Objective
This study was conducted to evaluate the role of methylation of adenylate cyclase activating peptide 1 (ADCYAP1), paired box gene 1 (PAX1), cell adhesion molecule 1 (CADM1), and T-lymphocyte maturation–associated protein (MAL) during carcinogenesis.
Methods
We evaluated the methylation of 4 genes by using the cervical carcinoma cell lines (CaSki, SiHa, HeLa, and C33A) and cervical neoplastic cells from 56 subjects with human papillomavirus 16 (HPV16)–infected low-grade squamous intraepithelial lesions (LSILs), 50 subjects with HPV16-infected high-grade squamous intraepithelial lesions (HSILs), and 24 subjects with HPV16-infected invasive cervical cancer who attended Seoul St. Mary’s Hospital. Methylation of the 4 genes was evaluated using quantitative bisulfate pyrosequencing.
Results
The ADCYAP1 promoter was hypermethylated in the 4 cell lines (CaSki, 97.40 ± 1.39; SiHa, 82.04 ± 17.02; HeLa, 96.14 ± 2.08; and C33A, 78 ± 10.18). PAX1 and CADM1 were hypermethylated in the HPV16/18-infected cell lines CaSki (PAX1, 91.18 ± 9.91; CADM1, 93.5 ± 7.33), SiHa (PAX1, 96.14 ± 2.08; CADM1, 93.15 ± 8.81), and HeLa (PAX1, 82.04 ± 17.02; CADM1, 92.43 ± 9.95). MAL was hypermethylated in the CaSki cell line (96.04 ± 4.74). Among human cervical neoplastic cells, the methylation indices of ADCYAP1 were 7.8 (95% confidence interval [95% CI], 7.0–8.6) in subjects with LSILs and 39.8 (95% CI, 29.0–54.7) in those with cervical cancer (P < 0.001); for PAX1, 7.2 (95% CI, 6.1–8.5) and 37.8 (95% CI, 27.1–52.7), respectively; for CADM1, 3.5 (95% CI, 3.0–4.0) and 17.7 (95% CI, 10.8–29.1), respectively; for MAL, 2.7 (95% CI, 2.5–3.0) and 13.0 (95% CI, 7.6–22.0), respectively (P < 0.001 for each). Immunohistochemical staining results were positive in the cytoplasm of subjects with low methylation of the 4 gene promoters; however, they were negative in the cytoplasm of those with hypermethylation of the 4 gene promoters.
Conclusions
The results of this study suggest that the methylation of ADCYAP1, PAX1, CADM1, and MAL may be highly associated with the development of cervical cancer, and that gene expression can be suppressed by gene promoter hypermethylation.
Key Words: DNA methylation, Cervical cancer, Human papillomavirus
Cervical cancer is the third most common cancer and the leading cause of cancer-related mortality in women worldwide.1 Development of cervical cancer is causally related to infection with high-risk human papillomavirus (types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68).2 Although high-risk HPVs can be detected in many women, progression of a high-risk HPV-positive premalignant lesion to invasive cancer is rare. Consistent with the multistep nature of human carcinogenesis, additive host cell alterations drive progression to invasive cancer.3 These events involve chromosomal alterations and epigenetic changes. The chromosomal alterations affect the structure and expression of oncogenes or tumor suppressor genes.4 The main mechanism by which HPV16 is involved in cervical carcinogenesis is the expression of the 2 early viral genes E6 and E7. E6 protein binds to the tumor suppressor gene p53 and promotes its degradation, and E7 inactivates the pRb gene. These events cause disruption of cell cycle regulation and immortalization.5
DNA methylation is a typical mechanism of epigenetic alterations, and it has been shown to suppress gene expression that controls tumor suppression and invasion-controlling genes.6–8 Recent studies have reported that hypermethylation of tumor suppressor genes induces the development of cancer.6,7 Cell adhesion molecule 1 (CADM1) is associated with the development of cervical cancer. CADM1 is hypermethylated in cervical cancer cell lines infected with HPV16 and HPV18.9 CADM1 promoter hypermethylation has also been detected in 40% of all lung cancer cases, 32% of all prostate cancer cases, 27% of all pancreatic adenocarcinoma cases, and 83% of all cervical squamous cell carcinoma cases.9,10 Paired box gene 1 (PAX1) is hypermethylated in the high-grade cervical lesion (cervical intraepithelial neoplasia, CIN 2-3) and cervical cancer but not in the low-grade cervical lesion or normal epithelium.11–13 T-lymphocyte maturation–associated protein (MAL) represses tumor suppressor activity by hypermethylation. Ninety percent of all patients with cervical squamous cell carcinomas and 93% of all patients with cervical adenocarcinomas are hypermethylated.6 The tumor suppressor activity of MAL has also been demonstrated in esophageal and breast cancers.14–16 Adenylate cyclase–activating polypeptide 1 (ADCYAP1) is hypermethylated in cervical cancer cell lines infected with HPVs, and the methylation level is increased from CIN 1 to CIN 3 and cervical cancer.17
Studies to date have been conducted to determine if various genes are hypermethylated in relation to the development of cancer and to analyze the methylation patterns of genes that are known to correlate with the development of cancer in patients with cervical cancer. Therefore, the aims of this study were to evaluate the methylation patterns of the aforementioned 4 genes according to cervical pathology in subjects infected with HPV16, to analyze the patterns of gene expression using immunohistochemical staining, and to elucidate the role of methylation of the aforementioned 4 genes in the development of cervical cancer.
