Abstract
In Myanmar, civil unrest and establishment of internally displaced persons (IDP) settlement along the Myanmar-China border have impacted malaria transmission. The growing IDP populations raise deep concerns about health impact on local communities. Microsatellite markers were used to examine the source and spreading patterns of Plasmodium falciparum between IDP settlement and surrounding villages in Myanmar along the China border. Genotypic structure of P. falciparum was compared over the past three years from the same area and the demographic history was inferred to determine the source of recent infections. In addition, we examined if border migration is a factor of P. falciparum infections in China by determining gene flow patterns across borders. Compared to local community, the IDP samples showed a reduced and consistently lower genetic diversity over the past three years. A strong signature of genetic bottleneck was detected in the IDP samples. P. falciparum infections from the border regions in China were genetically similar to Myanmar and parasite gene flow was not constrained by geographical distance. Reduced genetic diversity of P. falciparum suggested intense malaria control within the IDP settlement. Human movement was a key factor to the spread of malaria both locally in Myanmar and across the international border.
Keywords: Plasmodium falciparum, malaria transmission, border migration, genetic bottleneck, microsatellites, genetic diversity
Graphical Abstract
1. Introduction
Since the inception of the Roll Back Malaria Initiative by World Health Organization (WHO) in 1999, malaria control has been greatly intensified in endemic countries. In conjunction with better financial support and improved technology, malaria morbidity and mortality have been reduced tremendously worldwide (WHO 2008, 2013). However, the reductions in malaria are not universal. While some countries such as Morocco and Syria did eliminate malaria in the past decade, many others such as Thailand and China are still experiencing a sustained low-level transmission (Coker et al., 2011). In Myanmar, the situation is particularly concerning because of the resurgence of the disease and lack of sufficient, consistent information on malaria transmission (Cui et al., 2012a; WHO 2013). Myanmar has the highest malaria burden among other Southeast Asian countries with approximately 200,000 cases per year (Coker et al., 2011; WHO 2013). The number of malaria cases and malaria-induced mortality in Myanmar has been consistently high for the past three decades (Cui et al., 2012b). Factors such as the emergence/spread of P. falciparum resistance to artemisinins and P. vivax resistance to chloroquine, inadequate epidemiological data to assess malaria situations, complex vectorial systems, and above all civil unrest make malaria control very difficult in Myanmar (Coker et al., 2011; Cui et al., 2012a & b; Delacollette et al., 2009; Cheeseman et al., 2012; Phyo et al., 2012; Li et al., 2013).
Myanmar has been engaged in the world's longest-running civil unrest. Commonly known as Burma, Myanmar succumbed to ethnic civil unrest and turmoil since its independence in 1948, and this conflict remains unresolved today (Socheat et al., 2003). Political instability and military conflicts have driven hundreds of thousands of citizens into relocation camps known as Internally Displacement Persons (IDP) settlement scattered throughout the country's borders, specifically the Myanmar-China and Myanmar-Thailand borders. Large-scale human movement has led to intensive transmission of malaria in the IDP settlement (Delacollette et al., 2009; Archavanitkul et al., 2010; Kumar et al. 2012). In addition, the high proportion of ethnic minorities who live in remote countryside receives very little attention and healthcare resources from the central government (WHO 2008). For example, in Kachin State, the remote border region of northeast Myanmar, the estimated malaria incidence and morbidity and mortality rates have been shown higher than other parts of Myanmar (WHO 2008; Lee et al., 2006; Li et al., 2013). Regional seasonality coupled with population movement along country borders and countryside heavily impact malaria transmission in both Myanmar and China.
This study aimed to examine the sources and spreading patterns of P. falciparum between IDP settlement and surrounding villages in eastern Myanmar and western China along the international border. Moreover, we compared the parasite samples collected in the past three years to determine if there were changes in genotype structure over time within the IDP settlement and local community. In-depth knowledge and information on the extent of malaria spread are keys to target disease control efforts in high-risk areas. This is of particular relevance when most other parts of Southeast Asia are entering the malaria elimination phase.
2. Materials and methods
2.1. Blood sample collection
Nearly 300 samples that were diagnosed with P. falciparum infections were collected from clinics or hospitals located in two IDP settlements, Je Yang Hka (JYH) and Hpum Lum Yang (HLY), and four surrounding villages/towns including military base (CMH), Ja Htu Kawng (JHK), Laiza (LZCH), and Mai Sak Pa (MSP) in Myanmar along the international border of China, in addition to two town hospitals Tengchong (TC) and Yingjiang (YJ) in Yunnan, China (Figure 1; Supplemental Table S1). Among the village localities, LZCH is a major regional hospital that represents a larger catchment area of nearly 100,000 people from surrounding smaller villages. By contrast, CMH, JHK, and MSP are local clinics that represent a smaller catchment area of approximately 3,000 people. For the IDP settlement population size is variable; in 2012 the population size of HLY and JYH were about 1,600 and 8,600 respectively. Because of uneven sample size among localities, genetic variation was compared among three locality settings: regional hospital (LZCH), villages (CMH, JHK, and MSP), and IDP settlement (JYH and HLY). Samples from JYH and LZCH were collected in three consecutive years 2011, 2012, and 2013 for genetic composition comparison. All studied individuals showed fever or malaria-related symptoms at the time of sampling, and were detected with falciparum-infection by microscopic examination and PCR assays. For each individual, 30–50 µl of blood was blotted on Whatman 3MM filter papers. Filter papers were air-dried and stored in zip-sealed plastic bags with silica gel absorbent at room temperature until DNA extraction. Parasite DNA was extracted from dried blood spots by the Saponin/Chelex method (Bereczky et al., 2005).
