Abstract
Objective:
To compare cutaneous and nasal population levels between patients colonized and infected by multidrug-resistant organisms in two intensive care units.
Methods:
A prospective cohort study was performed in adult intensive care units of two hospitals in Belo Horizonte, Brazil (April 2012 to February 2013). Clinical and demographic data were first collected by reviewing patients’ charts. Then, samples collected with nasal, groin, and perineum swabs were cultivated in selective media for 48 h at 37°C. After isolation, determination of antimicrobial susceptibility and biochemical identification were performed.
Results:
A total of 53 cases of colonization were observed by the following bacteria in decreasing frequencies: imipenem-resistant Acinetobacter baumannii (50.9%), vancomycin-resistant Enterococcus faecalis (43.4%), extended-spectrum beta-lactamase–producing Klebsiella pneumoniae (37.7%), imipenem-resistant Pseudomonas aeruginosa (32.1%), oxacillin-resistant Staphylococcus aureus (7.5%), and imipenem-resistant Klebsiella pneumoniae (5.7%). Among these colonization cases, 26 (49.0%) were followed by infection with bacteria phenotypically similar to those of the colonization. A relation between high population levels of colonization by most of the multidrug-resistant organisms at anatomical sites and a subsequent infection was observed. After colonization/infection, bacterial population levels decreased progressively and spontaneously until disappearance by day 45 in all the anatomical sites and for all the multidrug-resistant organisms.
Conclusion:
There was a correlation between high population levels of colonization by multidrug-resistant organisms at anatomical sites and a subsequent infection. Reduction in multidrug-resistant organism populations after colonization at anatomical sites could be a preventive measure to reduce evolution to infection as well as transmission of these bacteria between patients in intensive care unit.
Keywords: Nosocomial infection, antimicrobial susceptibility, colonization, infection
Background
Alterations in the indigenous microbiota are observed in patients during their permanence in a hospital, especially when antimicrobials have being administered. After hospital discharge, a patient may carry multidrug-resistant organisms (MDROs), whether they are commensal, such as Staphylococcus epidermidis, or truly pathogenic, such as methicillin-resistant Staphylococcus aureus (MRSA).1
MDROs that commonly infect humans have multiple resistance mechanisms and include the following microorganisms: Enterococcus faecium, S. aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. Despite the fact that Gram-negative bacilli (BGN) are the most problematic drug-resistance bacteria in Brazil, there are no systematic data concerning its prevalence in colonization and/or infection in the country.2–4
Early detection of patients colonized with MDROs allows preventive measures to prevent progression and/or transmission of infection, such as isolation or selective decontamination. These measures can reduce pathogen transfer, infection rates, morbidity, mortality, and hospital costs, especially in intensive care units (ICUs). Among patients admitted to ICUs, various risk factors favor infection occurrence, such as antimicrobial use over long periods, time after ICU admission, immunosuppression, medical devices, microorganism virulence, and loss of skin integrity.3–5
As the identification of MDROs as well as the determination of their distribution among anatomical sites in colonized patients and of the evolution of their population levels (i.e. decreasing until elimination or progressing until infection) may contribute to the control of healthcare-associated infections (HCAI), these have been the objectives of this study performed in patients in ICU.
