Significance
A high-resolution structure was obtained for a drug candidate achieving pharmacological activity by inducing and stabilizing protein–protein interaction, a mechanism difficult to study in structural biology. We found that with poorly diffracting protein crystals, a protein stabilizing compound can improve crystal quality and enable the acquisition of a high-resolution structure. It also becomes apparent from this structure how improvements in pharmacologic potency can be achieved by improving protein–protein interaction stabilization and clear avenues for compound optimization are apparent from the data. The binding site observed in crystallography was biologically validated by mutational analysis, which also provides for the first time, to our knowledge, an understanding of a pathway by which viable, drug resistant virus variants may evolve against this drug class.
Keywords: HBV treatment, HBV inhibitor, core, capsid, protein–protein interaction
Abstract
The hepatitis B virus (HBV) core protein is essential for HBV replication and an important target for antiviral drug discovery. We report the first, to our knowledge, high-resolution crystal structure of an antiviral compound bound to the HBV core protein. The compound NVR-010–001-E2 can induce assembly of the HBV core wild-type and Y132A mutant proteins and thermostabilize the proteins with a Tm increase of more than 10 °C. NVR-010–001-E2 binds at the dimer–dimer interface of the core proteins, forms a new interaction surface promoting protein–protein interaction, induces protein assembly, and increases stability. The impact of naturally occurring core protein mutations on antiviral activity correlates with NVR-010–001-E2 binding interactions determined by crystallography. The crystal structure provides understanding of a drug efficacy mechanism related to the induction and stabilization of protein–protein interactions and enables structure-guided design to improve antiviral potency and drug-like properties.
Hepatitis B virus (HBV) infection is a major global health concern. It is estimated that 240 million people live with chronic hepatitis B (CHB) with a high risk of developing severe liver disease or liver cancer. Globally, 780,000 deaths per year are associated with HBV infection (1). Current approved treatment options (IFN alpha products or nucleoside analogs) are indicated for treatment of only a subset of CHB patients and are curative in only a very small proportion of patients, resulting in an urgent need for new types of therapies that can increase HBV cure rates.
The HBV core protein is a viral protein with no known related protein present in human cells. The core protein performs multiple essential roles at different stages of the virus life cycle; it interacts with other host and viral proteins and has to be able to form a capsid stable enough to protect viral RNA and DNA and be able to release viral DNA at the right time in the virus life cycle. HBV core protein is therefore an excellent target for the development of new, virus-selective, safe and effective antiviral agents to improve treatment options for this disease (2, 3). The HBV core protein consists of 183–185 amino acids that form an N-terminal (amino acids 1–149) capsid assembly domain and a C-terminal nucleic acid binding domain (amino acids 150–185). The viral capsid in infectious HBV particles is formed from 120 copies of assembled core protein dimers enclosing the viral DNA (4, 5). Small molecules that target the HBV core protein assembly domain can disrupt functional HBV capsid assembly and can be potent inhibitors of HBV replication (6–11). Most of these compounds have the ability to induce aggregation of the HBV core N-terminal assembly domain in vitro. Representative HBV core inhibitors from the heteroaryldihydropyrimidine (HAP) series of compounds, BAY 41–4109 and GLS4, have also shown antiviral activity in vivo, in mouse models of HBV replication (12–14). However, drug-like properties of current leads have not been optimized, and new classes of HBV core protein targeting compounds need to be developed to maximize antiviral efficacy as well as pharmacokinetic and safety profiles. Lead optimization has been hampered by the lack of high-resolution crystal structures. Crystal structures of assembled wild-type HBV capsid have only been obtained at resolutions above 3 Å, and structures including bound small-molecule inhibitors have been determined at 4.2- and 5.05-Å resolution. Considering these resolution limits together with the technical and time challenges in achieving these structures, the previously published methods are insufficient to support structure-guided lead optimization (5, 15, 16).
Here we present the structure of NVR-010–001-E2, a potent inhibitor of HBV replication from the HAP series of antiviral agents, bound to its target site on the HBV core protein at 1.95-Å resolution. The impact of mutations in the binding site on antiviral activity of NVR-010–001-E2 is consistent with the compound–protein interactions observed in the crystal structure and provides a starting point for the evaluation of possible pathways to drug resistance development. The data presented here provide a structural explanation for the ability of a small molecule to induce protein assembly and exemplify an efficient method to facilitate structure-guided optimization of HBV core inhibitors.
Results
HBV Replication Inhibitors Can Induce Assembly of Wild-Type and Y132A Mutant HBV Core Protein Assembly Domain.
The heteroaryldihydropyrimidine (HAP) series of compounds, represented by compound BAY 41–4109, was first described more than 12 years ago. It was shown that HAP compounds could be potent inhibitors of HBV replication, and they were found to target the N-terminal assembly domain of the HBV core protein (9, 13). Optimization of these early lead compounds has, however, been slow. In an effort to establish methods for structure-guided lead optimization we characterized the interaction of representative compounds from the HAP series (Fig. 1A) with the N-terminal assembly domain of the HBV core protein N-terminal domain (CoreND) and the CoreND protein variant with a single point mutation at position Y132A (CoreND-Y132A). The tyrosine residue at position 132 is a key amino acid to stabilize the interaction between HBV core protein dimers, and this interaction is essential for the formation of icosahedral capsid structures. When this tyrosine is mutated to alanine, the protein becomes deficient in capsid assembly, although it remains capable of coassembly with wild-type core protein (17, 18). Crystallization of the Y132A variant has been achieved and resulted in improved resolution compared with previous results with wild-type protein capsid (18, 19).
