forces suppress CpG and CpG-containing trinucleotides particularly when an A or U is next to the core CpG motif. These results are consistent with the avoidance of CpGs in AU contexts observed in influenza viruses replicating in humans (15, 22, 23). Given the apparent bias against CpG and UpA, we sought to determine if these motifs were linked. Pearson correlation between these forces across all GENCODE ncRNA in humans and mice showed no correlation between CpG and UpA biases (r = 0.0006; SI Appendix, Fig. S2). Therefore, the forces on CpG and UpA are likely independent. Moreover, every significant trimer across the GENCODE is correlated to CpG, UpA, or both. As a result, all significant trimers can be explained by their CpG or UpA motif use.
Cancer-Enriched Noncoding Repeat RNA May Have Anomalous Motif Use.
Prior work revealed aberrant expression of ncRNA across a spectrum of mouse and human cancers (8, 9). These sequences were found in the Repbase database of human and murine repetitive elements and the Functional Annotation of Mouse (FANTOM) database of murine noncoding elements (currently NONCODE) (24, 25). We also found high induction of GSAT in a murine testicular teratoma and liposarcoma tumor model (8, 9) (SI Appendix, Fig. S3). Focusing on these cancer-expressed repeats, we found a surprisingly significant enrichment of anomalous motif use patterns compared with other ncRNAs. In the Repbase database, we tested whether the bias on dinucleotide and trinucleotide motifs observed in repetitive element sequences fell outside the distribution obtained from GENCODE lncRNA. Remarkably, we found hundreds of sequences falling outside of this distribution. Many have high use of CpG dinucleotides, including a set of endogenous viruses (SI Appendix, Table S2) recently implicated in the innate immune response in tumors (13). We conclude that although the portions of the noncoding regions typically expressed as lncRNAs have motif use patterns similar to RNA from coding regions, there are many genomic regions with atypical motif use that are not transcribed in normal cells or tissues.
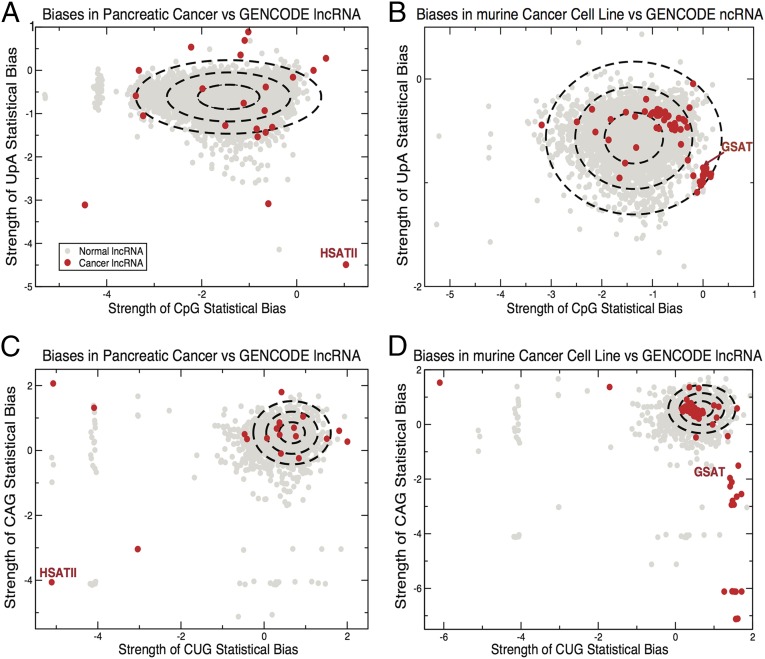
We use the forces that quantify the strength of the statistical bias on the often underrepresented CpG and UpA dinucleotides to differentiate between ncRNAs found preferentially in cancerous cells and the total lncRNA referenced in GENCODE for humans and mice, because these two dinucleotides essentially account for all significant trinucleotide motifs in this set. We use the distribution of forces on CpG and UpA to define a null hypothesis, which we approximate by a Gaussian distribution (Fig. 2). Many ncRNAs from cancerous cells are clearly outside the distribution, often to a large extent. In particular, HSATII, the main ncRNA up-regulated in human pancreatic cancers, is far outside the human distribution, and GSAT, the main murine ncRNA implicated in murine tumoral cell lines, is well outside the mouse distribution. Within our null hypothesis, the P values for all ncRNAs considered here are less than 10−61 for human pancreatic cancer data and less than 10−2 for murine cell line data.
