Abstract
The definitive sinoatrial node (SAN), the primary pacemaker of the mammalian heart, develops from part of pro-pacemaking embryonic venous pole that expresses both Hcn4 and the transcriptional factor Shox2. It is noted that ectopic pacemaking activities originated from the myocardial sleeves of the pulmonary vein and systemic venous return, both derived from the Shox2+ pro-pacemaking cells in the venous pole, cause atrial fibrillation. However, the developmental link between the pacemaker properties in the embryonic venous pole cells and the SAN remains largely uncharacterized. Furthermore, the genetic program for the development of heterogeneous populations of the SAN is also under-appreciated. Here, we review the literature for a better understanding of the heterogeneous development of the SAN in relation to that of the sinus venosus myocardium and pulmonary vein myocardium. We also attempt to revisit genetic models pertinent to the development of pacemaker activities in the perspective of a Shox2-Nkx2-5 epistatic antagonism. Finally, we describe recent efforts in deciphering the regulatory networks for pacemaker development by genome-wide approaches.
Keywords: pacemaker development, venous pole, atrial fibrillation, SAN, pulmonary vein
1. Introduction
The orchestrated contraction of the four-chambered mammalian heart is highly coordinated by the cardiac conduction system (CCS). The matured CCS of mammals contains the sinoatrial node (SAN), atrioventricular node (AVN), AV bundle and ventricle conduction networks [1]. The primary pacemaker, the SAN, is embedded in the junction between the right superior vena cava (RSVC) and the right atrium (RA). The SAN cells maintain highest automaticity and function to provide the primary impulse for each coordinated contraction. Dysregulation of SAN development and homeostasis causes sick sinus syndrome (SSS) [2].
1.1. The SAN Can Be Divided into Subdomains by Anatomical and Genetic Criteria
Initially noted as “a small condensed area of tissue located just where the cava sinks into the auricle” more than 100 years ago [3], the SAN is now recognized as a complex functional unit and the subdomains within the SAN have been defined both anatomically and genetically. Anatomically, a “head” region of the SAN can be identified as a histologically discrete structure that extends superiorly and wraps around the RSVC. The SAN also extends inferiorly toward the inferior vena cava (IVC) to form a “tail” structure [4,5,6,7].
Genetically, in the mouse, the SAN can be divided into a Tbx18+/Nkx2-5− head domain and a Tbx18−/Nkx2-5+ tail/peripheral domain. In addition, the entire SAN can be identified by the expression of Shox2, as shown in Figure 1, as well as Tbx3, Isl1, and Hcn4 [8,9,10,11]. A number of reporters/markers were also used to identify the developing SAN, such as the CCS-LacZ transgene [12] and the surface marker HNK-1 [13], though their specific SAN localization was not indicated. While the distinct functions of the SAN head and SAN tail/peripheral domain have not been well characterized, a recent study indicates that the Nkx2-5+ SAN tail/peripheral domain is essential for normal SAN function [9].
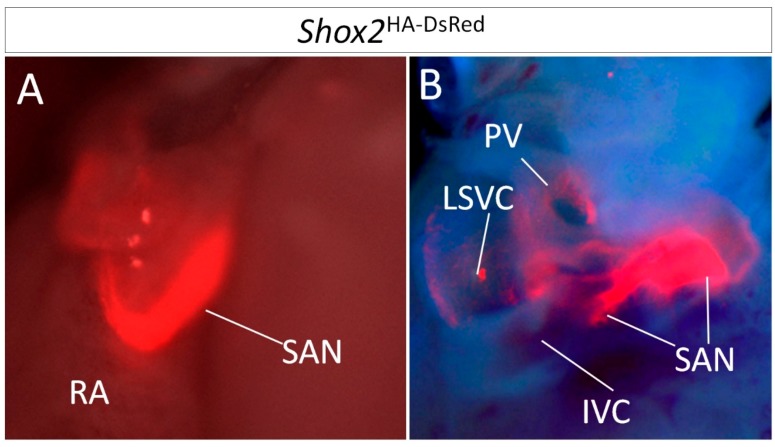
Figure 1.
Visualization of Shox2 expression in the SAN by Shox2HA-DsRed allele. (A) Frontal view of Shox2 expression in the SAN of a mouse carrying a Shox2HA-DsRed allele at Postnatal Day 8 (P8), as revealed by DsRed expression. (B) Dorsal view of Shox2 expression in the same mouse. PV, pulmonary vein; RA, right atrium; IVC, inferior vena cava; SAN, Sinoatrial node; LSVC, left superior vena cava.