Fig. 1.
Structure and activity of HBV core inhibitors. (A) Chemical structures. (B) Antiviral activity in HepG2.2.15 cells, NVR 010–001-E2 (squares), GLS4 (circles), BAY 41–4109 (triangles), and BAY 41–4109-IE (diamonds). (C) Thermal shift assay using wild-type CoreND protein in the absence of compound (DMSO, black line), in the presence of BAY 41–4109-IE (orange, wide dashed line), BAY 41–4109 (red, dotted line), GLS4 (NVR-010–002-E2, blue, dot-dashed line), and NVR-010–001-E2 (black, dashed line). (D) Antiviral activity in HepG2.2.15 cells. Data shown are mean values (± SD).
Recombinant CoreND and CoreND-Y132A proteins were purified from Escherichia coli. In agreement with previous reports, CoreND was a stable dimer in solutions of up to 150 mM NaCl at high pH (pH 9.6) and could assemble into capsids at higher salt concentrations and lower pH that could be visualized by electron microscopy (SI Appendix, Fig. S1 A and C). CoreND-Y132A was a stable dimer in solution up to at least 1 M NaCl and at neutral pH, consistent with the previously described deficiency in capsid assembly (SI Appendix, Fig. S1 B and D). The HAP compound BAY 41–4109 could induce the assembly of CoreND into larger than capsid structures, as observed by electron microscopy (SI Appendix, Fig. S1E). The (+) S enantiomer of BAY 41–4109, named BAY 41–4109-IE, did not show antiviral activity in cell culture and did not induce assembly of CoreND protein, whereas another HAP compound, NVR-010–001-E2, also induced assembly (SI Appendix, Fig. S1H) and the closely related compound GLS4 also showed the ability of inducing assembly of CoreND protein, as described below. BAY 41–4109, NVR-010–001-E2, and GLS4 were potent inhibitors of HBV replication in cell culture, as measured by the reduction of HBV DNA in the supernatant of HepG2.2.15 cells (Fig. 1 B and D). BAY 41–4109 inhibited HBV replication in HepG2.2.15 cells with a mean EC50 of 120 nM (range 85–170 nM), whereas its (+) S enantiomer BAY41-4109-IE did not inhibit HBV replication (EC50 > 30 µM). NVR-010–001-E2 and GLS4 were ∼10-fold more potent than BAY41-4109, with mean EC50 values of 11 nM (range 8.1–13 nM) and 15 nM (range 10–21 nM) (Fig. 1 B and D). In a thermal shift assay the dimeric CoreND protein showed temperature-induced unfolding, with a mean melting temperature (Tm) of 65 °C (Fig. 1C). The addition of the inactive enantiomer BAY 41–4109-IE did not affect the unfolding profile, whereas BAY 41–4109, NVR-010–001-E2, and GLS4 increased the Tm by mean 11 °C, 17 °C, and 17 °C, respectively (Fig. 1C). The active antiviral compounds could also increase the thermal stability of HBV capsid formed from recombinant CoreND protein. Capsid alone or in the presence of BAY 41–4109-IE showed a mean Tm of 79 °C. BAY 41–4109, NVR-010–001-E2, and GLS4 increased the Tm by mean 4 °C, 10 °C, and 11 °C, respectively (SI Appendix, Fig. S2A), and could therefore further stabilize HBV protein even from preformed capsid structures. The inactive enantiomer BAY 41–4109-IE did not affect the thermal stability of the capsid.
The active compounds BAY 41–4109, NVR 010–001-E2, and GLS4 could also induce assembly of the Y132A mutant protein (CoreND-Y132A), although this mutant protein was not amenable to salt-induced assembly, as observed by electron microscopy (SI Appendix, Fig. S1). The larger protein structures induced by BAY 41–4109 were sheets or tubes, whereas those induced by NVR 010–001-E2 were large, rounded open structures, similar to those induced with wild-type protein. A dose-dependent induction of CoreND-Y132A assembly by the compounds could also be observed with analytical size exclusion chromatography by the emergence of a defined new peak that eluted early, with an apparent molecular weight of ∼2.4 MDa. (SI Appendix, Fig. S3). NVR-010–001-E2 and BAY 41–4109 could fully convert the protein dimer into the larger molecular structures (SI Appendix, Fig. S3). The binding of NVR-010–001-E2 or GLS4 to CoreND-Y132A was associated with a significant increase in Tm, as determined by fluorescent thermal shift analysis (TSA) (SI Appendix, Fig. S2B). The CoreND-Y132A dimer protein unfolded with a mean Tm value of 61 °C, slightly lower than that of wild-type dimer. The temperature-induced unfolding was shifted by a mean 22 °C and 20 °C by NVR-010–001-E2 and GLS4, respectively. Interestingly, the protein structures induced from CoreND-Y132A by BAY 41–4109 (SI Appendix, Figs. S1 and S3) showed similar Tm compared with the dimer protein (SI Appendix, Fig. S2B).
Crystal Structure of NVR-010–001-E2 Bound to the HBV Core Protein.