Methods
Patients
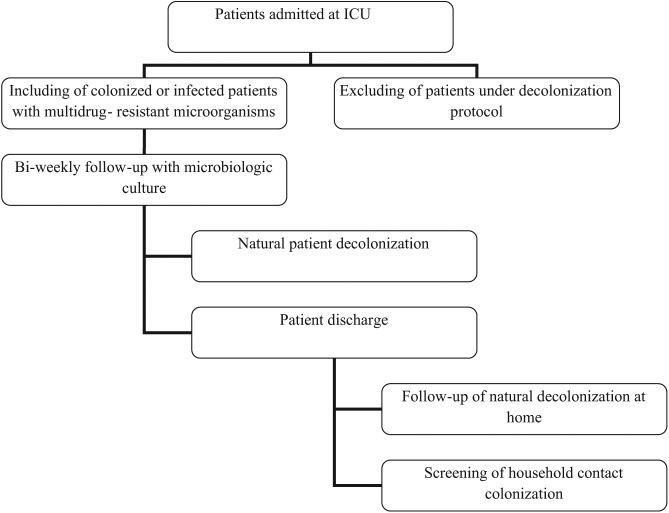
A prospective cohort study was performed in adult ICUs of two different hospitals in Belo Horizonte, Brazil, between April 2012 and February 2013. The research was approved by the Ethical Committee (0386.0.203.000-11). The ICUs showed the following characteristics: (1) ICU (18 beds): located in a large teaching hospital and admitting mainly surgical patients and (2) ICU (10 beds): located in a large philanthropic hospital and admitting both surgical patients and those from medical clinics. In both ICUs, the mean length of stay was 15 days. The study population consists of all patients identified as colonized or infected during the study period. Patients or their next of kin were requested to give informed written consent prior to sampling. The follow-up was carried out every 15 days at the hospital and every month at community after patient discharge (Figure 1). Patients were eligible if they were over 18 years old and were colonized with ampicillin- or vancomycin-resistant Enterococcus spp., oxacillin- or vancomycin-resistant S. aureus, imipenem- or third-generation cephalosporin-resistant K. pneumoniae, or ciprofloxacin-resistant and/or imipenem/meropenem-resistant A. baumannii and P. aeruginosa.6
Figure 1.
Follow-up flowchart of natural decolonization of admitted colonized or infected patients in the ICU.
For each selected patient, the following data were retrieved from patients’ charts: age, sex, monitoring by medical devices (i.e. mechanical ventilation, venous catheter, drains, and nasal and indwelling catheters), and Average Severity Index Score (ASIS). ASIS was classified into five levels (A–E) according to the severity range regarding the patients’ clinical conditions,7 antimicrobial use (i.e. class, previous use, use during the length of stay in the ICU, and time of use), and the occurrence of infection by the MDROs cited above. The information regarding infection occurrence was retrieved from patient chart review. In both ICUs, there was no protocol of decolonization, and antimicrobials were used in patient treatment only in cases of infection.
Sampling and treatment of samples
Each colonized or infected patient was followed up with surveillance culture every 15 days during the length of stay at the hospital as described. Three anatomical sites (nasal cavity, groin, and perineum)8 in the same patient were sampled with a sterile swab (Haste Plástico—J. P). Each swab was immersed and shaken in a tube containing 1 mL of sterile saline. From this initial suspension, the following dilutions were performed: nasal cavity and groin, 10−2 and 10−4; perineum, 10−2 and 10−6. Then, a volume of 0.1 mL of each dilution was plated onto the following selective media—MacConkey agar (K. pneumoniae and A. baumannii), cetrimide agar (P. aeruginosa), Baird–Parker agar, Mannitol salt agar (S. aureus), and Bile Esculin agar (Enterococcus spp.)—and incubated at 37°C for 48 h.
Microbiological analysis
Semi-quantitative counts of bacterial growth were performed using the adequate plate and the following formula: colony counts on Petri dishes × inverse of dilution factor × 10 = number of colony-forming units (CFU)/mL of swab initial suspension. Data were expressed as log10 CFU/mL.
After counts, each different phenotype was re-isolated on Brain Heart Infusion (BHI) or Mannitol salt agar (S. aureus) and then submitted to catalase test and Gram stain. Identification was performed using specific cards of the VITEK 2® system for Gram-positive and Gram-negative bacteria (bioMérieux, Marcy-l’Etoile, France). The repetitive extragenic palindromic sequence–based polymerase chain reaction (rep-PCR) using DiversiLab system (bioMérieux) was used according to the manufacturer’s recommendations to compare strain similarity.
