Significance
Activating mutations in the B-Raf proto-oncogene serine/threonine kinase (BRAF) gene occur in many tumor types, the highest incidence being in malignant melanoma and papillary thyroid carcinoma. In patients with BRAF mutations tumor progression is more rapid than in patients without these mutations. Therapeutic strategies presently aim at inhibiting BRAF resulting in slower tumor progression; however, lasting remission is rarely accomplished. In this paper we identify the oncomiR-3151 as a downstream effector of mutated BRAF. MicroRNA-3151 (miR-3151) targets TP53 and other members of the TP53 pathway resulting in its inhibition. Simultaneous inhibition of BRAF and miR-3151 potentiates the effects on tumor cell growth. These data establish a link between mutated BRAF and the TP53 pathway, allowing novel therapeutic approaches to be considered.
Keywords: microRNA, BRAF, TP53, PTC, melanoma
Abstract
The B-Raf proto-oncogene serine/threonine kinase (BRAF) gene is the most frequently mutated gene in malignant melanoma (MM) and papillary thyroid cancer (PTC) and is causally involved in malignant cell transformation. Mutated BRAF is associated with an aggressive disease phenotype, thus making it a top candidate for targeted treatment strategies in MM and PTC. We show that BRAF mutations in both MM and PTC drive increased expression of oncomiR-3151, which is coactivated by the SP1/NF-κB complex. Knockdown of microRNA-3151 (miR-3151) with short hairpin RNAs reduces cell proliferation and increases apoptosis of MM and PTC cells. Using a targeted RNA sequencing approach, we mechanistically determined that miR-3151 directly targets TP53 and other members of the TP53 pathway. Reducing miR-3151’s abundance increases TP53’s mRNA and protein expression and favors its nuclear localization. Consequently, knockdown of miR-3151 also leads to caspase-3–dependent apoptosis. Simultaneous inhibition of aberrantly activated BRAF and knockdown of miR-3151 potentiates the effects of sole BRAF inhibition with the BRAF inhibitor vemurafenib and may provide a novel targeted therapeutic approach in BRAF-mutated MM and PTC patients. In conclusion, we identify miR-3151 as a previously unidentified player in MM and PTC pathogenesis, which is driven by BRAF-dependent and BRAF-independent mechanisms. Characterization of TP53 as a downstream effector of miR-3151 provides evidence for a causal link between BRAF mutations and TP53 inactivation.
Despite intensive research, patients diagnosed with metastatic malignant melanoma (MM) have one of the poorest survival rates of all human cancers. The B-Raf proto-oncogene serine/threonine kinase (BRAF) gene encodes an oncogenic tyrosine kinase that is frequently mutated to a constitutively active form in MM (V600E; ∼50–60%) (1). When mutated, BRAF is associated with a more aggressive disease phenotype (1, 2). Direct inhibition of aberrantly activated BRAF (i.e., vemurafenib treatment) has shown some effectiveness, but only in a subset of patients and with the eventual development of resistance in most cases (3). A second key feature of many MMs is the inactivation of the tumor suppressor gene tumor protein p53 (TP53) (4). Pharmacologic restoration of TP53 activity (in combination with BRAF inhibition) represents a promising therapeutic strategy in MM and will likely be useful for treating a broader spectrum of patients once agents with higher effectiveness have been identified (5, 6). Therefore, a better understanding of the deregulated downstream signaling of BRAF and the inactivation of TP53 are major challenges in MM research. However, a direct link between mutated BRAF and reduced TP53 activity has not been described. Importantly, discoveries regarding the pathophysiology of mutated BRAF in MM may also advance our understanding of other human malignancies with frequent BRAF mutations.
MicroRNA miR-3151 was identified by small RNA sequencing of melanoma samples (7), and particularly high abundance was found in BRAF mutated (BRAFmut) tumors (8). Because miR-3151 has been shown to display oncogenic potential in acute myeloid leukemia (AML) by direct targeting of the tumor suppressor TP53 (8), we hypothesized that miR-3151 may also be causally implicated in melanoma carcinogenesis. Because its expression is higher in BRAFmut tumors, we speculated that BRAFmut itself may increase the abundance of miR-3151. If such a novel axis could be established, the proposed mechanism might also be applicable to other BRAFmut cancers.
Here, we studied the effects of knockdown and ectopic expression of miR-3151 in the two solid tumors with the highest frequency of BRAF mutations–MM and papillary thyroid cancer (PTC). Furthermore, we used a comprehensive targeted RNA sequencing approach to identify the functionally relevant downstream targets of miR-3151 in MM and PTC. Finally, we performed pilot in vitro experiments to test the effectiveness of combining inhibition of miR-3151 with the clinically approved BRAF inhibitor vemurafenib.
Results
Targeted Knockdown of miR-3151 Increases Apoptosis and Reduces Cell Viability of MM Cells.