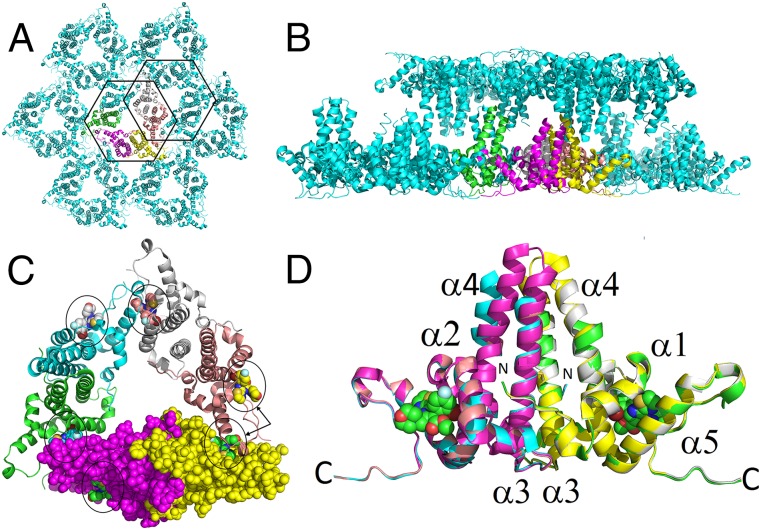
The available crystal structures of small molecules bound to HBV capsid indicate that HAP compounds bind to the interface between HBV core protein dimer assembly domains. However, the lengthy and inefficient process of capsid crystallization and the low resolution of the resulting structures prevent that method from being suitable for use in structure-guided drug design (5, 15, 16). After observing the ability of antiviral core inhibitors to stabilize HBV core protein, including the Y132A mutant protein, by inducing stabilized capsid-like structures, we investigated if such protein stabilization could increase the quality and resolution of HBV core protein crystals. Crystals of the CoreND-Y132A protein were obtained within 3 days from a 10 mg/mL protein solution. The presence of NVR-010–001-E2 could significantly enhance the quality of the protein crystals, achieving high-resolution diffraction up to 1.95 Å, whereas in the absence of the compound, resolution remained above 3 Å. Final data collection and refinement statistics are summarized in Table 1. The asymmetric unit was a closed trimer of dimers, representing the HBV capsid assembly intermediate or nucleation complex. In the crystal, these hexamers are positioned in a head-to-head arrangement. Fig. 2 shows the hexamers in the asymmetric units and an overlay of the three dimers that form the closed trimer of dimers. There are six compound binding sites in the hexamer (trimer of dimers), indicated by circles in Fig. 2C, three sites representing dimer–dimer interactions within the hexamer of the asymmetric unit, and three sites representing hexamer–hexamer interactions, indicated on one representative dimer by arrows in Fig. 2C. The structures of the three dimers are very similar, with differences only in the stalk region. Compared with dimer A-B, the ends of the stalks in dimers C-D and E-F were disordered, indicating structural flexibility at the stalk tips, and helix 4 in dimer C-D shows a kink.
Table 1.
Data collection and refinement statistics
| Parameters | CoreND-Y132A + NVR 010–001-E2 |
| Radiation source | APS 21-ID-f |
| Wavelength (Å) | 0.9787 |
| Resolution range (last shell) (Å) | 50–1.95 (2.0–1.95) |
| Space group | C2 |
| Unit cell (Å, deg) | 152.6, 88.2 102.25, 90, 131.5, 90 |
| Unique reflections | 95,947 (7,085) |
| Multiplicity | 4.7 (4.8) |
| Completeness (%) | 99.9 (100) |
| Mean I/ sigma(I) | 11.35 (3.1) |
| R-sym | 0.098 (0.506) |
| Reflections used for refinement | 95,947(6,696) |
| Reflections used for R-free | 4,810 (379) |
| R-work/R-free | 0.219/0.252 |
| Number of nonhydrogen atoms | |
| Protein | 3,544 |
| Ligand | 180 |
| Water/other | 441 |
| RMSD bonds (Å) | 0.015 |
| RMSD angles (°) | 1.70 |
| Ramachandran favored (%) | 96 |
| Ramachandran outliers (%) | 0 |
| Molprobity clashscore | 4.0 (99th) |
| Average B-factor | |
| Protein | 20.7 |
| Ligands | 20.2 |
| Water/other | 31.4 |
Fig. 2.
Crystal structure of CoreND-Y132A protein bound to NVR-010–001-E2. (A) The hexamer in the asymmetric unit is shown in color. (B) Packing of sheets derived from hexamers showing the head-to-head arrangement of the proteins. (C) Hexamer with the six bound NVR-010–001-E2 molecules highlighted by circles. Individual monomers are colored yellow (monomer A), magenta (monomer B), green (monomer C), cyan (monomer D), gray (monomer E), and salmon (monomer F). Monomers A and B are shown in space-filling mode. The arrows towards monomer A (yellow) and F (salmon) highlight the two different compound binding sites corresponding to intrahexamer (monomers A and F) and interhexamer (monomer F) interactions. (D) Overlay of the three dimers in the asymmetric unit. The structures vary only in the stalk region. For monomers C and D, the ends of the stalk helices are disordered, and helix 4 has a kink at amino acid 80. Two representative binding sites from monomers A and B are indicated by space-filling models of bound NVR-010–001-E2.
HAP Compound Interaction with HBV Core Protein.