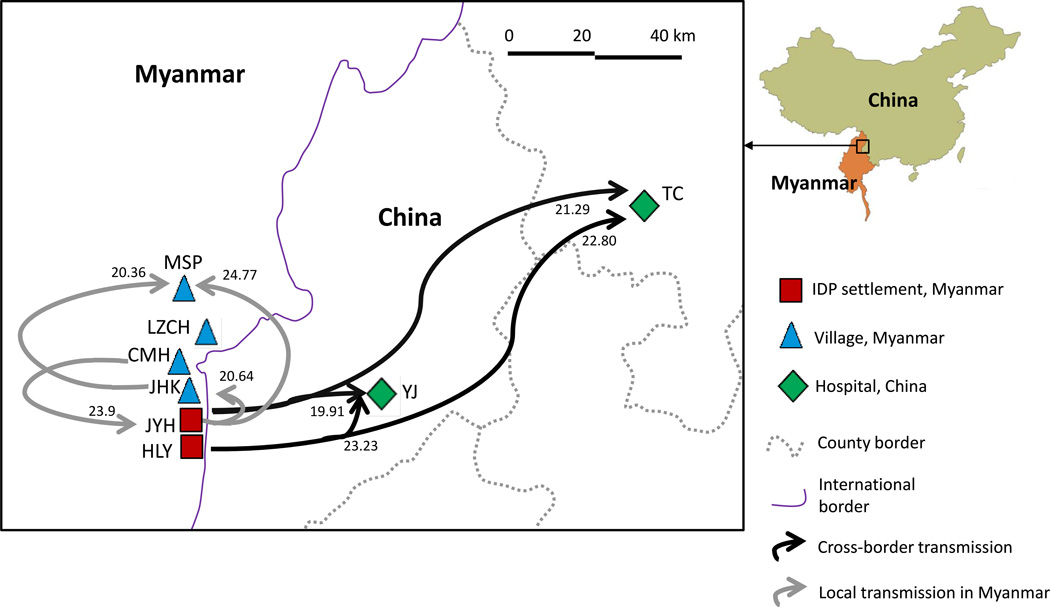
Figure 1.
Geographical distribution of the IDP settlement and surrounding villages in Myanmar and China along the international border. Arrows indicate the pathway of Plasmodium falciparum transmission and numbers on top of the arrows indicate the migration rate (M) as presented in Supplementary Table S3 (only M value of 20 or above was shown).
2.2. Microsatellite genotyping
Thirteen single-copy microsatellites (SSRs) with tri- or tetranucleotide repeats, which map to 14 chromosomes are typed for P. falciparum. Alleles were PCR-amplified with the published oligonucleotide primers following the published protocol (Su et al., 1999; Anderson et al., 1999). After PCR amplification, products were pooled as follows: POLY2+TAA124+POLYα+TAA81, TAA42+ TAA87+TAA109, PE87a+9735+PFPK2, TAA80+PfG377+TAA116 according to their sizes and fluorescent labels (Supplemental Table S2). The pooled products were separated on an ABI 3730 capillary sequencer and all allele sizes were determined and visualized in Peak Scanner.
2.3. Data analyses
2.3.1. Linkage disequilibrium genetic diversity, and clonal assessment
To examine whether the SSR loci represent an independent set of markers in the P. falciparum genome, genotyping linkage disequilibrium (LD) was tested by Fisher’s exact test for each pair of loci with GenePop version 4.2, using the Markov chain method with 100 batches and 10,000 iterations per batch (Raymond and Rousset, 1995). Significance values were adjusted by sequential Bonferroni correction for multiple comparisons.
Genotypic diversity and clonal variation were calculated in GenoDive version 2.0b4 (Meirmans and Van Tienderen, 2004) and GenClone version 2.0 (Amaud-Haond and Belkhir, 2007). We first calculated genetic distances using the method of Smouse & Peakall – a squared Euclidean distance based on the number of times a certain allele is found in the two individuals (Smouse and Peakall,1999). The minimal distance class was set as threshold to identify the follow: (i) the number of multilocus genotypes (G) where parasite samples with identical genotypes at all the examined microsatellite loci were identified as a single clone and thus the number of unique multilocus genotypes was referred to the number of unique clones; (ii) Simpson’s diversity index (D), also referred as clonal diversity corrected for sample size (a measure of the proportion of unique genotypes/clones in a population) that ranges from zero (where two randomly chosen individuals in a population represent a single clone) to one (where individuals all represent different clones); and (iii) genotype evenness (E) that ranges from zero (where one or a few clones dominate in a population) to one (where all clones are of equal frequency in a population). In addition, the number of effective alleles and expected heterozygosity were estimated with GENALEX for each of the localities (Peakall and Smouse, 2006).
2.3.2. Population structure and isolation-by-distance analyses
A model-based Bayesian method implemented in STRUCTURE (version 2.3.4) was performed to examine partitioning of individuals to genetic clusters (Pritchard et al., 2000). All samples from the different years were included in a single analysis to allow comparison of clustering patterns among sites as well as among years. The number of clusters (K) was determined by simulating a range of K values from 1 (no genetic differentiation among all localities) to 8 (all localities were genetically differentiated from one another). The posterior probability of each value was then used to detect the modal value of ΔK, a quantity related to the second order rate of change with respect to K of the likelihood function. Posterior probability values were estimated using a Markov Chain Monte Carlo (MCMC) method, and a burn-in period of 500,000 iterations followed by 106 iterations of each chain were performed to ensure convergence of the MCMC. Each MCMC chain for each value of K was run eight times with the ‘independent allele frequency’ option that allows individuals with ancestries in more than one group to be assigned into one cluster. Individuals were assigned into K clusters according to membership coefficient values (Q) ranged from 0 (lowest affinity to a cluster) to 1 (highest affinity to a cluster). The partitioning of clusters was visualized with the program DISTRUCT (Rosenberg 2004).
