Abstract
Objectives:
Previous studies found higher expression levels of DNA methyltransferase 1 in liver samples from smokers compared to those from non-smokers. In contrast, expression levels of DNA methyltransferase 3a and DNA methyltransferase 3b were similar in smokers and non-smokers. This study extends these studies to establish a causal linkage to cigarette smoke exposure by examining whether DNA methyltransferase expression is modulated by cigarette smoke condensate.
Methods:
These experiments were conducted in an in vitro system using HepG2 human liver cells. The dose range of cigarette smoke condensate was 0.1–120 µg/mL. The duration of exposure was up to 72 h.
Results:
In a 24-h exposure, DNA methyltransferase 1 expression was found to increase significantly in a dose-dependent manner (greater than threefold at 100 µg/mL cigarette smoke condensate). Expression levels of DNA methyltransferase 3a and DNA methyltransferase 3b were, however, not affected under these conditions. The effect of two cigarette constituents, nicotine and cotinine, on DNA methyltransferase 1 expression was also examined. Nicotine exposure significantly increased DNA methyltransferase 1 expression in a dose-dependent manner (greater than twofold at 50 µM). However, under these conditions, cotinine did not increase DNA methyltransferase 1 expression.
Conclusion:
These results clearly provide additional support of the modulating effect of cigarette smoke on DNA methyltransferase 1 expression. Given the potential of alterations in DNA methyltransferase expression to affect cellular function, this pathway may play a critical role in cigarette smoke-induced toxicity.
Keywords: Cigarette smoke condensate, DNA methyltransferase, human liver cells, nicotine
Introduction
Cigarette smoking is a major established environmental risk factor for numerous diseases, including cancer and diseases of the cardiovascular and respiratory systems.1 Several modes of action are involved in the toxicology of cigarette smoke, including epigenetic mechanisms.2 Epigenetic mechanisms resulting in changes in gene expression can lead to disruption in cellular function and cause a variety of diseases. Cytosine methylation is a major epigenetic modification of human DNA and has been shown to influence a number of cellular processes.3 Aberrant DNA methylation is causally implicated in human cancer,4,5 and there is increasing evidence of its involvement in diseases of the respiratory and cardiovascular systems, various autoimmune diseases, and multiple neuronal disorders.6,7 A global loss of DNA methylation (hypomethylation) and a gene-specific gain of DNA methylation (hypermethylation) are two distinct hallmarks of carcinogenesis.8,9 Numerous studies associate cigarette smoking with gene promoter hypermethylation in tobacco-related cancers.10–12 In two recent epigenome-wide association studies comparing smokers and never smokers, a number of the CpG sites identified as differentially methylated were, however, found to be hypomethylated in smokers.13,14 Several studies have also examined the association between cigarette smoke exposure and global DNA methylation.15,16
DNA methylation is the result of the activity of a family of DNA methyltransferase (DNMT) enzymes, including DNMT1, DNMT3a, and DNMT3b, that catalyze the transfer of a methyl group from the ubiquitous methyl donor S-adenosyl methionine to the 5-position of cytosines residing in the dinucleotide sequence cytosine-guanine.17 DNMTs 1, 3a, and 3b have been implicated to different extents in initiating gene silencing through de novo methylation and recruitment of chromatin remodeling proteins.18–20 DNMT1 has both maintenance and de novo methyltransferase activity, associates with chromatin, and is responsible for ~90% of methyltransferase activity in mammalian cells. DNMT3a and DNMT3b are ubiquitously expressed and can be detected in most adult tissues. DNMT3a and 3b are thought to function as de novo DNMTs. These enzymes were shown to have equal preferences in vitro for unmethylated and hemimethylated DNA. Studies have shown that DNMTs function in cooperation with each other to facilitate DNA methylation in both human and mouse systems.21
Elevated levels of DNMTs have been found in many cancer cells in vitro and in several types of human tumors in vivo, including hepatocellular carcinoma.20,22–25 In addition, increased DNMT levels are required to maintain the phenotype of fibroblasts transformed with ras and with the fos oncogene.26 Induced DNMT1 over-expression in cultured cell lines gradually induces CpG hypermethylation and cell transformation.27 Altered DNMT expression is related to changes in genome-wide DNA methylation patterns that can have potent effects on expression of the large number of genes that are controlled by promoter methylation.