Fig. 2.
ncRNA from cancer cells contains outliers from normal motif use. Distribution of UpA and CpG bias in lncRNA taken from human tumors (A) and murine cell lines (B) (indicated in red) plotted against lncRNA from GENCODE (indicated in gray). Each ellipse indicates 1 SD from the mean value in the GENCODE dataset. The forces on CAG and CUG are also shown for human tumors (C) and murine cell lines (D).
Many of the ncRNAs from the studies of Leonova et al. (8) and Ting, et al. (9) are outliers of at least three SDs with respect to at least one of the significant motifs implicated in the previous section, accounting for a median of 70.86% of the modulated Repbase RNA expression induced in pancreatic cancer, along with even higher percentages (73.95% and 85.74%, respectively) in the smaller sets of prostate and lung cancers. HSATII is the most differentially expressed (by a considerable margin) in the pancreatic cancer data, and HSATII and BSR are the highest in prostate and lung cancer data. In p53 KO murine cell lines treated with demethylation agents, around 68 ncRNAs are significantly modulated (8). Among those ncRNAs, 79.03% of the total expression comes from outliers as defined above, with the vast majority coming from GSAT and B2. Overall, we observed that repetitive sequences containing unusual motif use had varying degrees of conservation. However, the subset preferentially expressed in cancerous cells and tissues is encoded by sequences of more recent evolutionary origin. HSATII and GSAT are only conserved back to primates and mice, respectively, and 21 of the 22 ncRNAs from the study of Ting et al. (9) are conserved in humans and primates but extend no further back in evolution. Any function is likely to be species-specific.
ncRNAs with Unusual Motif Use Highly Expressed in Cancers Are Immunostimulatory.
Our analysis highlights that many ncRNAs up-regulated in cancer display abnormal nucleotide motif use that we had previously related to immunogenic properties in viruses. The innate immune system contains several effector cells that react to immunogenic nucleic acids, such as exogenous viral and bacterial nucleic acids, as well as endogenous nucleic acids that can be released upon cell death (6). Among those effectors, the mononuclear phagocytic system [macrophages, monocytes, and dendritic cells (DCs)] contains key regulators of innate immune activation and adaptive immunity (26–28). DCs efficiently sense and sample their environment to integrate information and mount a proper response, which may be tolerogenic or immunogenic. To test whether ncRNA with highly unusual motif use could be recognized as a DAMP by some nucleic acid-sensing pattern recognition receptors (PRRs), we studied the effect of human HSATII and murine GSAT following transfection in human monocyte-derived DCs (moDCs) and murine bone marrow-derived macrophages. Liposomal transfection was required for stimulation, whereas naked RNA had no effect, implying recognition is consistent with activation via an endosomal or intracellular sensor (SI Appendix, Fig. S4). The general sets of recognition pathways tested are indicated in the SI Appendix, Fig. S5.