An FST analysis was conducted using θ, an FST-estimator in SPAGeDi version 1.2e (Hardy and Vekemans, 2002). FST values were tested for significance using 10,000 permutations. Genetic differentiation among sites was displayed by multidimensional scaling plot based on the estimated DS values (an analog of FST). Furthermore, an analysis of molecular variance (AMOVA) was used to determine the hierarchical distribution of genetic variance within and among populations of the same year, as well as samples of different years but of the same population using GENALEX (Peakall and Smouse 2006). The relationships between genetic distances (DS values estimated from SPAGeDi) and traveling distances (estimated from the route that connects between two sites based on the road network of the region) were analyzed for the IDP settlement and village samples from different years (2011–2013), respectively, by Mantel tests (10,000 randomizations) and reduced major axis (RMA) regression in the Isolation By Distance (IBD) (version 1.52; Bohonak 2002).
2.3.3. Detection of bottlenecks and migration rates
We used BOTTLENECK (v1.2.02) to detect the signature of a genetic bottleneck (Piry et al., 1999). Two tests were performed on the samples from IDP settlement (JYH) and local community (LZCH) of which sample size was large and consistent in the past three years. First was the test on the overall distribution of allele frequency classes. Second was the Wilcoxon-signed rank test to compare the number of loci that present a heterozygosity excess to the number of such loci expected by chance only using three different mutation models: the infinite alleles model (IAM), the stepwise mutation model (SMM), and a combination of those two extreme hypotheses, the two-phase model (TPM). In the TPM, the proportion of IAM was set to 10% and the variance of those mutations to 12. These are generic values typical for many microsatellite markers (Busch et al., 2007; Hundertmark and Van Daele, 2010).
Gene flow among IDP settlement and village populations was estimated for each year by performing a maximum-likelihood analysis implemented in Migrate-N (v2.4.2; Beerli 2004; Beerli and, Felsenstein, 2001). Parameters including Θ (defined as 4Neµ, where Ne is the effective population size and µ is the mutation rate per generation and site), M (m/µ, where m is the immigration rate scaled by mutation rate), and the number of effective migrants per generation Nem (ΘM/2 for haploid individuals) were estimated. Four independent runs were conducted with the Brownian motion model using 10 short chains with 5,000 sampled genealogies and 3 long chains with 50,000 sampled genealogies to obtain the mean and range of Θ, M, and Nem values.
2.4. Ethics statement
Scientific and ethical clearance was given by the institutional scientific and ethical review boards of Kunming Medical University, China; University of California at Irvine, USA; Pennsylvania State University, USA; and the Ministry of Health of Kachin State, Myanmar. Written informed consent/assent for study participation was obtained from all consenting heads of households or parents/guardians (for minors under age 18) and each individual who was willing to participate in the study.
3. Results
3.1. Genetic diversity in IDP settlement and local community
No significant linkage disequilibrium was detected for all pairwise combinations of the 13 microsatellite loci (Bonferroni corrected P<0.05) among the P. falciparum samples. All samples revealed a single, sharp peak for each of the microsatellite loci without ambiguity in allele scoring. In general, genotypic diversity and expected heterozygosity were shown to be significantly higher for the community samples collected from the regional hospital and village clinics than samples from the IDP settlement (Table 1; P<0.01, two-tailed t-test). Among the IDP samples, a drastic decline in genetic diversity was observed over the past three years where genotypic diversity and expected heterozygosity were found to be lowest in 2013, indicative of a reduced proportion of unique clones/genotypes within the population. On the other hand, only little differences were observed in genetic diversity among the community samples in the past three years (Table 1), suggesting that the proportion of unique clones remained similar in these populations.
Table 1.
Comparison of genetic diversity measures based on 13 microsatellite loci.
| Genotypic diversity | Gene diversity | ||||||
|---|---|---|---|---|---|---|---|
|
Locality setting |
Year |
Sample size |
G | D | E | Ne | He |
| Regional hospital |
|||||||
| 2011 | 33 | 18.00 | 0.94 | 0.90 | 2.68 (±0.43) |
0.46 (±0.08) |
|
| 2012 | 50 | 29.35 | 0.97 | 0.82 | 2.60 (±0.42) |
0.49 (±0.06) |
|
| 2013 | 42 | 14.53 | 0.87 | 0.44 | 1.81 (±0.16) |
0.45 (±0.06) |
|
| Village | |||||||
| 2011 | 39 | 20.59 | 0.98 | 0.93 | 2.81 (±0.45) |
0.46 (±0.06) |
|
| 2012 | 26 | 15.91 | 0.93 | 0.80 | 2.45 (±0.40) |
0.58 (±0.08) |
|
| 2013 | 5 | 3.57 | 0.90 | 0.89 | 1.59 (±0.13) |
0.39 (±0.08) |
|
| IDP settlement |
|||||||
| 2011 | 31 | 3.45 | 0.71 | 0.86 | 1.96 (±0.23) |
0.42 (±0.07) |
|
| 2012 | 54 | 10.56 | 0.67 | 0.54 | 1.72 (±0.12) |
0.37 (±0.06) |
|
| 2013 | 40 | 5.19 | 0.54 | 0.73 | 1.08 (±0.05) |
0.23 (±0.04) |
|
G: Number of multilocus genotypes corrected for sample size
D: Simpson’s diversity index corrected for sample size
E: Genotypic evenness
Ne: Number of effective alleles (Nielsen et al. 2003)
He: Expected heterozygosity corrected for sample size (Nei 1978)
AMOVA indicated that most of the genetic variation (>75%; Table 2) was within populations in the 2011, 2012, and 2013 samples, respectively. Very little variation was found between IDP settlements and villages in Myanmar as well as between Myanmar and China. For the 2012 samples, a greater proportion (22%) of the variation was detected among populations. When samples were compared across years for each of the locality settings, we found a contrasting distribution of variation (Table 2). For the community samples, most of the variation (88–100%; Table 3) was within the same year and only a relatively small proportion of variation between years. However, for the IDP samples up to 41% of variation was between years (Table 2), indicative of considerable genetic differentiation over time.
