Abstract
Emerging and reemerging human viral pathogens pose great public health concerns since therapeutics against these viruses are limited. Thus, there is an urgent need to develop novel drugs that can block infection of either a specific virus or a number of viruses. Viral entry is thought to be an ideal target for potential therapeutic prevention. One of the challenges of developing antivirals is that most of these viruses are highly pathogenic and therefore require high biosafety-level containment. In this study, we have adopted a comparative high-throughput screening protocol to identify entry inhibitors for three enveloped viruses (Marburg virus, influenza virus H5N1, and Lassa virus) using a human immunodeficiency virus–based pseudotyping platform. We demonstrate the utility of this approach by screening a small compound library and identifying putative entry inhibitors for these viruses. One major advantage of this protocol is to reduce the number of false positives in hit selection, and we believe that the protocol is useful for inhibitor screening for many enveloped viruses.
Keywords: phenotypic drug discovery, antiviral drugs, high-throughput screening, cell-based assays, comparative HTS
Introduction
Viral infection can devastate human populations either globally, like influenza pandemics,1 or locally, such as outbreaks of Ebola and Marburg viruses (filoviruses) in Africa.2 One of the major challenges in combating these viral diseases is that the vaccine and therapeutic options against them are usually limited. Furthermore, emergence of highly pathogenic and new viruses and drug resistance viruses render the limited antiviral vaccines and therapeutic treatments inadequate. Thus, there is an urgent need to discover and develop antiviral therapeutic treatments, either targeting one particular virus or broadly targeting multiple viruses.
Viral entry is the first essential step in the viral replication cycle; consequently, blocking viral entry into the target cells will lead to blockage of viral infection and is an attractive antiviral strategy. For many enveloped viruses, such as influenza virus, human immunodeficiency virus (HIV), filoviruses, and arenaviruses, entry to the target cells is dictated by a single viral surface glycoprotein (GP), which is responsible for receptor recognition and binding (attachment) and for mediating viral/cell membrane fusion.3–7 Successful development of anti-HIV entry drugs to block gp120mediated CCR5 receptor binding8,9 and gp41-mediated fusion (T-20)10,11 has convincingly demonstrated in principle the feasibility and promise of targeting other enveloped viruses at the entry step for therapeutics.
One challenge in research and drug discovery for highly pathogenic viruses is that high-containment facilities (biosafety level 3 or level 4, or BSL-3/4) are required to handle these viruses. However, for many enveloped viruses, this obstacle can often be circumvented by a surrogate system called viral pseudotyping,12 which has been widely used by many researchers, including us, to study the entry mechanisms of highly infectious enveloped viruses, such as filoviruses and H5N1 “bird flu” influenza virus, and to identify and develop antiviral therapeutics.13–22 We and others have identified numerous antiviral entry inhibitors using a robust HIV-1–based pseudotyping assay,20,21,23 and importantly, the efficacy of these inhibitors has been validated with infectious viruses, demonstrating the screening power of this approach.
In this study, we have adopted this HIV-based pseudotyping assay to develop a high-throughput screening (HTS) protocol to screen entry inhibitors for three different viruses: influenza virus H5N1, the filovirus Marburg virus (MARV), and the arenavirus, Lassa virus (LASV). By screening a small chemical library of 1200 compounds, we demonstrate that this protocol allows one to identify specific and shared entry inhibitors for different viruses with a greatly reduced number of false positives, alleviating a major problem in HTS. This protocol can provide a powerful screen strategy for identifying entry inhibitors for other enveloped viruses. In addition, the concept of comparative approaches described in this report can be used in other biological HTS assays.
Materials and Methods
Cell Culture and Plasmids
Human 293T embryonic kidney cells and A549 human lung epithelial cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Cellgro, Manassas, VA) supplemented with 10% fetal bovine serum (GIBCO, Carlsbad, CA), 100 μg/mL streptomycin, and 100 U penicillin (Invitrogen, Carlsbad, CA). The three types of pseudoviri ons for HTS were created by the following plasmids: hem agglutinin (HA), isolated from a highly pathogenic avian influenza virus, A/Goose/Qinghai/59/05 (H5N1) strain18; Marburg virus glycoprotein (GP)24; LASV envelope GP25; and the HIV-1 proviral vector pNL4-3.Luc.R−E−,26,27 which was obtained through the National Institutes of Health (NIH) AIDS Research and Reference Reagent program.