Previous studies in our laboratory examined the potential role of cigarette smoking in influencing DNMT expression in human tissue by comparing expression levels in liver samples from smokers and non-smokers.28,29 Cigarette smoking is causally associated with liver cancer.30 Higher expression levels of DNMT1 were found in smokers compared to non-smokers. In contrast, expression levels of DNMT3a and DNMT3b were similar in smokers and non-smokers. This study extends these studies to establish a causal linkage to cigarette smoke exposure by examining whether DNMT expression is modulated by cigarette smoke condensate (CSC) in an in vitro system using HepG2 human liver cells. The effects of nicotine and cotinine, as constituents of cigarette smoke, were also investigated.
Material and methods
Cell lines and treatment conditions
The human liver cell line, HepG2, was obtained from the American Type Culture Collection (Manassas, VA). HepG2 cells were grown in William’s E medium, supplemented with 5% fetal bovine serum (FBS; Atlanta Biologicals, Norcross, GA), 15,000 U penicillin, 15,000 U streptomycin, 2 mM l-glutamine (Life Technologies, Carlsbad, CA). Cells were routinely maintained at 37°C in a humidified 5% CO2 atmosphere. CSC was purchased from Murty Pharmaceuticals (Lexington, KY) and was prepared using a smoking machine designed for Federal Trade Commission testing. The particulate matter from Kentucky standard cigarettes (1R3F; University of Kentucky, Lexington, KY) was collected on Cambridge glass fiber filters and the amount of CSC obtained was determined by weight increase on the filter. CSC was prepared by dissolving the collected smoke particulates in dimethyl sulfoxide (DMSO) to yield a 4% solution (w/v). The CSC was diluted into DMSO and aliquots were stored at −80°C. For smoke condensate exposure experiments, cells (400,000 cells per plate) were cultured in 100 mm dishes in appropriate media with or without CSC (0.1, 1.0, 10, 100, 120 µg/mL; in DMSO; final vol., 0.1%) for 24, 48, or 72 h. Nicotine and cotinine were purchased from Sigma-Aldrich (St. Louis, MO). Cells were treated with or without nicotine or cotinine (0.1, 1.0, 10.0, 20.0, 50.0 µM) for 24, 48, or 72 h. Cell proliferation was assessed by MTT techniques using the Cell Titer 96 Non-Radioactive Cell Proliferation Assay (Promega, Madison, WI).
RNA isolation and quantitative real-time polymerase chain reaction analysis
Qiagen RNeasy isolation kit (Valencia, CA) was used to isolate total RNA. A Clontech cDNA synthesis kit (Mountain View, CA) was used for cDNA synthesis. Quantitative real-time polymerase chain reaction (QRT-PCR) was performed to determine expression levels of the DNMTs. The primers for real-time polymerase chain reaction (PCR) (synthesized by Sigma-Genosys; Woodlands, TX) were DNMT1 (forward: GCACAAACTGACCTGCTTCA and reverse: GCCTTTTCACCTCCATCAAA), DNMT3a (forward: 5′CGTCTCCGAACCACATGAC and reverse: 5′-CGTCTCCGAAC CACATGAC), DNMT3b (forward: 5′-CCAGCTGAAGCCCATGTT and reverse: 5′-ATTTGTCTTGAACGCTTG), GAPDH (forward: 5′GAAGGTGA AGGTCGGAGTC and reverse: 5′-GAAGATGGTGATGGG ATTTC) . QRT-PCR reactions were performed in a volume of 50 µL containing 50 ng cDNA, 400 nM of each primers, deionized water, and 25 µL SYBR Green Master Mix (Stratagene, Cedar Creek, TX) using the following conditions: 50°C for 2 min and denaturing at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Optical data were collected during the 60°C step. PCRs were carried out in 96-well thin-wall PCR plates covered with optically clear sealing film (Applied Biosystems, Forest City, CA). Amplification, detection, and data analysis were performed using the ABI PRISM 7000 Sequence Detector system (Applied Biosystems). Results were expressed using the comparative threshold method after validation, following the recommendations of the manufacturer (Applied Biosystems). The threshold cycle number (CT) value for DNMT was normalized against GAPDH and calculated as ΔCT = CTDNMT − CTGAPDH. Thereafter, the relative mRNA levels of these genes after treatment were calculated using the ΔΔCT method: ΔCT (treatment) − ΔCT (vehicle) = ΔΔCT (treatment). The fold changes of mRNA levels were expressed as 2−ΔΔCT. All PCR reactions were performed in triplicate in three independent experiments.
