Significance
Legumes form a root structure, the nodule, in which nitrogen-fixing bacteria (rhizobia) reside. In this symbiotic relationship, the bacteria provide nitrogen to the plant and in return obtain fixed carbon from the host. Once released into the cytoplasm of the host cell, the rhizobia undergo a remarkable transformation, including genome amplification and cell elongation, before reaching the differentiated nitrogen-fixing state. Small plant-derived peptides with antimicrobial activities have been known to play critical roles in the differentiation of rhizobia in legumes that form indeterminate nodules. By studying the Medicago truncatula dnf4 mutant, we discovered that an antimicrobial peptide, NCR211, plays a critical role in the survival and function of differentiated rhizobia in host cells for successful symbiotic nitrogen fixation.
Keywords: nitrogen-fixing symbiosis, Sinorhizobium meliloti, Medicago truncatula, legume, NCR antimicrobial peptides
Abstract
In the nitrogen-fixing symbiosis between legume hosts and rhizobia, the bacteria are engulfed by a plant cell membrane to become intracellular organelles. In the model legume Medicago truncatula, internalization and differentiation of Sinorhizobium (also known as Ensifer) meliloti is a prerequisite for nitrogen fixation. The host mechanisms that ensure the long-term survival of differentiating intracellular bacteria (bacteroids) in this unusual association are unclear. The M. truncatula defective nitrogen fixation4 (dnf4) mutant is unable to form a productive symbiosis, even though late symbiotic marker genes are expressed in mutant nodules. We discovered that in the dnf4 mutant, bacteroids can apparently differentiate, but they fail to persist within host cells in the process. We found that the DNF4 gene encodes NCR211, a member of the family of nodule-specific cysteine-rich (NCR) peptides. The phenotype of dnf4 suggests that NCR211 acts to promote the intracellular survival of differentiating bacteroids. The greatest expression of DNF4 was observed in the nodule interzone II-III, where bacteroids undergo differentiation. A translational fusion of DNF4 with GFP localizes to the peribacteroid space, and synthetic NCR211 prevents free-living S. meliloti from forming colonies, in contrast to mock controls, suggesting that DNF4 may interact with bacteroids directly or indirectly for its function. Our findings indicate that a successful symbiosis requires host effectors that not only induce bacterial differentiation, but also that maintain intracellular bacteroids during the host–symbiont interaction. The discovery of NCR211 peptides that maintain bacterial survival inside host cells has important implications for improving legume crops.
Many legume plants satisfy their nitrogen needs by interacting with nitrogen-fixing bacteria (rhizobia) to form a specialized symbiotic organ, the root nodule. As rhizobia penetrate root hair cells through invaginations of host membrane called infection threads, the cortical cells underneath start dividing and eventually build the nodule in which the invading rhizobia are internalized to form intracellular organelles known as symbiosomes.
Among legumes, Medicago truncatula belongs to a group that forms indeterminate nodules, where a meristem continuously produces new nodule cells. An indeterminate nodule can be divided into four zones harboring different cell types (1). The meristem of indeterminate nodules is located in the apical region (zone I), which constantly supplies new cells to the nodule. These new cells then become infected with rhizobia in zone II, where the rhizobial cells are released from the infection thread into the host cytoplasm to form symbiosomes. On release into the host cytoplasm, the bacteroids are encapsulated with a host-derived membrane (the peribacteroid membrane), blocking the rhizobia from directly contacting the cytoplasm. The released bacteroids multiply and gradually colonize the host cell. In interzone II-III, rhizobial nif genes are turned on as the bacteroids expand, primarily by elongation, occupying a majority (∼65%) of the host cell volume. The volume of vacuoles decreases dramatically, down to ∼30% of the cell volume, which is correlated with the suppression of HOPS (homotypic fusion and vacuole protein sorting complex) gene expression in infected cells (2). In zone III, as the infected cells stop expanding, bacteroids become terminally differentiated and actively convert atmospheric nitrogen into ammonia, which can be readily transferred to the host plant for assimilation into amino acids. In older nodules, zone IV (also called the senescence zone) is present, where the host cells and bacteroids degenerate.
Although in certain legumes the nitrogen-fixing bacteroids remain morphologically similar to free-living bacteria and are capable of reverting back to the nonsymbiotic lifestyle, bacteroids in nodules formed on the inverted-repeat lacking clade (IRLC) of legumes, such as M. truncatula, undergo remarkable transformations to differentiate terminally, such that they are no longer able to survive independent of their host. Features of bacterial differentiation include genome amplification and cell elongation, and the host cell also expands by endoreduplicating its own genome (3, 4). Studies have shown that the differentiation of bacteroids requires the delivery of host effectors, such as nodule-specific cysteine-rich (NCR) peptides, through the endoplasmic reticulum secretory system (5, 6). Present only in IRLC legumes, NCRs are defensin-like antimicrobial peptides, some of which drive the differentiation of bacteroids (5, 7, 8). They are now recognized as major players in the interaction between the legume host and rhizobia. The importance of this family is underscored by their massive expansion in the IRLC lineage, with more than 500 members encoded in the M. truncatula genome.
The best evidence for the requirement of the NCR peptides to date has been obtained by disrupting the nodule-specific protein secretory pathway, where intracellular rhizobia no longer differentiate (6). However, blocking protein secretion in the nodule indiscriminately is a blunt instrument that offers no insight into the specificity of individual NCR peptides. The large size of the NCR family and the limited sequence homology among its members hinder efforts to generalize their role in the symbiosis. Their sequence diversity and distinct temporal and spatial expression patterns have led to the suggestion that they may perform diverse functions, with different bacterial targets and modes of action (9), but direct proof of functional diversity is still lacking.
Our best understanding is of the subset of peptides that induce bacterial differentiation. NCR247 has been studied in particular detail, and has been reported to enter bacteroids and interact with a plethora of proteins (10). In addition, NCR247 treatment causes massive transcriptome changes in the bacteria (∼15% of the Sinorhizobium meliloti genome), affecting critical cell cycle regulators and cell division genes (11). Genetically, the only in planta evidence supporting the role of NCR peptides in bacterial differentiation comes from ectopic expression of NCR035 in Lotus japonicus (5). Although loss-of-function genetic results would be desirable, it is generally assumed that within such a large group, the contribution from any single NCR peptide will not be sufficient for its absence to cause a significant phenotype.
Here, by studying the defective in nitrogen fixation mutant, dnf4, we report that an individual NCR peptide can be indispensable for the nitrogen-fixing symbiosis. The symbiotic mutant phenotype indicates that the M. truncatula DNF4 gene is required for the survival and function of differentiating bacteroids. Map-based cloning followed by whole genome sequencing identified the DNF4 gene as encoding NCR211. Our results indicate that while some NCRs such as NCR247 have been suggested to induce bacterial differentiation, DNF4/NCR211 on the other hand acts on protecting differentiating bacteroids in symbiosomes from degeneration.
Results
dnf4 Is Defective in Maintaining Differentiating Bacteroids Inside the Host Cell.