MATERIALS AND METHODS
Cervical Carcinoma Cell Lines and Human Tissue Samples
Four cervical cancer cell lines were used in this study. CaSki (HPV16-positive), SiHa (HPV16-positive), HeLa (HPV18-positive), and C33A (HPV-negative, p53 mutation) cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, Va). Each cell line was grown as follows: SiHa, HeLa, and C33A cells in Dulbecco modified Eagle medium (WelGENE, Korea) and CaSki cells in RPMI-1640 medium (Gibco-BRL, San Francisco, Calif). All media were supplemented with 10% fetal bovine serum (Gibco-BRL) and 1% Antibiotic-Antimycotic (Gibco-BRL). All these cells were cultured at 37°C in a humidified atmosphere of 95% air and 5% CO2. A total of 130 human cervical cells were identified from Seoul St. Mary’s Hospital, The Catholic University of Korea after ethical approval. Each patient gave signed informed consent, and the study protocol was approved by our institutional review board (KC12SISI0130). We evaluated the methylation patterns of the 4 genes in cervical neoplastic cells obtained from 56 subjects with low-grade squamous intraepithelial lesions (LSILs), 50 subjects with high-grade squamous intraepithelial lesions (HSILs), and 24 subjects with cervical cancer.
HPV Genotyping Test
For HPV genotyping, we used the HPV DNA Chip (Mygene, Seoul, Korea), a polymerase chain reaction (PCR) base microarray system. The genotyping experiments, including the preparation and testing of specimens, were performed according to the manufacturer’s instructions. The HPV DNA chip contains 22 type-specific probes that consist of 15 high-risk groups (types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68, and 69) and 7 low-risk groups (types 6, 11, 34, 40, 42, 43, and 70). The PCR product was hybridized onto the chip at 40°C for 2 hours and washed with 3× SSPE and with 1× SSPE for 2 minutes each. Hybridized signals were visualized with a DNA Chip Scanner (GSI Lumpnics Scanarray Lite, Ottawa, Ontario, Canada).
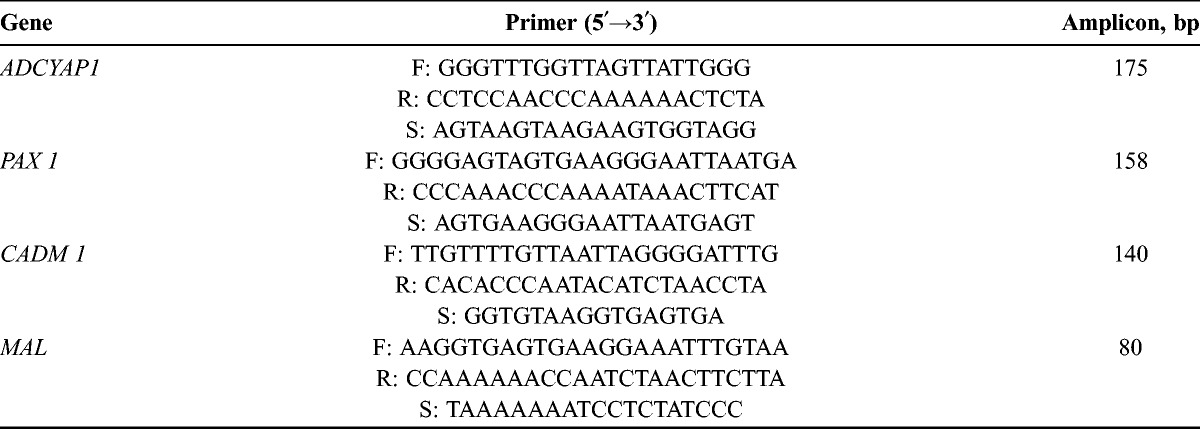
Pyrosequencing for Methylation Analysis
To allow quantitative determination of methylation at the selected CpG sites of CADM1, PAX1, MAL, and ADCYAP1 genes, cytosine percentage methylation was determined by pyrosequencing using a PyroMark Q96 ID pyrosequencer. A genomic DNA sequence corresponding to −1.0 to +1.0 kb from the transcription start site of these genes was downloaded from NCBI Reference Sequence Database Build 37.2 and predicted putative CpG islands in the 5′ regulatory region using the MethPrimer software (http://www.urogene.org/methprimer). Specific bisulfite PCR and pyrosequencing primers were designed to amplify 175 bp of 5′ untranslated regions, including the 4 CpG sites of these genes by PSQ Assay Design software version 1.0 (Qiagen, Valencia, Calif). Two hundred nanograms of genomic DNA was modified by sodium bisulfite PCR using the EZ DNA Methylation kit (ZYMO Research) according to the manufacturer’s instructions. Bisulfite-modified DNA was amplified in a 25-μL reaction with the primer set and Taq polymerase (Solgent, Daejeon, Korea) (Table 1). Pyrosequencing was performed using the PyroGold kit and PyroMark 96ID instrument (Qiagen) as instructed by the manufacturer. Briefly, 20 μL of each biotinylated PCR product was immobilized on streptavidin-coated Sepharose HP beads (Amersham Biosciences, Mass) and then subjected to sequencing using automatically generated nucleotide dispensation order for “sequence to analyze” corresponding to each reaction. The degrees of methylation (methylation index) for each of the targets and samples were calculated as the mean percentage of methylated cytosine over the sum of total cytosine for all CpGs examined. Non-CpG cytosine residues were used as built-in controls to verify bisulfite conversion. Each assay also included controls for self-annealing of sequencing primers.