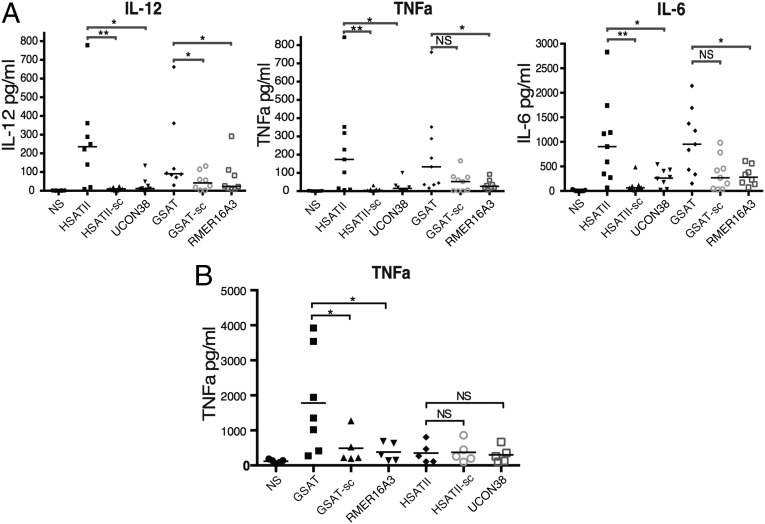
We generated different ncRNAs by in vitro transcription using minigenes coding for the two main candidate outliers computationally predicted to have immunogenic motif use (HSATII and GSAT). As controls, we derived RNA from minigenes encoding scrambled (sc) versions with the same nucleotide content but having normal motif use (labeled HSATII-sc and GSAT-sc) and repetitive elements of comparable length but having normal motif use patterns (RMER16A3 and UCON38), as described in SI Appendix. In human moDCs, liposomal transfection of HSATII induced significant production of IL-6, IL-12, and TNF-alpha relative to both endogenous controls and their scrambled versions (Fig. 3A). A similar profile of cytokines was elicited by moDCs in response to selected Toll-like receptor (TLR) agonists (SI Appendix, Fig. S6A). The candidate murine immunogenic ncRNA, GSAT, had less pronounced immunogenic properties but still induced IL-12 (Fig. 3A). Upon liposomal transfection of the same ncRNA into immortalized murine bone marrow-derived macrophages (imBMs), the immunogenic properties of HSATII were strongly attenuated, whereas the murine GSAT induced high levels of TNF-alpha (Fig. 3B) and monocyte chemotactic protein 1 (MCP-1), but not IFN-gamma, IL-6, or IL-12. The imBM almost exclusively regulates TNF-alpha in response to PRR agonists (SI Appendix, Fig. S6B).
Fig. 3.
i-ncRNA stimulates human moDC cytokine production. Quantification of inflammatory cytokine production in human moDCs (A) and murine imBMs (B) upon liposomal transfection of human i-ncRNA (HSATII) and murine i-ncRNA (GSAT) vs. their scrambled and endogenous controls. Each point represents the mean value of the experimental replicates for each individual condition; the bar represents the median. The significance of i-ncRNA stimulation is analyzed by the nonparametric Mann–Whitney test to compare their effect vs. their scrambled and endogenous controls. NS, not significant. *P < 0.05; **P < 0.01.
HSATII and GSAT ncRNA induced IL-12 in human moDCs similar to the TLR3 ligand poly-IC (a synthetic dsRNA mimic; SI Appendix, Fig. S5). The absence of an effect by ncRNA with normal motif use [i.e., the scrambled forms (Fig. 3 A and B)] suggests specific sequence patterns within the RNA, such as CpG and UpA motifs, regulate immunostimulatory activity. Such motif use could also influence secondary conformations that may contribute to immunogenic properties, although we checked that the scrambled sequences did not lower the RNA minimum folding energy. Based upon these observations, we refer to HSATII and GSAT as immunogenic ncRNA or i-ncRNA. Interestingly, our study corroborates previous findings by Leonova et al. (8) that ncRNA, such as GSAT, can induce an innate response, although in those studies, the type I IFN pathway was also activated. Our initial investigations into this pathway were inconclusive (SI Appendix, Fig. S6C).
Dissection of the Immunostimulatory Properties of i-ncRNA.