Statistical analysis
Prism IV software (GraphPAD Software, Inc., La Jolla, CA) was used for graphical analyses. Data were analyzed for statistical significance using Student’s t test. Differences with p values of <0.05 were considered statistically significant.
Results
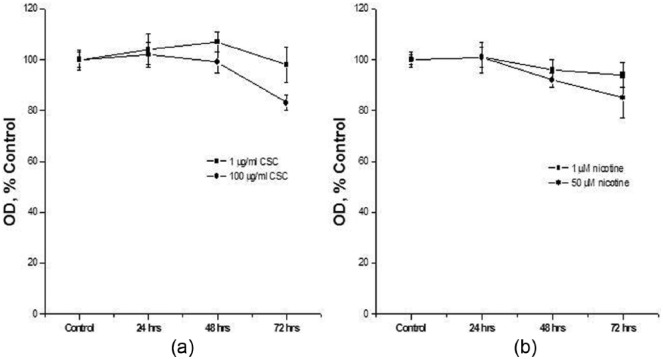
The effect of CSC and nicotine on cell growth was assessed. In these experiments, the dose of CSC was 1 µg/mL or 100 µg/mL. The duration of exposure was up to 72 h. Under these conditions, there was a modest inhibition of cell growth that did not exceed 20% (Figure 1(a)). Similar results were observed with nicotine (1 and 50 µM; Figure 1(b)).
Figure 1.
Effect of CSC and nicotine on cell proliferation. HepG2 cells were treated with (a) 1 or 100 µg/mL CSC for 24, 48, or 72 h or (b) 1 or 50 µM nicotine for 24, 48, or 72 h and assayed by MTT techniques. Data are presented as mean ± SD of at least three determinations.
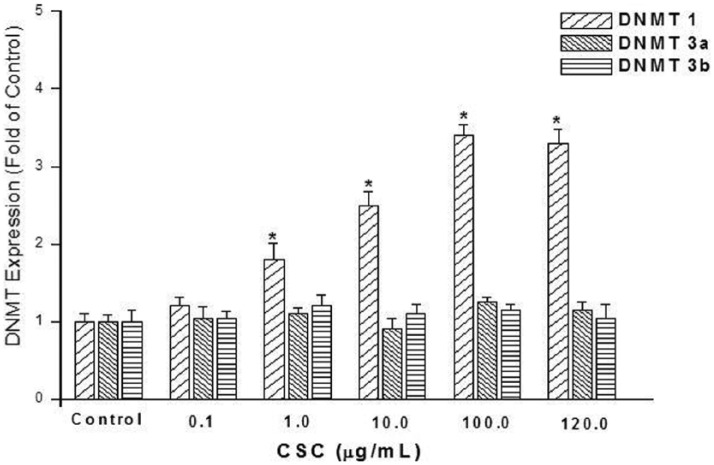
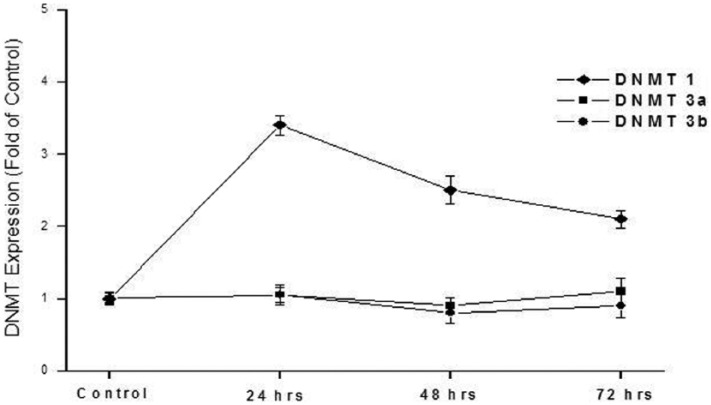
The effect of CSC on expression of DNMT1, DNMT3a, and DNMT3b was assessed in HepG2 cells. In these experiments, the dose range of CSC was 0.1–120 µg/mL. The duration of exposure was up to 72 h. In a 24-h exposure, DNMT1 expression was found to significantly increase (up to 3.4-fold) in a dose-dependent manner (1–100 µg/mL), as shown in Figure 2. Induction of DNMT1 expression was greatest at 24-h exposure compared to exposure durations of 48 or 72 h (Figure 3). Cells in these experiments were exposed to 100 µg/mL CSC. Under the conditions included in this study, expression levels of DNMT3a and DNMT3b were, however, not affected (Figures 2 and 3).