TABLE 1.
Primers for bisulfate PCR and pyrosequencing
Immunohistochemistry
The experiments were performed with 0.4-μm-thick sections obtained from formalin-fixed and paraffin-embedded tissues. Tissue sections were deparaffinized in xylene and rehydrated in graded alcohol. After antigen retrieval, tissue sections were sequentially treated with 3% hydrogen peroxide and goat serum. Sections were then incubated overnight at 4°C with anti-CADM1 antibody (ab3910; Abcam, Cambridge, Mass), anti-MAL antibody (ab52911; Abcam), anti-PAX1 antibody (ab111752; Abcam), and anti-ADCYAP1 antibody (ab104154; Abcam) that were diluted 1:200 (CADM1), 1:250 (MAL), and 1:50 (PAX1 and ADCYAP1) in phosphate buffer solution. After being rinsed in phosphate buffer solution, the sections were incubated with peroxidase-conjugated goat antimouse immunoglobulin G for 1 hour at room temperature. The sections were rinsed and visualized using the 3,3-diaminobenzidine kit. Finally, all sections were counterstained with hematoxylin and eosin and covered with a coverslip. Staining was evaluated by percentages of tumor cell positivity and staining intensity. All sections were also immunohistochemically stained.
Statistical Analysis
The cutoff point of gene hypermethylation was determined by receiver operating characteristic curve analysis. The Fisher exact test and analysis of variance were used to compare between subjects with LSILs, those with HSILs, and those with cervical cancer. The odds ratios (ORs) of methylation indices of various combinations of the 4 genes were obtained by penalized likelihood logistic regression analysis. All statistical analyses were conducted using SPSS software, version 11.5 (SPSS Inc, Chicago, Ill).
RESULTS
Methylation of the 4 Gene Promoters in the Cervical Carcinoma Cell Lines
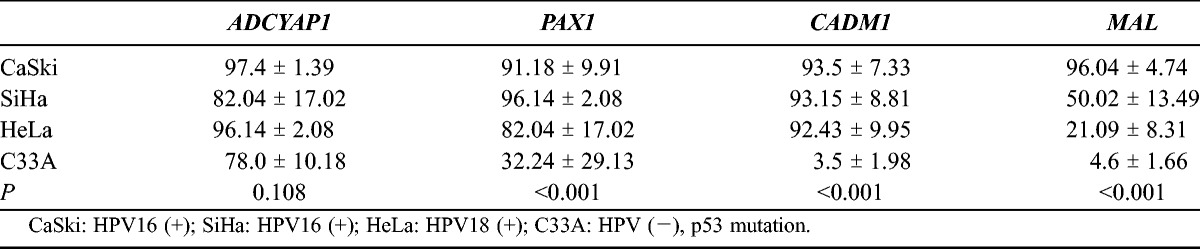
Table 2 shows the methylation degrees of the 4 genes in the cervical carcinoma cell lines. ADCYAP1 promoter was hypermethylated in the 4 cell lines. PAX1 and CADM1 promoters were hypermethylated in the CaSki and SiHa cell lines infected with HPV16 and the HeLa cell line infected with HPV18, whereas they were not in the C33A cell line. MAL promoter was hypermethylated in the CaSki and SiHa cell lines, whereas it was not in the HeLa and C33A cell lines. Based on these results, hypermethylation of PAX1 and CADM1 was associated with high-risk HPV infection. Hypermethylation of MAL was also associated with high-risk HPV infection, although the methylation indices of HeLa and C33A cell lines were not statistically significantly different (P = 0.148). However, methylation of ADCYAP1 was noted in all the cervical cancer cell lines, and the methylation level of ADCYAP1 was higher in the HPV-infected cervical cancer cell lines than in the HPV-negative cervical cancer cell lines, although it was not statistically significant.