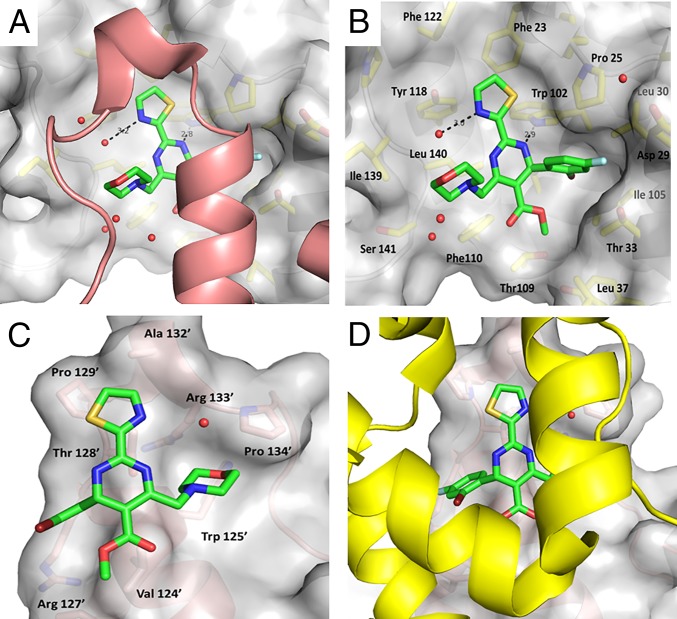
The electron density for NVR-010–001-E2 clearly shows the position of the ligand in the binding site at the dimer–dimer interface (Fig. 3). The compound binding site is composed of a binding surface and a hydrophobic pocket provided by one protein subunit, whereas the C-terminal helix (α5) and loop of the neighboring protein subunit forms a “lid” for additional interaction with the compound. The central pyrimidine of the NVR-010–001-E2 compound lies at the floor of the binding surface and anchors to the protein via a hydrogen bond between one of the nitrogens and the side chain of Trp102. Each substitution around the central pyrimidine then contributes additional binding affinity. The bromofluoro-substituted phenyl group fits into a well-defined, largely hydrophobic pocket created by Pro25, the hydrophobic face of Asp29, Leu30, and Thr33 from helix 4 and Trp102, Ile105, and Ser106 from helix 2, with the bromine pointing toward the protein and deep into the hydrophobic pocket. The fluoro-edge of the phenyl group interacts with the lid of the adjacent protein provided by residues Val124’, the alkyl chain of Arg127’, and Thr128’ (note that amino acid residues of the lid are numbered with apostrophes). The thiazole moiety of NVR-010–001-E2 also sits in a hydrophobic environment created by aromatic residues Trp102, Phe23, Phe122, and Tyr118. The thiazole nitrogen makes a water-mediated hydrogen bond to the backbone of Leu140. The thiazole also interacts with Thr128’ of the lid. The ester moiety of NVR-010–001-E2 sits at the apex between the stalk and the dimer interaction domains against a wall formed by Thr109 and Phe110 on one side and Thr33 and Leu37 on the other side. Interaction with the lid is through Val124’, whereas the free oxygen of the ester group remains solvent-exposed. The primary interactions of the morpholino group of NVR-010–001-E2 are with the lid of the neighboring dimer, which is formed by helix 5 and the loop following helix 5, in particular residues Val124’, Trp125’, Thr128’, and Pro134’. The multiple interactions of NVR-010–001-E2 with the binding surface and the lid are consistent with the potent ability of the compound to induce dimer–dimer interactions and HBV core protein assembly into large protein multimer structures. A 2D view of the compound interactions with the A (lid) and F (contact domain) monomers is shown in SI Appendix, Fig. S6.
Fig. 3.
Binding site of NVR-010–001-E2. (A) The ligand sits in the groove of the contact domain (gray surface) on one dimer and is covered by a lid formed by helix 5 from the adjacent dimer, colored in salmon. (B) Binding surface to the contact domain. The lid is removed for clarity. (C and D) Binding surface to the lid. The contact domain is removed for clarity in C and shown in yellow in D.
Mutational Analysis of the HAP Binding Site.
To test the role of specific compound–protein interactions in the core inhibitor binding site, we first investigated the conservation of amino acids and viability of binding site mutants for HBV replication. A total of 2,800 public core sequences across all eight HBV genotypes (A-H) were collected into an HBV core protein sequence database. The conservation and variability of the 24 amino acids that are located within 5 Å of the ligand in the binding pocket formed by protein monomers A and F of the core hexamer are shown in SI Appendix, Tables S1 and S2. Among the amino acids located within 5 Å of the ligand, only positions 105 and 109 showed a conservation of <99% when all 2,800 sequences were considered (SI Appendix, Table S1). When the amino acid frequencies were analyzed within individual HBV genotypes, positions 102 and 118 also showed a low-level amino acid variability (SI Appendix, Table S2).
The replication competence of these polymorphisms was evaluated using a transient transfection phenotyping assay based on a total of nine different core variants generated in the context of the HBV genotype B strain: W102G, W102R, I105L, I105T, I105V, T109I, T109M, T109S, and Y118F. Replication competence was determined by measuring intracellular encapsidated HBV DNA levels in HepG2 cells 3 days after transfection. As shown in SI Appendix, Fig. S4, compared with wild-type HBV, the two variants for Trp102 were both incompetent for HBV replication (<2%), whereas the other mutants showed sufficient replication capacity for further analysis.