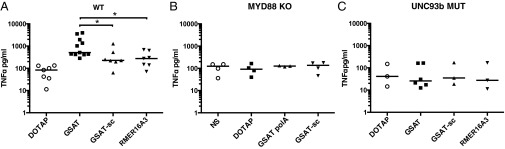
Pathogen-associated molecular patterns and DAMPs activate innate immune cells through PRRs. To characterize better the mechanisms involved in sensing i-ncRNA, we studied the immunomodulatory properties of HSATII and GSAT on a panel of imBMs that lack specific PRRs or effector molecules in their downstream signaling pathways (SI Appendix, Fig. S5). Whereas GSAT induced a TNF-alpha response, HSATII did not induce differential cytokine expression in these immortalized cells, indicating that there is either a species-specific effect, because the cells are murine, or a cell type-specific effect, because these cells are macrophages. This result is perhaps unsurprising, because different species and cell types express different PRRs, and HSATII and GSAT have different sequence compositions. Significantly, the absence of two key adaptor and regulatory proteins, MYD88 and UNC93B1:UNC93B3d (UNC93b), respectively, eliminated the differential response to GSAT in imBMs (Fig. 4).
Fig. 4.
MYD88 and UNC93b control GSAT i-ncRNA stimulation. Genetic screen of the innate immune pathway related to i-ncRNA function in murine imBMs. The imBM cells of different genotypes (WT, MYD88 KO, and UNC93b3d/3d MUT) have been stimulated by liposomal transfection (DOTAP liposomal transfection reagent) of the murine i-ncRNA (GSAT). TNF-alpha (TNF-a) production in the supernatant has been quantified, and each point represents the mean value of the experimental replicates for each individual condition; the bar represents the median. *P < 0.05.
MYD88 is a key cytosolic adaptor protein that is used by all TLRs except TLR3 to activate the transcription factor NF-κB. Similarly, the mutated form of UNC93b essentially eliminated inflammatory responses in imBMs. Although less well characterized than MYD88, this protein is known to interact with several endosomal TLRs (TLR3, TLR7, and TLR9) and has been implicated in TLR trafficking between the endoplasmic reticulum and endosomes, and their resultant maturation (29–31). We tested the requirement for TLR3, TLR7, and TLR9, which are known to recognize dsRNA, ssRNA, and CpG DNA, respectively (32–34) (SI Appendix, Fig. S7A and S8). None of these receptors were required for GSAT to activate TNF-alpha production from imBMs. Additional pathways investigated, including the stimulator of IFN genes (STING) and inflammasome pathways, are discussed in SI Appendix and did not contribute to i-ncRNA stimulatory activity. Altogether, our data are consistent with a requirement for i-ncRNA activation through signaling pathways that rely upon MYD88 and UNC93b. The precise receptor involved in initial recognition remains to be determined.
Discussion
There is a surprising similarity to be drawn between foreign viral nucleotide sequences and select ncRNAs silent in normal cells, yet transcribed in cancer cells, activating innate immunity (23, 29, 35–37). We determined that ncRNAs expressed predominantly in normal cells from humans and mice reflect patterns of nucleotide sequence motif avoidance, such as underrepresentation of CpG-containing sequences and reduced UpA, similar to protein-coding RNA. Such patterns often include a many-fold underrepresentation of CpG-containing sequences and reduced UpA motif use compared with expected levels. However, the genome also harbors repetitive elements, which often have abnormal use of CpG and UpA motifs compared with the use of CpG and UpA motifs observed in RNA expressed in normal cells and tissues. Sets of these ncRNAs, typically newer genome entries over evolutionary time scales, can be expressed at very high levels in cancerous cells and tumors. As a result, human and mouse elements expressed in cancer cells can have different sequences but can share high CpG content and are not generally observed in the human or mouse transcriptome in normal cells.