Compound Library and Control
The Prestwick Chemical Library contains 1200 drugs approved by the Food and Drug Administration. The active compounds were selected for high chemical and pharmacological diversity as well as for bioavailability and safety in humans. Three hundred twenty unique compounds were arrayed in a 384-well plate at a 10-mM concentration in DMSO, leaving columns 1, 2, 23, and 24 with DMSO. The positive control drug for this assay, azidothymidine (AZT; Sigma, St. Louis, MO), was solubilized at 10 mM in DMSO. The stock solution was diluted to a final concentration of 25 μM for the screens.
Production of Pseudovirions
All three types of pseudovirions—MARV/HIV, avian influenza virus (AIV)/HIV, and LASV/HIV—were produced by transient cotransfection of human 293T cells using a polyethylenimine–based transfection protocol.28 Plasmids encoding MARV GP, HA/neuraminidase (NA), LASV GP, and replication-defective HIV vector (pNL4-3.Luc.R−E−) were used for transient cotransfection into 293T-producing cells. Five hours after transfection, cells were washed with phosphate-buffered saline, and 40 mL of fresh medium was added to each plate (150 mm). Forty-eight hours after transfection, the supernatants were collected and filtered through a 0.45-μm pore size filter (Nalgene, Rochester, NY). The pseudovirion stocks were stored at 4 °C prior to use.
High-Throughput Screen
Target A549 cells were seeded into white, flat-bottom, 384-well plates (CulturPlate; PerkinElmer, Waltham, MA) at a density of 1000 cells/well in a 30-μL assay medium using JANUS liquid handler MDT (Modular Dispense Technology; PerkinElmer) and incubated at 37 °C, 5% CO2 , with high humidity. Twenty-four hours later, 0.2 μL of each compound was added to 80 μL pseudovirions per well in an intermediate 384-well plate through a pin tool (V&P Scientific, San Diego, CA) and mixed thoroughly using a JANUS liquid handler MDT. This resulted in a final concentration of 25 μM (0.25% DMSO) for all compounds. Then, 30 μL compound-pseudovirus mixture per well was transferred to the target cell plate in duplicate. Plates were incubated at 37 °C, 5% CO2 for 48 h. After incubation, 15 μL of neolite (PerkinElmer) was added to each well using the JANUS liquid handler MDT, and plates were incubated at room temperature for 5 to 10 min. Luciferase activity was measured by an EnVision plate reader (PerkinElmer).
Data Analysis
The luminescence signal in each well was measured on the EnVision plate reader. The data were exported as commaseparated values files and analyzed using the statistical programming language R.29 An R script was employed to calculate the median of luminescence signals of all samples in each plate, and then the signal in each well was normalized by the plate sample median.
The Z′ factor was calculated from the normalized signals from DMSO and AZT control wells on each plate with the following equation: 1 – 3(StdDMSO + StdAZT)/(MeanDMSO – MeanAZT).
The coefficient of variation (%) of each compound was calculated from the normalized signals in corresponding wells in the duplicate plates with the following equation: 100 × Stdduplicate wells/Meanduplicate wells.
Percentage of inhibition (% inhibition) was calculated as 100 × (1 – normalized signal). Hit compound lists were generated separately from screens of three viruses by applying various % inhibition cutoffs, and shared hits and virus-specific hits were picked by comparing these lists.