Figure 2.
Effect of CSC dose on expression levels of DNMTs 1, 3a, and 3b. HepG2 cells were exposed to 0.1, 1.0, 10, 100, or 120 µg/mL CSC for 24 h. Expression levels were determined by quantitative real-time RT-PCR. Data are presented as mean ± SD of at least three determinations. The symbol “*” Denotes a significant difference (p < 0.05) compared to controls.
Figure 3.
Effect of CSC exposure duration on expression levels of DNMTs 1, 3a, and 3b. HepG2 cells were exposed to 100 µg/mL CSC for 24, 48, or 72 h. Expression levels were determined by quantitative real-time RT-PCR. Data are presented as mean ± SD of at least three determinations.
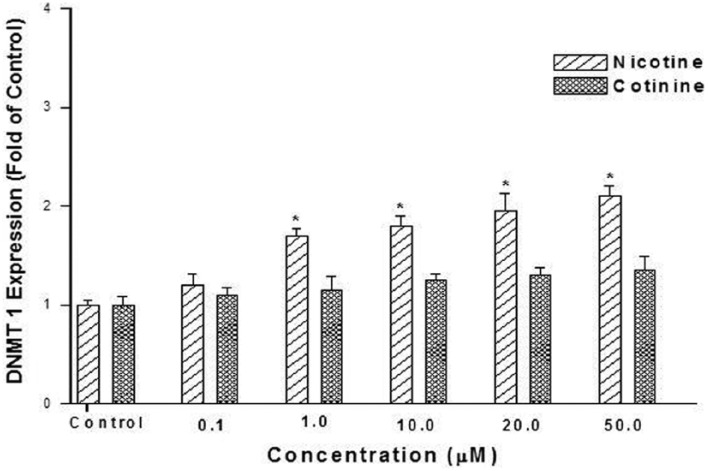
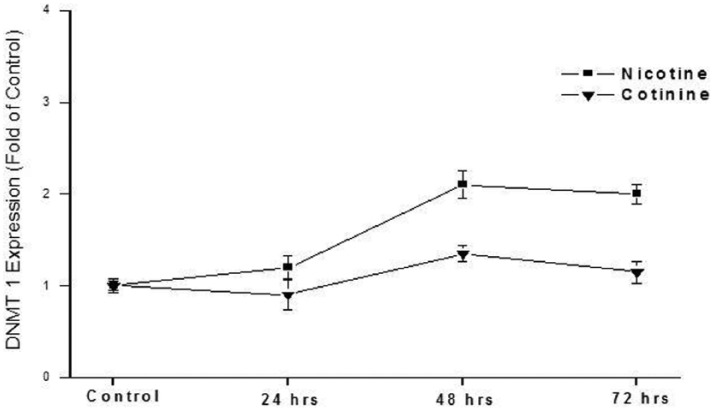
The effect of two cigarette constituents, nicotine and cotinine, on DNMT1 expression was also examined in this in vitro system. Cotinine is also a metabolite of nicotine. Nicotine exposure significantly increased DNMT1 expression (up to 2.1-fold) in a dose-dependent manner (1–50 µM; Figure 4). Cotinine was, however, not shown to increase DNMT1 expression under the conditions employed in this study. Cells were exposed for 48 h because the highest increase in DNMT1 expression was shown at this exposure duration at a dose level of 50 µM (Figure 5).
Figure 4.