We previously proposed that immunostimulatory and proinflammatory properties of highly inflammatory influenza and other RNA viruses derive, in part, from RNA containing CpGs in AU-rich contexts, which are avoided in RNA viruses circulating in humans. Experimental evidence has supported this hypothesis (23, 38, 39). Recently we recast our analysis in the language of statistical physics in a way that is theoretically insightful and computationally efficient (15). In this language, the evolution and optimization of nucleotide sequence motifs are driven by the interplay between selective and entropic forces. The latter randomize motif frequencies in a genome under constraints, whereas the former are largely Darwinian, optimizing for functions enhancing viral replication and spread. However, ncRNAs transcribed mostly in cancerous cells would not be exposed to the same selective and entropic forces as coding RNAs and ncRNAs transcribed in normal cells. Based on motif use patterns, we predicted many ncRNAs may have immunogenic properties, presenting DAMPs.
We focused experimentally on HSATII and murine GSAT, because they are preferentially and highly expressed in carcinogenic processes and exhibit abnormal patterns of motif use. In particular, human HSATII is enriched in CpG motifs in AU-rich contexts avoided in genomes of humans and human-adapted viruses. We demonstrate that their computationally predicted immunogenic properties lead to the induction of inflammatory cytokines in human and murine innate cells (Fig. 3 A and B). Our observations, together with previous work by Leonova et al. (8), strongly suggest that these endogenous i-ncRNAs are recognized as DAMPs by cellular nucleic acid PRRs.
We identified a key role for MYD88 and UNC93b as regulators of GSAT immunogenicity, but without evidence for the common endosomal nucleic acid sensors typically regulated by UNC93b or associated with the MYD88 adaptor (TLR2, TLR4, TLR7, and TLR9). Our results indicate that in the murine imBM background, there is potent induction of TNF-alpha. Further studies will be required to elucidate whether TLR13, which has been identified in murine cells and recognizes ribosomal bacterial and viral RNA, is involved, or whether there exist intracellular sensors of i-ncRNA associated with MYD88 (40–42), as there are for dsDNA (DHX-9 or DHS-36) (43). Interestingly, we find alignment of GSAT contains a subsequence conserved in immunogenic RNA isolated from bacterial ribosomal RNA, which specifically activates murine TLR13 (41).
Activation of innate immune signaling can contribute to either carcinogenesis or antitumoral immunity. TLR signaling and MYD88 have been associated with tumor development (44). Given that HSATII and GSAT expression has been found to be pervasive in many tumor types and induces responses that differ by species or cell type, the role of i-ncRNA in tumorigenesis is likely dependent on the particular RNA expressed and other properties of the tumor microenvironment. For instance, HSATII activates macrophages and monocytes in our study, suggesting it may be a mechanism for attraction and retention of tumor-associated macrophages. These macrophages have consistently been shown to be a poor prognostic in cancer, leading to increased tumorigenesis, metastasis, and immunoevasion (45). Under this hypothesis, HSATII is used by the tumor to keep macrophages in the tumor microenvironment while driving out T cells. Interestingly, the viral-like behavior of HSATII transcripts is found not only in the immune response to these elements but also in their ability to reverse-transcribe in cancer cells, akin to retroviruses (46).