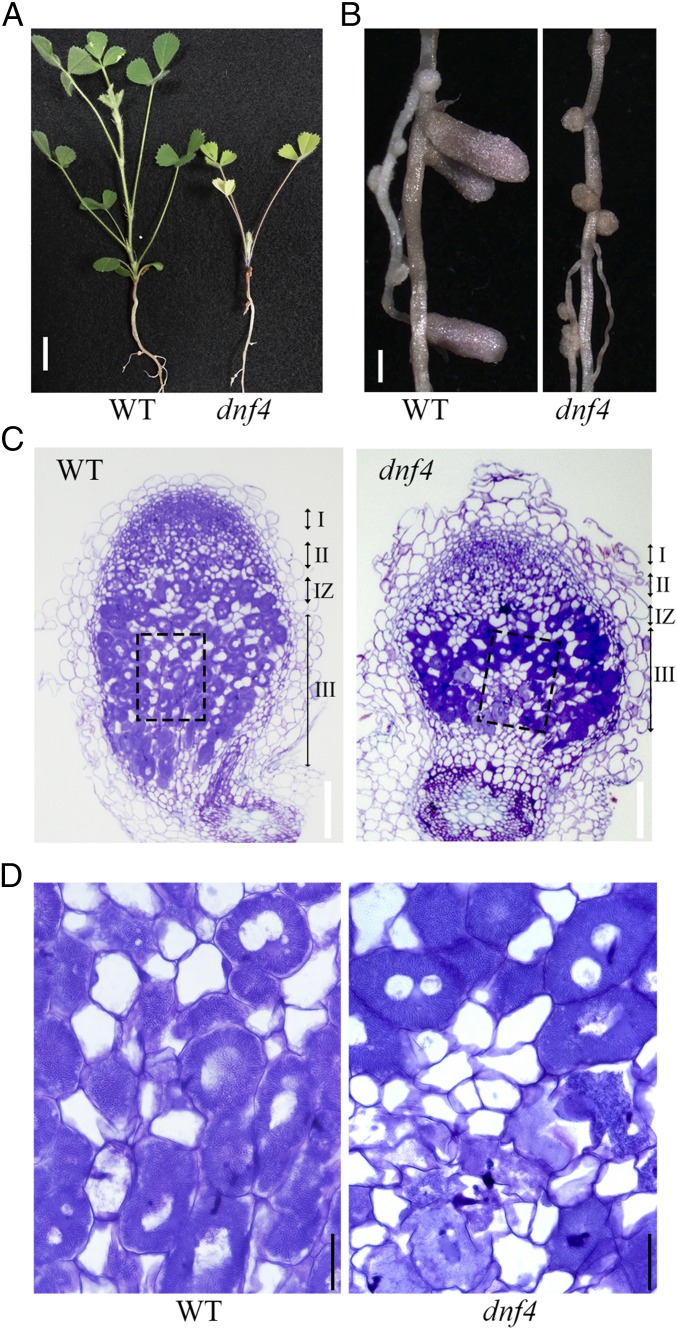
The dnf4 mutant was isolated from a screen for M. truncatula mutants defective in nitrogen fixation (12). In growth medium lacking exogenous nitrogen, the inability of dnf4 mutant plants to use atmospheric nitrogen resulted in growth retardation and chlorotic leaves (Fig. 1A). The nodules of dnf4 also remained small and white, whereas wild type nodules elongated and developed a pink color, which derives from the accumulation of leghemoglobin (Fig. 1B). Toluidine blue-stained nodules of dnf4 at 10 d postinoculation (dpi) appeared normal in zones I and II, as well as in interzone II-III, where the dark color suggests that the host cells were fully occupied by bacteria (Fig. 1C); however, in the nitrogen-fixing zone III, bacteroids appeared to disappear from the central part of some host cells proximal to the root. Under higher magnification, fully infected dnf4 cells exhibited a morphology similar to that of WT cells, with elongated bacteroids arranged around the central vacuole; however, cells that may have once contained bacteroids were clearly degenerated (Fig. 1D).
Fig. 1.
dnf4 is unable to develop functional nodules. (A) Plants were inoculated with S. medicae ABS7 and grew for 4 wk. Yellow leaves are the sign of nitrogen deficiency. (Scale bar: 10 mm.) (B) dnf4 nodules remain small and white, whereas the wild type nodules elongate and show pink color. (Scale bar: 1 mm.) (C) Toluidine blue staining of 10-dpi nodules inoculated with ABS7. (Scale bar: 100 μm.) (D) Magnification of boxed areas in C. Elongated bacteroids stain dark blue. dnf4 nodules show degradation of symbiosomes and host cells at the nitrogen fixation zone proximal to the root. (Scale bar: 20 μm.)
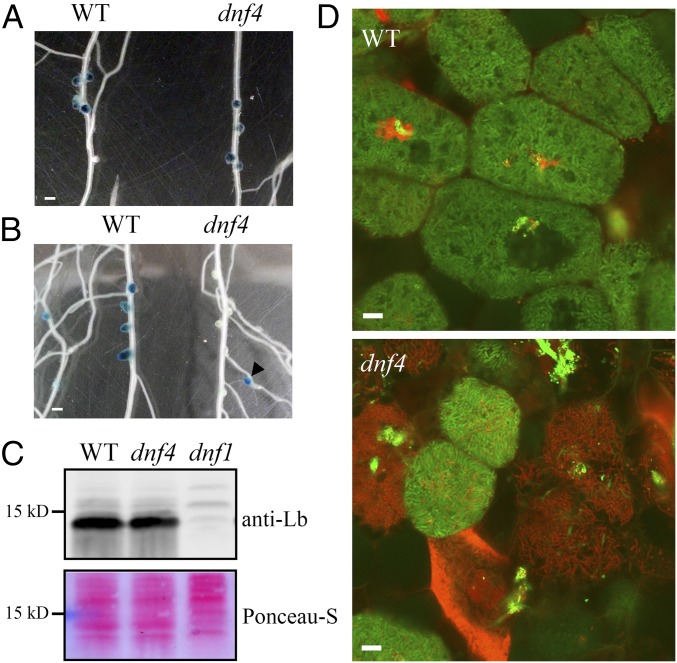
To further analyze the nature of the defect in dnf4, we examined the activity of the nifH bacterial gene using a promoter fusion construct with the GUS gene that encodes β-glucuronidase (12). The nifH gene encodes one of the subunits of the nitrogenase enzyme complex, and thus is required for nitrogen fixation. At 10 dpi, nodules of wild type and dnf4 plants showed similar nifH expression (Fig. 2A); however, in dnf4 nodules, nifH expression was lost by 14 dpi (Fig. 2B), whereas younger nodules that formed later on the lateral roots (arrowhead in Fig. 2B) still displayed nifH expression. These data indicate that the nifH gene was expressed in young dnf4 nodules, but its expression was abolished in older ones.
Fig. 2.
dnf4 nodules express fixation genes but fail to sustain differentiated bacteroids. (A and B) Activity of a nifH::GUS reporter in WT and dnf4 nodules at 10 dpi (A) and 14 dpi (B). The arrowhead in B denotes a young dnf4 nodule on a lateral root. (Scale bar: 1 mm.) (C) Accumulation of leghemoglobin in 10-dpi nodules. Ponceau S-stained membrane is shown as a loading control. (D) Live/dead staining of bacteroids in dnf4 compared with WT nodules (14 dpi). Live bacteroids have a green signal (GFP) and dying bacteroids have a red signal from propidium iodide (PI). PI can only enter bacteroids with compromised membranes. (Scale bar: 10 µm.)