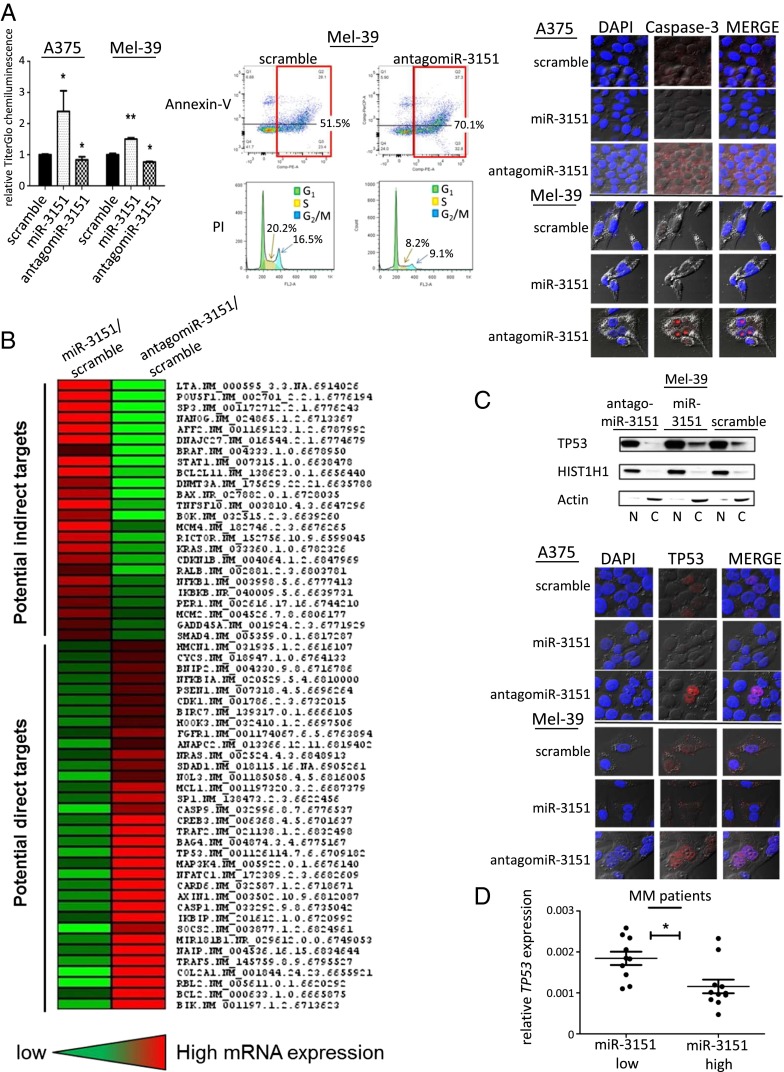
To assess the effects of miR-3151’s abundance on MM cells, we stably infected Mel-39 and A375 cells with miR-3151, antagomiR-3151, or scramble control, and then determined the cell viability with chemiluminescent TiterGlo assays. Whereas increasing the abundance of miR-3151 increased the proliferation of both cell lines compared with scramble control, antagomiR-3151 led to reduced proliferation (Fig. 1A). To test whether the reduction in cell number is solely due to a lowered rate of cell division, or additionally potentiated by an increase in cell death and apoptosis, we performed flow cytometric annexin-V apoptosis assays, and propidium iodide cell cycle analysis in antagomiR-3151–infected Mel-39 cells. Indeed, knockdown of miR-3151 increased the percentage of apoptotic cells and also reduced the percentage of cells in both S phase and G2 phase (Fig. 1A). Additionally, we used confocal microscopy to compare the expression and cellular localization of caspase-3, a major determinant of apoptotic activity. As seen in Fig. 1A, antagomiR-3151 increased caspase-3 expression.
Fig. 1.
Effect of antagomiR-3151 on cell viability and discovery of miR-3151 downstream targets in MM. (A, Left) Viability of A375 and Mel-39 cells after forced miR-3151 expression or knockdown in chemiluminescent assays. Three experiments, bars, mean ± SD; *P < 0.05, **P < 0.005, two-tailed t test. In both A375 and Mel39 cells, overexpression of miR-3151 led to increased chemiluminescent activity and, thus, increased proliferation, whereas knockdown of miR-3151 led to decreased proliferation. (A, Middle) Example plots of annexin-V apoptosis assays and example histograms of propidium iodide cell cycle analysis. Green area is G1 phase, yellow is S phase, and blue is G2/M phase. Assays were performed in Mel-39 cells after miR-3151 knockdown. Infection with antagomiR-3151 led to an increase in the number of cells undergoing apoptosis and a decrease in cells undergoing cell division. (A, Right) Confocal microphotographs show A375 and Mel-39 cells transfected with scramble, miR-3151, or antagomiR-3151 stained for Caspase-3 (red). DAPI nuclear staining is shown in blue. In both cell lines, infection with antagomiR-3151 increased the expression of Caspase-3. (B) Heatmap of gene expression changes concordantly affected by forced miR-3151 expression or miR-3151 knockdown based on targeted RNA sequencing. Genes with concordant decreased expression upon miR-3151 overexpression and decreased expression after miR-3151 knockdown were considered potential direct targets of miR-3151. (C, Upper) Western blot with nuclear fractionation lysates to determine the relative nuclear (N) and cytoplasmic (C) abundance of TP53 in Mel-39 cells after manipulation of miR-3151 expression. Infection with antagomiR-3151 led to increased nuclear localization of TP53. (C, Lower) Confocal microphotographs show A375 and Mel-39 cells transfected with scramble, miR-3151, or antagomiR-3151 stained for TP53 (red). DAPI nuclear staining is shown in blue. When infected with antagomiR-3151, cells displayed increased nuclear localization of TP53. (D) TP53 expression in high miR-3151 and low miR-3151 expressing MM patients, *P < 0.05, two-tailed t test. Patients with high expression of miR-3151 had relatively lower TP53 expression compared with low miR-3151 expressers (as defined by median cut).
AntagomiR-3151 Activates the TP53 Pathway in MM and Increases the Nuclear Localization of TP53.
To explore the downstream targets—and thus the effectors—of miR-3151 in MM, we took a broad screening approach for the identification of possible target genes. Using the TruSeq RNA platform (Illumina), we custom designed a panel of 361 genes and assessed their expression in Mel-39 MM cells with knockdown or forced expression of miR-3151. A total of 34 genes showed ≥20% down-regulation in the cells ectopically expressing miR-3151 and concordant ≥20% up-regulation in the antagomiR-3151 cells compared with scramble. Therefore, these genes were considered as potential direct miR-3151 targets (Fig. 1B). In addition, 24 genes showed concordant ≥20% up-regulation by miR-3151 and down-regulation by antagomiR-3151 (Fig. 1B), suggesting indirect regulatory mechanisms.
In line with previous reports about the effects of miR-3151 on leukemic cells (8), TP53 was down-regulated by forced miR-3151 expression and up-regulated by antagomiR-3151 (Fig. 1B). In addition, several TP53 pathway members were also down-regulated, including: BCL2, RBL2, CASP1, CASP9, MAP3K4, and CDK1 (Fig. 1B). Eleven of the 34 down-regulated genes were predicted to harbor at least one binding site for miR-3151 in their 3′-UTR and may therefore be potential direct miR-3151 targets (Table S1).
Table S1.