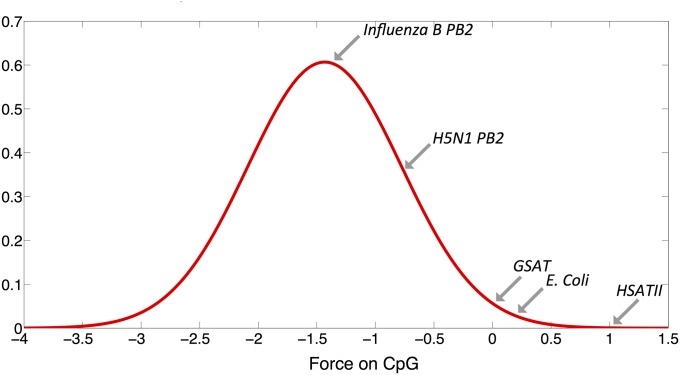
The i-ncRNA, not subject to the same forces as ncRNA transcribed in steady state, may retain or evolve to mimic features of foreign RNA, as seen by comparing HSATII and GSAT with typical human ncRNA and foreign genomic material in Fig. 5 (15, 47). Indeed, HSATII and GSAT cluster more closely, in terms of motif use patterns, with bacterial rather than human RNA. Such RNA may have been selected to identify and eliminate cells when their epigenetic state is disrupted. Essentially self-“junk” RNA may have been maintained or may have evolved to mimic non–self-pathogen–associated patterns to create a danger signal. We propose that such a mechanism would be a previously unidentified aspect of “genetic mimicry,” where the host is, for all practical purposes, mimicking pathogen-associated nucleic acid patterns. HSATII and GSAT emanate from the pericentromeres, which harbor new repetitive elements with no known function (48). This region, unlike centromeres or regions critical for structure or regulation, may dynamically produce unusual repetitive elements that can adapt to a particular organism’s PRRs. Our studies indicate that under the “extraordinary” circumstances where these repetitive elements are expressed, they could play a critical role in the regulation of immune responses against cancer.
Fig. 5.
Motif use in HSATII and GSAT clusters with foreign RNA. A comparison of the forces on CpG dinucleotides is plotted against the distribution of forces on all GENCODE lncRNA relative to a sequences nucleotide bias. The force on CpG dinucleotides for HSATII and GSAT is shown on the distribution, along with the average values for the longest gene (PB2) in human influenza B and avian H5N1 and all Escherichia coli coding regions.
Materials and Methods
We consider an RNA sequence of length L, hereafter called , and a motif m [a series of contiguous nucleotides (e.g., CpG)]. Our objective is to define a probabilistic model over the set of the sequences, , such that the average value of the number, , of occurrences of the motif m in coincides with the number, , of occurrences of that motif in . To do so, we consider a random-nucleotide model, where nucleotides are independently distributed according to the frequencies , with , found in . We then introduce the weakest bias that allows us to reproduce on average.
The probability of a sequence in this least-constrained, maximum entropy model is
| [1] |
where
| [2] |
ensures the probability is correctly normalized. Parameter referred to as a selective force (or just force) on the motif m, introduces a statistical bias over P (15). The force quantifies the strength of statistical bias, which may be due to selection on a motif. In the absence of bias (, the probability of simplifies to the product of its nucleotide frequencies, and the number of motifs is what one would expect in a typical sequence with nucleotide frequencies given by . Positive values for push the distribution toward sequences with larger than what one would expect, whereas negative values for favor sequences with a smaller than expected.
The value of the force, , is computed by maximizing the probability of the sequence over . This calculation is equivalent to finding the value of such that the average number of motifs,
| [3] |
equals By scanning the sequences in the GENCODE database, we obtain the forces shown in Fig. 2.
The logarithm of the number of sequences having repetitions of m is bounded from above by the entropy of the random-nucleotide model; the equality is reached in the absence of bias only . The difference between those entropies is the entropy cost corresponding to the constraint on the average number of occurrences of m, and is denoted by . It is the Legendre transform of (Eqs. 2 and 3):
| [4] |
Efficient computational techniques allow us to calculate the sum over the sequences in Eq. 2 in a time growing only linearly with L.