Host leghemoglobin proteins bind free oxygen to create a microoxic environment in nitrogen-fixing cells, protecting the nitrogenase enzyme from inactivation by oxygen. nifH is expressed only in low-oxygen conditions, implying that leghemoglobin is present in dnf4 nodules. We observed that 10–20% of dnf4 nodules were slightly pink, suggesting that these nodules express leghemoglobin proteins. This proposition was confirmed by immunoblot analyses of proteins extracted from 10-dpi nodules. The 10-dpi nodules of dnf4 also accumulated leghemoglobin proteins at a level near that measured in wild type, in contrast to the early nodulation mutant dnf1, in which leghemoglobin protein was not detected (12) (Fig. 2C). Thus, dnf4 nodules are deficient in the symbiosis even though both partners express genes characteristic of mature nitrogen-fixing nodules. These results are also consistent with a previous report of the dnf4 mutant being defective in later stages of nodule development, unlike mutants affected in earlier stages of nodule development, such as dnf1, dnf2, and dnf5 (12).
To observe microscopic details of dnf4 defects, mutant nodules were inoculated with GFP-expressing S. meliloti Rm1021 and stained with propidium iodide (PI), a fluorescence dye that can penetrate only compromised biological membranes. Wild type nodules at 14 dpi had few PI-positive bacteroids. In contrast, dnf4 nodule cells showed widespread PI staining in bacteroids. Furthermore, dnf4 bacteroids, including ones staining positive for PI, appeared elongated and differentiated (Fig. 2D), unlike other early-stage mutants (6, 13), although the degree of bacteroid differentiation was unclear. Coupling PI staining with SYTO9, a commonly used viability dye, yielded a similar pattern. In WT nodules, most nitrogen-fixing bacteroids were green (Fig. S1A). In dnf4, most cells containing differentiated bacteroids were red, suggesting that the bacteria in these cells were dead; however, bacteria in recently infected cells were still alive, and in some nodules, these newly infected cells formed a sharp band in the infection zone, suggesting that the infection process itself was normal (Fig. S1B). Some bacteroids in dnf4 cells appeared to reach the final stage of differentiation, when they are radially aligned (Fig. S1 C and D). In summary, the phenotypic characterization of dnf4 nodules strongly suggests that this mutation causes defects in the maintenance of differentiating bacteroids within host cells.
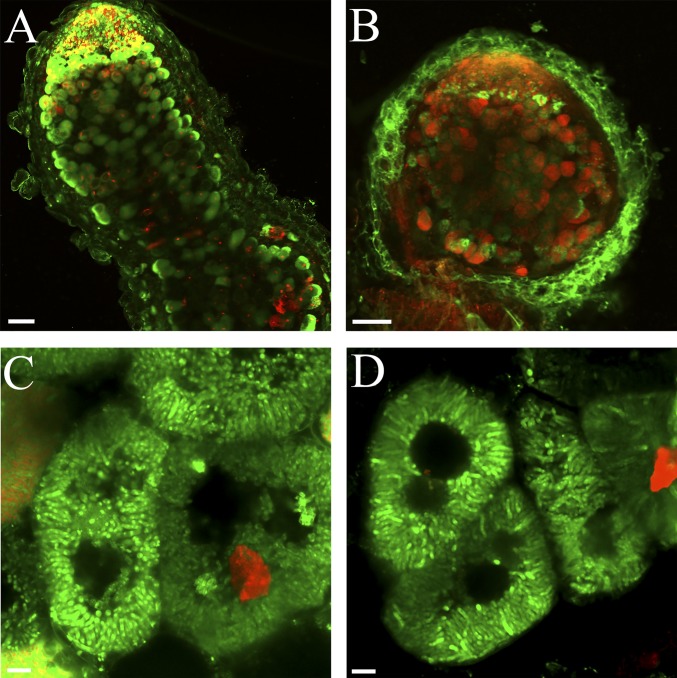
Fig. S1.
Live-dead staining of whole nodules with SYTO9 and PI. The procedures are similar to that used for Fig. 2D, except that the bacteria do not contain fluorescent proteins. (A and B) WT (A) and dnf4 (B) whole nodule sections. (Scale bar: 100 µm.) (C and D) Images of individual WT (C) and dnf4 (D) cells. (Scale bar: 10 µm.)
Deletion of NCR211 Is Responsible for the dnf4 Phenotype.
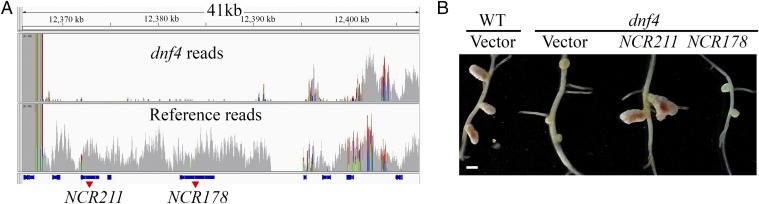
To identify the causal mutation of the dnf4 phenotype, we crossed the dnf4 mutant (in the Jemalong background) with the A20 ecotype. Using map-based cloning, we narrowed the region to the upper arm of chromosome 4, between molecular markers 003F07 and h2_133k2b. Genomic DNA pooled from phenotypically mutant plants was sequenced. An unrelated line in the A17 background (derived from Jemalong) was also sequenced as a reference. Alignment of sequence reads to the A17 reference genome identified a deletion of ∼35 kb unique to dnf4 close to the mapping interval (Fig. 3A). This deletion, verified by PCR amplification (Fig. S2), eliminated five predicted genes. Two of these five genes encode NCR211 and NCR178, both of which are expressed in nodules, whereas the other three genes encode hypothetical proteins not expressed in nodules according to available RNA-seq data (www.jcvi.org/medicago/index.php).
Fig. 3.
Identification of the DNF4 gene. (A) Read density plot of dnf4 mutant (pool of mapping population) and an unrelated EMS mutant as a reference, showing that the deletion is unique to dnf4. The blue bars on the bottom represent gene models. (B) Nodule morphology on hairy roots transformed with empty vector or NCR211 or NCR178 genomic constructs. (Scale bar: 1 mm.)
Fig. S2.
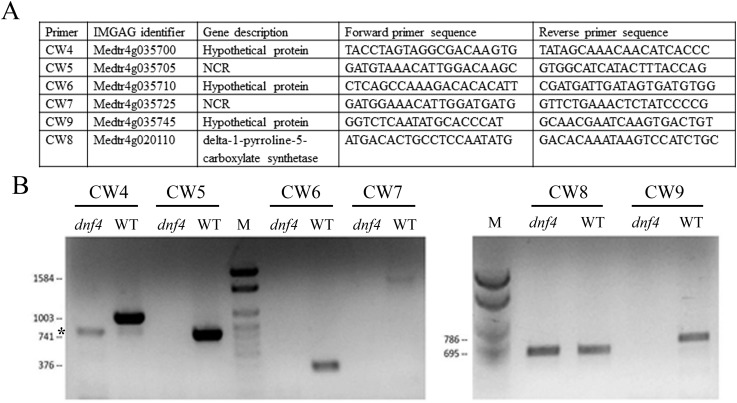
Verification of a deletion in dnf4. (A) Sequences of the primers used for PCR verification. CW8 amplifies a distant gene near the deletion site as a positive control. (B) DNA agarose gel picture of PCR products. Expected sizes of PCR products are shown to the left of the gels. The asterisk indicates a nonspecific band. M, size marker.