Results
Strategy of Cell-Based Comparative HTS
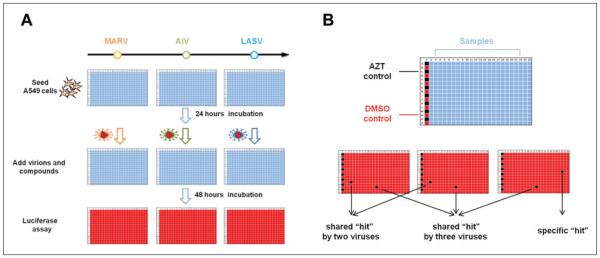
We have adopted a cell-based comparative HTS protocol to identify entry inhibitors for enveloped viruses. The basic screening protocol is based on the following principles. First, for the enveloped viruses, the entry is mediated by the viral GP projecting from the viral envelope (lipid). Thus, the GP of a virus dictates its entry tropism. For many enveloped viruses such as HIV, filoviruses, and influenza viruses, a single GP is necessary and sufficient to mediate viral entry.6,30 Second, viral GPs usually can be incorporated onto another virus to generate “chimeric” viral particles, commonly referred to as pseudovirions.12 Such pseudoviral particles, in general, retain the entry property dictated by the incorporated GPs on the virions. Thus, the generated pseudovirions can be used as a reliable surrogate system to study entry mechanism and to screen entry inhibitors of an enveloped virus of interest with alleviated safety concerns since the research can be carried out in a biosafety level 2 (BSL-2) facility even for highly pathogenic viruses, which require BSL-3 or BSL-4 facilities. Third, three types (or more) of pseudovirions were employed here sequentially (or at the same time) to screen the same chemical library so that these screens were used as counterscreens to each other, allowing us to identify virus-specific and “shared” hit compounds with greatly reduced false positives, a major problem with HTS. In the current study, the following three types of pseudovirions—MARV (HIV with Marburg glycoprotein GP incorporated on the surface), AIV (HIV with avian influenza virus H5N1 HA), and LASV (HIV with Lassa arenavirus GP)—were produced and used to evaluate a small chemical library (Prestwick library; see below) sequentially (Fig. 1A), and the effect of each compound on three types of pseudovirions was directly compared, and virus-specific and “shared” hit compounds were easily identified (Fig. 1B). For example, since all three types of pseudovirions share the same HIV core, a compound that can inhibit the AIV but not MARV or LASV is most likely a specific inhibitor to influenza viral entry. Therefore, in principle, one should be able to identify the compounds that specifically inhibit HA-mediated entry from those that specifically inhibit MARV or LASV GP-mediated entry through a single round of HTS.
Figure 1.
Experimental design of the comparative high-throughput screening protocol. (A) The comparative compound screen workflow. A549 cells are seeded into 384-well plates and infected by pseudovirions 24 h later in the presence of diverse compounds. After a 48-h incubation, the virus infection is detected by luciferase activity assay. (B) Compound library plate design and data interpretation. Azidothymidine (AZT) controls (black) and DMSO controls (red) are arranged on the first column of the 384-well plate; compounds are arranged in the columns 3 through 22. Compound only showing low signal (black) in assay plate of one type of virus is regarded as this virus’ specific hit; compound showing low signals in assay plates of two different viruses is regarded as the shared hit by these two viruses; compound showing low signals in the assay plates of all three viruses is regarded as a shared hit by three viruses. AIV, avian influenza virus; LASV, Lassa virus; MARV, Marburg virus.
Experimental Protocols
The experimental protocols are depicted in Figure 2. The two flowcharts show two key stages of the HTS screens: (1) production of HIV pseudovirions (Fig. 2A) and (2) compound screen (Fig. 2B).
Figure 2.
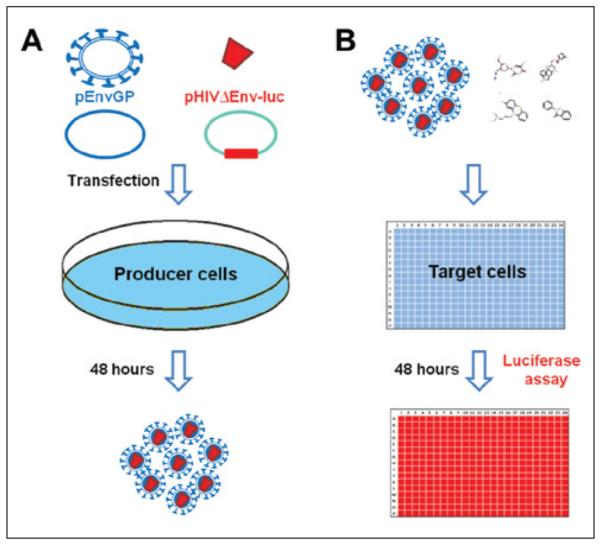
Experimental procedure flowcharts. (A) Generation of pseudotyped virions. 293T producer cells are cotransfected with plasmid-containing virus envelope protein gene (pEnvGP) and plasmid-containing human immunodeficiency virus (HIV) genome with the luciferase reporter gene but lacking the HIV envelope protein gene (pHIVΔEnv-luc). Forty-eight hours later, pseudovirions in the supernatant are collected for infection assay. (B) Virus infection assay. Pseudovirions and compounds are applied to A549 target cells, and infection is determined 48 h later by luciferase activity assay.