The effect of the replication-competent single-point mutations on the sensitivity of HBV replication to inhibition by the HAP compounds NVR 010–001-E2 and BAY 41–4109 was determined by comparative dose–response analysis in the phenotyping assay (Table 2 and SI Appendix, Fig. S5). HBV genome containing the core Y118F variant was less susceptible to inhibition by both core inhibitors (Table 2). The antiviral activities of the HAP compounds were similar to wild type across the mutants at position 105. Mutations T109M and T109I were associated with decreased susceptibility, whereas the T109S variant was susceptible to inhibition by both HAP compounds (Table 2). The observed impact of mutations on the antiviral activity of the HAP compounds is consistent with the binding interactions determined by crystallography.
Table 2.
Antiviral activities of NVR 010–001-E2 and BAY 41–4109 against the replication of HBV variants with amino acid mutations in the HBV core protein sequence compared with wild-type virus
| HBV Variant | NVR-010–001-E2 EC50 (µM)* | NVR-010–001-E2 EC50 fold change† | BAY 41–4109 | BAY 41–4109 |
| EC50 (µM) | EC50 fold change† | |||
| Wild type | 0.038 ± 0.015 | 1 | 0.13 ± 0.023 | 1 |
| Y118F | 0.14 ± 0.034 | 3.7 | 1.1 ± 0.39 | 8.2 |
| I105V | 0.048 ± 0.015 | 1.3 | 0.17 ± 0.0015 | 1.3 |
| I105L | 0.019 ± 0.005 | 0.49 | 0.058 ± 0.013 | 0.44 |
| I105T | 0.047 ± 0.006 | 1.2 | 0.20 ± 0.037 | 1.5 |
| T109S | 0.016 ± 0.001 | 0.43 | 0.053 ± 0.013 | 0.40 |
| T109M | 0.11 ± 0.030 | 2.8 | 0.49 ± 0.051 | 3.7 |
| T109I | 0.35 ± 0.029 | 9.2 | 2.8 ± 0.89 | 21 |
Mean EC50 values and SDs from at least three independent studies determined in HepG2 cells.
Ratio of the mean EC50 value of core variants over wild-type HBV.
Discussion
The HBV core protein is an important target for the development of new therapies for improved treatment of HBV infection. The acquisition of high-resolution structural information of small-molecule binding to HBV core protein or capsid has proven difficult, progress in compound optimization has been very slow, and only recently have the first HBV core inhibitors entered clinical studies (20). The determination of the crystal structures of assembled capsids with or without small-molecule inhibitors is hampered by many technical hurdles, such as long purification protocols, slow-growing crystals, low-resolution diffraction limits, and challenging refinement (5, 15). Low-resolution (>4 Å) crystal structures with the assembly activators HAP-1 (15) and AT-130 (16) have been reported; however, at these low resolutions, the binding interactions of the small molecules are difficult to definitively model and cannot enable effective structure-guided lead optimization. The Y132A mutant of HBV core protein is unable to assemble into capsid by itself, but can form capsids when coassembled with wild-type protein (17). This protein remains dimeric in solution even under conditions that normally induce capsid assembly, such as high salt. This property has allowed for the generation of crystal structures of mutant HBV core protein in hexameric arrangements (trimers of dimers) that reflect the basic capsid building blocks (18, 19).
The CoreND-Y132A protein used in this study crystallized in a different crystal form compared with the previously reported structures 3KXS and 4BMG (18, 19). This could be due to the specific construct used for protein expression or differences in the protein purification or crystallization protocols. Whereas the crystals of the apoprotein diffracted to ∼3-Å resolution, we found that the addition of NVR-010–001-E2 could significantly stabilize the structure and increase resolution. When the CoreND-Y132A crystals were soaked with ligand NVR-010–001-E2, the diffraction improved dramatically to provide better than 2-Å resolution.
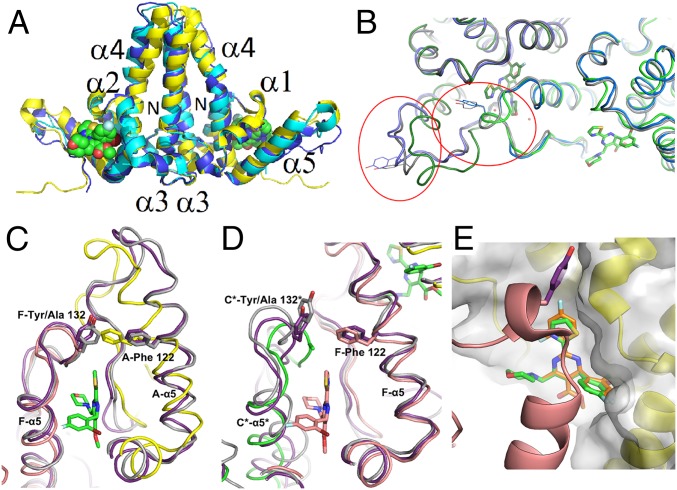
The CoreND-Y132A protein crystallized as a trimer of dimers, consistent with previous low-resolution structures. Fig. 4A shows an overlay of the A-B dimer from the current NVR 010–001-E2 bound structure with the two previously reported apoprotein structures. The backbones from the three dimers are similar in structure (RMSD 1.95 Å and 2.62 Å compared with 3KXS and 4BMG, respectively). The backbone of the NVR-010–001-E2 bound structure of the closed hexamer is most similar to 4BMG, which also crystallized as a closed hexamer (RMSD 2.42 Å across all six protein chains in the hexamer), compared with the open hexamer in 3KXS (18, 19). Structural differences between the A-B dimers are apparent in the stalk helices and around the C terminus where the interdimer interactions occur (Fig. 4A). This movement of the C terminus allows for the binding site to accommodate the inhibitor. The structures of the C-terminal regions are also different between the two apoprotein structures, 4BMG and 3KXS (RMSD 3.86 Å across all six chains), consistent with inherent flexibility of the core protein in this region. Because HBV capsid can assemble in both T = 4 and T = 3 forms, and two distinct hexameric forms of the protein are required for the formation of an icosahedral capsid structure, significant flexibility in the interdimer contact region is a functional requirement of the protein (4, 5, 15, 21). Structural flexibility has also been shown to extend to the stalk region, consistent with the structural disorder at the stalk tips in the crystal structure and with the high solvent content of this area of the structure (21).