Potential direct target genes of miR-3151
| Gene | No. of predicted binding sites | mirSVR score | PhastCons score |
| BIK | 1 | −0.1048 | 0.5207 |
| CARD6 | 1 | −0.0107 | 0.6 |
| NFATC1 | 1 | −0.0006 | 0.3967 |
| TP53 | 2 | −0.003 | 0.648 |
| −0.0063 | 0.4148 | ||
| SP1 | 2 | −0.0041 | 0.653 |
| −0.0022 | 0.6501 | ||
| MCL1 | 2 | −0.0011 | 0.4804 |
| −0.2373 | 0.6276 | ||
| NOL3 | 1 | −0.0093 | 0.5414 |
| NRAS | 2 | −0.0023 | 0.5623 |
| −0.0036 | 0.5216 | ||
| ANAPC2 | 1 | −0.0026 | 0.475 |
| FGFR1 | 1 | −0.0018 | 0.5134 |
| HOOK3 | 1 | −0.0038 | 0.5079 |
Genes that showed concordant ≥20% down-regulation in their mRNA expression by miR-3151 and ≥20% up-regulation by antagomiR-3151 in the RNA sequencing approach were considered potential direct miR-3151 targets. Their 3′-UTRs were tested for predicted miR-3151 binding sites using a computational prediction approach (www.microRNA.org). Listed are the genes with predicted miR-3151 binding sites including their affinity scores (miRSVR score and PhastCons score).
To gain first insights into the pathways affected by miR-3151, we performed a pathway analysis of the up- and down-regulated genes (n = 58) by using the Ingenuity platform. The top scoring molecular and cellular functions affected by miR-3151 were cell death and survival; cellular development; and DNA replication, recombination, and repair (Table S2).
Table S2.
Molecular and cellular functions predicted to be influenced by miR-3151 (Ingenuity)
| Pathway name (Ingenuity) | P | No. of molecules |
| Cell death and survival | 7.11−30 to 4.65−07 | 48 |
| Cellular development | 2.62−17 to 4.54−07 | 42 |
| DNA replication, recombination, and repair | 5.65−17 to 4.54−08 | 20 |
| Cell cycle | 7.09−17 to 2.75−07 | 30 |
| Cellular growth and proliferation | 1.35−16 to 3.36−07 | 46 |
Listed are the top ranking components of the Global Canonical Pathway (GCP) category after pathway analysis (Ingenuity), pathway names and involved molecules defined by Ingenuity) of the miR-3151 associated gene expression signature. The P value associated with a pathway in GCP is a measure of the likelihood that the association between a set of focus genes in the analyzed experiment and a given pathway is due to random chance. The P value identifies statistically significant overrepresentation of focus genes in a given process. It is automatically calculated by the ingenuity pathway analysis program (ingenuity.com) using the right-tailed Fisher’s exact test.
To distinguish between nuclear (transcriptionally active) TP53 and cytoplasmic TP53, we performed subcellular fractionation of Mel-39 cells after either stable introduction or knockdown of miR-3151 compared with scramble control. Expression of antagomiR-3151 led to a preferred nuclear localization of TP53 (Fig. 1C). We used confocal microscopy to validate the changes in the subcellular localization caused by antagomiR-3151 in A375 and Mel-39 cells. The knockdown of miR-3151 resulted in a dramatic nuclear accumulation of TP53 thereby suggesting an increased transcriptional potential of the tumor suppressor gene caused by antagomiR-3151 (Fig. 1C).
Finally, to strengthen the evidence that miR-3151 is an important regulator of TP53 expression in MM in vivo, we determined miR-3151 and TP53 expression by using tumor RNA from a cohort of MM patients (n = 21) and assessed a possible association of the expression levels of the two genes. Patients with high expression of miR-3151 had lower expression of TP53 (Fig. 1D).
miR-3151 Expression Can Be Increased by BRAF Mutations and the SP1/NF-κB Transactivating Complex.
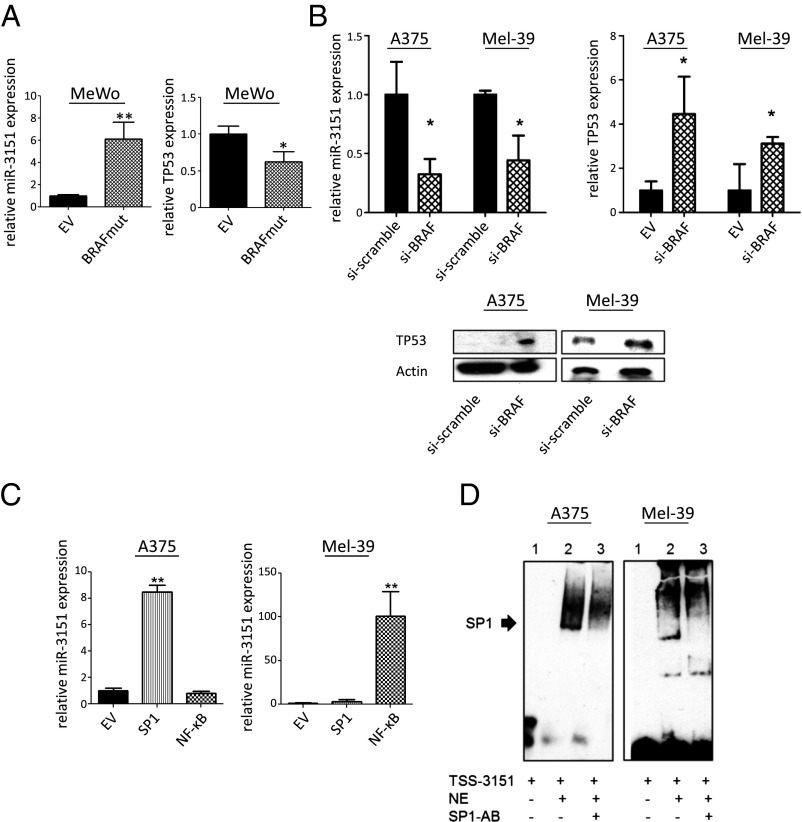
Because BRAFmut MM samples have on average fivefold higher expression of miR-3151 compared with BRAFwt samples (8), we wanted to elucidate whether miR-3151 may be a direct downstream effector of aberrantly activated BRAF and whether it contributes to the increased disease aggressiveness associated with the BRAF mutation. If miR-3151 is downstream of BRAF, an introduction of the mutation into BRAFwt cells should increase miR-3151 expression and, consequently, lead to a further reduction of TP53, whereas knockdown of BRAFmut allele expression should reduce miR-3151s abundance and elevate TP53 expression. In line with our hypothesis, transfection of MeWo cells (BRAFwt) with a BRAFmut expression construct increased miR-3151 expression and reduced TP53 expression (Fig. 2A). siRNA-mediated knockdown of BRAFmut in A375 and Mel-39 cells reduced miR-3151 expression and increased TP53 at both the mRNA and protein levels (Fig. 2B).