Production of HIV Pseudovirions
Production of high quality of the three types of HIV pseudovirions, each carrying a particular viral GP, was an integral part of a robust HTS. To accomplish this, an HIV vector, pNL4-3.Luc. R.E., was cotransfected with the viral GP plasmids of interest (MARV glycoprotein GP, avian influenza virus H5N1 HA, or LASV glycoprotein GP) to the 293T producer cells. This HIV vector is replication deficient since the HIV is Env- and Vpr-.26,27 Importantly, this HIV vector carries a luciferase gene as a reporter, and thus the level of viral infection can be measured by the luciferase activity of the infected cells. Forty-eight hours after transfection, the supernatants were collected, filtered, and titered to ensure the quality of the produced pseudovirions. It was found that the prepared pseudovirions could be stored at 4 °C for up to at least 2 weeks prior to use without reducing the infectivity of the pseudovirions.
Determination of the Viral Infectivity
The infectivity of each stock of the collected pseudovirions was determined by the following method. Target A549 cells in a 384-well plate were infected with aliquots (30 μL/well) of the collected pseudovirions, and the luciferase activity (relative light units, RLUs) of the infected cells was measured 48 h postinfection. A stock of pseudovirion preparation with at least 106 RLUs (normally within the range of 106 to 2 × 106 RLUs) was used for HTS.
Compound Screening
Each of the 1200 compounds from the Prestwick library (25 μM final concentration) was robotically dispensed into plates with A549 cells in duplicate (384-well plates). Based on the parameters optimized in our pilot experiments, approximately 1000 A549 cells were seeded for each well. To each set of the 384-well plates with A549 cells, one type of the HIV pseudovirions (MARV, AIV, or LASV) was individually added to each well with the predetermined amounts, the cells were incubated for 48 h, and luciferase substrate was directly added to the wells and the luciferase activity of each well measured. Although it seemed feasible to screen with the three types of pseudovirions simultaneously in a parallel HTS format, we chose to perform the screening sequentially in this study.
Parameter Optimization and Quality Control
HTS assay quality is crucial in HTS experiments. In our screens, we use three parameters to assess the screening quality.
Signal-to-Noise Ratio
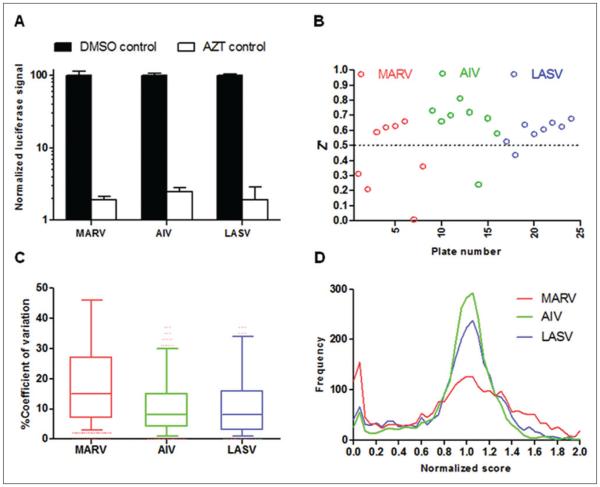
Selection of effective positive and negative controls is critical for assay development. In our screens, we used eight DMSO (0.25%) control wells as negative controls and eight AZT (5 μM in 0.25% DMSO) control wells as positive controls in each plate. AZT was used as a positive control because it is an HIV reverse transcriptase inhibitor, and it inhibits the infection of HIV-based pseudotype viruses. As shown in Figure 3A, in our three separate screens, the AZT controls gave only ~2% of signals compared with DMSO controls, giving a more than 50-fold signal-to-noise ratio.
Figure 3.
Quality control of the comparative high-throughput screening. (A) The signals in DMSO or azidothymidine (AZT) control wells were normalized by plate median and are shown in black columns (DMSO) and white columns (AZT). Results are the means ± standard deviation. (B) The Z′ factors were calculated from normalized signals of DMSO and AZT control wells and are plotted for each plate from Marburg virus (MARV; red circle), avian influenza virus (AIV; green circle), and Lassa virus (LASV; blue circle) screens. (C) Coefficient of variation was used to assess screen reproducibility, and it was calculated for each compound from its duplicate wells by the following equation: 100 × standard deviation of duplicate wells/mean of duplicate wells. Box-and-whisker plots are employed to represent the range of coefficients of variation for each screen. (D) For each compound, the luciferase signal was normalized by the plate median to get the normalized score, which was used to plot the distribution of signals for each screen.