The two NCR genes were introduced into the dnf4 mutant by hairy root transformation to test their ability to rescue the dnf4 phenotype. Only the NCR211-encoding gene (Medtr4g035705) proved capable of complementing the dnf4 mutant, confirming that it corresponds to DNF4 (Fig. 3B). The rescue of dnf4 phenotype by NCR211 was observed in all transgenic plants (n ≥10) selected by the red fluorescence of the DsRED1 marker gene.
The DNF4 gene is predicted to produce a 58-aa-long preprotein. Cleavage of the signal peptide [the first 24 residues, as predicted by SignalP (14)] would yield an anionic (pI = 5.38) NCR211 peptide 34 aa long, with four conserved cysteines. A BLASTp search identified NCR178 (Medtr4g035725) as the closest homolog of the mature DNF4 (61.8% identity), suggesting that these two NCR genes may have arisen from a recent local duplication event. Our phenotypic analysis of the dnf4 mutant suggests that NCR211’s function is necessary to maintain the viability of differentiating bacteroids.
dnf4 Is Allelic to esn1.
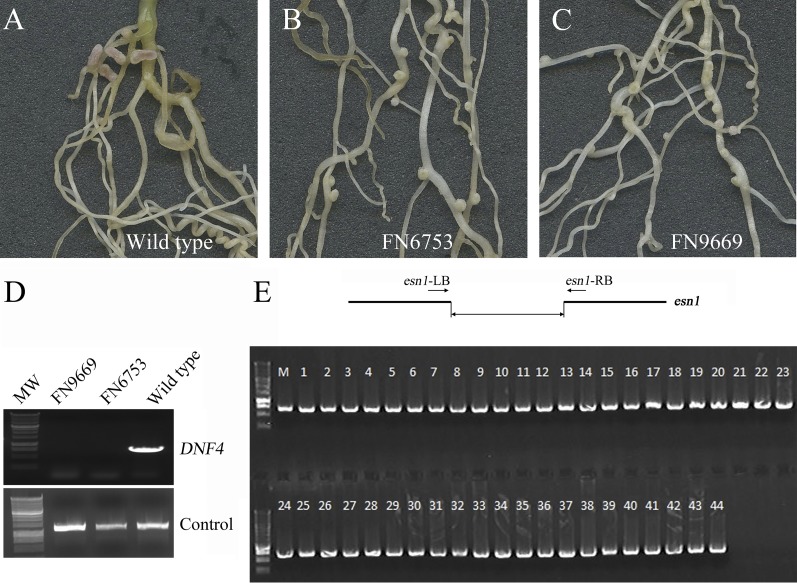
We noticed that the defects in dnf4 are similar to two M. truncatula symbiotic mutants deficient in nitrogen fixation, FN6753 and FN9669 (named early senescent nodule1, or esn1) (15) from a different irradiation-generated mutant collection. Under nitrogen-replete conditions, FN6753 and FN9669 are indistinguishable from wild type (15). Inoculated with S. meliloti Rm1021 under nitrogen depletion, FN6753 and FN9669 exhibited symbiotic phenotypes resembling the phenotype of the dnf4 mutant (Fig. S3 A–C) (15). To assess whether the symbiotic defects of FN6753 and FN9669 result from mutations in NCR211, we first examined whether NCR211 is deleted in these mutants. PCR results showed that NCR211 is absent in both mutants (Fig. S3D). We next carried out a linkage analysis using a backcrossed F2 population of the esn1 mutant. Wild type and esn1 mutants segregated at a ratio close to 3:1 (95 wild type and 44 mutant), and the deletion in NCR211 was linked to the symbiotic phenotype (Fig. S3E). Based on these results, we conclude that FN9669 and FN6753 represent additional alleles of the dnf4 mutant.
Fig. S3.
FN6753 and FN9669 (esn1) are allelic to dnf4. (A–C) Symbiotic nitrogen fixation-deficient phenotypes of FN6753 (B) and FN9669 (C) compared with wild type A17 (A). (D) PCR analysis showing deletion of DNF4 in both mutants. (E) Genetic linkage analysis showing a linkage between the dnf4/esn1 deletion as illustrated at the top to the mutant phenotype in a backcrossed F2 population. All 44 samples are esn1 mutants. The PCR analysis was designed to amplify across the deletion interval.
DNF4 Is Highly Expressed in Nodules and Localizes to the Symbiosome.
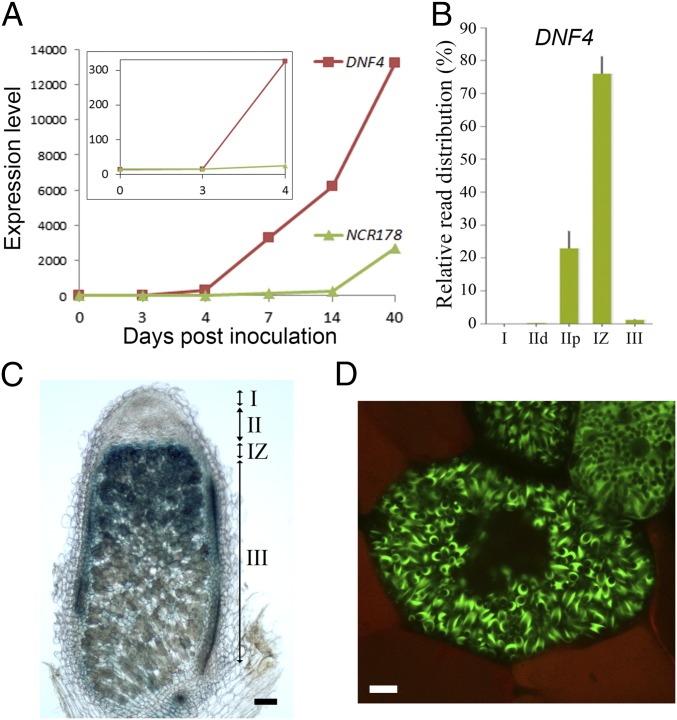
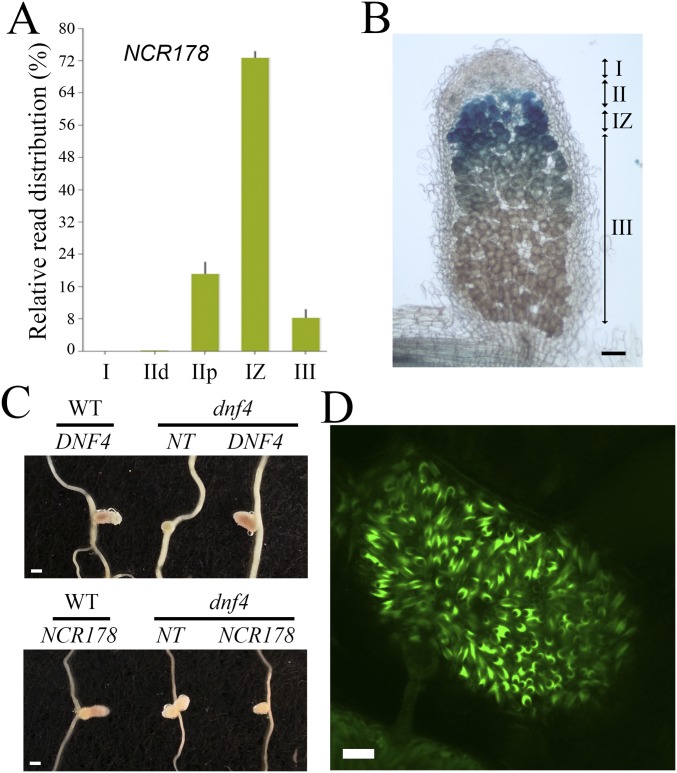
Microarray analysis of NCR genes during nodule development allowed us to search for the expression pattern of both DNF4 (NCR211) and NCR178 on Rm1021 inoculation (16). As shown in Fig. 4A, both genes were induced starting at 4 dpi, and their expression level continued to increase up to 40 dpi; however, the expression was much higher (by ∼22-fold at 14 dpi) for DNF4 compared with NCR178. Spatial gene expression profiling of Sm2011-inoculated nodules was recently performed with laser-capture microdissection (LCM) followed by RNA-seq (17). Data from this study indicate that expression of DNF4 was high in the proximal half of zone II and reached its highest level in interzone II-III (Fig. 4B). NCR178 exhibited a similar expression pattern (Fig. S4A). To confirm this pattern derived from LCM, promoter-GUS constructs of DNF4 and NCR178 were introduced into M. truncatula with hairy root transformation. Consistent with the LCM data, the promoter activity of DNF4 was highest in interzone II-III (Fig. 4C). GUS activity was also present in zone III, as well as in proximal zone II. The promoter of NCR178 showed an expression pattern similar to that of DNF4 (Fig. S4B). The promoter-GUS expression analyses were performed with nodules from more than five independent transgenic roots, with similar results.