Table 2.
Results of analyses of molecular variance (AMOVA) of Plasmodium falciparum samples across years of different locality settings.
| Samples | Source of variation | Percentage of variation | P-value |
|---|---|---|---|
| Partition by site | |||
| 2011 | Between IDP and villages in Myanmar | 3 | ns |
| Among populations | 9 | ns | |
| Within populations | 88 | - | |
| 2012 | Between China and Myanmar | 2 | ns |
| Between IDP and villages in Myanmar | 0 | ns | |
| Among populations | 22 | * | |
| Within populations | 76 | - | |
| 2013 | Between IDP and villages in Myanmar | 3 | ns |
| Among populations | 0 | * | |
| Within populations | 97 | - | |
| Partition by year | |||
| Regional hospital | |||
| Among years | 12 | ** | |
| Within years | 88 | - | |
| Village | |||
| Among years | 0 | ns | |
| Within years | 100 | - | |
| IDP settlement | |||
| Among years | 41 | ** | |
| Within years | 59 | - |
‘ns’ denotes P-value>0.05;
P-value≤0.05;
P-value≤0.01;
‘-’ denotes significance test not applicable.
Table 3.
Summary of the parameters and results based on BOTTLENECK analyses.
| Locality setting | Year of collection | Mode shift | Mutation model | Heterozygote excess |
|---|---|---|---|---|
| Regional hospital | ||||
| 2011 (n=33) | ||||
| Normal L-shaped | IAM | P = 0.3 | ||
| SMM | 0.34 | |||
| TPM | 0.48 | |||
| 2012 (n=50) | ||||
| Normal L-shaped | IAM | 0.22 | ||
| SMM | 0.07 | |||
| TPM | 0.54 | |||
| 2013 (n=42) | ||||
| Normal L-shaped | IAM | 0.31 | ||
| SMM | 0.003** | |||
| TPM | 0.06 | |||
| IDP settlement | ||||
| 2011 (n=29) | ||||
| Shifted mode | IAM | 0.002** | ||
| SMM | 0.02* | |||
| TPM | 0.005** | |||
| 2012 (n=47) | ||||
| Normal L-shaped | IAM | 0.83 | ||
| SMM | 0.21 | |||
| TPM | 0.46 | |||
| 2013 (n=40) | ||||
| Shifted mode | IAM | 0.008* | ||
| SMM | 0.03* | |||
| TPM | 0.05* |
Village samples were not included because of the small sample size in 2013;
P-value≤0.05;
P-value≤0.01
3.2. Genetic clustering of samples
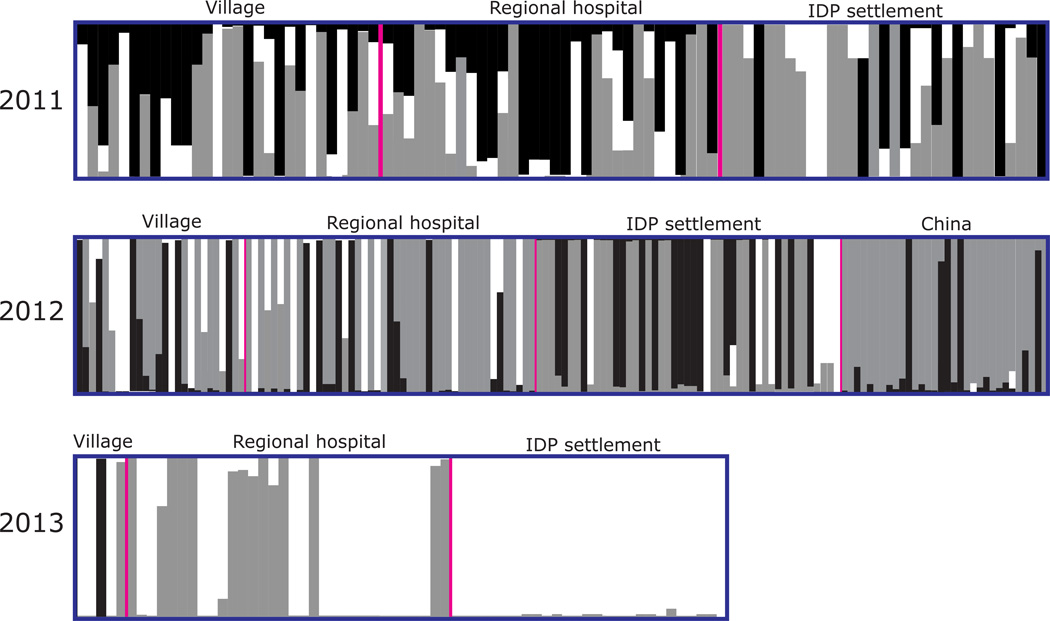
The optimal partitioning of all samples is obtained for K=3 (Figure 2) by STRUCTURE analyses. For the 2011 samples, individuals from the villages, regional hospital, and IDP settlement constituted mixed genetic clusters (Figure 2). Almost all individuals had Q-value <0.9, indicative of a resemblance to at least two genetic clusters. The genetic composition of the 2012 samples for the most part was similar to that of 2011. Individuals from China (TC and YJ) were more genetically uniform and they shared the same genetic clusters (gray and black) with samples from Myanmar (Figure 2). Interestingly, the genetic composition of the 2013 samples was markedly different from those of 2011 and 2012 (Figure 2). For the village and regional hospital samples, it was apparent that nearly half of the individuals constituted the gray cluster similar to individuals of previous years, whereas the remaining individuals (except one that was assigned to the black cluster) constituted the white cluster based on a set of pre-existing alleles (Figure 2). The change was most substantial in the IDP settlement where samples changed from an admixture of alleles resembling three genetic clusters (2011 and 2012) to homogenously belonging to one cluster with almost no admixture of alleles (white for 2013; Figure 2).