Antimicrobial susceptibility test was performed using Bauer–Kirby method with the following antimicrobials: vancomycin (Enterococcus spp.); imipenem (P. aeruginosa, K. pneumoniae, and A. baumannii); third generation of cephalosporin, ceftazidime, cefotaxime, ceftriaxone, and aztreonam (K. pneumoniae); and cefoxitin (S. aureus). Agar dilution with vancomycin was used for S. aureus according to Clinical and Laboratory Standards Institute (CLSI).6
Statistical analysis
Statistical analysis was performed using Student’s t test, chi-square test for media, and Fisher’s exact test for proportions. The Spearman correlation–associated Bonferroni test was used for the analysis of association between bacterial population levels at anatomical sites and colonization. The Spearman correlation was performed regarding the variable “colonization” treated as dichotomy and the abnormal distribution of bacterial population levels. The logistical regression was used in the analysis of variables associated with colonization status. A p-value less than 0.05 was considered significant, and 95% confidence intervals were reported. The analysis was performed using the STATA™ software, version 11.0.
Results
Demographic data
A total of 53 patients were colonized with MDROs. The patients’ average age was 55 years, and the proportions of male and female were quite similar (52% and 48%, respectively). The most frequent levels of severity (ASIS) were C (81%—physiological stability, requiring nursing intensive care) and D (physiological instability, requiring medical and nursing intensive care and constant evaluation). Their primary diagnoses were in decreasing order: cardiovascular disease (27.0%), malignancies (20.0%), infectious diseases (7.0%), and external causes (5.0%). Among the 53 follow-up patients, 16.0% died. The mean length of stay in the ICU was 22.3 days, and after ICU discharge, the mean time in the wards was 21.2 days. The majority of patients (64.0%) underwent surgery, and among the 53 colonized patients, 30.0% had a pressure ulcer.
Distribution of colonization by MDRO
A total of 74.0% (39/53) of the patients were colonized by more than one MDRO, including Gram-negative and Gram-positive bacteria. Among the patients colonized with only one MDRO (14/53), Gram-negative bacteria (9/14) and vancomycin-resistant E. faecalis (VRE) (5/14) were identified during the follow-up. The bacteria identified in colonized patients were, in decreasing order of frequency, as follows: imipenem-resistant A. baumannii (50.9%), VRE (43.4%), extended-spectrum beta-lactamase (ESBL)–producing K. pneumoniae (37.7%), imipenem-resistant P. aeruginosa (32.1%), oxacillin-resistant S. aureus (ORSA; 7.5%), and imipenem-resistant K. pneumoniae (5.7%) (Table 1).
Table 1.
Frequencies of colonized and infected patients with MDRO in ICU, main colonization sites, and factors associated with colonization.
| MDRO | Number of patients colonized by each MDRO/total number (%) | Main anatomical sites of colonization | Factors associated with colonization | Infection (% in relation to total number of infected patients—26) |
|---|---|---|---|---|
| Imipenem-resistant Acinetobacter baumannii | 27/53 (50.9) | Nasal cavity; groin | Pressure ulcer; carbapenem | 35 |
| Vancomycin-resistant Enterococcus | 23/53 (43.4) | Perineum | Drain; glycopeptide | 19 |
| Klebsiella pneumoniae ESBL | 20/53 (37.7) | Groin | Fluoroquinolone | 27 |
| Imipenem-resistant Pseudomonas aeruginosa | 17/53 (32.1) | Nasal cavity; groin | Fluoroquinolone; mechanical ventilation | 11 |
| Oxacillin-resistant Staphylococcus aureus | 4/53 (7.5) | Nasal cavity | No factor associated | 0 |
| Imipenem-resistant K. pneumoniae | 3/53 (5.7) | Groin | Fluoroquinolone | 8 |
MDRO: multidrug-resistant organism; ICU: intensive care unit; ESBL: extended-spectrum beta-lactamase.
Nasal cavity (Spearman correlation: ρ = 0.7; p < 0.05) and groin (ρ = 0.6; p < 0.05) were important sites for imipenem-resistant A. baumannii as well as for imipenem-resistant P. aeruginosa (nasal cavity: ρ = 0.8; p < 0.05 and groin: ρ = 0.7; p < 0.05), whereas groin was an important site for K. pneumoniae ESBL (ρ = 0.7; p < 0.05) and perineum for VRE (ρ = 0.6; p < 0.05). ORSA was recovered mainly from the nasal cavity (p < 0.05) (Table 1).