Fig. 4.
DNF4 (NCR211) is highly expressed in interzone II-III, and the GFP fusion protein localizes to the peribacteroid space. (A) Temporal expression pattern of DNF4 and NCR178 after inoculation with Rm1021. (Inset) Expression at early time points. (B) Spatial expression pattern of DNF4. I, zone I; IId, zone II distal; IIp, zone II proximal; IZ, interzone II-III; III, zone III. (C) DNF4 promoter GUS activity. (Scale bar: 100 µm.) (D) Localization of DNF4-GFP translational fusion in the peribacteroid space. (Scale bar: 10 µm.)
Fig. S4.
NCR178 is highly expressed in the interzone II-III, and the GFP fusion protein localizes to the peribacteroid space. (A) Spatial expression pattern of NCR178. I, zone I; IId, zone II distal; IIp, zone II proximal; IZ, interzone II-III; III, zone III. (B) NCR178 promoter GUS activity. (Scale bar: 100 µm.) (C) Complementation of dnf4 by C-terminal GFP fusion constructs. NCR211-GFP transformed nodules recover the wild type-like nodule morphology, whereas NCR178-GFP transformed nodules do not. The transgenic nodules were selected by DsRED1 fluorescence from hairy root-transformed roots. NT, nontransgenic root. (Scale bar: 1 mm.) (D) Localization of NCR178-GFP translational fusion in the peribacteroid space. (Scale bar: 10 µm.)
Previous studies of some NCR peptides showed localization inside bacteroids or on the bacteroid membrane (5, 10). Here the genomic sequences of DNF4 and NCR178 were fused in frame with a GFP sequence to create translational fusion constructs for localization studies. The constructs were introduced into both wild type and dnf4 plants by hairy root transformation. The DNF4-GFP construct was able to fully rescue the defective nodule phenotype of dnf4 in all transgenic plants recovered (n >10), indicating that the GFP fusion does not interfere with the function of DNF4 (Fig. S4C). DNF4-GFP was observed in the peribacteroid space between the host and the bacteroid membranes (Fig. 4D). An NCR178-GFP fusion protein (in wild type background) also localized to the peribacteroid space (Fig. S4D). The localization of DNF4 to the symbiosome suggests that the NCR211 peptide may directly or indirectly interact with bacteroids.
Exogenously Supplied NCR211 Does Not Reverse Membrane Permeability of the Symbiosome.
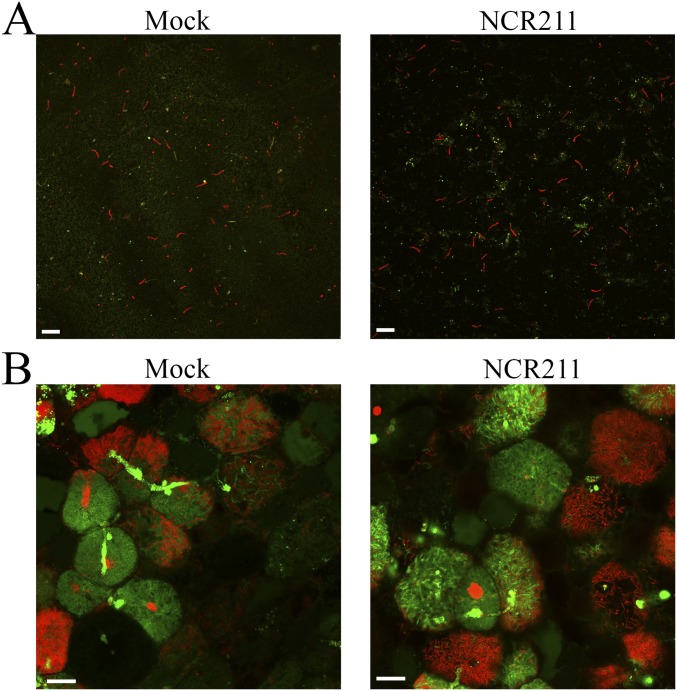
The mature DNF4 peptide has certain sequence similarities with the scorpion toxin BmKK2, which acts as a blocker of eukaryotic potassium channels embedded in the cell membrane (18). As the DNF4 peptide localizes to the symbiosome, it could be in close proximity to three membranes: the host peribacteroid membrane, the bacterial outer membrane, and the bacterial inner membrane. Bacteroids in dnf4 nodules readily take up PI, suggesting that all three membranes are permeabilized. To test whether DNF4 acts on the membranes of differentiated symbiosomes to block membrane permeabilization, we applied a synthetic NCR211 peptide to dnf4 symbiosomes either extracted from nodules or in situ, and then proceeded with PI staining. In neither case did NCR211 prevent the uptake of the PI dye (Fig. S5). Along with later tests demonstrating that the synthetic NCR211 peptide is biologically active (next section and Fig. 5), these results suggest that NCR211 might not block membrane permeability in the symbiosome.