TABLE 2.
Methylation status in the cervical carcinoma cell lines
Methylation of Human Cervical Neoplastic Tissues
Table 3 shows the methylation indices of the 4 genes from the study subjects. For ADCYAP1, methylation indices were 7.8 (95% confidence interval [95% CI], 7.0-8.6) in subjects with LSILs, 12.3 (95% CI, 9.8–15.3) in those with HSILs, and 39.8 (95% CI, 29.0–54.7) in those with cervical cancer (P < 0.001); for PAX1, 7.2 (95% CI, 6.1–8.5), 10.9 (95% CI, 8.4–14.3), and 37.8 (95% CI, 27.1–52.7), respectively (P < 0.001); for CADM1, 3.5 (95% CI, 3.0–4.0), 5.6 (95% CI, 4.0–4.7), and 17.7 (95% CI, 10.8–29.1), respectively (P < 0.001); for MAL, 2.7 (95% CI, 2.5–3.0), 3.7 (95% CI, 3.0–4.6), and 13.0 (95% CI, 7.6–22.0), respectively (P < 0.001). Methylation indices were significantly more increased in subjects with cervical cancer than in those with preinvasive cervical lesions (P < 0.001). Table 4 shows the methylation frequencies of the 4 genes above the cutoff points. For ADCYAP1, methylation frequencies above the cutoff point were 7.1% in those with LSILs, 34% in those with HSILs, 91.7% in subjects with cervical cancer (P < 0.001); for PAX1, 14.3%, 34%, and 87.5%, respectively (P < 0.001); for CADM1, 73.2%, 90%, and 95.8%, respectively (P = 0.016); for MAL, 23.2%, 32%, and 83.3%, respectively (P <0.001). For the 4 genes, methylation frequencies above the cutoff point tended to increase as pathologic grade became higher, and they were significantly more increased in subjects with cervical cancer. The ORs of methylation indices of various combinations of the 4 genes are shown in Table 5. A combination of ADCYAP1 and PAX1 showed the highest OR in subjects with invasive cervical cancer (OR, 40.87; 95% CI, 9.88–169.10), which suggests that this combination could be useful for discriminating the preinvasive lesion from invasive cancer, whereas a combination of CADM1 and MAL showed the lowest OR in those with invasive cervical cancer (OR, 13.33; 95% CI, 3.60–49.28). Figure 1 shows the correlation between the 4 genes using log transformation. ADCYAP1 and PAX1 had the highest correlation coefficient (r = 0.82, P < 0.001), whereas MAL and CADM1 had the lowest correlation coefficient (r = 0.58, P < 0.001).
TABLE 3.
Mean methylation frequencies of the study subjects
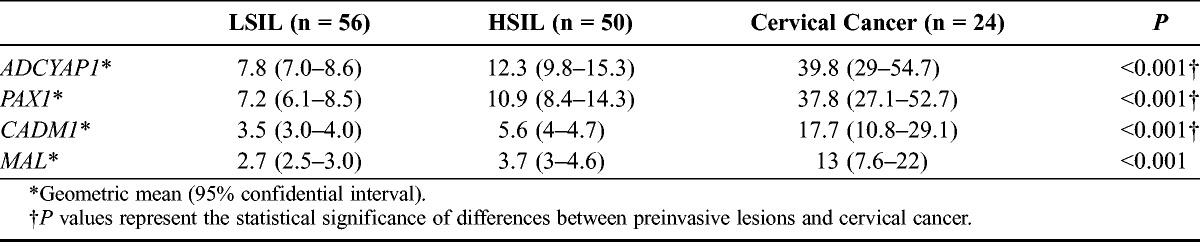
TABLE 4.