Our aim is to find anomalous motif use in a sequence where the number of motif occurrences is different from what is expected by chance in the random-nucleotide model (i.e., associated with a significant nonzero force). We express the likelihood of observing the natural sequence with a given motif count as
| [5] |
This likelihood is therefore directly related to the entropic cost: The larger the cost, the more likely is the motif to be statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. K. Fitzgerald (University of Massachusetts Medical School), Dr. R. Vance (University of California, Berkeley), Dr. G. Barton (University of California, Berkeley), and the Biodefense and Emerging Infections Research Resources Repository [American Type Culture Collection/National Institute of Allergy and Infectious Diseases (NIAID)] for helping us collect murine immortalized macrophages. We also thank Dr. N. Vabret for many helpful discussions and A. Munk for all of his assistance. B.D.G. was supported by NIH [National Cancer Institute (NCI)] Grant 5P01CA087497-13; N.B. was supported by NIH (NIAID) Grants 5R01AI081848-05 and 5R01AI081848-05, NCI Grant 1R01CA180913-01A1, and the Cancer Research Institute; D.T.T. was supported by NIH (NCI) Grant K12CA087723-11A1, Department of Defense (US Army) Grant W81XWH-13-1-0237, and the Burroughs Welcome Fund; and R.M. and S.C. were supported by L’Agence Nationale de la Recherche Grant ANR-13-BS04-0012-01.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 15008.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1517584112/-/DCSupplemental.
References
- 1.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrow J, et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012;22(9):1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. 2011;12(10):671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- 5.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atianand MK, Fitzgerald KA. Molecular basis of DNA recognition in the immune system. J Immunol. 2013;190(5):1911–1918. doi: 10.4049/jimmunol.1203162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang K, et al. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene. 2014;547(1):1–9. doi: 10.1016/j.gene.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 8.Leonova KI, et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci USA. 2013;110(1):E89–E98. doi: 10.1073/pnas.1216922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ting DT, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331(6017):593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293(5532):1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 11.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4(2):143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 12.Yi L, Lu C, Hu W, Sun Y, Levine AJ. Multiple roles of p53-related pathways in somatic cell reprogramming and stem cell differentiation. Cancer Res. 2012;72(21):5635–5645. doi: 10.1158/0008-5472.CAN-12-1451. [DOI] [PubMed] [Google Scholar]
- 13.Zeng M, et al. MAVS, cGAS, and endogenous retroviruses in T-independent B cell responses. Science. 2014;346(6216):1486–1492. doi: 10.1126/science.346.6216.1486. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Greenbaum BD, Levine AJ, Bhanot G, Rabadan R. Patterns of evolution and host gene mimicry in influenza and other RNA viruses. PLoS Pathog. 2008;4(6):e1000079. doi: 10.1371/journal.ppat.1000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenbaum BD, Cocco S, Levine AJ, Monasson R. Quantitative theory of entropic forces acting on constrained nucleotide sequences applied to viruses. Proc Natl Acad Sci USA. 2014;111(13):5054–5059. doi: 10.1073/pnas.1402285111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ulitsky I, Bartel DP. lincRNAs: Genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levine AJ, Greenbaum B. The maintenance of epigenetic states by p53: The guardian of the epigenome. Oncotarget. 2012;3(12):1503–1504. doi: 10.18632/oncotarget.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coleman JR, et al. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320(5884):1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mueller S, et al. Live attenuated influenza virus vaccines by computer-aided rational design. Nat Biotechnol. 2010;28(7):723–726. doi: 10.1038/nbt.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mueller S, Papamichail D, Coleman JR, Skiena S, Wimmer E. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J Virol. 2006;80(19):9687–9696. doi: 10.1128/JVI.00738-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlin S, Doerfler W, Cardon LR. Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? J Virol. 1994;68(5):2889–2897. doi: 10.1128/jvi.68.5.2889-2897.