Effect of nicotine and cotinine dose on expression levels of DNMT1. HepG2 cells were exposed to 0.1, 1.0, 10.0, 20.0, and 50.0 µM nicotine or cotinine for 48 h. Expression levels were determined by quantitative real-time RT-PCR. Data are presented as mean ± SD of at least three determinations. The symbol “*” denotes a significant difference (p < 0.05) compared to controls.
Figure 5.
Effect of nicotine and cotinine exposure duration on expression of DNMT1. HepG2 cells were exposed to 50.0 µM nicotine or cotinine for 24, 48, or 72 h. Expression levels were determined by quantitative real-time RT-PCR. Data are presented as mean ± SD of at least three determinations.
Discussion
In this study, an in vitro model system was employed to provide direct evidence that cigarette smoke induces the expression of DNMT1 in human liver cells. No effect was demonstrated with DNMT3a or DNMT3b. These results are consistent with our previous observations in human liver tissue samples in which higher levels of DNMT1 were found in smokers while levels of DNMT3a and DNMT3b were similar in smokers and non-smokers.28,29 Reports of other in vitro studies of the effects of CSC on DNMT expression are limited, and the results have been varied. Similar to the findings in the current study, CSC exposure in T-24 bladder cancer cells induced a significant time-dependent increase in the level of DNMT1, but had no effect on DNMT3a and DNMT3b.31 The time dependence did, however, differ with the current study given the decrease in DNMT1 expression after 24 h that was shown in this study. In another study, increased DNMT1 expression was observed in B2B cells treated with 1% CSC for 12 weeks.32 However, in an examination of expression levels of DNMTs 1, 3a, and 3b in A549 lung cancer cells, a time-dependent (12–72 h) decrease in the levels of DNMT3b by CSC exposure was detected.33 There was no effect on DNMT3a expression. In human bronchial epithelial cells (HBEC) exposed to CSC, after 5 months, decreased DNMT1 and increased DNMT3B expression were observed.34 Exposures of shorter durations were found not to affect the expression of either DNMT. Reasons for these variations in results are unknown but may reflect different cell types, doses, or durations of exposure.
Only limited data appear to be available on the effects of cigarette smoke constituents on DNMT expression. Our examination of the effect of nicotine and cotinine on DNMT1 expression explored the contribution of these constituents. At least one other report describes the effect of nicotine on DNMT1. Results were, however, in contrast to those found in this study. Down-regulation of DNMT1 expression was demonstrated in the frontal cortex of mice injected with nicotine.35 The effect of other cigarette smoke constituents has also been investigated. Exposure of HBEC lung cells to methylnitrosourea and benzo(a)pyrene-diolepoxide increased significantly levels of DNMT1.36 Additional research will be needed to develop this area of study.
The underlying mechanism(s) for the observed influence of CSC on DNMT expression remain to be completely established. Earlier studies suggest that one of the mechanisms involved in increased DNMT1 in smokers may be the ras pathway since smoking has been found to be associated with increased ras expression.37–39 The JUN/FOS transcription factors have been shown to activate murine DNMT1 transcription through the AP-1 target motifs.40 Furthermore, oncogenic RAS-stimulated signal transduction pathways increase murine DNMT1 expression40 and expression in human T cells41 and alter DNA methylation patterns.42 More recent studies have identified several other signaling pathways involved in the regulation of DNMT expression that may have a role in CSC-induced expression of DNMT1. These include transforming growth factor-β (TGF-β),43 glioma-associated oncogene family zinc finger 1 (GLI1),44 a transcriptional factor in Hedgehog signaling pathway, and Sp1/NF-κB.45 Levels of each of these transcriptional factors have been shown to respond to cigarette smoke.46–48 DNMT1 can also be regulated by microRNAs, as demonstrated with miR-148a and miR-152.49,50 Although shown for other microRNAs, whether cigarette smoke affects these microRNAs has, however, not been reported.