Methylation frequencies of the 4 genes above the cutoff points
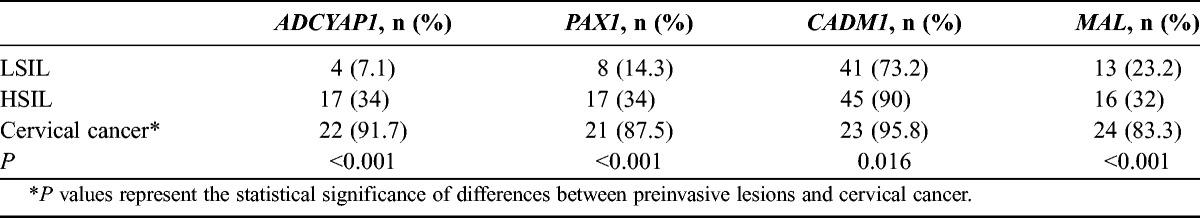
TABLE 5.
Diagnostic value of combination pairs of the 4 gene methylation frequencies in distinguishing between preinvasive lesions (low-grade intraepithelial and high-grade intraepithelial lesions) and cervical cancer
FIGURE 1.
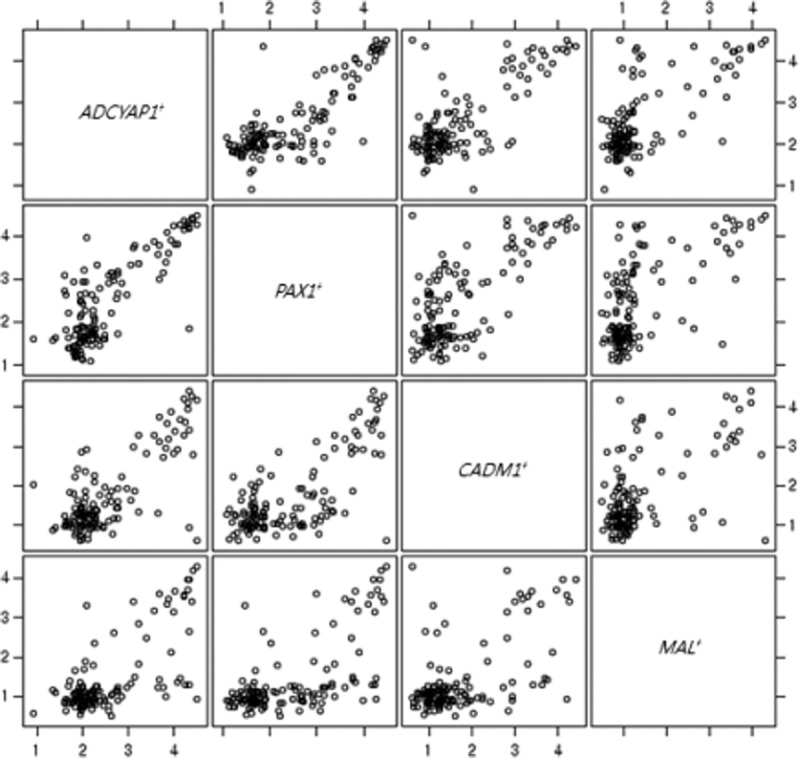
Relationship between the 4 genes ADCYAP1, PAX1, CADM1, and MAL.
Association of Reduced Gene Expression With Promoter Hypermethylation
To examine the correlation between methylation patterns and gene silencing, we assessed 4 protein expressions in cervical preinvasive cervical lesions and cervical cancers (Fig. 2). Panels A to C of Figure 2 show immunohistochemical staining of the cervical neoplastic tissues from subjects with LSILs and HSILs below the cutoff point of CADM1 methylation, which is positive for CADM1 in the cytoplasm. Figure 2D shows immunohistochemical staining of the cervical neoplastic tissue from a subject with an HSIL below the cutoff point of MAL methylation, which is positive for MAL in the cytoplasm. Figure 2E shows immunohistochemical staining of the cervical cancer tissue with increased methylation of ADCYAP1, which is negative for ADCYAP1 in the cytoplasm. Figure 2F shows immunohistochemical staining of the cervical cancer tissue with increased CADM1 methylation, which is negative for CADM1 in the cytoplasm.
FIGURE 2.