1.2. Lineage Development of the SAN in Relation to Other Venous Pole Structures
During early development, after the formation of a linear heart tube through bilateral cardiogenic plate fusion (the first heart field, FHF), additional progenitor cells are continuously recruited from the surrounding mesenchyme to the outflow tract and the venous pole (inflow tract) where the SAN resides [14,15,16,17]. A morphologically distinct SAN structure can be identified as early as Embryonic Day 9.5 (E9.5) in the mouse. The initial function of the SAN was thought to start at around E12.5 [18] in the mouse, with typical SAN-like action potential (AP) configurations being identified at E14.5 [9,19]. Although the SAN is the dominant pacemaker in the venous pole, cells with pacemaking activities can be found not only in the SAN, but also in the myocardial sleeves of the systemic venous return and the pulmonary vein (PV) myocardium. It was proposed that these cells with pacemaking activities can act as ectopic pacemaker to trigger atrial fibrillation (A-Fib) [20,21], making the characterization of the lineage origin of these sites in relation to the SAN an interesting subject. Multiple lineages have been shown to contribute to the development of the venous pole based on marker expression and genetic fate mapping studies, such as the Tbx18+/Nkx2-5− sinus venosus (SV) cells that develop into the myocardial sleeves of the systemic venous return [14,22], the posterior second heart field (SHF) cells mapped by Mef2C-AHF-Cre that develop into the primary atrial septum [16,23], and the cells labeled by Hcn4CreERT2 induced from E9.5 onward [24]. Based on the observation that Tbx18+ lineage also maps to the SAN, it was assumed that the SAN is derived mainly from the Tbx18+/Nkx2-5− SV myocardial cells [14,22]. Interestingly, the lineage and genetic attribution of the myocardial sleeves around the PV have been under heavy debate regarding whether it is a SV-like lineage. Similar to the SV myocardium, the PV myocardial cells are CCS-LacZ positive and HNK-1 positive as well [25,26]. However, unlike the SV myocardium, the PV myocardium is derived from a Tbx18−/Nkx2-5+ lineage [27], arguing against the hypothesis that the PV myocardium and SV myocardium share a common origin. Our recent study showed that the PV myocardium, SV myocardium, as well as SAN are all derived from Shox2+ cells [9], offering a new perspective for understanding the lineage attribution of the PV myocardium. Interestingly, by using the expression of Shox2 and cTnT as a marker for myocardial cells, a Shox2+/cTnT+/Hcn4+ myocardial continuation was clearly identified in the proximal junction of the left superior vena cava (LSVC) and the PV as early as E10.5, supporting a common origin of the SV and PV myocardium [9]. In adult mice, this group of cells remains positive for Hcn4 (Figure 2), suggesting a potential etiology for A-fib if the counterpart of these cells is present in humans. Moreover, Shox2 is expressed in the PV side of the left atrial-PV structural continuum [9], providing a molecular marker for distinguishing the PV derived structure from the atrial-PV confluence structural continuum. Thus, although some genes, such as Tbx3 and Isl1, are the SAN specific markers, other genes that mark the SAN also serve as lineage markers for the venous pole such as Tbx18 that labels the SV myocardium and Shox2 that labels the SV myocardium along with the PV myocardium. Such correlation coincides with the knowledge that the SV and PV myocardial cells are prone to develop foci induced A-Fib [20,21], and that the Shox2+ SV and PV myocardial cells display pacemaker-like characteristics at early embryonic stages [9,20,21]. Given that pacemaker properties are shared among Shox2+ cells at early developmental stages regardless of the expression of Tbx3 and Isl1 that are predominantly expressed in the SAN [9,18], an interesting questions is raised: is the genetic program that controls pacemaker properties in the venous pole independent from the one that controls the development of the histologically discrete SAN?
Figure 2.
Hcn4 expression in the proximal junction of the PV and LSVC myocardium of an adult mice. (A) Dorsal view of EGFP expression (arrowheads) in an adult Hcn4CreERT2; ROSAmTmG mouse seven days after tamoxifen induction. (B) Hcn4 positive cells (arrowheads) in the junction between the PV and LSVC of the same mouse. LV, left ventricle; PV, pulmonary vein; TR, Trachea; LAA, left atrial appendage; LSVC, left superior vena cava.
2. Genetic Models for SAN Development
To further understand SAN development in relation to the SV and PV myocardium, we sought to revisit literature describing mouse models generated for studying SAN development. We categorized these models by whether or not mutations would cause phenotypes in both the venous pole and SAN.