Fig. S5.
Exogenous application of NCR211 does not prevent the uptake of PI dye. (A) Symbiosomes were extracted from the dnf4 nodules and incubated for 4 h with (Right) or without (Left) 20 µM NCR211 peptide. SYTO9 (green, 5 µM) and PI (red, 30 µM) staining was done afterward. (Scale bar: 5 µm.) (B) Hand-sectioned dnf4 nodules were incubated for 4 h with (Right) or without (Left) 10 µM NCR211 peptide. SYTO9 (green, 5 µM) and PI (red, 30 µM) staining was done afterward. (Scale bar: 5 µm.)
Fig. 5.
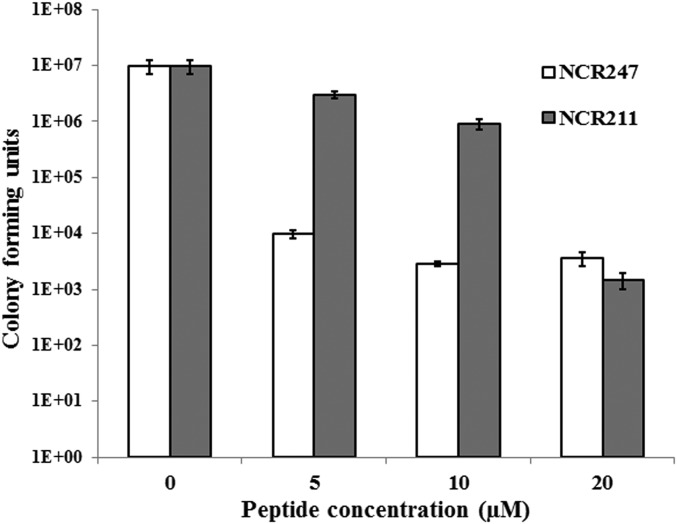
A synthetic NCR211 peptide blocks the proliferation of free-living S. meliloti Rm1021. Bacteria were treated with either NCR211 or NCR247 for 3 h at the indicated concentrations, and spotted on solid media. The number of colonies was recorded 2 d later. Error bars indicate SDs (n = 3). The graph represents a typical outcome from multiple (>3) replications.
DNF4 Blocks the Proliferation of Free-Living Rhizobia.
Although the synthetic NCR211 peptide does not appear to repair compromised symbiosome membranes, the localization pattern of DNF4 nonetheless suggests that it may interact with the bacteroids in some fashion. To test this hypothesis, we applied synthetic NCR211 and a different NCR peptide, NCR247, to free-living S. meliloti Rm1021. NCR247 treatments blocked subsequent colony formation, as reported previously (5). NCR211 treatments also blocked colony formation (Fig. 5). Although the kinetics of the two NCR peptides appeared different, at the highest concentration the inhibitory effect of NCR211 was comparable to that of NCR247 (Fig. 5).
Discussion
Approximately 600 genes encoding NCR peptides are predicted in the M. truncatula genome (19). The lack of symbiotic mutants in this gene family to date has been attributed to gene redundancy (16). Here we report that removing a single NCR peptide (NCR211) leads to defects in nitrogen fixation. Phenotypic characterization suggests that dnf4 causes problems in the symbiosis at stages later than other dnf mutants, such as dnf1, dnf2, and dnf5, because a number of late symbiotic genes from bacteria (nodF, bacA and nifH) and plants (LB1, CAM1 and Nodulin 31) are still expressed in dnf4 (12). This classification is supported by our microscopic analyses demonstrating that differentiating bacteroids are present in dnf4 mutant nodules, in contrast to those of dnf1 and dnf2, in which the bacteroids barely differentiate (6, 13). Furthermore, PI staining of differentiated bacteroids in the dnf4 mutant suggests that NCR211 functions to maintain the viability of differentiating bacteroids. The loss of bacterial viability in dnf4 is further supported by the lack of signals from the bacterial GFP reporter in PI-stained bacteroids (Fig. 1D), because doubly fluorescent bacteroids are rarely observed.
The deletion in the dnf4 mutant includes two NCR genes, NCR211 and NCR178. Although these are the closest homologs based on mature peptide sequences, only NCR211 can rescue the dnf4 phenotype. The promoter-GUS activity, LCM RNA-seq, and microarray data show similar temporal and spatial expression patterns for NCR211 and NCR178; however, the expression level of NCR211 is much higher than that of NCR178. Therefore, the lack of complementation of dnf4 by NCR178 might be related to its lower transcript level. An alternative explanation is that the differences in sequences of the two peptides might be sufficient to confer different biochemical activities and distinct biological functions. Promoter swapping analysis of the two genes could answer this question.
Structurally similar to defensins, some NCR peptides have bactericidal activities at high concentrations. At lower concentrations, certain NCR peptides induce membrane permeabilization, genome amplification, and cell elongation, which are features of terminal bacteroid differentiation in IRLC legumes (5). One of the NCR peptides, NCR247, was recently shown to bind many bacterial proteins, including ribosomal proteins and GroEL, indicating that this peptide may control bacteroids through multiple mechanisms (10). Massive transcriptome changes (∼15% of the genome) by NCR247 are likely the result of numerous compromised cellular targets (10).
DNF4 is very different from NCR247 in terms of amino acid composition, charge, and length. Furthermore, whereas NCR247 has been reported to penetrate bacteroid membranes, DNF4-GFP is observed in the peribacteroid space (Fig. 4D), although this observation requires further verification using additional methods. The fact that nodules without DNF4 contain differentiating bacteroids indicates that DNF4 acts after the bacteroids have initiated the differentiation program. Based on these observations, we propose that DNF4 functions to promote the survival of intracellular bacteroids during their differentiation. The expression of DNF4 is high in the older parts of zone II and the highest in interzone II-III, where bacteroids undergo the final steps of differentiation before reaching maximum size. It appears that DNF4 may be maximally expressed in these cells to protect terminally differentiating bacteria from otherwise lethal conditions. In the absence of DNF4, these bacteria perish either before or after reaching full differentiation. The expression pattern of DNF4 is similar to that of NCR247 and also overlaps with the pattern of NCR035 in interzone II-III (5, 10); therefore, DNF4 could be in a position to balance the differentiation induced by other NCRs, such as NCR247 and NCR035. Finally, our finding that synthetic NCR211 is active on S. meliloti in culture suggests that this effect may be direct.
At first sight, it may appear perplexing that DNF4/NCR211 supports the survival of differentiating bacteroids in planta while also blocking free-living bacteria from forming colonies in culture. These two activities may reflect the same mode of action by NCR211 on bacterial biology, however. Differentiating bacteroids and free-living bacteria have different physiologies. The manipulation of the same bacterial target by DNF4 in these two very different physiological contexts could manifest itself in distinct outcomes. For bacteria, differentiation can be stressful, or even lethal if left unprotected. The induction of DNF4 could provide a timely intervention to establish a sublethal, stable state in bacteroids. DNF4’s action may be detrimental to free-living and proliferating bacteria, however. Uncontrolled proliferation of the intracellular bacterial population is clearly a risk to the host. The dual effect of DNF4/NCR211 may reflect a mechanism to ensure that the rhizobia stay in a properly differentiated state.