Z′ Factor
Z′ factor is the most commonly used statistical parameter to assess screen quality.31 Normalized signals from both DMSO and AZT control wells were used for Z′ factor calculation. Figure 3B shows the Z′ factors of 24 plates from the three separate screens, with most Z′ factors above 0.5, indicating that the overall quality of HTS was excellent.
Variation of Duplicate Wells
To increase the reliability of HTS, we performed each HTS with duplicate 384-well plates. In addition, by comparing the signals in duplicate plates, we could also assess the reproducibility of the screens. Coefficient of variation (CV) was calculated from duplicate wells and used as an indicator of assay reproducibility. As shown in Figure 3C, the median CVs in three screens ranged from 10% to 15%, indicating that screen reproducibility was excellent. We also observed that different viruses had different responses to the Prestwick compound library. MARV had the widest CV range and correspondingly the broadest signal distribution, and AIV had a narrow CV range and the sharpest signal distribution (Fig. 3C,D).
Data Analysis and Identification of Putative Entry Inhibitors
Analysis of the HTS data from the three sequential screens allowed us to directly identify and classify entry inhibitors for MARV, AIV, and LASV in the Prestwick chemical library since the three screens could be used as counterscreens for each another. Using 70%, 80%, or 90% inhibition as arbitrary cutoffs, putative entry inhibitors for individual viruses as well as “shared inhibitors” for two or three viruses could be easily identified (Table 1). For example, if we used 80% inhibition as the cutoff, 177, 53, and 84 hit compounds were identified for MARV, AIV, and LASV based on the three individual screens, respectively. However, pairwise and three-way comparisons revealed that only 19 and 10 compounds could be classified as putative MARVor LASV-specific entry inhibitors, respectively, whereas there was no AIV-specific entry inhibitor. Furthermore, two compounds were classified as MARV/ AIV specific since they did not inhibit LASV (less than 20% inhibition), four compounds as MARV/LASV specific, and only one compound as AIV/LASV specific (Table 1 and Fig. 4), demonstrating the power of this approach.
Table 1.
Summary of Putative Entry Inhibitors for Three Viruses from the Prestwick Library.
| Inhibition, % | MARV | AIV | LASV | MARV/AIV | MARV/LASV | AIV/LASV | “Shared” | Nonclassified (MARV/AIV/ LASV) |
|---|---|---|---|---|---|---|---|---|
| >90 | 140 (12)a | 35 (0) | 51 (7) | 28 (0)b | 34 (1) | 25 (1) | 22c | 88/4/7d |
| >80 | 177 (19) | 53 (0) | 84 (10) | 44 (2) | 63 (4) | 37 (1) | 36 | 87/8/10 |
| >70 | 195 (25) | 66 (0) | 115 (14) | 53 (3) | 84 (10) | 46 (1) | 42 | 75/9/13 |
The number of compounds showing >90% inhibition in a screen is listed. Here 140 compounds were identified in the Marburg virus (MARV) screen. However, only 12 compounds, shown in parentheses, were identified as putative entry inhibitors of MARV after comparison with two other virus screens (>90% inhibition in MARV screen but <20% inhibition in the other two screens).
The number of compounds showing >90% inhibition in two screens is listed. Here 28 compounds were identified in the MARV and avian influenza virus (AIV) screens. However, no compound, shown in parentheses, was identified as the putative entry inhibitor for MARV and AIV (>90% inhibition in MARV and AIV screens but <20% inhibition in the Lassa virus [LASV] screen).
The number of compounds showing >90% inhibition in all three screens is listed as “shared.” See text for the explanation.
The number of compounds that cannot be classified as the a, b, or c group in the screens is listed (88 for MARV, 4 for AIV, and 7 for LASV).
Figure 4.
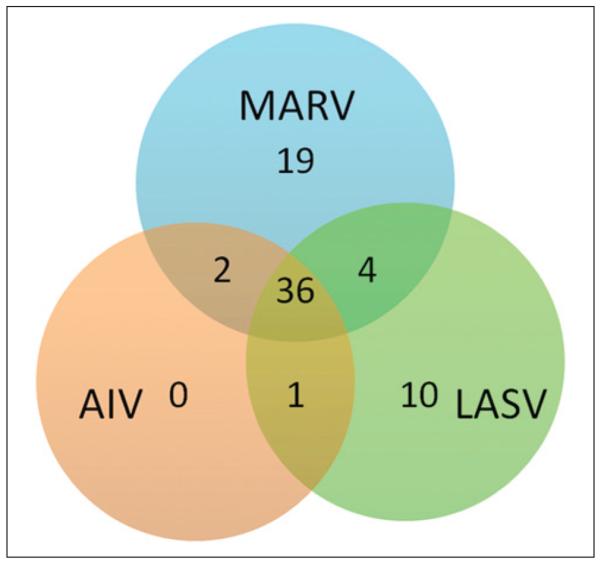
Venn diagram of three viruses’ hit selection. All hit selection criteria were based on an inhibition rate greater than 80% on the target virus compared with an inhibition rate lower than 20% on the other viruses. In total, 19, 0, and 10 hits are specific to Marburg virus (MARV), avian influenza virus (AIV), and Lassa virus (LASV), respectively; 2, 4, and 1 hits are shared by two of the three viruses, and 36 hits are “shared hits” for three viruses.