A limitation of this study is noted. CSC is widely used in model systems to study in vitro effects of tobacco smoke.31–34,51–55 A disadvantage is that cell culture models often do not exhibit all the differentiated and functional characteristics of the corresponding native epithelium or the entire organ. A concern can also be the inherent instability, especially on long-term culture. Treatment with CSC may not exactly replicate in vivo responses to smoke exposures. However, in in vitro research, cellular and subcellular functions can be studied with more ease in a simplified, direct biological model system using guidelines for good cell culture practice, which allows the prediction of mechanisms that may be relevant to in vivo situations. HepG2 are widely used and are useful as a model system in vitro for human hepatic cells. These cells are highly differentiated and display many of the genotypic features of normal liver cells.56 This cell line is, however, known to have low metabolic capacities.57 Despite the potential limitation, our experiments yielded several interesting and potentially relevant findings.
In summary, a full understanding of the harmful effects of cigarette smoke requires consideration of its epigenetic action. Results from this study clearly provide additional support of the modulating effect of cigarette smoke on DNMT1 expression. Although the underlying mechanisms are not yet known, given the potential of alterations in DNMT expression to affect cellular function, this pathway may play a critical role in cigarette smoke-induced diseases.
Footnotes
Declaration of conflicting interests: The views presented in this article do not necessarily reflect those of the Food and Drug Administration.
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
References
- 1. Office on Smoking and Health. The health consequences of smoking: a report of the surgeon general. Atlanta, GA: Centers for Disease Control and Prevention, 2004. [PubMed] [Google Scholar]
- 2. Hammons G, Lyn-Cook B. Epigenetics in tobacco smoke toxicology. Curr Top Toxicol 2011; 7: 63–77. [Google Scholar]
- 3. Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 2002; 16: 6–21. [DOI] [PubMed] [Google Scholar]
- 4. Esteller M. Epigenetics in cancer. New Engl J Med 2008; 358: 1148–1159. [DOI] [PubMed] [Google Scholar]
- 5. Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 2008; 9: 465–476. [DOI] [PubMed] [Google Scholar]
- 6. Baccarelli A, Rienstra M, Benjamin EJ. Cardiovascular epigenetics: basic concepts and results from animal and human studies. Circ Cardiovasc Genet 2010; 3: 567–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schwarz DA. Epigenetics and environmental lung disease. Proc Am Thorac Soc 2010; 7: 123–125. [DOI] [PubMed] [Google Scholar]
- 8. Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer 2003; 3: 253–266. [DOI] [PubMed] [Google Scholar]
- 9. Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007; 128: 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim DH, Nelson HH, Wiencke JK, et al. p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res 2001; 61: 3419–3424. [PubMed] [Google Scholar]
- 11. Russo AL, Thiagalingam A, Pan H, et al. Differential DNA hypermethylation of critical genes mediates the stage-specific tobacco smoke-induced neoplastic progression of lung cancer. Clin Cancer Res 2005; 11: 2466–2470. [DOI] [PubMed] [Google Scholar]
- 12. Vaissiere T, Hung RJ, Zaridze D, et al. Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res 2009; 69: 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zeilinger S, Kuhnel B, Klopp N, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS ONE 2013; 8(5): e63812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Besingi W, Johansson A. Smoke-related DNA methylation changes in the etiology of human disease. Hum Mol Genet 2014; 23: 2290–2297. [DOI] [PubMed] [Google Scholar]
- 15. Smith IM, Mydlarz WK, Mithani SK, et al. DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. Int J Cancer 2007; 121: 1724–1728. [DOI] [PubMed] [Google Scholar]