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenbaum BD, Rabadan R, Levine AJ. Patterns of oligonucleotide sequences in viral and host cell RNA identify mediators of the host innate immune system. PLoS One. 2009;4(6):e5969. doi: 10.1371/journal.pone.0005969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jimenez-Baranda S, et al. Oligonucleotide motifs that disappear during the evolution of influenza virus in humans increase alpha interferon secretion by plasmacytoid dendritic cells. J Virol. 2011;85(8):3893–3904. doi: 10.1128/JVI.01908-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110(1-4):462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 25.Xie C, et al. NONCODEv4: Exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014;42(Database issue):D98–D103. doi: 10.1093/nar/gkt1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guilliams M, et al. Dendritic cells, monocytes and macrophages: A unified nomenclature based on ontogeny. Nat Rev Immunol. 2014;14(8):571–578. doi: 10.1038/nri3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kroemer G, Galluzzi L, Kepp O, Zitvogel L. Immunogenic cell death in cancer therapy. Annu Rev Immunol. 2013;31:51–72. doi: 10.1146/annurev-immunol-032712-100008. [DOI] [PubMed] [Google Scholar]
- 28.Sabado RL, Bhardwaj N. Dendritic cell immunotherapy. Ann N Y Acad Sci. 2013;1284:31–45. doi: 10.1111/nyas.12125. [DOI] [PubMed] [Google Scholar]
- 29.Casrouge A, et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314(5797):308–312. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- 30.Lee BL, et al. UNC93B1 mediates differential trafficking of endosomal TLRs. eLife. 2013;2:e00291. doi: 10.7554/eLife.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tabeta K, et al. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7(2):156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- 32.O’Neill LA, Golenbock D, Bowie AG. The history of Toll-like receptors - redefining innate immunity. Nat Rev Immunol. 2013;13(6):453–460. doi: 10.1038/nri3446. [DOI] [PubMed] [Google Scholar]
- 33.Broz P, Monack DM. Newly described pattern recognition receptors team up against intracellular pathogens. Nat Rev Immunol. 2013;13(8):551–565. doi: 10.1038/nri3479. [DOI] [PubMed] [Google Scholar]
- 34.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14(10):1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bogunovic D, et al. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc Natl Acad Sci USA. 2009;106(48):20429–20434. doi: 10.1073/pnas.0905139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kayagaki N, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479(7371):117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 37.Cosset É, et al. Comprehensive metagenomic analysis of glioblastoma reveals absence of known virus despite antiviral-like type I interferon gene response. Int J Cancer. 2014;135(6):1381–1389. doi: 10.1002/ijc.28670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Atkinson NJ, Witteveldt J, Evans DJ, Simmonds P. The influence of CpG and UpA dinucleotide frequencies on RNA virus replication and characterization of the innate cellular pathways underlying virus attenuation and enhanced replication. Nucleic Acids Res. 2014;42(7):4527–4545. doi: 10.1093/nar/gku075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vabret N, et al. The biased nucleotide composition of HIV-1 triggers type I interferon response and correlates with subtype D increased pathogenicity. PLoS One. 2012;7(4):e33502. doi: 10.1371/journal.pone.0033502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li XD, Chen ZJ. Sequence specific detection of bacterial 23S ribosomal RNA by TLR13. eLife. 2012;1:e00102. doi: 10.7554/eLife.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oldenburg M, et al. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science. 2012;337(6098):1111–1115. doi: 10.1126/science.1220363. [DOI] [PubMed] [Google Scholar]
- 42.Shi Z, et al. A novel Toll-like receptor that recognizes vesicular stomatitis virus. J Biol Chem. 2011;286(6):4517–4524. doi: 10.1074/jbc.M110.159590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim T, et al. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc Natl Acad Sci USA. 2010;107(34):15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang JQ, Jeelall YS, Ferguson LL, Horikawa K. Toll-like receptors and cancer: MYD88 mutation and inflammation. Front Immunol. 2014;5:367. doi: 10.3389/fimmu.2014.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noy R, Pollard JW. Tumor-associated macrophages: From mechanisms to therapy. Immunity. 2014;41(1):49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bersani F, et al. Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proc Natl Acad Sci. 2015;112:15148–15153. doi: 10.1073/pnas.1518008112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maumus F, Quesneville H. Ancestral repeats have shaped epigenome and genome composition for millions of years in Arabidopsis thaliana. Nat Commun. 2014;5:4104. doi: 10.1038/ncomms5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.