Fig. 4.
Compound binding site comparisons. (A) Overlay of the dimer A-B (yellow, NVR-010–001-E2 shown in space-filling mode) with the A-B dimer of 3KXS (cyan, [18]) and 4BMG (blue, [19]). (B) Overlay of the NVR-010–001-E2 bound structure with wild-type capsid (2G33, blue, [15]) and wild-type capsid with compound HAP-1 putatively bound (3G34, gray, [15]). Helix 5 and the C-terminal tail are circled in red for the two corresponding dimers, with Tyr132 shown for the wild-type structures. (C and D) Overlay of the NVR 010–001-E2 bound structure with wild-type capsid (2G33, gray, [15]) and wild-type capsid with compound HAP-1 putatively bound (3G34, purple, [15]). (C) In the A-F interface, helix 5 of monomer A shifts so that Phe122 can fill the space normally occupied by Tyr132’ of monomer F (monomer A, yellow; monomer F, salmon). (D) The interface in the capsid between hexamers with monomers F (salmon) and C* from the next hexamer (green). (E) Overlay of the model of BAY 41–4109 (orange) on the structure of NVR-010–001-E2 (green).
The Tyr132 residue at the C-terminal end of the core protein assembly domain is an important mediator of interdimer interaction to facilitate core protein assembly in the process of capsid formation. The mutation of Tyr132 to Alanine renders the protein assembly deficient. NVR 010–001-E2, GLS4, and BAY 41–4109 could compensate for the lack of Tyr132 and induce assembly of Y132A mutant HBV core protein in solution. Fig. 4B illustrates the extensive interface that is formed between NVR 010–001-E2 and the two protein dimers to facilitate interdimer interaction. NVR 010–001-E2 provides an increased binding surface compared with Tyr132, consistent with the lack of requirement for Tyr132 and the indeed more effective induction of protein assembly and increased stabilization of the assembled protein compared with wild-type protein in the absence of NVR 010–001-E2.
Fig. 4 C and D shows an overlay of the compound-free and the HAP-1 bound capsid structures with the NVR 010–001-E2 bound structure. As an example for the intrahexamer dimer–dimer interaction, the overlay of the protein interface of monomers A (yellow) and F (salmon) with the same interface from the wild-type capsid structures 2G33 (gray) and 3G34 (purple) is shown in Fig. 4C. Helix 5 of the Y132A mutant protein is shifted and allows Phe-122 to occupy a similar location to that of Tyr132 in the lid to close the binding pocket above the thiazole moiety of NVR 010–001-E2. There was no apparent density of HAP-1 in this site in the capsid. As an example of the hexamer–hexamer interface, the overlay of monomers F (salmon) with monomer C* (green) from the next hexamer is shown in Fig. 4D. Helix 5 of the F monomer remains similar between the three structures, whereas the C* protein C terminus with Ala132 closed in toward the compound binding site. The differences between these structures may be related to the impact of curvature in the capsid, to compound induced adaptation (induced fit) of binding site structure, or a combination of both.
BAY41-4109 differs from NVR 010–001-E2 in three aspects (Fig. 1A): The bromine is replaced by a chlorine substitution, the thiazole is replaced by a difluoropyridine, and the morpholino group is missing. Fig. 4E shows the binding model of BAY 41–4109 based on the NVR 010–001-E2 structure. The most dramatic difference in binding site interaction is caused by the lack of the morpholino group and the resulting reduction in binding interaction with the “lid” of the binding site. The reduction in interdimer binding interaction of BAY 41–4109 compared with NVR 010–001-E2 is consistent with the reduced stability of BAY 41–4109 induced HBV core protein assemblies. The crystal structure also clearly explains the requirement for the correct stereochemistry of the pyrimidine substitution to allow the halogenated phenyl group to bind into the critical hydrophobic binding pocket on the core protein. The S(+) enantiomer BAY 41–4109-IE cannot effectively bind into this binding site, consistent with the lack of core protein binding, lack of assembly induction, and lack of antiviral activity of this compound.