Fig. 2.
Regulation of miR-3151 by BRAF-dependent and BRAF-independent mechanisms. (A) Effects of BRAFmut on miR-3151 expression and TP53 expression in BRAFwt MeWo cells. Upon introduction of BRAFmut, cells had increased miR-3151 expression and decreased TP53 expression. Three experiments, bars, mean ± SD; *P < 0.05, two-tailed t test. (B) Effects of BRAF knockdown (si-BRAF) on miR-3151 expression and TP53 expression in BRAFmut A375 and Mel-39 cells. In both cell lines, when BRAF was silenced, miR-3151 expression decreased, whereas TP53 expression increased at both the RNA and protein levels. Three experiments, bars, mean ± SD; *P < 0.05, **P < 0.005, two-tailed t test. Inset shows an example immunoblot of TP53 protein expression after BRAF knockdown. (C) Effects of forced SP1 and NF-ĸB expression on miR-3151 expression in A375 and Mel-39 cells. A375 cells responded preferentially to SP1 transfection, whereas Mel-39 cells responded to NF-ĸB transfection with miR-3151 overexpression. Three experiments, bars, mean ± SD; *P < 0.05, **P < 0.005, two-tailed t test. (D) Visualization of SP1 binding to miR-3151s transcription start site (TSS-3151) by using EMSA. Nuclear extracts (NE) were used from A375 and Mel-39 cells, shifting performed with SP1-antibody (SP1-AB). The SP1/ NF-ĸB transactivating complex was bound to TSS-3151 in A375 and Mel-39 cells.
In AML, miR-3151 can be regulated by the Sp1 transcription factor/nuclear factor kappa-B (SP1/NF-κB) transactivating complex (8), which binds to the regulatory region of miR-3151 (TSS-3151). To test whether this activation also regulates miR-3151 in MM, we transfected A375 and Mel-39 cells with expression constructs for SP1, NF-κB (p65), or both. Whereas A375 cells showed a greater response to SP1, Mel-39 cells reacted preferentially to transfection with NF-κB (Fig. 2C). Finally, electrophoretic mobility shift assays with nuclear extracts harvested from both A375 and Mel-39 cells indicated a direct interaction of SP1/NF-κB with TSS-3151 (Fig. 2D).
Thus, aberrant expression of miR-3151 in melanoma seems to be a combined effect of BRAFmut and transcriptional activation by the SP1/NF-κB transactivating complex.
The BRAF–miR-3151–TP53 Axis Is Also Present in Papillary Thyroid Cancer.
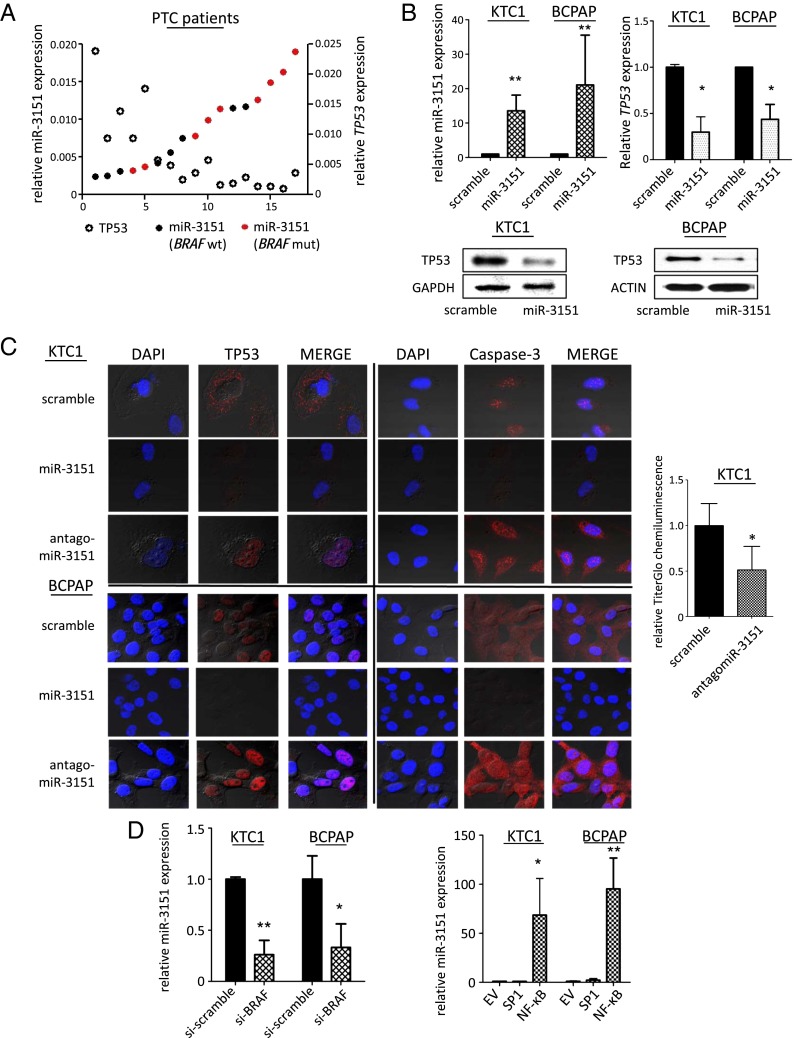
In addition to MM, BRAF mutations are frequently found in PTC (9) and are associated with a more aggressive disease phenotype (10). To test whether the newly identified axis among BRAF mutations, miR-3151 expression, and reduced TP53 levels is also present in PTC, we first determined the BRAF mutation status and the endogenous miR-3151 expression in a cohort of PTC tumor samples (n = 16). As seen in MM, patients with BRAFmut tumors had higher endogenous miR-3151 expression (Fig. 3A). In addition, patients with higher miR-3151 expression had lower endogenous TP53 expression levels (Fig. 3A). To validate that miR-3151 affects TP53 expression, we stably introduced miR-3151 into two PTC cell lines (KTC1 and BCPAP). Forced miR-3151 expression reduced TP53 expression at both the mRNA and protein levels (Fig. 3B) and also resulted in a reduction of nuclear TP53 (Fig. 3C). Conversely, antagomiR-3151 infection strongly enhanced the expression and nuclear localization of TP53 (Fig. 3C). The changes in TP53 expression/subcellular localization were accompanied by alterations in caspase-3 expression levels, indicating that knockdown of miR-3151 enhances cell death in PTC (Fig. 3C). This observation was further supported by TiterGlo assays, which showed decreased viability in cells infected with antagomiR-3151 (Fig. 3C). Finally, the effects of BRAFmut knockdown on miR-3151 expression (Fig. 3D) and the transcriptional activation of miR-3151 by the SP1/NF-ĸB complex was also validated in PTC cells, with NF-ĸB being the main transcriptional driver (Fig. 3D).