Figure 2.
Bayesian inferences of the K clusters estimated by STRUCTURE among Plasmodium falciparum samples collected from IDP settlement and local community in Myanmar and from China in 2011–2013. The three identified clusters are presented as three different colors (black, gray, and white), and individuals are represented as columns. Within each column (individual), the extent of the component colors indicates the magnitude of the membership coefficient (Q) corresponding to each cluster.
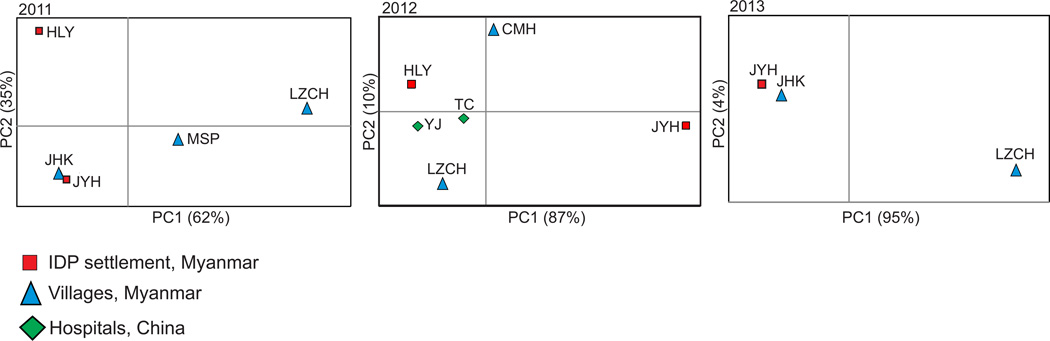
Mantel tests indicated no significant associations between geographical and genetic distances among sites with respect to different years (2011: R2 = 0.08, P > 0.05; 2012: R2 = 0.16, P > 0.05; 2013: R2 = 0.41, P > 0.05). Although the two IDP settlements (JYH and HLY) were located in close geographical proximity (Fig. 1), they were not genetically close to each other (Figure 3). Likewise, TC and YJ (China) were geographically distant from most of the Myanmar sites; however, they were genetically close to the IDP settlement (HLY; Figure 3). These findings suggested that parasite gene flow via human movement was not limited by physical distance.
Figure 3.
Scatter plots of pairwise DS values (an analog of FST) showing the genetic relatedness of Plasmodium falciparum among localities with respect to years. The first two axes that contain over 95% of the total variation are shown. Locations of the studied sites are presented in Figure 1 and Table 1.
3.3. Demographic change and migrations
BOTTLENECK analyses detected excess heterozygosity in the IDP settlement samples from 2011 and 2013 under all the three mutation models (Table 3). The mode-shift test also indicated a deviation from the normal L-shape distribution in allele frequency as an evidence of a bottleneck in these samples. For the regional hospital samples, although excess heterozygosity was detected under the SMM model in the 2013 samples, mode-shift test did not show any signature of bottleneck (Table 3).
Migrate-N analyses revealed that all populations had relatively small effective population size (Θ), suggesting the effect of drift was unequivocally as significant as migration (Supplementary Table S3). Given that most values of M (m/µ) were >1, the effect of migration (m) was larger than the effect of mutation (µ). For the 2011 samples, the effective number of migrants per generation Nem ranged from 0.08–1.36. The greatest migration was observed from IDP settlement JYH to village MSP (Nem=1.36, M=24.77; Supplementary Table S3). For the 2012 samples, the greatest migration was observed from village CMH to IDP settlement JYH (Nem=0.72, M=23.9; Supplementary Table S3) followed by migration from HLY to China (TC: Nem=0.68, M=22.88; YJ: Nem=0.70, M=23.23). For the 2013 samples, the greatest migration was found from IDP settlement JYH to village JHK (Nem=0.31, M=20.64; Supplementary Table S3). Overall, these results indicated that gene flow between IDP settlement and villages occurred in both directions (Figure 1) and that the combined effects of migration and drift influenced genetic variability of the parasite populations. Plasmodium falciparum infections detected in China were migrated or introduced from Myanmar IDP settlements and such migration was likely asymmetrical (Figure 1).
4. Discussion
According to Refugees International published in January 2012, an estimated 500,000 people were displaced by internal conflict in eastern Myanmar. The settlement of displaced populations has raised concerns of health impact on local residents (Cui et al., 2012a & b; Delacollette et al., 2009). Malaria is one of the most commonly reported causes of death among refugees and displaced persons in disease endemic countries (Connolly et al., 2004). Several studies have shown that disease incidence rates in refugee/IDP settlements are generally high because of the overcrowding and substandard living conditions (CDC, 1992; Toole and Waldman, 1997; Salama et al., 2001; Connolly et al., 2004). For example, civil wars and conflicts have occurred in the Sahel region of Africa for many decades and malaria epidemics are common in refugee camps (Rowland and Nosten, 2001; Bayoh et al., 2001; Burns et al., 2012).
While it is generally thought that displaced populations are more vulnerable to infectious diseases, our data indicated an overall lower and reduced genetic diversity in the parasite populations from the IDP settlement than the community samples in Myanmar. These findings lend support to the observation of a lower malaria annual incidence rate in the IDP settlements (54 cases/1,000 population) as compared to local villages (134 cases/1,000 population; Zhou et al., unpublished data). In the IDP settlements, malaria incidence was found to be relatively low during the first 21 months since the settlements established in July 2011 without seasonal peaks. By contrast, in local villages peak malaria incidence was consistently observed during May-August in 2011–2013, and the peak incidence rates increased from year to year (Zhou et al., unpublished data).