The mutational analysis of the binding site was consistent with the structure of the binding site determined by crystallography. The largest impact on antiviral activity of both compounds was conferred by mutation T109I, which is a rare polymorphism present in 1.2% of the genotype B sequences in our assembled database of 2,800 core protein sequences. Thr109 forms a polar binding subsite to accommodate the polar ester group of NVR 010–001-E2 and BAY 41–4109. The replacement by the larger nonpolar Ile significantly affects ester binding in this area. Similarly, the replacement of Thr109 with the larger and less polar Met was associated with a resistance shift for both compounds. In contrast, the replacement of Thr109 with Ser resulted in a 2.5-fold improvement in antiviral activity for both compounds, consistent with the importance of polarity in this subsite. The conservative mutation of Tyr118 to Phe also affected the potency of both compounds. Tyr118 is involved in the formation of a hydrophobic environment with Trp102, Phe23, and Phe122 and is involved in a direct stacking interaction with the aromatic thiazole and pyridine moieties of the inhibitors. The change to Phe may change the geometry toward a less favorable interaction with the compounds in this subsite. These mutations occur as natural polymorphisms and show replication competence in HepG2 cells. They therefore provide a possible pathway for resistance development in patients treated with compounds of this chemical class. However, the HepG2 phenotypic assay system does not reflect all structural and functional requirements of the HBV core protein. For example, the HBV core protein functions that enable nuclear entry of HBV DNA, modulation of host gene expression, and cccDNA function are not all visible in this assay. It is therefore unknown at this time whether a virus with these mutations would be viable in vivo with sufficient fitness to enable chronic HBV infection and resistance development in patients treated with HAP compounds.
The high-resolution structure of NVR 010–001-E2 bound to CoreND-Y132A protein provides clear rational targets for efficacy optimization within the class of HAP compounds. Compared with BAY 41–4109, NVR 010–001-E2 has achieved an ∼10-fold improvement in antiviral activity. These improved antiviral properties are provided mostly by the presence of an additional structural moiety, a morpholino group, which adds significant interactions of the compound with the “lid” structure of another core protein dimer and therefore increases the ability of the compound to stabilize core protein dimer–dimer interactions and thus the ability to more effectively induce protein assembly into highly stable protein structures compared with BAY 41–4109 (Fig. 4E). The morpholino group is, however, not an optimal structural moiety at this position, and lead optimization in this compound class will therefore focus on replacing this group for further improved protein interaction and improved drug-like properties, such as increased metabolic stability and solubility. In summary, we have established a fast and robust method to allow the generation of the first to our knowledge high-resolution crystal structure of HBV core protein bound to small-molecule inhibitors. The structure will allow faster rational optimization of antiviral leads into development candidates with improved antiviral and pharmacologic properties.
Materials and Methods
Detailed information on methods is provided in SI Appendix.
Compounds.
GLS4, NVR-010–001-E2, BAY 41–4109, and the inactive enantiomer of BAY41-4109 (BAY41-4109-IE) were synthesized by WuXi AppTec. All compounds were dissolved in DMSO to generate 20-mM stock solutions.
Protein Expression and Purification.
HBV core protein N-terminal domain (amino acids 1–149) of HBV genotype D strain adyw was expressed in E. coli. The protein used for crystallography was expressed with a C-terminal His6 tag, which was cleaved with TEV protease, leaving additional C-terminal amino acid residues, KLENLYFQ. The proteins used for biochemical experiments were expressed with N-terminal His6_SUMO tags that could be removed by cleavage with Ulp-1 protease (22).
Electron Microscopy.
Electron microscopy with HBV capsid formed from recombinant HBV core protein was performed as described (23).
Thermal Shift Analysis.
Assays were performed at a final concentration of 4 μM HBV core protein monomer, 25 μM ligands, 1% DMSO. Melting temperatures were measured after 1 h incubation.
Analytical Size Exclusion Chromatography.
HPLC experiments were carried out at 4 °C using a Superdex 200 10/300 GL column.
Crystallization.
CoreND-Y132A protein at 10mg/mL in 50mM Tris pH 9.0, 2 mM dithiothreitol DTT was combined with reservoir solution containing 100 mM ammonium citrate/citric acid pH 6.5, 9% (vol/vol) isopropanol, 10% (wt/vol) PEG 3350, supplemented with 10% (vol/vol) 2-Methyl-2,4-pentanediol at a 2:1 volume ratio at 20 °C. Crystals were soaked overnight with compound NVR-010–001-E2. Final data collection and refinement statistics are summarized in Table 1 (PDB code 5E0I).
Antiviral Assays.
Antiviral activity was determined in HepG2.2.15 cells.
Transient Transfection Phenotyping.
Plasmid DNA containing a 1.1x genotype B HBV genome under the control of a cytomegalovirus (CMV) promoter was previously cloned from serum of an HBV-infected patient (GenBank AY220698, Fudan University, China) (24). HBV core variants W102G/R, I105V/L/T, T109S/M/I, and Y118F were generated by site-directed mutagenesis. HepG2 cells were transfected with HBV plasmids.
Supplementary Material
Acknowledgments
This work was sponsored by Novira Therapeutics. We thank Donald Lorimer for his contribution to crystallization trials and crystallography peer review, Adel Naylor-Olsen for scientific discussion and help with figure preparations, and Stefan Becker for crystallography peer review, scientific discussion, and critical reading.
Footnotes
Conflict of interest statement: The authors are employees of Novira Therapeutics (K.K., A.M.L., R.V., S.R., C.E., G.H., and O.A.F.), Beryllium (C.L., R.B., K.A., and J.A.), and Vista Informatics Corporation (G.L.).
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5E0I).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1513803112/-/DCSupplemental.