The main characteristics related to colonization with MDROs were the use of medical devices and antimicrobials during the length of stay in the ICU. In a bivariate analysis, the use of drains and glycopeptides was associated with VRE colonization, fluoroquinolone use with imipenem-resistant K. pneumoniae or K. pneumoniae ESBL colonization, and presence of pressure ulcers and carbapenem use with imipenem-resistant A. baumannii colonization. For imipenem-resistant P. aeruginosa, some variables were highlighted in the final logistic regression model, such as use of mechanical ventilation and fluoroquinolones (Table 1).
Distribution of infection by MDRO
Among the 26 infection cases observed in this study, phenotypical identification of the pathogenic agent indicated bacterial species similar to those identified in previous colonization of anatomical sites showing the following decreasing distribution: imipenem-resistant A. baumannii (35.0%), K. pneumoniae ESBL (27.0%), VRE (19.0%), imipenem-resistant P. aeruginosa (11.0%), and imipenem-resistant K. pneumoniae (8.0%). There was no registry of infection by ORSA (Table 1).
Overall, there was a correlation between high population levels of colonization by MDRO at anatomical sites and a subsequent infection. This was particularly observed for infections with K. pneumoniae ESBL or imipenem-resistant K. pneumoniae (6.0 log10 CFU/mL for colonization followed by infection and 3.0 log10 CFU/mL in patients who remained only colonized), VRE (6.3 log10 CFU/mL and 5.2 log10 CFU/mL, respectively), and imipenem-resistant P. aeruginosa (5.0 log10 CFU/mL and 4.3 log10 CFU/mL, respectively) (Table 2). This correlation was not observed for imipenem-resistant A. baumannii. The main sites of infection were the respiratory tract (16 cases), bloodstream (10 cases), and urinary tract (5 cases).
Table 2.
Mean population levels (log CFU/mL) of colonized patients who evolved to infection with MDRO or not.
| MDRO | Colonization levels in patients who evolved to infection (26) | Colonization levels in patients who did not evolve to infection (27) |
|---|---|---|
| Imipenem-resistant Acinetobacter baumannii | 6.0 | 6.5 |
| Vancomycin-resistant Enterococcus | 6.3 | 5.2 |
| Klebsiellae pneumoniae | 6.0 | 3.0 |
| Imipenem-resistant Pseudomonas aeruginosa | 5.0 | 4.3 |
MDRO: multidrug-resistant organism; CFU: colony-forming units.
The evolution of population levels in each anatomical site was recorded by bacterial counts at four time points with bi-weekly sampling intervals. Figure 2 shows that population levels decreased until disappearance by day 45 for all the MDROs and all the anatomical sites. These decreased kinetics were quite similar between most of the MDROs, except for VRE which presented a slower elimination profile.
Figure 2.