However, classification of one group of compounds, labeled as “shared” in Table 1 (36 compounds with 80% inhibition as the cutoff), was less straightforward since these compounds could belong to any of the following three groups: (1) they indeed represented shared inhibitory compounds—that is, some of these compounds could block viral entry for all three viruses, MARV, AIV, and LASV. (2) Some of the compounds could block HIV replication after entry. In fact, one of the compounds in the Prestwick library is AZT, a reverse transcriptase inhibitor for HIV, which was identified as a shared hit in our screens, as expected. (3) Some of the compounds were simply toxic to the target cells at the tested concentration (25 μM). Further evaluation of this class of compounds will be discussed below.
Discussion
In this report, we describe a cell-based comparative HTS protocol to identify entry inhibitors for enveloped viruses, particularly highly pathogenic ones. The utility and power of this protocol are demonstrated by screening a small library of 1200 compounds (Prestwick library) for three different viruses—MARV, AIV, and LASV—and putative entry inhibitory compounds, both virus specific and broad spectrum, were identified for these viruses by comparing the three separate screens.
Several aspects of the comparative HTS protocol described in this report make it an attractive approach for antiviral drug discovery targeting the entry step for the highly pathogenic enveloped viruses such as HIV, filoviruses (Ebola and Marburg viruses), avian influenza virus H5N1, and arenaviruses, which require high-containment facilities (BSL-3 or BSL-4) for handling infectious viruses. Thus, using a defective HIV-based pseudotyping platform as a surrogate system in the screens reduces the safety concerns, allowing one to carry out the cell-based HTS in a BSL-2 facility, making the HTS highly practical and amenable for most researchers. In addition, the HIV vector used in this study, pNL4.3. Luc,17,26,27 which carries a luciferase gene as the reporter, was advantageous over other HIV vectors, which use green fluorescent protein (GFP) as the reporter gene, since we observed that using luciferase as the reporter typically gave a much greater signal-to-background ratio (routinely more than 100 to 1000) than that of GFP (roughly around 10) in the pilot experiments (data not shown). Such a greater signal-to-background ratio is desirable since it is generally thought that a ratio more than 10 is required for a robust HTS.
The most important advantage of the comparative HTS protocol working with several viruses, either simultaneously (parallel screens) or sequentially as in the current study (comparative screens), instead of performing one screen for one virus as routinely done by different researchers, is its power in reducing the number of the false-positive hits and quickly identifying putative entry inhibitors for different viruses. This point is well illustrated by the results presented in the Results section (see Table 1 and Figure 4). For example, a screen with MARV indicated that 140 compounds in the Prestwick library (1200 compounds total) displayed more than 90% inhibition on infection of the HIV pseudovirions expressing Marburg GP. However, comparative analysis of the data sets with the other two screens (AIV and LASV) suggested that only 12 compounds are putative entry inhibitors specifically for MARV. Similarly, only 7 of 51 compounds can be classified as LASV-specific inhibitors (using 90% inhibition as the cutoff; see Table 1). In essence, these three screens were used as counterscreens against one another, allowing us to identify virus-specific entry inhibitory compounds without any other assay.
However, it should be noted that there are certain limitations associated with using the aforementioned HIV pseudotyping platform as a surrogate HTS protocol. The most important one is whether a pseudotyping entry assay can accurately mimic the authentic entry pathway(s) of an infectious virus. Therefore, it is imperative to validate putative entry inhibitors using assays with virus-like particles32,33 and eventually infectious viruses. Another limitation of the current protocol, although not a major one, is that it is not always easy to distinguish two groups of compounds, shared and nonclassified (see Table 1), by comparing the results from the screens. Therefore, additional experiments such as entry assays with a different core (such as murine leukemia virus, instead of HIV) of pseudovirions14 and cytotoxicity assays will be needed to determine the nature of the shared hit and nonclassified compounds. In fact, it can be highly beneficial to identify entry inhibitors with broad-spectrum activity for antiviral drug discovery and development because broad-spectrum antiviral therapeutics are highly desirable. Furthermore, some of the shared hit compounds may block viral entry of viruses and instead may target HIV replication.34 Therefore, the comparative screening protocol using an HIV pseudotyping platform described in this study, as well as identification of the shared hit compounds to target HIV replication, can be used for anti-HIV drug discovery as well.