- 16. Guerrero-Preston R, Goldman LR, Brebi-Mieville P, et al. Global DNA hypomethylation is associated with in utero exposure to cotinine and perfluorinated alkyl compounds. Epigenetics 2010; 5: 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet 2000; 9: 2395–2402. [DOI] [PubMed] [Google Scholar]
- 18. Okano M, Bell DW, Haber DA, et al. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999; 99: 247–257. [DOI] [PubMed] [Google Scholar]
- 19. Pradhan S, Bacolla A, Wells RD, et al. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J Biol Chem 1999; 274: 33002–33010. [DOI] [PubMed] [Google Scholar]
- 20. Robertson KD, Uzvolgyi E, Liang G, et al. The human DNA methyltransferases (DNMTs) 1, 3a, and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res 1999; 27: 2292–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rhee I, Bachman KE, Park BH, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 2002; 416: 552–556. [DOI] [PubMed] [Google Scholar]
- 22. Belinsky SA, Nikula KJ, Baylin SB, et al. Increased cytosine DNA-methyltransferase activity is target-cell-specific and an early event in lung cancer. Proc Natl Acad Sci USA 1996; 93: 4045–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nagai M, Nakamura A, Makino R, et al. Expression of DNA (5-cytosin)-methyltransferases (DNMTs) in hepatocellular carcinomas. Hepatol Res 2003; 26: 186–191. [DOI] [PubMed] [Google Scholar]
- 24. Oh BK, Kim H, Park HJ, et al. DNA methyltransferase expression and DNA methylation in human hepatocellular carcinoma and their clinicopathological correlation. Int J Mol Med 2007; 20: 65–73. [PubMed] [Google Scholar]
- 25. Zhang JJ, Zhu Y, Wu JL, et al. Association of increased DNA methyltransferase expression with carcinogenesis and poor prognosis in pancreatic ductal adenocarcinoma. Clin Transl Oncol 2012; 14: 116–124. [DOI] [PubMed] [Google Scholar]
- 26. Bakin AV, Curran T. Role of DNA 5-methylcytosine transferase in cell transformation by fos. Science 1999; 283: 387–390. [DOI] [PubMed] [Google Scholar]
- 27. Vertino PM, Yen RW, Gao J, et al. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-)-methyltransferase. Mol Cell Biol 1996; 16: 4555–4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xiao Y, Word B, Starlard-Davenport A, et al. Age and gender affect DNMT3a and DNMT3b expression in human liver. Cell Biol Toxicol 2008; 24: 265–272. [DOI] [PubMed] [Google Scholar]
- 29. Hammons GJ, Yan Y, Lopatina NG, et al. Increased expression of hepatic DNA methyltransferase in smokers. Cell Biol Toxicol 1999; 15: 389–394. [DOI] [PubMed] [Google Scholar]
- 30. Sasco AJ, Secretan MB, Straif K. Tobacco smoking and cancer: a brief review of recent epidemiological evidence. Lung Cancer 2004; 45(Suppl. 2): S3–S9. [DOI] [PubMed] [Google Scholar]
- 31. Yang W, Cui S, Ma J, et al. Cigarette smoking extract causes hypermethylation and inactivation of WWOX gene in T-24 human bladder cancer cells. Neoplasma 2012; 59: 216–223. [DOI] [PubMed] [Google Scholar]
- 32. Tennis MA, VanScoyk MM, Wilson LA, et al. Methylation of Wnt7a is modulated by DNMT1 and cigarette smoke condensate in non-small cell lung cancer. PLoS ONE 2012; 7(3): e32921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu H, Zhou Y, Boggs SE, et al. Cigarette smoke induces demethylation of prometastatic oncogene synuclein-γ in lung cancer cells by downregulation of DNMT3B. Oncogene 2007; 26: 5900–5910. [DOI] [PubMed] [Google Scholar]
- 34. Liu F, Killian JK, Yang M, et al. Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene 2010; 29: 3650–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Satta R, Maloku E, Zhubi A, et al. Nicotine decreases DNA methyltransferase 1 expression and glutamic acid decarboxylase 67 promoter methylation in GABAergic interneurons. Proc Natl Acad Sci USA 2008; 105: 16356–16361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Damiani LA, Yingling CM, Leng S, et al. Carcinogen-induced gene promoter hypermethylation is mediated by DNMT1 and causal for transformation of immortalized bronchial epithelial cells. Cancer Res 2008; 68: 9005–9014. [DOI] [PubMed] [Google Scholar]
- 37. Chowdhury P, Mantague DC, Rayford PL, et al. Nicotine alters pancreatic gene (H-ras) expression and induces point mutation and exocrine pancreatic injury. In: XVI international cancer congress (ed Rao RS, Deo MG, Sanbhvi LD, et al.), New Delhi, India, 1994, pp. 369–373. [Google Scholar]