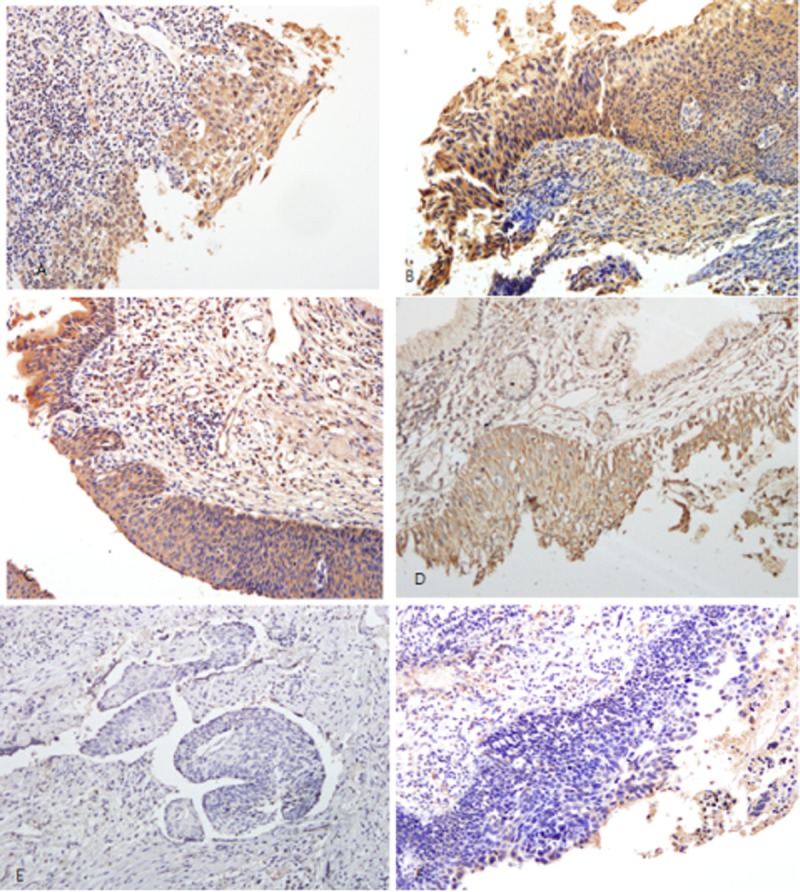
Representative immunohistochemical staining. A, LSIL without hypermethylation. It shows positivity for CADM1 in the cytoplasm (×100). B and C, HSIL without hypermethylation. It shows positivity for CADM1 in the cytoplasm (×100). D, HSIL without hypermethylation. It shows positivity for MAL (×200) in the cytoplasm. E, Squamous cell carcinoma with ADCYAP1 hypermethylation. It shows negativity for ADCYAP1 (×200) in the cytoplasm. F, Squamous cell carcinoma with CADM1 hypermethylation. It shows negativity for CADM1 in the cytoplasm (×200).
DISCUSSION
Our results suggested that silencing of some genes may be a frequent event in cervical cancer and that CADM1, MAL, PAX1, and ADCYAP1 promoter hypermethylation could be the main mode of their inactivation. Thus, silencing of these genes may be associated with the acquisition of a more advanced stage of disease during HPV-mediated malignant transformation, and hypermethylation frequencies of these gene promoters may be increased with the severity of disease. Based on these findings, methylation patterns can be used to detect aggressive precancer lesions earlier.
There have been many studies on the relationship between CADM1/MAL hypermethylation and the development of cervical cancer. Hypermethylation frequency increases as the duration of high-risk HPV exposure becomes longer (≥5 years).18 A combination of CADM1/MAL hypermethylation frequency and cytologic findings can help predict those who are at high risk of HPV infection.19
CADM1 plays an important role in both the invasion of tumors and the avoidance of immunity.9 Loss of CADM1 function is related to decreased cell adhesion and occurs before tumor formation, invasion, and anchorage-independent growth.10 CADM1 gene silencing is involved in the early phases of tumorigenesis.20 Both the frequency and degree of CADM1 promoter methylation are high in cervical cancer.9 In our study, immunohistochemistry showed that hypermethylation may be associated with decreased expression of the CADM1 protein. Previous studies have demonstrated that the density of CADM1 methylation is proportional to the degrees of anchorage-independent growth and gene silencing in HPV-transformed keratinocytes.9,10
Overexpression of MAL reduces the proliferation rate and suppresses tumor cell characteristics, such as migration and anchorage-independent growth.14,21 Analysis of a large series of cervical biopsies has revealed that both the frequency and level of MAL promoter methylation increase with the severity of disease.22 In our study, the degree of methylation increased with the pathologic grade, which is consistent with the results of previous studies.