In summary, the cell-based comparative screening protocol using an HIV pseudotyping platform described in this report can be used as a powerful and robust HTS protocol to identify entry inhibitors for many enveloped viruses, particularly those highly pathogenic viruses. We suggest that, in addition to identification of novel entry inhibitors for different viruses, such high-throughput screens with numerous viruses may be insightful in understanding entry pathways of different viruses, revealing the shared pathways as well as the divergence for different viruses.
Acknowledgments
We thank Dr. Michael Farzan (Scripps Florida) for the Lassa virus glycoprotein (GP) plasmid.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was partially supported by National Institutes of Health grant UO1 AI77767 to L.R., and we acknowledge support from the UIC Center for Clinical and Translational Science through a grant from the National Institutes of Health, National Center for Advancing Translational Sciences, UL1TR000050. J.W. was supported by the China Scholarship Council.
Footnotes
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Wright PF, Newumann G, Kawaoka Y. Orthomyxoviruses. In: Fields Virology; Knipe DM, Howley PM, editors. Vol. 2. Wolters Kluwer, Lippincott Williams & Wilkins; Philadelphia, PA: 2007. pp. 1691–1740. [Google Scholar]
- 2.Sanchez A, Geiger TW, Feldmann H. fIloviridae. Marburg and Ebola viruses. In: Knipe DM, Howley PM, editors. Fields Virology. Vol. 1. Wolters Kluwer, Lippincott Williams & Wilkins; Philadelphia, PA: 2007. pp. 1409–1448. Ch. 40. [Google Scholar]
- 3.White JM. Viral and Cellular Membrane Fusion Proteins. Annu. Rev. Physiol. 1990;52:675–697. doi: 10.1146/annurev.ph.52.030190.003331. [DOI] [PubMed] [Google Scholar]
- 4.Doms RW, Moore JP. HIV-1 Membrane Fusion: Targets of Opportunity. J. Cell. Biol. 2000;151:F9–F14. doi: 10.1083/jcb.151.2.f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckert DM, Kim PS. Mechanisms of Viral Membrane Fusion and Its Inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 6.Skehel JJ, Wiley DC. Receptor Binding and Membrane Fusion in Virus Entry: The Influenza Hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 7.Rojek JM, Kunz S. Cell Entry by Human Pathogenic Arenaviruses. Cell Microbiol. 2008;10:828–835. doi: 10.1111/j.1462-5822.2007.01113.x. [DOI] [PubMed] [Google Scholar]
- 8.Abel S, Back DJ, Vourvahis M. Maraviroc: Pharmacokinetics and Drug Interactions. Antivir. Ther. 2009;14:607–618. [PubMed] [Google Scholar]
- 9.Levy JA. HIV Pathogenesis: 25 Years of Progress and Persistent Challenges. AIDS. 2009;23:147–160. doi: 10.1097/QAD.0b013e3283217f9f. [DOI] [PubMed] [Google Scholar]
- 10.Lalezari JP, Henry K, O'Hearn M, et al. Enfuvirtide, an HIV-1 Fusion Inhibitor, for Drug-Resistant HIV Infection in North and South America. N. Engl. J. Med. 2003;348:2175–2185. doi: 10.1056/NEJMoa035026. [DOI] [PubMed] [Google Scholar]
- 11.Kilby JM, Hopkins S, Venetta TM, et al. Potent Suppression of HIV-1 Replication in Humans by T-20, a Peptide Inhibitor of gp41-Mediated Virus Entry. Nat. Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 12.Hunter E, Swanstrom R. Retrovirus Envelope GPs. Cur. Topic Microbiol. Immun. 1990;157:187–253. doi: 10.1007/978-3-642-75218-6_7. [DOI] [PubMed] [Google Scholar]