Fig. 3.
Downstream effects and upstream regulation of miR-3151 in PTC. (A) Endogenous miR-3151 (indicated by filled circles) and TP53 expression levels (indicated by open circles) in tumor samples from BRAFwt and BRAFmut PTC patients. Patients with high expression of miR-3151 had lower TP53 expression (P < 0.05, two-tailed Student’s t test). BRAFmut patients (indicated by red color) had higher expression of miR-3151 compared with BRAFwt patients (indicated by black color; P < 0.05, two-tailed Student’s t test). (B, Upper Left) miR-3151 expression achieved by stable overexpression with the lentiviral miR-3151 expression construct in KTC1 and BCPAP cell lines compared with scramble control. Three experiments, bars, mean ± SD; **P < 0.005, two-tailed t test. (B, Upper Right) Effects of forced miR-3151 expression on TP53 mRNA in KTC1 and BCPAP cells. Three experiments, bars, mean ± SD; *P < 0.05, two-tailed t test. (B, Lower) Effects of forced miR-3151 expression on TP53 protein levels in KTC1 and BCPAP cells. Forced expression of miR-3151 decreased TP53 expression at both the RNA and protein levels in both PTC cell lines. (C, Left) Confocal microphotographs show A375 and Mel-39 cells transfected with scramble, miR-3151, or antagomiR-3151 stained for TP53 or Caspase-3 (red). DAPI nuclear staining is shown in blue. Knockdown of miR-3151 led to increased expression of TP53 and Caspase-3 in PTC cells. (C, Right) Cell viability of KTC1 cells after knockdown of miR-3151, as measured by TiterGlo assay. Knockdown of miR-3151 led to decreased chemiluminescent activity and, therefore, a decreased proliferation rate. Three experiments, bars, mean ± SD; *P < 0.05, two-tailed t test. (D, Left) Effects of BRAF knockdown (si-BRAF) on miR-3151 expression and TP53 expression in KTC1 and BCPAP cells. Silencing BRAF in both cell lines decreased miR-3151 expression. Three experiments, bars, mean ± SD; *P < 0.05,**P < 0.005, two-tailed t test. (D, Right) Effects of SP1 and NF-ĸB on miR-3151 expression in KTC1 and BCPAP cells. Overexpression of NF-ĸB increased miR-3151 expression in both cell lines. Three experiments, bars, mean ± SD; *P < 0.05, **P < 0.005, two-tailed t test.
Combined Knockdown of miR-3151 and Presence of BRAFmut in MM and PTC Cells Increases Sensitivity to Vemurafenib Treatment.
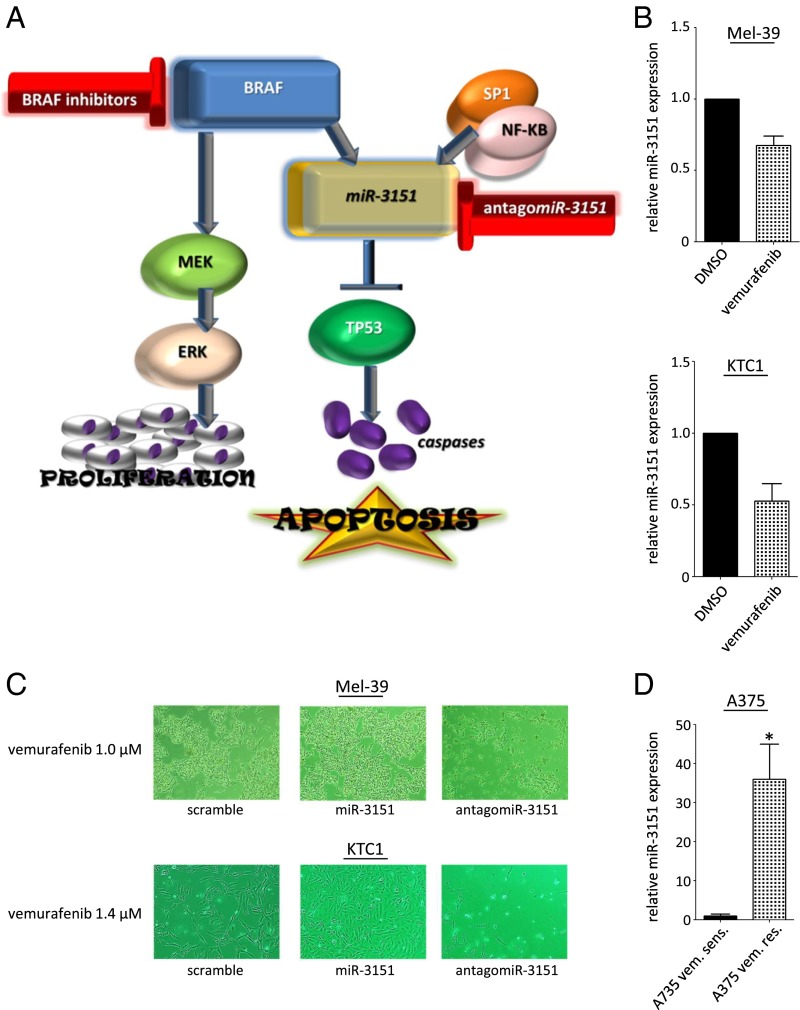
Although we identified miR-3151 as a previously unidentified downstream effector of BRAF, it is only one of many known BRAF targets. Perhaps the most important genes known to be affected by aberrantly activated BRAF are mitogen-activated protein kinase kinase 7 (MEK) and mitogen-activated protein kinase 1 (ERK), which facilitate signal transduction and ultimately lead to increased gene transcription and translation (11). Of note, above we demonstrated that miR-3151 is not only activated by BRAF, but also by the SP1/NF-κB transactivation complex. Thus, a simultaneous inhibition of BRAFmut (to stop activation of the MEK/ERK cascade and also reduce miR-3151 expression) and additional targeting of miR-3151 (to stop its activation by SP1/NF-κB), either with proteasome inhibitors to limit SP1/NF-κB’s binding activity or by direct miR-3151 inhibition with antagomiR-3151, may be an effective combined treatment approach (Fig. 4A). As a proof of principle, we first tested the effects of the BRAF inhibitor vemurafenib on miR-3151 expression. Indeed, treatment of BRAFmut Mel-39 (MM) and KTC1 cells (PTC) led to a reduction of miR-3151 expression (Fig. 4B). Next, we treated Mel-39 and KTC1 cells stably expressing miR-3151, antagomiR-3151, or scramble control. Whereas treatment with 1.0 μM (Mel-39) and 1.4 μM (KTC1) vemurafenib had no effect on cell proliferation in miR-3151–infected cells, treatment reduced the growth in scramble-infected cells and completely eradicated antagomiR-3151–infected cells (Fig. 4C). Finally, we created a vemurafenib-resistant A375 cell line and compared its endogenous miR-3151 expression level to the parental (nonresistant) A375 cells. The vemurafenib-resistant A375 cells had a 35-fold higher endogenous miR-3151 expression (Fig. 4D). Taken together, these pilot experiments may encourage in vivo tests of this combined targeted treatment approach.