2.1. Genes and Genetic Models for SAN Dysgenesis without Complications in the SV and PV Myocardium
Tbx3: Tbx3, encoding a T-Box transcription factor, has been well characterized for its essential role in embryonic development and postnatal function of the SAN and AVN. Null mutation or hypomorphism of Tbx3 causes sick sinus syndrome and atrial-ventricular (A-V) conduction block [8,28,29,30,31,32]. Tbx3 acts predominantly as a molecular repressor on the expression of working myocardial specific genes in the SAN [1,33]. It has been proposed that Tbx3 functions to compete with Tbx5 for binding with Nkx2-5, thus repressing the activation of the target genes of Nkx2-5 and Tbx5 such as Scn5α (encoding Nav1.5) and Gja5 (encoding Cx40) [1,33]. Tbx3 may also repress Nkx2-5 expression directly as conditional inactivation of Tbx3 elicits ectopic activation of Nkx2-5 in the adult SAN [32]. Interestingly, although it has been established that Hcn4 is a repressive target of Nkx2-5 [9,34], null mutation of Tbx3 does not cause downregulation of Hcn4, suggesting that Tbx3 is only partially required for the maintenance of the SAN program. The fact that Tbx3 is expressed in a manner more restricted than that of Shox2 (Figure 3) in the SAN region at E10.5 when the dominant pacemaker activity is not yet confined to the SAN [18] suggests that the definitive SAN cells are already specified before the establishment of the dominant pacemaker activity and require a distinctive genetic program likely centering around Tbx3.
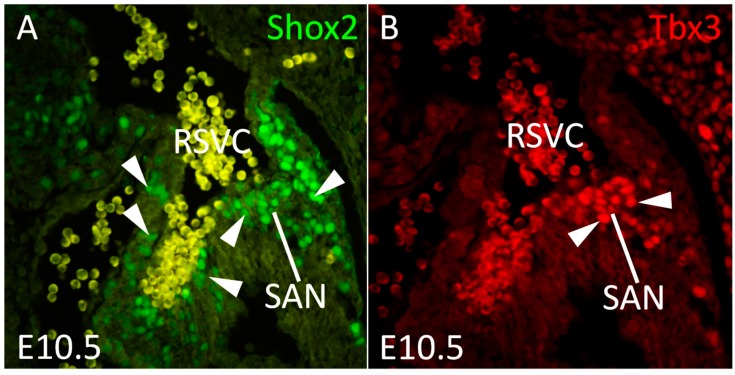
Figure 3.
Expression of Shox2 and Tbx3 in the SAN at E10.5. Immunofluorescence shows the expression of Shox2 (arrowheads) in the SAN and other tissues around the SAN (A) and the restricted expression of Tbx3 (B) (arrowheads) in the SAN. SAN, sinoatrial node; RSVC, right superior vena cava.
Isl1: Isl1 encodes a LIM-domain homeodomain transcription factor. Although Isl1-Cre maps to the whole SHF derivatives of the mouse heart, Isl1 is expressed later on only in a small subpopulation of cells in the heart including the SAN [35,36]. Recently, the essential function of Isl1 in the development and function of the SAN was revealed by genetic and transcriptome studies in an Isl1F/F; Hcn4ERT2Cre/+ model in which the deletion of Isl1 is induced at stages beyond E10.5 at which point the role of Isl1 in maintaining the progenitor state of SHF cells becomes relatively limited and its deletion is less likely to complicate the analysis of Isl1 function in the SAN [10,11]. As it was expected, inactivation of Isl1 causes bradycardia associated with reduced size of the SAN and down-regulation of a number of SAN specific genes including Hcn4, Shox2, and Tbx3 [10,11].
2.2. Gene and Genetic Models Affecting Both the SAN and Other Venous Pole Components
Tbx18: Tbx18 is the first gene that was shown to be able to convert non-pacemaker cardiomyocytes into pacemaker-like cells when it was forcedly expressed [37,38]. Tbx18 is expressed in the pro-epicardial organ (PEO), developing SV myocardium, and SAN head in the venous pole [39]. Tbx18-Cre could also map to part of the SAN tail/peripheral domain, but not to the PV myocardium [39]. Accordingly, inactivation of Tbx18 causes dysgenesis of both the SAN head and the SV myocardium [39]. Interestingly, Tbx18 is never expressed in the PV myocardium, arguing for a different lineage origin of the PV myocardium versus the SV myocardium. Although the SAN head undergoes severe dysgenesis in Tbx18 mutants, a functional SAN tail/peripheral domain is retained, implying the existence of an unidentified mechanism that controls pacemaker development in the SAN tail/peripheral domain and possibly in the PV myocardium as well.