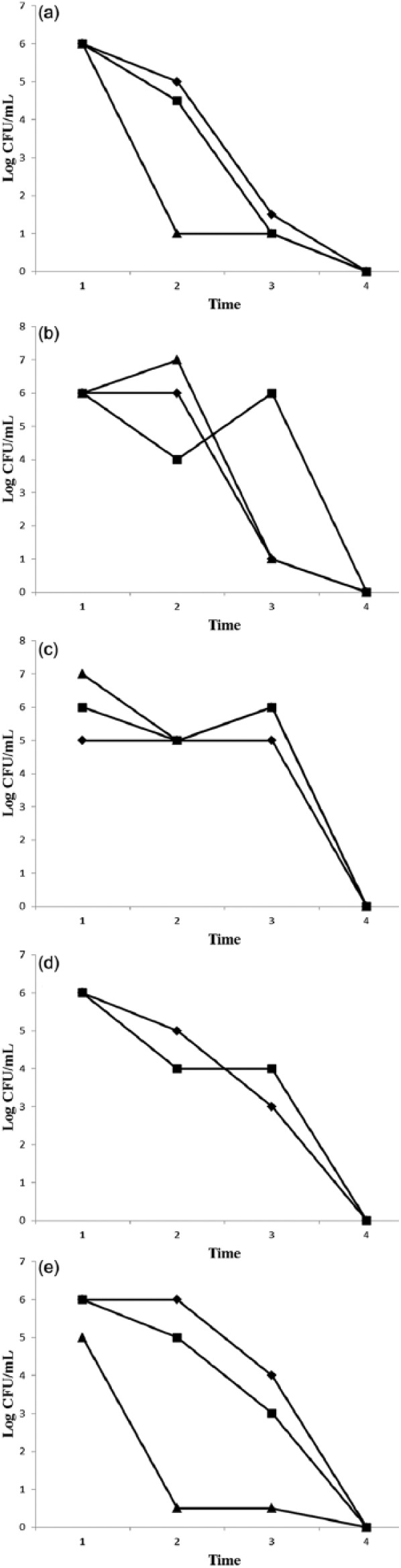
Evolution of population levels (log CFU/mL) in the nasal cavity (♦), perineum (◆), and groin (■) of MDRO from patient in ICU after 1 (1), 15 (2), 30 (3), and 45 (4) days. (a) Imipenem-resistant Acinetobacter baumannii, (b) imipenem-resistant or ESBL Klebsiellae pneumoniae, (c) vancomycin-resistant Enterococcus, (d) imipenem-resistant Pseudomonas aeruginosa, and (e) oxacillin-resistant Staphylococcus aureus.
Using the DiversiLab technique, different patterns of relatedness were revealed between the strains isolated from colonization cases, as well as between bacteria isolated from colonization and those from infection. In a comparison of the similarity among isolates of imipenem-resistant K. pneumoniae, three distinct patterns were noted (Figure 3). High similarity (>90%) was observed between the isolates Kp7 and Kp19. These bacteria were isolated from two distinct patients in the same ICU (ICU II). Another similarity (80%) was observed between isolate Kp4 (resistant to imipenem) isolated from a patient of ICU II and Kp30 (ESBL) from a patient of ICU I. Concerning imipenem-resistant A. baumannii from ICU II, only one pattern was observed, showing 95% of similarity between the isolates.
Figure 3.
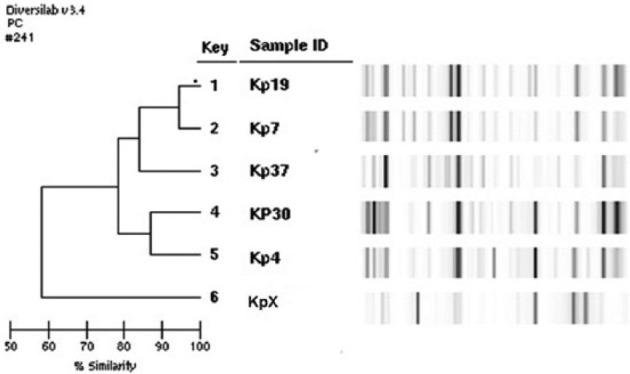
Patterns of similarity among isolates of Klebsiella pneumoniae associated with patient colonization in ICU.
Discussion
In temporal evaluation of microbiota in a healthy population, the nasal cavity and groin were generally among the most important sites in terms of composition and structure.9 In the microbial community of these anatomical sites, MDROs are frequently found. Among the patients who were evaluated in this study, the groin represented an important and unique site of colonization for K. pneumoniae ESBL and imipenem-resistant K. pneumoniae. In a cross-sectional study involving a long-term treatment facility in Chicago, correlation analysis between bacterial colonization and anatomical sites showed that rectum and groin were the most important sites where K. pneumoniae–producing carbapenemase has been detected.10 For ORSA, the main site of colonization was the nasal cavity, and this was also observed in this study. As expected, perineum was the main site in which VRE was identified. Enterococcus spp. are components of gut microbiota, and under the selective pressure of antimicrobials, resistant populations can be selected. Generally, microbiological evaluation for VRE detection is performed by rectal or peri-anal swabs.11–14
In equilibrated microbial ecosystems with a well-established and rich variety of bacterial species, the colonization resistance offered by this microbiota provides protection against pathogen invasion. However, environmental alterations due to antimicrobial use or presence of mechanical devices can stress the composition and function of the microbial ecosystem, allowing an initial colonization by potential pathogenic microorganisms which can be followed by infection.15 In this study, factors such as antimicrobial use (glycopeptide, carbapenem, and fluoroquinolone) and presence of mechanical devices (drain and mechanical ventilation) were associated with colonization for most of the MDROs. However, factors associated with colonization by ORSA were not identified here, probably due to the low number of colonization by this bacterium.