Fig. 4.
Effects of combined BRAFmut and miR-3151 inhibition on MM and PTC cells. (A) Schematic depiction of the proposed BRAF–miR-3151–TP53 axis. Targeting miR-3151 and BRAF through antagomiR-3151 and BRAF inhibitors, respectively, is a potential treatment strategy for both MM and PTC. (B) Effects of vemurafenib treatment on miR-3151 expression in Mel-39 (1.0 μM vemurafenib; Upper) and KTC1 cells (1.4 μM vemurafenib; Lower) compared with vehicle. Vemurafenib treatment led to decreased miR-3151 expression in both MM and PTC cells. Three experiments, bars, mean ± SD. (C) Representative images of the differential effect of vemurafenib treatment on Mel-39 and KTC1 cells after forced miR-3151 expression or knockdown. Overexpression of miR-3151 decreased the sensitivity to vemurafenib treatment, whereas knockdown of miR-3151 increased the sensitivity to vemurafenib treatment. (D) Changes in miR-3151 expression after development of vemurafenib resistance (A375 vem. res.) compared with the parental cell line (A375 vem. sens.). Vemurafenib resistant cells showed increased miR-3151 expression compared with vemurafenib sensitive cells. Resistant A375 cells were maintained in 2 μM vemurafenib. Three experiments, bars, mean ± SD; *P < 0.05, two-tailed t test.
Discussion
MM is the most deadly skin cancer, accounting for 75% of all skin cancer deaths with numbers continuously increasing (2). When diagnosed at an advanced stage (stage IV) the 5-y survival rates of the patients are only ∼15% (2), emphasizing the urgent need for better treatment options for these patients. PTC is the most common type of thyroid cancer accounting for ∼80% of all thyroid cancers (9, 10). Although the prognosis of PTC is generally favorable, the prognosis of patients diagnosed at an advanced disease stage is still poor (9, 10). BRAF mutations are the most frequent mutations in MM and PTC and are associated with increased disease aggressiveness and poor outcome (1). Therefore, targeting BRAF has become one of the most promising treatment options in BRAF-mutated MM and—in clinical trials—also PTC patients (3, 12–14). Still, many questions regarding the downstream signaling of mutated BRAF remain open.
As a second crucial event, the tumor suppressor TP53 is inactivated in many MM cases and plays a critical role in melanoma progression. The mechanisms of TP53 inactivation are not yet fully understood, but it has been demonstrated that restoration of TP53 activity represents an attractive treatment strategy in MM (4, 5). In fact, disruption of the TP53 pathway via short hairpin RNA in benign nevi with BRAF mutations may promote malignant transformation of the cells (6). Thus, the identification of a causal link between these two important players would be a major contribution to our understanding of MM pathophysiology.
Vemurafenib is an orally administered, small molecule that selectively inhibits BRAF. It has been approved by the FDA for the treatment of unresectable or metastatic MM with the presence of the BRAF V600E mutation (12). In an international multicenter trial (NCT01006980), treatment with vemurafenib was superior to treatment with chemotherapy (dacarbazine), but not all patients benefited from treatment with vemurafenib (13, 14).
Deregulation of specific miRs has been implicated in the disease initiation and progression of virtually every human cancer, including MM and PTC (15, 16). Specific targeting of aberrantly activated miRs, either indirectly via blockage of their upstream activation or directly via antagomiRs, represents a promising new therapeutic approach (17, 18).
We show that miR-3151 is up-regulated in BRAFmut MM and PTC and provide the first evidence to our knowledge that miR-3151 leads to increased cell growth by direct down-regulation of its target TP53. We propose that miR-3151 is a crucial link between BRAF mutations and the observed down-regulation of TP53.
Additional up-regulation of miR-3151 by BRAF-independent mechanisms (SP1/NF-κB transactivating complex) may explain why some patients show tumor progression under vemurafenib treatment and indicates that combining vemurafenib with other chemotherapeutics may be a more effective strategy.