- 38. Kuo MY, Chang HH, Hahn LJ, et al. Elevated ras p21 expression in oral premalignant lesions and squamous cell carcinoma in Taiwan. J Oral Pathol Med 1995; 24: 255–260. [DOI] [PubMed] [Google Scholar]
- 39. Anderson D, Hughes JA, Cebulska-Wasilewska A, et al. Biological monitoring of workers exposed to emissions from petroleum plants. Environ Health Perspect 1996; 104(Suppl. 3): 609–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rouleau J, MacLeod AR, Szyf M. Regulation of the DNA methyltransferase by the Ras-AP-1 signaling pathway. J Biol Chem 1995; 270: 595–601. [DOI] [PubMed] [Google Scholar]
- 41. Yang J, Deng C, Hemati N, et al. Effect of mitogenic stimulation and DNA methylation on human T cell DNA methyltransferase expression and activity. J Immunol 1997; 159: 1301–1309. [PubMed] [Google Scholar]
- 42. MacLeod AR, Rouleau J, Szyf M. Regulation of DNA methylation by the Ras signaling pathway. J Biol Chem 1995; 270: 11327–11337. [DOI] [PubMed] [Google Scholar]
- 43. Zhang Q, Chen L, Helfand BT, et al. TGF-β regulates DNA methyltransferase expression in prostate cancer, correlates with aggressive capabilities, and predicts disease recurrence. PLoS ONE 2011; 6: e25168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. He S, Wang F, Yang L, et al. Expression of DNMT1 and DNMT3a are regulated by GLI1 in human pancreatic cancer. PLoS ONE 2011; 6(11): e27684. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45. Liu S, Liu Z, Xie Z, et al. Bortezomib induces DNA hypomethylation and silenced gene transcription by interfering with Sp1/NF-κB-dependent DNA methyltransferase activity in acute myeloid leukemia. Blood 2008; 111: 2364–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang RD, Wright JL, Churg A. Transforming growth factor-β1 drives airway remodeling in cigarette smoke-exposed tracheal explants. Am J Resp Cell Mol 2005; 33: 387–393. [DOI] [PubMed] [Google Scholar]
- 47. Lemjabbar-Alaoui H, Dasari V, Sidhu SS, et al. Wnt and Hedgehog are critical mediators of cigarette smoke-induced lung cancer. PLoS ONE 2006; 1(1): e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hasnis E, Bar-Shai M, Burbea Z, et al. Mechanisms underlying cigarette smoke-induced NF-κB activation in human lymphocytes: the role of reactive nitrogen species. J Physiol Pharmacol 2007; 58(Suppl. 5): 275–287. [PubMed] [Google Scholar]
- 49. Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology 2010; 51: 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pan W, Zhu S, Yuan M, et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol 2010; 184: 6773–6781. [DOI] [PubMed] [Google Scholar]
- 51. Hellermann GR, Nagy SB, Kong X, et al. Mechanism of cigarette smoke condensate-induced acute inflammatory response in human bronchial epithelial cells. Respir Res 2002; 3: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nagathihalli NS, Massion PP, Gonzalez AL, et al. Smoking induces epithelial-to-mesenchymal transition in non-small cell lung cancer through HDAC-mediated downregulation of E-cadherin. Mol Cancer Ther 2012; 11: 2362–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nagaraj NS, Beckers S, Mensah JK, et al. Cigarette smoke condensate induces cytochromes P450 and aldo-keto reductases in oral cancer cells. Toxicol Lett 2006; 165: 182–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xu H, Ferro TJ, Chu S. Cigarette smoke condensate inhibits ENaC α-subunit expression in lung epithelial cells. Eur Respir J 2007; 30: 633–642. [DOI] [PubMed] [Google Scholar]
- 55. Shizu M, Itoh Y, Sunahara R, et al. Cigarette smoke condensate upregulates the gene and protein expression of proinflammatory cytokines in human fibroblast-like synoviocyte line. J Interferon Cytokine Res 2008; 28: 509–522. [DOI] [PubMed] [Google Scholar]
- 56. Sassa S, Sugiata O, Galbraith RA, et al. Drug metabolism by the human hepatoma cell, Hep G2. Biochem Biophys Res Commun 1987; 143: 52–57. [DOI] [PubMed] [Google Scholar]
- 57. Xu JJ, Diaz D, O’Brien PJ. Applications of cytotoxicity assays and pre-lethal mechanistic assays for assessment of human hepatotoxicity potential. Chem Biol Interact 2004; 150: 115–128. [DOI] [PubMed] [Google Scholar]