Few studies evaluate the role and correlation between population levels in colonization with MDRO and a subsequent infection, and most of them are focused on ORSA. In these analyses of colonization by ORSA, higher bacterial populations are associated with a dominant tissue tropism and can constitute risk factors for infection.16,17 In this study, infection episodes by most of the MDROs were associated with higher population levels previously detected in various anatomical sites 1–10 days before the infection when compared to population levels of patients who did not evolve to infection. However, K. pneumoniae ESBL was the only MDRO whose previous colonization levels were statistically associated with the occurrence of posterior infection. This observation highlights the importance of such type of determination to evaluate the risk of both MDRO transmission and evolution to infection. However, more analyses are required concerning the role of the population levels in colonization and infection.16,17
Variation in population levels of MDROs in the various anatomical sites sampled was verified in colonized patient, and this evolution was marked by a progressive decrease throughout the course of patient evaluation with an elimination by day 45 observed at the fourth time point. This apparently natural decolonization could be associated with the reestablishment of the composition and protective function of the local indigenous microbiota. However, the bacterial population decrease at anatomical sites may have also been influenced by antimicrobial use in cases of infection. As an example, the use of polimyxin E in the therapeutic regimen during infection could be correlated with the decrease in imipenem-resistant P. aeruginosa and K. pneumoniae ESBL. Anyway, microbial population determination and its variation below detectable levels contribute to an evaluation of the time required for the maintenance of precautionary measures, such as isolation. High microbial populations are probably associated with longer periods of colonization and an increased risk of infection.16,17 This point is still unclear in the literature, and there are no defined recommendations. In some studies, the possibility of decolonization is assumed when the bacterium is absent in three consecutive evaluations.9,11,14
In this study, three different patterns of similarity among the isolated strains were identified for K. pneumoniae ESBL and imipenem-resistant K. pneumoniae, ranging from 80% to 90%. The greatest similarity was verified between two strains isolated from the case of patient colonization in ICU II. However, a significant percent (80%) similarity was also registered between strains recovered from a case of patient colonization in ICU I and one in ICU II, which may be due to the possibility of patient readmission after discharge from the other hospital. The patterns identified for imipenem-resistant A. baumannii isolates showed high similarity, but they were restricted to ICU II and exhibited temporal variation.
In this study, the results suggest a correlation between infection occurrence and high bacterial population during the colonization period as well as a spontaneous decrease in colonization status. However, some limitations for these conclusions must be highlighted such as the possibility of low population levels has not been detected in microbiological culture medium, the relatively short period of follow-up, and the sample size of colonization/infection cases. Anyway, a prophylactic measure to reduce the microbial load related to MDROs in anatomical sites may be important in clinical practice regarding the possibility of decreasing bacterial spread and infection occurrence. This aspect in clinical practice could imply a possible decrease in mean time of permanence under strict isolation after identification of colonization with MDROs.
Acknowledgments
INSERM U914 Emerging Resistance to Antibiotics, Faculté de Médecine et Université Paris-Sud, Kremlin-Bicêtre, France, is acknowledged for the similarity determinations.
Footnotes
Declaration of conflicting interests: The authors declare no conflict of interest.
Funding: This study was supported by grant from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq)—No. 472823/2011-6.