Determination of miR-3151 and TP53 expression in BRAFmut tumors during vemurafenib therapy will provide insight into the predictive value of miR-3151 expression. Connecting BRAF mutations with TP53 down-regulation via increased expression of miR-3151 is a testable approach ready to be assessed in preclinical trials.
Materials and Methods
Patient Samples.
PTC samples (n = 16) were obtained from the Human Cancer Genetics Tissue Bank at Ohio State University (OSU). All patients provided written informed consent according to the Declaration of Helsinki to store and use their tissue for discovery studies according to OSU institutional guidelines under protocols approved by the OSU Institutional Review Board. Total RNA samples from MM patients (n = 21) were purchased from Asterand.
Tissue Culture Conditions.
For studying miR-3151 in MM A375, Mel-39 and MeWo cells were provided by the laboratory of W.E.C. [obtained from American Type Culture Collection (ATCC)]. Cells were cultured in DMEM supplemented with 20% (vol/vol) FBS and 1% Antibiotic-Antimycotic (Gibco). For studying miR-3151 in PTC, KTC1 and BCPAP cells were obtained from the ATCC. Cells were cultured in RPMI-1640 medium supplemented with 20% (vol/vol) FBS and 1% Antibiotic-Antimycotic (Gibco).
For transcriptional luciferase reporter assays, HEK293 cells, obtained from ATCC, were cultured in DMEM culture medium supplemented with 10% FBS, l-glutamine (200 mM), and antibiotic/antimycotic agent (all Life Technologies Corporation/Gibco) and grown at 37 °C with 5% (vol/vol) CO2.
To create vemurafenib-resistant A375 cells, the cells were cultured in increasing concentrations of vemurafenib (0–10 μM) over a period of 8 wk with twofold dosage increases once a week. After confirmation of resistance by using MTT assays, the cells were cultured in 2 μM vemurafenib.
Overexpression of miR-3151 and miR-3151 Knockdown.
For stable expression, the stem loop of miR-3151 with 200-bp flanking sequence was cloned into an HIV-based lentiviral dual promoter vector as described (8) (pCDH-CMV-MCS-EF1-copGFP+Puro cDNA; System Biosciences). For targeted knockdown of miR-3151, a custom-made antagomiR-3151 was purchased from System Biosciences. As a control, lentiviral scramble miR was used according to the manufacturer’s instructions (miRZiP000; System Biosciences). Lentiviral construct (4,500 ng) was transfected into 293TN cells by using 45 μL of pPACKH1 and 55 μL of PureFection (System Biosciences). After 48 h and 72 h, the supernatant containing the pseudoviral particles was collected and the virus precipitated overnight at 4 °C by using 5 mL of PEG-IT virus precipitation solution (System Biosciences). We used 200 μL of PBS and 25 μM Hepes buffer for resuspension of the pelleted virus. We infected 200,000 cells per mL in triplicate with 20 IU virus, using 5 μL of Transdux Infection Reagent (System Biosciences).
cDNA Synthesis and miR-3151 mRNA Analysis.
To check for successful overexpression of miR-3151 and to analyze the effect of forced miR-3151 expression on the predicted target genes, RNA from 1 million cells was harvested on day 14 after infection and reverse transcribed to cDNA by using the TaqMan MicroRNA Reverse Transcription Kit (Life Technologies Corporation/Applied Biosystems) or the SuperScript III First-Strand cDNA Synthesis Kit (Life Technologies Corporation/Invitrogen). Both kits were used according to the manufacturer’s instructions. Simultaneously, protein (from 4 million cells) was harvested and used for Western blotting. miR-3151 abundance was determined by qRT-PCR as described (8).
Transcriptional Luciferase Reporter Assays for the miR-3151 Promoter Analysis.
Luciferase reporter constructs (∼50-bp genomic sequence) containing the predicted TSS for miR-3151 were cloned into the multiple cloning site of a promoterless luciferase reporter vector (pGL4.11; Promega) by using the KpnI and SacI restriction sites: TSS-3151 clon F (KpnI; −487 bp of stemloop) gcacggtaccGTAGTCAGAGCGGTGGGATG, TSS-3151 clon R (SacI; −290 bp of stemloop) gtgcgagctcCAGAATGAGACAGACCTGAG. Expression constructs for the potentially activating transcription factors SP1 and NF-κB were constructed as described (8). HEK293 cells were transfected in triplicate with 250 ng of luciferase reporter construct and 100 ng of control construct (pGL4.74; Promega) and cotransfected with 50 ng of the different expression constructs or empty pIRES2-EGFP vector control. Transfected cells were incubated for 24 h at 37 °C with 5% CO2 in Opti-MEM II medium containing the Lipofectamine and plasmid combination. Protein lysates were assessed for firefly luciferase and Renilla luciferase activities according to the recommendations detailed in the Dual-Luciferase Reporter Assay System (Promega). Relative expression was normalized by using the activity of cotransfected Renilla luciferase.
Transient Overexpression of BRAFmut, SP1, and NF-κB and BRAF Knockdown.
For transient overexpression, 3 μg of the overexpression constructs of BRAFmut, SP1, NF-κB (all cloned in pIRES2-EGFP vector; Clontech), and/or the p65 subunit of NF-κB (cloned as pCMV-p65) were transfected in triplicate into 3 million Mel-39 and A375 cells by using Purefection transfection reagent according to the manufacturer’s instructions (System Biosciences). Knockdown of BRAF was performed by using BRAF siRNA pools (sc-36368; Santa Cruz) and compared with siRNA scramble pools (Santa Cruz). Efficient knockdown was validated by Western blot.
TruSeq-Targeted RNA Analysis.
A customized add-on panel comprised of 361 genes using the backbone of the Illumina apoptosis panel and the Illumina stem cell panel was designed by using DesignStudio (Illumina) for the TruSeq Targeted RNA expression analysis. Library preparations using 100 ng of total RNA (miR-3151+scramble: harvest 3 h after transfection, antagomiR-3151/scramble: harvest 24 h after transfection), and the Miseq run were performed according to the manufacturer’s instructions. MiSeqReporter software was used to estimate target hits for each transcript after aligning reads against references specified by Targeted Oligo Pool, using banded Smith-Waterman alignment. The raw count data were then normalized by using the R library DESeq (V 1.14.0), built based on negative binomial distribution, with variance and mean linked by local regression (19). Percent relative changes of mRNA expression of miR-3151 and antagomiR-3151 compared with scramble were estimated.