Host control of terminal bacteroid differentiation has evolved in multiple lineages of legumes, indicating a possible fitness benefit to the host plant (20). Furthermore, nodules with differentiated bacteroids returned more benefit to the host (20). In an accompanying report, Horváth et al. (28) identified M. truncatula DNF7 encoding NCR169, suggesting that more than one NCR peptide can be indispensable for the nitrogen-fixing symbiosis. In another companion study, Price et al. (29) recovered a rhizobial peptidase capable of degrading host NCR peptides. This collection of discoveries demonstrates the evolving nature in controlling bacterial differentiation in classical host–microbe mutualism.
Materials and Methods
Plant Materials and Growth Conditions.
M. truncatula Jemalong ecotype and dnf4 mutant plants were grown in vermiculite in a growth chamber (22/18 °C, 16-h/8-h day/night) under fluorescent lamps (∼100 µmol·m−2·s−1). Plants were inoculated with Sinorhizobium medicae strain ABS7 carrying pHemA::lacZ (21), S. meliloti Rm1021 carrying pNifH::uidA (12), or Rm1021 carrying pTrp::GFP (22).
Plasmid Constructs.
For promoter-GUS fusion constructs, genomic DNA was amplified with the primers DNF41-F1 (5′-ggatccCGGAGTGTAGGGGTGATGTT-3′) and DNF41-R3 (5′-CAAACTTGAGAATTTCAGCCA-3′) for NCR211, as well as DNF42-F10 (5′-ggatccCCTC-TTAAATTGATTAAAGGC-3′) and DNF42-R5 (5′-TTTTTTTTTTAACTTTGCCTCT-3′) for NCR178. The products were cloned into the pCR8/GW/TOPO vector (Life Technologies), checked for sequence errors, and transferred to the pMDC163 binary vector (23) by LR recombination. For the translational fusions of GFP to the C-termini of NCRs, genomic DNA was amplified with primers DNF41-F1 and DNF41-R4 (5′-CTTGGGATAGCTCACA-CAATT-3′) for NCR211 and DNF42-F10 and DNF42-R6 (5′-GTGGGTACGACAAT-CACAA-3′) for NCR178, and then cloned into the pCR8/GW/TOPO vector. The Gateway-compatible GFP vector pK7FWG2-R (24) was cut with HindIII/SpeI, blunted with Klenow enzyme, and self-ligated to remove the 35S promoter in front of the Gateway cassette, resulting in pMK77. The inserts in the pCR8/GW/TOPO vector were checked for sequence errors and then transferred to the pMK77 binary vector by LR recombination.
Microscopy.
For toluidine blue staining, nodules were fixed in 2.5% (vol/vol) glutaraldehyde in 0.05 M cacodylate buffer, washed, dehydrated, and embedded in Technovit 7100 (Heraeus Kulzer), according to the manufacturer’s instructions. Then 5-µm sections were cut on a microtome (Reichert-Jung) and stained with 0.05% toluidine blue solution in 1× PBS buffer. Photographs were taken with an Eclipse TE2000-S microscope (Nikon).
Transgenic nodules were selected based on DsRED1 expression using an Olympus SZ61 binocular fitted with an ET590lp optical filter (Chroma Technology). A Nikon NI-150 illuminator fitted with an ET560/40x optical filter cube (Chroma Technology) was used for excitation of DsRED1.
PI staining was done with hand-sectioned nodules at 30 µM for 20 min. Sections were then briefly washed with water and observed under a confocal microscope (Olympus FluoView FV1000).
The GFP/SYTO9 (Life Technologies) signal was detected by excitation with a 473-nm laser and emission with a 490- to 540-nm bandpass filter. The PI/DsRED1 signal was detected by excitation with a 559-nm laser and emission with a 575- to 675-nm bandpass filter.
SDS/PAGE and Immunoblot Analysis.
Nodule samples were harvested from 21-dpi plants. Total protein extracts were prepared by grinding the samples in a 1.5-mL microtube with SDS sample buffer [60 mM Tris⋅HCl pH 6.8, 60 mM DTT, 2% (wt/vol) SDS, 15% (wt/vol) sucrose, 5 mM ε-amino-N-caproic acid, and 1 mM benzamidine]. Protein concentration was measured with a Coomassie blue binding assay (25). Here 10 μg of total nodule proteins were separated by SDS/PAGE and blotted onto a nitrocellulose membrane for immunoblot analysis. The membrane was probed with an antibody against alfalfa leghemoglobin provided by Dr. Carroll Vance (University of Minnesota). A secondary antibody conjugated with horseradish peroxidase was incubated with the membrane. The signal was visualized by enhanced chemiluminescence (Thermo Scientific).
Identification of the DNF4 Gene.
The dnf4 mutant was isolated from mutagenized M. truncatula Gaertn. cv Jemalong seeds created by fast neutron bombardment (12). The dnf4 mutant was crossed with the A20 ecotype for rough mapping. The location was narrowed down to the upper arm of chromosome 4 between markers 003F07 and h2_133k2b. Then genomic DNA was extracted from a pool of 40 mutant plants isolated from the mapping population and sent to the J. Craig Venter Institute for whole-genome sequencing using an Illumina HiSeq. Visualization of sequence reads was done with the Integrative Genomics Viewer (26). The primers used to amplify across the deletion break point in esn1 are esn1-LB (5′-GGTCCTGAAGTACCATATCTTAGT-3′) and esn1-RB (5′-GATTAAGTGCAAGTATACAT-TGTCC-3′).
Complementation of the dnf4 Mutant.
Genomic DNA sequences encoding NCR211 (Medtr4g035705) and NCR178 (Medtr4g035725) were amplified and cloned into pCR8/GW/TOPO with primers DNF41-F1 and DNF41-R1 (5′-ggatccGGAGGGGGTCTAT-GAGAGCA-3′) for NCR211 and DNF42-F1 (5′-ggatccGCAGGTTTGACATCCTCACC-3′) and DNF42-R1 (5′- CCTCACTAATCACTGACGGACC-3′) for NCR178. The cloned inserts were checked for errors and transferred to a binary vector, pKGW-R (24). Hairy root transformation was performed with Agrobacterium rhizogenes strain ARqua1 harboring the constructs according to the method outlined by Boisson-Dernier et al. (27). More than 10 independent transgenic roots were selected by red fluorescence of the DsRED1 marker gene for scoring of nodule phenotype.
GUS Staining.
Roots with nodules were fixed in cold 90% (vol/vol) acetone for 30 min and then rinsed with cold water. Samples were then immersed in 50 mM sodium phosphate buffer (pH 7.2) containing 2 mM 5-bromo-4-chloro-3-indoxyl-β-d-glucuronide cyclohexylammonium salt (X-Gluc), 0.2% Triton X-100, and 2 mM each potassium ferricyanide and ferrocyanide under vacuum for 30 min, followed by further incubation at 37 °C for 2–5 h. The nodules were embedded in 6% (wt/vol) low-melting agarose and sectioned in 70-µm slices with a vibratome (Vibratome 100 Plus). Nodules from more than five independently transformed roots showed the same GUS staining pattern.