References
- 1. Thompson-Chagoyán OC, Maldonado J, Gil A. Colonization and impact of disease and other factor son intestinal microbiota. Dig Dis Sci 2007; 52: 2069–2077. [DOI] [PubMed] [Google Scholar]
- 2. Rice LB. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis 2008; 197: 1079–1081. [DOI] [PubMed] [Google Scholar]
- 3. Vikram HR, Dumigan DG, Kohan C, et al. Discontinuation of contact precautions for patients no longer colonized with methicillin-resistant Staphylococcus aureus. Infect Control Hosp Epidemiol 2010; 31: 541–543. [DOI] [PubMed] [Google Scholar]
- 4. Sader HS, Jones RN, Gales AC, et al. SENTRY antimicrobial surveillance program report: Latin American and Brazilian results for 1997 through 2001. Braz J Infect Dis 2004; 8: 25–79. [DOI] [PubMed] [Google Scholar]
- 5. Schechner V, Kotlovsky T, Kazma M, et al. Asymptomatic rectal carriage of blaKPC producing carbapenem-resistant Enterobacteriaceae: who is prone to become clinically infected? Clin Microbiol Infect 2013; 19: 451–456. [DOI] [PubMed] [Google Scholar]
- 6. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial disk susceptibility tests; approved standard—tenth edition (CLSI document M02-A10). Wayne, PA: Clinical and Laboratory Standards Institute, 2011. [Google Scholar]
- 7. Mccusker ME, Périssé ARS, Roghmann MC. Severity-of-illness markers as predictors of nosocomial infection in adult intensive care unit. Am J Infect Control 2002; 30: 139–144. [DOI] [PubMed] [Google Scholar]
- 8. Mermel LA, Cartony JM, Covington P, et al. Methicillin-resistant Staphylococcus aureus (MRSA) colonization at different body sites: a prospective, quantitative analysis. J Clin Microbiol 2011; 49: 1119–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grice EA, Kong HH, Conlan S, et al. Topographical and temporal diversity of the human skin microbiome. Science 2009; 324: 1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thurlow CJ, Prabaker K, Lin MY, et al. Anatomic sites of patient colonization and environmental contamination with Klebsiella pneumoniae carbapenemase-producing Enter-obacteriaceae at long-term acute care hospitals. Infect Control Hosp Epidemiol 2013; 34: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pacio GA, Visintainer P, Maguire G, et al. Natural history of colonization with vancomycin-resistant enterococci, methicillin-resistant Staphylococcus aureus, and resistant Gram negative bacilli among long-term-care facility residents. Infect Control Hosp Epidemiol 2013; 24: 246–250. [DOI] [PubMed] [Google Scholar]
- 12. Babady NE, Gilhuley K, Cianciminio-Bordelon D, et al. Performance characteristics of the Cepheid Xpert vanA Assay for rapid identification of patients at high risk for carriage of vancomycin-resistant Enterococci. J Clin Microbiol 2012; 50: 3659–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim YJ, Kim S, Kim YR, et al. Risk factors for vancomycin-resistant enterococci infection and mortality in colonized patients on intensive care unit admission. Am J Infect Control 2012; 40: 1018–1019. [DOI] [PubMed] [Google Scholar]
- 14. Sohn KM, Peck KR, Joo EJ, et al. Duration of colonization and risk factors for prolonged carriage of vancomycin-resistant enterococci after discharge from the hospital. Int J Infect Dis 2013; 17: 240–246. [DOI] [PubMed] [Google Scholar]
- 15. Robinson CJ, Bohannan BJM, Young VB. From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev 2010; 74: 453–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stone ND, Lewis DR, Lowery HK, et al. Importance of bacterial burden among methicillin-resistant Staphylococcus aureus carriers in a long-term care facility. Infect Control Hosp Epidemiol 2008; 29: 143–148. [DOI] [PubMed] [Google Scholar]
- 17. Siegel JD, Rhinehart E, Jackson M, et al. Guideline for isolation precautions: preventing transmission of infectious agents in healthcare settings, http://www.cdc.gov/ncidod/dhqp/pdf/isolation2007.pdf (2007, accessed 15 November 2009). [DOI] [PMC free article] [PubMed]