The role of PAX1 silencing in carcinogenesis is still unknown. However, hypermethylation and gene silencing are frequently observed in cervical cancer.11,12 The PAX gene family has been classified into 4 groups. PAX1, which belongs to class 1, is less frequently overexpressed in cancer.23 It has been suggested that the PAX1 gene is a tumor suppressor in cervical cancer and has a normal function in the development of other organs. In patients with equivocal cytology (atypical squamous cells), measurement of PAX1 hypermethylation frequency shows higher sensitivity and specificity than hybrid capture II (87.5% vs 62.5% for sensitivity, 98.0% vs 86.0% for specificity).13
The ADCYAP1 gene encodes an adenylate cyclase–activating peptide 1 that belongs to a member of the secretin/glucagon/vasoactive intestinal peptide family. It is associated with both cell proliferation and apoptosis in normal cells17,24–26 and is known to regulate the immune system.27 Although the role of ADCYAP1 in carcinogenesis has not yet been completely elucidated, overexpression or repression of ADCYAP1 has been reported in various cancers.24,28 It has been demonstrated that, in the cervical cancer line, ADCYAP1 expression is suppressed by hypermethylation and is reactivated by demethylation.17 In our study, ADCYAP1 was also hypermethylated in the cervical cancer cell lines and human cervical tissues. Moreover, hypermethylation of this gene was associated with inhibition of the gene expression. The frequency of ADCYAP1 hypermethylation was very low in low-grade cervical lesions but increased with carcinoma development, which suggests that ADCYAP1 hypermethylation may suppress the apoptotic effects of ADCYAP1.
Based on these results, gene hypermethylation may be associated with progression of preinvasive lesions to invasive types of cancer. Of the 4 genes, ADCYAP1 and PAX1 seem to strongly correlate with the development of invasive types of cancer, which can be used to predict invasive types of cancer (Table 5; Fig. 1).
CONCLUSIONS
The degree of gene methylation tended to increase with the pathologic grade in subjects infected with HPV16, and hypermethylation was frequently observed in those with invasive cervical cancer, which suggested that gene hypermethylation may decrease protein expression and can be involved in carcinogenesis. In conclusion, the results of this study suggest that silencing of CADM1, MAL, PAX1, and ADCYAP1 genes may be functionally involved in HPV-mediated transformation and that promoter hypermethylation of these genes, which are predictive of decreased gene expression in tissues, may be significantly associated with the development of cervical cancer. In addition, DNA methylation status can be used as a biomarker for detecting cervical cancer.
Footnotes
This study was supported by funding for research (no. 2013-E1005-02) of the Korea Centers for Disease Control and Prevention.
The authors declare no conflicts of interest.
REFERENCES
- 1. Wisman GB, Nijhuis ER, Hoque MO, et al. Assessment of gene promoter hypermethylation for detection of cervical neoplasia. Int J Cancer. 2006; 119: 1908– 1914. [DOI] [PubMed] [Google Scholar]
- 2. zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002; 2: 342– 350. [DOI] [PubMed] [Google Scholar]
- 3. Snijders PJ, Steenbergen RD, Heideman DA, et al. HPV-mediated cervical carcinogenesis: concepts and clinical implications. J Pathol. 2006; 208: 152– 164. [DOI] [PubMed] [Google Scholar]
- 4. Esteller M, Herman JG. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol. 2002; 196: 1– 7. [DOI] [PubMed] [Google Scholar]
- 5. Piyathilake CJ, Mascaluso M, Alvares RD, et al. A higher degree of methylation of the HPV 16 E6 gene is associated with a lower likelihood of being diagnosed with cervical intraepithelial neoplasia. Cancer. 2011; 117: 957– 963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Momparler RL, Bovenzi V. DNA methylation and cancer. J Cell Physiol. 2000; 183: 145– 154. [DOI] [PubMed] [Google Scholar]
- 7. Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005; 2(suppl 1): S4– S11. [DOI] [PubMed] [Google Scholar]
- 8. Esteller M. Epigenetics in cancer. N Eng J Med. 2008; 358: 1148– 1159. [DOI] [PubMed] [Google Scholar]
- 9. Steenbergen RD, Kramer D, Bakhuis BJ, et al. TSLC1 gene silencing in cervical cancer cell lines and cervical neoplasia. J Natl Cancer Inst. 2004; 96: 294– 305. [DOI] [PubMed] [Google Scholar]