References
- 1.Cowie BC, Carville KS, MacLachlan JH. Mortality due to viral hepatitis in the Global Burden of Disease Study 2010: New evidence of an urgent global public health priority demanding action. Antivir Ther. 2013;18(8):953–954. doi: 10.3851/IMP2654. [DOI] [PubMed] [Google Scholar]
- 2.Klumpp K, Crépin T. Capsid proteins of enveloped viruses as antiviral drug targets. Curr Opin Virol. 2014;5:63–71. doi: 10.1016/j.coviro.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Seeger C, Zoulim F, Mason WS. Hepadnaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Vol II. Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 2185–2221. [Google Scholar]
- 4.Dryden KA, et al. Native hepatitis B virions and capsids visualized by electron cryomicroscopy. Mol Cell. 2006;22(6):843–850. doi: 10.1016/j.molcel.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 5.Wynne SA, Crowther RA, Leslie AG. The crystal structure of the human hepatitis B virus capsid. Mol Cell. 1999;3(6):771–780. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- 6.Bourne C, et al. Small-molecule effectors of hepatitis B virus capsid assembly give insight into virus life cycle. J Virol. 2008;82(20):10262–10270. doi: 10.1128/JVI.01360-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campagna MR, et al. Sulfamoylbenzamide derivatives inhibit the assembly of hepatitis B virus nucleocapsids. J Virol. 2013;87(12):6931–6942. doi: 10.1128/JVI.00582-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho MH, Jeong H, Kim YS, Kim JW, Jung G. 2-amino-N-(2,6-dichloropyridin-3-yl)acetamide derivatives as a novel class of HBV capsid assembly inhibitor. J Viral Hepat. 2014;21(12):843–852. doi: 10.1111/jvh.12214. [DOI] [PubMed] [Google Scholar]
- 9.Deres K, et al. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science. 2003;299(5608):893–896. doi: 10.1126/science.1077215. [DOI] [PubMed] [Google Scholar]
- 10.Feld JJ, et al. The phenylpropenamide derivative AT-130 blocks HBV replication at the level of viral RNA packaging. Antiviral Res. 2007;76(2):168–177. doi: 10.1016/j.antiviral.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Wang XY, et al. In vitro inhibition of HBV replication by a novel compound, GLS4, and its efficacy against adefovir-dipivoxil-resistant HBV mutations. Antivir Ther. 2012;17(5):793–803. doi: 10.3851/IMP2152. [DOI] [PubMed] [Google Scholar]
- 12.Brezillon N, et al. Antiviral activity of Bay 41-4109 on hepatitis B virus in humanized Alb-uPA/SCID mice. PLoS One. 2011;6(12):e25096. doi: 10.1371/journal.pone.0025096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber O, et al. Inhibition of human hepatitis B virus (HBV) by a novel non-nucleosidic compound in a transgenic mouse model. Antiviral Res. 2002;54(2):69–78. doi: 10.1016/s0166-3542(01)00216-9. [DOI] [PubMed] [Google Scholar]
- 14.Wu G, et al. Preclinical characterization of GLS4, an inhibitor of hepatitis B virus core particle assembly. Antimicrob Agents Chemother. 2013;57(11):5344–5354. doi: 10.1128/AAC.01091-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bourne CR, Finn MG, Zlotnick A. Global structural changes in hepatitis B virus capsids induced by the assembly effector HAP1. J Virol. 2006;80(22):11055–11061. doi: 10.1128/JVI.00933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katen SP, Tan Z, Chirapu SR, Finn MG, Zlotnick A. Assembly-directed antivirals differentially bind quasiequivalent pockets to modify hepatitis B virus capsid tertiary and quaternary structure. Structure. 2013;21(8):1406–1416. doi: 10.1016/j.str.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bourne CR, Katen SP, Fulz MR, Packianathan C, Zlotnick A. A mutant hepatitis B virus core protein mimics inhibitors of icosahedral capsid self-assembly. Biochemistry. 2009;48(8):1736–1742. doi: 10.1021/bi801814y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Packianathan C, Katen SP, Dann CE, 3rd, Zlotnick A. Conformational changes in the hepatitis B virus core protein are consistent with a role for allostery in virus assembly. J Virol. 2010;84(3):1607–1615. doi: 10.1128/JVI.02033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexander CG, et al. Thermodynamic origins of protein folding, allostery, and capsid formation in the human hepatitis B virus core protein. Proc Natl Acad Sci USA. 2013;110(30):E2782–E2791. doi: 10.1073/pnas.1308846110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gane EJ, et al. Phase 1a safety and pharmacokinetics of NVR 3-778, a potential first-in-class HBV core inhibitor. Hepatology. 2014;60:1279A. [Google Scholar]
- 21.Böttcher B, Vogel M, Ploss M, Nassal M. High plasticity of the hepatitis B virus capsid revealed by conformational stress. J Mol Biol. 2006;356(3):812–822. doi: 10.1016/j.jmb.2005.11.053. [DOI] [PubMed] [Google Scholar]
- 22.Zlotnick A, et al. In vitro screening for molecules that affect virus capsid assembly (and other protein association reactions) Nat Protoc. 2007;2(3):490–498. doi: 10.1038/nprot.2007.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zlotnick A, Ceres P, Singh S, Johnson JM. A small molecule inhibits and misdirects assembly of hepatitis B virus capsids. J Virol. 2002;76(10):4848–4854. doi: 10.1128/JVI.76.10.4848-4854.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang JM, et al. High replicative full-length lamivudine-resistant hepatitis B virus isolated during acute exacerbations. J Med Virol. 2005;77(2):203–208. doi: 10.1002/jmv.20453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.