Significant bottlenecks were detected in the IDP settlement samples collected in 2011 and 2013, respectively, with markedly reduced genetic variation. Comparatively, there was only little support for heterozygosity excess or signature of bottleneck in the samples from the regional hospital, suggesting that the local parasite populations were relatively stable over the years and that genetic diversity was sustained high under the mutation-drift equilibrium. There are four possible explanations for such contrasting patterns. First, the regional hospital (LZCH) covers a relatively large catchment area and associated parasite pools. Our previous studies of parasite genetic diversity using the polymorphic antigenic markers indicated that parasites in this area were genetically diverse (Yuan et al., 2013). Thus, a smaller founder population of the parasite in the IDP settlement may in part explain the lower genetic diversity compared to that in the local community. Second, the reduced genetic diversity particularly in the 2013 samples may imply effective health management and disease control within the IDP settlements. For example, there were clinics/hospitals within all IDP settlements that provide free malaria diagnosis and treatment. However, in most of the local villages and regional hospitals health infrastructure is poor and is often short of antimalarial supplies (Zhou et al., 2014). Furthermore, in the IDP settlement, indoor and outdoor insecticide sprays were routine. Every household received free bed nets and approximately 60% of these bed nets were long-lasting insecticide-treated whereas 35% were non-insecticide-treated. By contrast, in the local villages only 0.4% of the nets were long-lasting insecticide-treated and as much as 43.3% were non-insecticide-treated (Zhou et al., unpublished data). In addition, the IDP settlements used deep-well pumped water and the drainage ditches were connected to the river that could reduce standing water habitat, whereas the local villages used open-well, shallow water that could serve as breeding sites for vector mosquitoes. These control measures in the IDP may altogether contribute to reduced parasite diversity. Third, the observed genetic bottlenecks in the parasite populations in IDP settlements may suggest greater effectiveness or usage of antimalarial drug in clearing the parasites compared to local villages. This notion remains to be investigated further whether P. falciparum in local villages are more resistant to antimalarial drug in use than the parasites that circulated in the IDP settlement. Fourth, the reduced proportion of unique parasite genotypes in the IDP settlements may reflect specialization or selection of clones that were adapted to the IDP habitats probably based on biting patterns of the vector mosquitoes, human behavior, as well as the physical environments.
The IDP settlement in Myanmar was first set up in August 2011 to accommodate residents from central and northeast Myanmar who were affected by civil unrest. Due to internal conflicts, tens of thousands of Burmese have fled their home villages and displaced in resettlement or refugee camps located along international borders, while many others have migrated to neighboring countries such as China and Thailand for jobs. The displacement of large numbers of people and their circulation can favor malaria transmission and disseminate the disease to other countries (Martens and Hall, 2000). Our data showed that parasite gene flow via human migration between IDP settlements and villages was bidirectional and frequent, contributing to the vast genetic variation within the parasite populations but very little differentiation among populations. Such migration was alarming because malaria spread not only occurred at the local scale but also stretched over 100 km away from Myanmar to China without much constraint in physical distance or political boundary. Infected cases in Yunnan (TC, YJ) were evidently introduced from Myanmar based on their genetic resemblance. These results were also supported by the traveling record of the studied subjects indicating their recent visits to Myanmar prior to the diagnosis and confirmation of malaria (Zhou et al., 2014). Similar situation was seen between Myanmar and Thailand where border migration played a key role in malaria epidemics in Thailand (Wiwanitkit et al., 2002; Zhou et al., 2005; Archavanitkul et al., 2010; Phyo et al., 2012).
Although China has demonstrated a decline in malaria morbidity and mortality, control efforts are hampered by the continuous influx of migrants with travel histories to Myanmar (Xu and Liu, 1997; Moore et al., 2008; Li et al., 2009). In the past years high malaria incidence were reported in areas along the international border of Myanmar and China (Clements et al., 2009; Liu et al., 2009; Li et al., 2013; Zhou et al., 2014). In 2010, over 90% of falciparum infections detected in China were imported cases and these cases were largely concentrated along the Yunnan/Myanmar border (Zhou et al., 2011). Border areas represent the largest reservoirs for malaria and frequent malaria introductions by migratory human populations could be difficult to monitor (Jitthal et al., 2013). The movement of infected people from malaria-endemic areas to areas where the disease had been or near eliminated (usually with non-immune people) can heighten epidemic risk in local communities leading to resurgence of the disease. More importantly, population movement is increasingly implicated in the spread of drug resistance in malaria (Socet al., 2003; Cheeseman et al., 2012; Cui et al., 2012b; Yuan et al., 2013). Hence, parasite reintroduction by human travel presents a major challenge in malaria elimination and coordinated international efforts are needed to tighten existing border patrol and prevent malaria reintroduction.
5. Conclusions
High malaria incidence in Myanmar represents a major challenge for malaria control and elimination in the GMS region especially neighboring countries such as China and Thailand. Because of the continuous conflict in Myanmar, the current magnitude and diversity of population movement are unprecedented, and thus the impact of population movement on malaria transmission can be complex and chronic. This study provided genetic information that shed light on malaria transmission in part of the GMS region with the goal to improve existing preventive measures and control efforts. Notwithstanding that displaced populations are vulnerable to infectious diseases like malaria, it is more concerning that the local community around displaced populations could be at greater increased risks by human movement and poor health infrastructure. The findings of low genetic diversity and bottlenecks in the parasite populations in IDP settlements support the view that influx of parasites through IDP settlements is low compared to local transmission, suggestive of better control effort in the IDP settlements. Thus, current control policy implemented in the IDP settlement e.g., by increasing indoor/outdoor insecticide spray to contain parasite spread and minimize local transmission, and by ensuring regular supply of effective antimalarial drug should be expanded and reinforced in local villages. In addition, areas at risk for epidemics through influx of infected people should be identified and local community should be provided with increased efforts and resources in disease surveillance and guidance. In long term, the costs involved in the handling malaria outbreaks and the widespread treatment of infections would far outweigh the costs involved in implementing preventive measures if no additional actions are taken.