TiterGlo and Caspase-3/7 Assays.
Cell viability and apoptosis changes in Mel-39, A375, and KTC1 cell lines infected with lentiviral miR-3151, antagomiR-3151, or scramble control were analyzed by using the chemiluminescent Capase-3/7 and Titer Glo assays (both Promega) 72 h after Puromycin selection using 20,000 cells in duplicate of three biological replicates according to the manufacturer’s instructions.
Western Blotting Assays.
Western blotting was performed according to standard procedures. Antibodies used were p53 (sc-126; Santa Cruz), Histone H3 (ab 32107; Abcam), and Actin (sc-1616; Santa Cruz).
Electrophoretic Mobility Shift Assay.
Nuclear proteins were extracted from Mel-39 and A375 cells by using the Nuclear Extract Kit (Active Motif) according to the manufacturer’s instructions. The oligonucleotide sequences used for the EMSA analysis are as follows: miR3151 promoter SP1 F EMSA, 5′/5Biosg/GCAGTGGGGTGGGGTTTGGA, miR3151 promoter SP1 R EMSA, and 5′/5Biosg/TCCAAACCCCACCCCACTGC. For EMSA, the Thermo Scientific LightShift Chemiluminescent EMSA Kit (Pierce/Thermo Fisher Scientific) was used according to the manufacturer’s instructions. The antibody used was SP1 (sc-59; Santa Cruz).
Confocal Staining and Microscopy.
Confocal staining was performed 24 h after transfection by standard procedures using the following antibodies: p53 (sc-126; Santa Cruz), Caspase-3 (9665S; Cell Signaling), Alexa Flour 647 goat anti-mouse, and Alexa Fluor 546 donkey anti-rabbit (both BD Biosciences). Confocal micrographs were taken by using the FV1000 Confocal Laser Scanning Microscope (Olympus) with a UPLFLN 40× Oil, N.A. 1.3 lens.
Statistical Methods.
Data were represented as mean ± SD of at least three independent experiments unless otherwise indicated and analyzed by the two-tailed or one-tailed Student’s t test. The means and SD were calculated and displayed in bar graphs as the height and the corresponding error bar, respectively. A P < 0.05 was considered statistically significant.
Acknowledgments
We thank Jan Lockman for technical support, the Human Cancer Genetics Tissue Bank at OSU for sample processing and storage services, and The Ohio State University Comprehensive Cancer Center’s Nucleic Acid and Microarray Shared Resources for technical support. This work was supported in part by National Cancer Institute Grants P30 CA16058 and P01 CA124570, the Coleman Leukemia Research Foundation (A.-K.E. and A.d.l.C.), the Pelotonia Fellowship Program (A.-K.E., R.P., S.E.M., C.J.W., and J.M.), and the Pelotonia IDEA Grant (to A.-K.E., A.d.l.C., and W.E.C.).
Footnotes
Conflict of interest statement: A.d.l.C. and A.-K.E. hold a US patent on miR-3151 in the diagnosis and treatment of cancer.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1520390112/-/DCSupplemental.
References
- 1.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Finn L, Markovic SN, Joseph RW. Therapy for metastatic melanoma: The past, present, and future. BMC Med. 2012;10:23. doi: 10.1186/1741-7015-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nat Med. 2013;19(11):1401–1409. doi: 10.1038/nm.3392. [DOI] [PubMed] [Google Scholar]
- 4.Houben R, et al. High-level expression of wild-type p53 in melanoma cells is frequently associated with inactivity in p53 reporter gene assays. PLoS One. 2011;6(7):e22096. doi: 10.1371/journal.pone.0022096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Box NF, Vukmer TO, Terzian T. Targeting p53 in melanoma. Pigment Cell Melanoma Res. 2014;27(1):8–10. doi: 10.1111/pcmr.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu H, et al. The role of BRAF mutation and p53 inactivation during transformation of a subpopulation of primary human melanocytes. Am J Pathol. 2009;174(6):2367–2377. doi: 10.2353/ajpath.2009.081057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stark MS, et al. Characterization of the melanoma miRNAome by deep sequencing. PLoS One. 2010;5(3):e9685. doi: 10.1371/journal.pone.0009685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisfeld AK, et al. Intronic miR-3151 within BAALC drives leukemogenesis by deregulating the TP53 pathway. Sci Signal. 2014;7(321):ra36. doi: 10.1126/scisignal.2004762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jo YS, et al. Influence of the BRAF V600E mutation on expression of vascular endothelial growth factor in papillary thyroid cancer. J Clin Endocrinol Metab. 2006;91(9):3667–3670. doi: 10.1210/jc.2005-2836. [DOI] [PubMed] [Google Scholar]
- 10.Xing M, et al. Association between BRAF V600E mutation and mortality in patients with papillary thyroid cancer. JAMA. 2013;309(14):1493–1501. doi: 10.1001/jama.2013.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solit DB, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439(7074):358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garbe C, Abusaif S, Eigentler TK. Vemurafenib. Recent Results Cancer Res. 2014;201:215–225. doi: 10.1007/978-3-642-54490-3_13. [DOI] [PubMed] [Google Scholar]
- 13.Chapman PB, et al. BRIM-3 Study Group Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sosman JA, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366(8):707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SK, Calin GA. Non-coding RNAs and cancer: New paradigms in oncology. Discov Med. 2011;11(58):245–254. [PubMed] [Google Scholar]
- 16.Mueller DW, Bosserhoff AK. The evolving concept of ‘melano-miRs’-microRNAs in melanomagenesis. Pigment Cell Melanoma Res. 2010;23(5):620–626. doi: 10.1111/j.1755-148X.2010.00734.x. [DOI] [PubMed] [Google Scholar]
- 17.Monroig PdelC, Chen L, Zhang S, Calin GA. Small molecule compounds targeting miRNAs for cancer therapy. Adv Drug Deliv Rev. 2015;81:104–116. doi: 10.1016/j.addr.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12(11):847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]