- 13.Chan SY, Speck RF, Ma MC, et al. Distinct Mechanisms of Entry by Envelope GPs of Marburg and Ebola (Zaire) Viruses. J. Virol. 2000;74:4933–4937. doi: 10.1128/jvi.74.10.4933-4937.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wool-Lewis R, Bates P. Characterization of Ebola Virus Entry by Using Pseudotyped Viruses: Identification of Receptor Deficient Cell Lines. J. Virol. 1998;72:3155–3160. doi: 10.1128/jvi.72.4.3155-3160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeffers SA, Sanders DA, Sanchez A. Covalent Modifications of the Ebola Virus GP. J. Virol. 2002;76:12463–12472. doi: 10.1128/JVI.76.24.12463-12472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takada A, Robison C, Goto H, et al. A System for Functional Analysis of Ebola Virus GP. Proc. Natl. Acad. Sci. U. S. A. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manicassamy B, Wang J, Jiang H, et al. Comprehensive Analysis of Ebola Virus GP1 in Viral Entry. J. Virol. 2005;79:4793–4805. doi: 10.1128/JVI.79.8.4793-4805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Y, Rumschlag-Booms E, ang J, et al. Analysis of Hemagglutinin-Mediated Entry Tropism of H5N1 Avian Influenza. Virol. J. 2009;6:39. doi: 10.1186/1743-422X-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Y, Tisoncik J, McReynolds S, et al. Identification of a New Region of SARS-CoV S Protein Critical for Viral Entry. J. Mol. Biol. 2009;394:600–605. doi: 10.1016/j.jmb.2009.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basu A, Li B, Mills DM, et al. Identification of a Small-Molecule Entry Inhibitor for Filoviruses. J. Virol. 2011;85:3106–3119. doi: 10.1128/JVI.01456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yermolina MV, Wang J, Caffrey M, et al. Discovery, Synthesis, and Biological Evaluation of a Novel Group of Selective Inhibitors of Filoviral Entry. J. Med. Chem. 2011;54:765–781. doi: 10.1021/jm1008715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez O, Johnson J, Manicassamy B, et al. Zaire Ebola Virus Entry into Human Dendritic Cells Is Insensitive to Cathepsin L Inhibition. Cell Microbiol. 2010;12:148–157. doi: 10.1111/j.1462-5822.2009.01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, Rothwangl K, Mesecar AD, et al. Lamiridosins, Hepatitis C Virus Entry Inhibitors from Lamium album. J. Nat. Prod. 2009;72:2158–2162. doi: 10.1021/np900549e. [DOI] [PubMed] [Google Scholar]
- 24.Manicassamy B, Wang J, Rumschlag E, et al. Characterization of Marburg Virus GP in Viral Entry. Virology. 2007;358:79–88. doi: 10.1016/j.virol.2006.06.041. [DOI] [PubMed] [Google Scholar]
- 25.Radoshitzky SR, Abraham J, Spiropoulou CF, et al. Transferrin Receptor 1 Is a Cellular Receptor for New World Hemorrhagic Fever Arenaviruses. Nature. 2007;446:92–96. doi: 10.1038/nature05539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He J, Choe S, Walker R, et al. Human Immunodeficiency Virus Type 1 Viral Protein R (Vpr) Arrests Cells in the G2 Phase of the Cell Cycle by Inhibiting p34cdc2 Activity. J. Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Connor RI, Chen BK, Choe S, et al. Vpr Is Required for Efficient Replication of Human Immunodeficiency Virus Type-1 in Mononuclear Phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 28.Tisoncik JR, Guo Y, Cordero KS, et al. Identification of Critical Residues of Influenza Neuraminidase in Viral Particle Release. Virol. J. 2011;8:14. doi: 10.1186/1743-422X-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Development Core Team . R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing; Vienna, Austria: 2011. http://www.R-project.org/ [Google Scholar]
- 30.White JM. Membrane Fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 31.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 32.Manicassamy B, Rong L. Expression of Ebolavirus GP on the Target Cells Enhances Viral Entry. Virol. J. 2009;6:75. doi: 10.1186/1743-422X-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tscherne DM, Manicassamy B, Garcia-Sastre A. An Enzymatic Virus-Like Particle Assay for Sensitive Detection of Virus Entry. J. Virol. Methods. 2010;163:336–343. doi: 10.1016/j.jviromet.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rumschlag-Booms E, Zhang H, Soejarto DD, et al. Development of an Antiviral Screening Protocol: One-Stone-Two-Birds. J. Antivir. Antiretrovir. 2011;3:1. doi: 10.4172/jaa.1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]