Supplementary Material
Highlights.
Falciparum in the IDP showed reduced genetic diversity over the past three years
Genetic diversity of P. falciparum was lower in the IDP than in local community
Falciparum infections detected in China were introduced from Myanmar IDP settlements
Malaria spreads locally in Myanmar and across international border via human movement
Acknowledgments
The authors thank the field team for their technical assistance. We are grateful to the communities and hospitals for their support and willingness to participate in this research. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the National Institute of Health. This work was funded by the National Institute of Health (U19 AI089672).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data
Supplementary Table S1. Locality information and sampling size of studied sites in the Myanmar-China border area.
Supplementary Table S2. Information of microsatellite loci used in the present study.
Supplementary Table S3. Frequency of migration between sampling locations of Plasmodium falciparum with respect to years.
All authors declared no conflict of interest.
References
- Arnaud-Haond S, Belkhir K. Genclone: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol. Ecol. Notes. 2007;7:15–17. [Google Scholar]
- Anderson TJC, Su XZ, Bockarie M, Lagog M, Day KP. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology. 1999;119:113–125. doi: 10.1017/s0031182099004552. [DOI] [PubMed] [Google Scholar]
- Archavanitkul K, Vajanasara K. Health of Migration Workers from Myanmar, Cambodia and Laos. In: Kanchanachitra C, Podhisita C, Archavanikul K, et al., editors. Thai Health 2010: Capitalism in crisis, opportunity for society? Bangkok: Amarin Pringting; 2010. pp. 31–32. [Google Scholar]
- Bayoh MN, Akhwale W, Ombo kM, Sang D, Engoki SC, Koros D, Hamel MJ. Malaria in Kakuma refugee camp, Turkana, Kenya: facilitation of Anopheles arabiensis vector populations by installed water distribution and catchment systems. Malar. J. 2011;10:149. doi: 10.1186/1475-2875-10-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P. Migrate: Documentation and program, part of LAMARC. Version 2.4.4. 2004 Available at: http://evolution.gs.washington.edu/lamarc.html.
- Beerli P, Felsenstein J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. PNAS. 2001;98:4563–4568. doi: 10.1073/pnas.081068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereczky S, Martensson A, Gil JP. Short report: Rapid DNA extraction from archive blood spots on filter paper for genotyping of Plasmodium falciparum. Am J. Trop. Med. Hyg. 2005;72:249–251. [PubMed] [Google Scholar]
- Bohonak AJ. IBD (Isolation By Distance): a program for analyses of isolation by distance. J. Hered. 2002;93:153–154. doi: 10.1093/jhered/93.2.153. [DOI] [PubMed] [Google Scholar]
- Burns M, Rowland M, N’Guessan R, Carneiro I, Beeche A, Ruiz SS, Kamara S, Takken W, Carnevale P, Allan R. Insecticide-treated plastic sheeting for emergency malaria prevention and shelter among displaced populations: an observational cohort study in a refugee setting in Sierra Leone. Am. J. Trop. Med. Hyg. 2012;87:242–250. doi: 10.4269/ajtmh.2012.11-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch JD, Waser PM, DeWoody A. Recent demographic bottlenecks are not accompanied by a genetic signature in banner-tailed kangaroo rats (Dipodomys spectabilis) Mol. Ecol. 2007;16:2450–2463. doi: 10.1111/j.1365-294X.2007.03283.x. [DOI] [PubMed] [Google Scholar]
- Cheeseman IH, Miller BA, Nair S, Nkhoma S, Tan A, Tan JC, Anderson TJ. A major genome region underlying artemisinin resistance in malaria. Science. 2012;336:79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centres for Disease Control and Prevention. Famine affected, refugee and displaced populations: recommendations for public health issues. MMWR. 1992;41:RR-1. [PubMed] [Google Scholar]
- Clements AC, Barnett AG, Cheng ZW, Snow RW. Space-time variation of malaria incidence in Yunnan province, China. Malar. J. 2009;8:180. doi: 10.1186/1475-2875-8-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker RJ, Hunter BM, Rudge JW, Liverani M, Hanvoravongchai P. Emerging infectious diseases in southeast Asia: regional challenges to control. Lancet. 2011;377:599–609. doi: 10.1016/S0140-6736(10)62004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly MA, Gayer M, Ryan MJ, Salama P, Spiegel P, Heymann DL. Communicable diseases in complex emergencies: impact and challenges. Lancet. 2004;364:1974–1983. doi: 10.1016/S0140-6736(04)17481-3. [DOI] [PubMed] [Google Scholar]
- Cui L, Yan G, Sattabongkot J, Chen B, Cao Y, Fan Q, Parker D, Sirichaisinthop J, Su XZ, Yang H, Yang Z, Wang B, Zhou G. Malaria in the Greater Mekong Subregion: Heterogeneity and complexity. Acta Trop. 2012;121:227–239. doi: 10.1016/j.actatropica.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Yan G, Sattabongkot J, Chen B, Cao Y, Fan Q, Parker D, Sirichaisinthop J, Su XZ, Yang H, Yang Z, Wang B, Zhou G. Challenges and prospects for malaria elimination in the Greater Mekong Subregion. Acta Trop. 2012;121:240–245. doi: 10.1016/j.actatropica.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacollette C, D’Souza C, Christophe lE, Thimasarn K, Abdur R, Bell D, Ehrenberg J. Malaria trends and challenges in the Greater Mekong Subregion. Southeast Asian J. Trop. Med. Public Health. 2009;40:674–691. [PubMed] [Google Scholar]
- Hardy OJ, Vekemans X. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Note. 2002;2:618–620. [Google Scholar]