Peptide Treatment of Free-Living S. meliloti.
S. meliloti Rm1021 cultures were grown to an OD600 of 0.3–0.6 in LB media and washed with 5 mM Mes-KOH buffer at pH 5.8. Then 200 µL of bacteria (diluted to an OD600 of 0.1) were treated with peptides (synthesized by GenScript) at the indicated concentrations and incubated for 3–4 h at 30 °C. After the peptide treatment, bacterial suspensions were serially diluted and plated out in triplicate on selective media. Colony-forming units per milliliter were measured after 2 d of incubation at 30 °C.
Acknowledgments
We thank Erik Limpens for the binary vectors with a fluorescence marker, Carroll Vance for the anti-leghemoglobin antisera, Siyeon Rhee for technical assistance with toluidine blue staining of nodules, Alice Cheung for access to a microtome, Tobias Baskin for access to a vibratome, and Jeanne Harris and Peter Chien for insightful discussions of the manuscript. Funding was provided by the University of Massachusetts (D.W.) and by the Samuel Roberts Noble Foundation and the National Science Foundation (Grant IOS-1127155, to R.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1500123112/-/DCSupplemental.
References
- 1.Vasse J, de Billy F, Camut S, Truchet G. Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J Bacteriol. 1990;172(8):4295–4306. doi: 10.1128/jb.172.8.4295-4306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gavrin A, et al. Adjustment of host cells for accommodation of symbiotic bacteria: Vacuole defunctionalization, HOPS suppression, and TIP1g retargeting in Medicago. Plant Cell. 2014;26(9):3809–3822. doi: 10.1105/tpc.114.128736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cebolla A, et al. The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J. 1999;18(16):4476–4484. doi: 10.1093/emboj/18.16.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kondorosi E, Kondorosi A. Endoreduplication and activation of the anaphase-promoting complex during symbiotic cell development. FEBS Lett. 2004;567(1):152–157. doi: 10.1016/j.febslet.2004.04.075. [DOI] [PubMed] [Google Scholar]
- 5.Van de Velde W, et al. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science. 2010;327(5969):1122–1126. doi: 10.1126/science.1184057. [DOI] [PubMed] [Google Scholar]
- 6.Wang D, et al. A nodule-specific protein secretory pathway required for nitrogen-fixing symbiosis. Science. 2010;327(5969):1126–1129. doi: 10.1126/science.1184096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fedorova M, et al. Genome-wide identification of nodule-specific transcripts in the model legume Medicago truncatula. Plant Physiol. 2002;130(2):519–537. doi: 10.1104/pp.006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mergaert P, et al. A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol. 2003;132(1):161–173. doi: 10.1104/pp.102.018192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maróti G, Kondorosi E. Nitrogen-fixing Rhizobium-legume symbiosis: Are polyploidy and host peptide-governed symbiont differentiation general principles of endosymbiosis? Front Microbiol. 2014;5:326. doi: 10.3389/fmicb.2014.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farkas A, et al. Medicago truncatula symbiotic peptide NCR247 contributes to bacteroid differentiation through multiple mechanisms. Proc Natl Acad Sci USA. 2014;111(14):5183–5188. doi: 10.1073/pnas.1404169111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Penterman J, et al. Host plant peptides elicit a transcriptional response to control the Sinorhizobium meliloti cell cycle during symbiosis. Proc Natl Acad Sci USA. 2014;111(9):3561–3566. doi: 10.1073/pnas.1400450111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Starker CG, Parra-Colmenares AL, Smith L, Mitra RM, Long SR. Nitrogen fixation mutants of Medicago truncatula fail to support plant and bacterial symbiotic gene expression. Plant Physiol. 2006;140(2):671–680. doi: 10.1104/pp.105.072132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourcy M, et al. Medicago truncatula DNF2 is a PI-PLC-XD–containing protein required for bacteroid persistence and prevention of nodule early senescence and defense-like reactions. New Phytol. 2013;197(4):1250–1261. doi: 10.1111/nph.12091. [DOI] [PubMed] [Google Scholar]
- 14.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 15.Xi J, Chen Y, Nakashima J, Wang SM, Chen R. Medicago truncatula esn1 defines a genetic locus involved in nodule senescence and symbiotic nitrogen fixation. Mol Plant Microbe Interact. 2013;26(8):893–902. doi: 10.1094/MPMI-02-13-0043-R. [DOI] [PubMed] [Google Scholar]
- 16.Nallu S, et al. Regulatory patterns of a large family of defensin-like genes expressed in nodules of Medicago truncatula. PLoS One. 2013;8(4):e60355. doi: 10.1371/journal.pone.0060355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roux B, et al. An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 2014;77(6):817–837. doi: 10.1111/tpj.12442. [DOI] [PubMed] [Google Scholar]
- 18.Zhang N, et al. Solution structure of BmKK2, a new potassium channel blocker from the venom of Chinese scorpion Buthus martensi Karsch. Proteins. 2004;55(4):835–845. doi: 10.1002/prot.20117. [DOI] [PubMed] [Google Scholar]
- 19.Zhou P, et al. Detecting small plant peptides using SPADA (Small Peptide Alignment Discovery Application) BMC Bioinformatics. 2013;14:335. doi: 10.1186/1471-2105-14-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oono R, Denison RF. Comparing symbiotic efficiency between swollen versus nonswollen rhizobial bacteroids. Plant Physiol. 2010;154(3):1541–1548. doi: 10.1104/pp.110.163436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leong SA, Williams PH, Ditta GS. Analysis of the 5′ regulatory region of the gene for delta-aminolevulinic acid synthetase of Rhizobium meliloti. Nucleic Acids Res. 1985;13(16):5965–5976. doi: 10.1093/nar/13.16.5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gage DJ. Analysis of infection thread development using Gfp- and DsRed-expressing Sinorhizobium meliloti. J Bacteriol. 2002;184(24):7042–7046. doi: 10.1128/JB.184.24.7042-7046.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133(2):462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smit P, et al. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science. 2005;308(5729):1789–1791. doi: 10.1126/science.1111025. [DOI] [PubMed] [Google Scholar]
- 25.Hong SW, Vierling E. Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high-temperature stress. Proc Natl Acad Sci USA. 2000;97(8):4392–4397. doi: 10.1073/pnas.97.8.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boisson-Dernier A, et al. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact. 2001;14(6):695–700. doi: 10.1094/MPMI.2001.14.6.695. [DOI] [PubMed] [Google Scholar]
- 28.Horváth B, et al. Loss of the nodule-specific cysteine rich peptide, NCR169, abolishes symbiotic nitrogen fixation in the Medicago truncatula dnf7 mutant. Proc Natl Acad Sci USA. 2015;112:15232–15237. doi: 10.1073/pnas.1500777112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price PA, et al. Rhizobial peptidase HrrP cleaves host-encoded signaling peptides and mediates symbiotic compatibility. Proc Natl Acad Sci USA. 2015;112:15244–15249. doi: 10.1073/pnas.1417797112. [DOI] [PMC free article] [PubMed] [Google Scholar]