- 10. Overmeer RM, Henken FE, Snijders PJ, et al. Association between dense CADM1 promoter methylation and reduced protein expression in high-grade CIN and cervical SCC. J Pathol. 2008; 215: 388– 397. [DOI] [PubMed] [Google Scholar]
- 11. Kan YY, Liou Y, Wang HJ, et al. PAX1 methylation as a potential biomarker for cervical cancer screening. Int J Gynecol Cancer. 2014; 24: 928– 934. [DOI] [PubMed] [Google Scholar]
- 12. Wang ZM. PAX1 methylation analysis by MS-HRM is useful in triage of high-grade squamous intraepithelial lesions. Asian Pac J Cancer Prev. 2014; 15: 891– 894. [DOI] [PubMed] [Google Scholar]
- 13. Huang TH, Lai HC, Liu HW, et al. Quantitative analysis of methylation status of the PAX1 gene for detection of cervical cancer. Int J Gynecol Cancer. 2010; 20: 513– 519. [DOI] [PubMed] [Google Scholar]
- 14. Marazuela M, Alonso MA. Expression of MAL and MAL2, two elements of the protein machinery for raft-medicated transport, in normal and neoplastic human tissue. Histol Histopathol. 2004; 19: 925– 933. [DOI] [PubMed] [Google Scholar]
- 15. Horne HN, Lee PS, Murphy SK, et al. Inactivation of the MAL gene in breast cancer is a common event that predicts benefit from adjuvant chemotherapy. Mol Cancer Res. 2009; 7: 199– 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mimori K, Shiraishi T, Mashino K, et al. MAL gene expression in esophageal cancer suppresses motility, invasion and tumorigenicity and enhances apoptosis through the Fas pathway. Oncogene. 2003; 22: 3463– 3471. [DOI] [PubMed] [Google Scholar]
- 17. Jung S, Yi L, Jeong D, et al. The role of ADCYAP1, adenylate cyclase activating polypeptide 1, as a methylation biomarker for the early detection of cervical cancer. Oncol Rep. 2010; 25: 245– 252. [PubMed] [Google Scholar]
- 18. Bierkens M, Hesselink A, Meijer CJ, et al. CADM1 and MAL promoter methylation levels in hrHPV-positive cervical scrapes increase proportional to degree and duration of underlying cervical disease. Int J Cancer. 2013; 133: 1293– 1299. [DOI] [PubMed] [Google Scholar]
- 19. Verhoef VM, van Kemenade FJ, Rozendaal L, et al. Follow-up of high-risk HPV-positive women by combined cytology and bi-marker CADM1/MAL methylation analysis on cervical scrapes. Gynecol Oncol. 2015; 137: 55– 59. [DOI] [PubMed] [Google Scholar]
- 20. Chen TM, Pecoraro G, Defendi V. Genetic analysis of in vitro progression of human papillomavirus–transfected human cervical cells. Cancer Res. 1993; 53: 1167– 1171. [PubMed] [Google Scholar]
- 21. Hatta M, Okino K, Onda M, et al. Down-regulation of members of glycolipid-enriched membrane raft gene famuly, MAL amd BENE, in cervical squamous cell cancers. J Obstet Gynaecol Res. 2004; 30: 53– 58. [DOI] [PubMed] [Google Scholar]
- 22. Hesselink AT, Heideman DA, Steenbergen RD, et al. Combined promoter methylation analysis of CADM1 and MAL: an objective triage tool for high-risk human papillomavirus DNA-positive women. Clin Cancer Res. 2011; 17: 2459– 2465. [DOI] [PubMed] [Google Scholar]
- 23. Robson EJ, He SJ, Eccles MR. A PANorama of PAX genes in cancer and development. Nat Rev Cancer. 2006; 6: 52– 62. [DOI] [PubMed] [Google Scholar]
- 24. Garcia-Fernandez MO, Bodega G, Ruiz-Villaespesa A, et al. PACAP expression and distribution in human breast cancer and healthy tissue. Cancer Lett. 2004; 205: 189– 195. [DOI] [PubMed] [Google Scholar]
- 25. Castorina A, Tiralongo A, Guinta S, et al. PACAP and VIP prevent apoptosis in schwannoma cells. Brain Res. 2008; 1241: 29– 35. [DOI] [PubMed] [Google Scholar]
- 26. Li M, Cortez S, Nakamachi T, et al. Pituitary adenylate cyclase-activating polypeptide is a potent inhibitor of the growth of light chain-secreting human multiple myeloma cells. Cancer Res. 2006; 66: 8796– 8803. [DOI] [PubMed] [Google Scholar]
- 27. Ganea D, Delgado M. Vasoactive intestinal peptide (VIP) and pituitary adenylate cyclase-activating polypeptide (PACAP) as modulators of both innate and adaptive immunity. Crit Rev Oral Biol Med. 2002; 13: 229– 237. [DOI] [PubMed] [Google Scholar]
- 28. Isobe K, Kaneko M, Kaneko S, et al. Expression of mRNAs for PACAP and its receptor in human neuroblastomas and their relationship to catecholamine synthesis. Regul Pept. 2004; 123: 29– 32. [DOI] [PubMed] [Google Scholar]