- Hundertmark KJ, Van Daele LJ. Founder effect and bottleneck signatures in an introduced, insular population of elk. Conserv. Genet. 2010;11:139–147. [Google Scholar]
- Jitthal N. Migration and malaria. Southeast Asian J. Trop. Med. Public Health. 2013;44:166–200. [PubMed] [Google Scholar]
- Kumar A, Chery L, Biswas C, et al. Malaria in South Asia: prevalence and control. Acta Trop. 2012;121:246–255. doi: 10.1016/j.actatropica.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TJ, Mullany LC, Richards AK, Kuiper HK, Maung C, Beyrer C. Mortality rates in conflict zones in Karen, Karenni, and Mon states in eastern Burma. Trop. Med. Int. Health. 2006;11:1119–1127. doi: 10.1111/j.1365-3156.2006.01651.x. [DOI] [PubMed] [Google Scholar]
- Li N, Parker DM, Yang Z, Fan Q, Zhou G, Ai G, Wang Y. Risk factors associated with slide positivity among febrile patients in a conflict zone of northeastern Myanmar along the China-Myanmar border. Malar. J. 2013;12:361. doi: 10.1186/1475-2875-12-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Nie RH, Li CF, Sun YH, Li GS. Active detection of malaria cases in Myanmar Wa ethnical villages of China-Myanmar border. Parasites Infect. Dis. 2009;7:6–9. [Google Scholar]
- Martens P, Hall L. Malaria on the move: human population movement and malaria transmission. Emerg. Infect. Dis. 2000;6:103–109. doi: 10.3201/eid0602.000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirmans PG, Van Tienderen PH. GenoType and GenoDive: two programs for the analysis of genetic diversity of asexual organisms. Mol. Ecol. Note. 2004;4:792–794. [Google Scholar]
- Moore SJ, Min X, Hill N, Jones C, Zaixing Z, Cameron MM. Border malaria in China: knowledge and use of personal protection by minority populations and implications for malaria control: a questionnaire-based survey. BMC Public Health. 2008;8:344. doi: 10.1186/1471-2458-8-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Note. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, McGready R, Nosten F. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet. 2012;379:1960–1966. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piry S, Luikart G, Cornuet JM. BOTTLENECK: a computer program for detecting recent reductions in effective size using allele frequency data. J. Hered. 1999;90:502–503. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2): a population genetic software for exact test and ecumenicism. J. Hered. 1995;86:248–249. [Google Scholar]
- Rosenberg NA. Distruct: a program for the graphical display of population structure. Mol. Ecol. Note. 2004;4:137–138. [Google Scholar]
- Rowland M, Nosten F. Malaria epidemiology and control in refugee camps and complex emergencies. Ann. Trop. Med. Parasit. 2001;95:741–754. doi: 10.1080/00034980120103405. [DOI] [PubMed] [Google Scholar]
- Salama P, Spiegel P, Brennan R. No less vulnerable: the internally displaced in humanitarian emergencies. Lancet. 2001;357:1430–1431. doi: 10.1016/S0140-6736(00)04570-0. [DOI] [PubMed] [Google Scholar]
- Socheat D, Denis MB, Fandeur T, et al. Mekong malaria. II. Update of malaria, multi-drug resistance and economic development in the Mekong region of Southeast Asia. Southeast Asian J. Trop. Med. Public Health. 2003;34:1–102. [PubMed] [Google Scholar]
- Smouse PE, Peakall R. Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity. 1999;82:561–573. doi: 10.1038/sj.hdy.6885180. [DOI] [PubMed] [Google Scholar]
- Su XZ, Gerdig MT, Huang Y, Huynh CQ, Liu A, You J, Wootton JC, Wellems TE. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science. 1999;286:1351–1353. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]
- Toole MJ, Waldman RJ. The public health aspects of complex emergencies and refugee situations. Annu. Rev. Publ. Health. 1997;18:283–312. doi: 10.1146/annurev.publhealth.18.1.283. [DOI] [PubMed] [Google Scholar]
- World Health Organization South-East Asia Region. Malaria in the Greater Mekong Subregion: regional and country profiles. New Delhi: WHO press; 2008. [Accessed 19 September 2014]. http://www.searo.who.int/myanmar/documents/malariainthegreatermekongsubregion.pdf. [Google Scholar]
- World Health Organization. World Malaria Report. Geneva: WHO press; 2013. [Accessed 19 September 2014]. http://www.who.int/malaria/publications/world_malaria_report_2013/report/en/ [Google Scholar]
- Wiwanitkit V. High prevalence of malaria in Myanmar migrant workers in a rural district near Thailand-Myanmar border. Scand. J. Dis. 2002;34:236–237. doi: 10.1080/00365540110077272. [DOI] [PubMed] [Google Scholar]
- Xu J, Liu H. Border malaria in Yunnan, China. Southeast Asian J. Trop. Med. Public Health. 1997;28:456–459. [PubMed] [Google Scholar]
- Yuan L, Zhao H, Wu L, Li X, Parker D, Xu S, Cui L. Plasmodium falciparum populations from northeastern Myanmar display high levels of genetic diversity at multiple antigenic loci. Acta Trop. 2013;125:53–59. doi: 10.1016/j.actatropica.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Sirichaisinthop J, Sattabongkot J, Jones J, Bjørnstad ON, Yan G, Cui L. Spatio-temporal distribution of Plasmodium falciparum and P. vivax malaria in Thailand. Am. J. Trop. Med. Hyg. 2005;72:256–262. [PubMed] [Google Scholar]
- Zhou G, Sun L, Xia R, et al. Clinical malaria along the China- Myanmar border, Yunnan Province, China, January 2011-August 2012. Emerg. Infect. Dis. 2014;20:675–678. doi: 10.3201/eid2004.130647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SS, Wang Y, Li Y. Malaria situation in the People’s Republic of China in 2010 [in Chinese] Chin. J. Parasitol. Parasitic Dis. 2011;29:401–403. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.