Shox2: Shox2 belongs to the family of short stature homeobox genes and is expressed in the developing SV and PV myocardium, and the SAN in both mice and humans [9], as well as in a small subdomain of the dorsal mesenchymal protrusion [19]. Previous studies showed that Shox2 is essential for SAN development partially by preventing Nkx2-5 expression in the SAN head [40,41]. Interestingly, Tbx3 is also essential for repressing the Nkx2-5 expression in the SAN head [32], raising the possibility that Shox2 and Tbx3 repress Nkx2-5 synergistically to ensure normal SAN development. This hypothesis is supported by the observation that mice carrying compounded Shox2 and Tbx3 hypomorphic alleles have significant higher level of Nkx2-5 in the SAN compared to the littermate controls [42]. Actually, Shox2 and Nkx2-5 are co-expressed extensively in the SAN tail/peripheral region that is also Tbx3 positive and in a group of cells surrounding the SAN. In addition, Shox2 and Nkx2-5 are also co-expressed in the other venous pole components, such as the PV myocardium that is Hcn4 positive in early embryonic stages and possesses pacemaker properties, suggesting an unidentified mechanism for Shox2 to control pacemaker development [9]. It was proposed that the Shox2-Bmp pathway may function to regulate SAN development [19,43]. However, Shox2-Cre mediated site specific inactivation of either Bmp4, the major BMP ligand that is highly expressed in the SV and SAN myocardium, or Smad4, which is required for the execution of canonical BMP signaling, results in a normal functional SAN [44]. These observations indicate that although Shox2 is required for the developmental expression of Bmp4 in the venous pole, the Shox2-Bmp4 pathway does not sufficiently account for the function of Shox2 in SAN development.
Nkx2-5: Numerous studies have pinpointed to the importance of Nkx2-5 in early cardiac development, maturation of cardiomyocytes, and A-V conduction axis. The role of Nkx2-5 in the venous pole, however, was underappreciated, partially due to embryonic lethality of Nkx2-5 null mice around E10.5, precluding functional analysis of Nkx2-5 at later stage of venous pole morphogenesis. Nkx2-5 is initially absent from the SV myocardium and the SAN head [9,20] but is acquired in the SV myocardium at around E14.5. The PV myocardium is Nkx2-5+ and expresses a relatively weak level of Hcn4 expression compared to the SV myocardium [9,45]. In an Nkx2-5 hypomorphism model in which the expression level of Nkx2-5 is reduced to about 25% of wide type level [45,46], the PV myocardium acquires strong Hcn4 expression and losses the expression of Gja5 (Cx40) partially, mimicking the SV myocardial phenotype [45]. Furthermore, hypomorphism of Nkx2-5 also results in an “invasion” of SAN phenotype to the surrounding atrial tissue, suggesting that the PV myocardium and the atrial tissues peripheral to the SAN are “default” to a pacemaker-like cell fate and such a “default” fate is suppressed by the presence of Nkx2-5 [45]. It was reported recently that this pacemaker “default” fate is primed and sustained by the presence of Shox2 [9]. Nevertheless, it is generally accepted that Nkx2-5 exerts a repressive effect on pacemaker program by facilitating the maturation of cardiomyocytes in the developing venous pole. Such notion is further supported by the observation that venous pole specific deletion of Nkx2-5 in the Sln-Cre; Nkx2-5F/F mouse model results in an enlarged SAN [47].
Pitx2: Components of the four-chambered heart, including venous pole structures, undergo left-right asymmetric development at early stage. It has been demonstrated that Pitx2, encoding a homeodomain transcription factors, confers left sided patterning information [48,49,50]. It is noted that the myocardial sleeves of the LSVC and PV are positive for Pitx2 and were considered “left sided” structures [51,52]. Pitx2 deficiency leads to the emergence of right-sided structures including the SAN and venous valves (VV) in the left-side SV-atrial junction [18]. Moreover, it was shown that haploinsufficiency of Pitx2 caused A-Fib and ectopic expression of SAN program including Tbx3 and Shox2 in the left-side of the heart [53,54]. Although the loss of either Nkx2-5 or Pitx2 results in the ectopic pacemaker phenotype, inactivation of Pitx2 leads to an acquirement of the complete pacemaker program in the left SV-atrial junction, whereas haploinsufficiency of Nkx2-5 causes only an upregulation of Hcn4 in the Pitx2+ left sided structure, suggesting that these two genes repress pacemaker program through independently functional mechanisms [53,54]. Such hypothesis is further supported by an observation that in some Pitx2− right sided structures such as left venous valve, haploinsufficiency of Nkx2-5 elicits an activation of a relatively complete SAN program including Tbx3 and Hcn4 [42].
3. Shox2-Nkx2-5 Antagonistic Mechanism in the Regulation of Pacemaker Properties
3.1. Shox2-Nkx2-5 Antagonistic Mechanism
To date, genetic, biochemical, and electrophysiological studies have suggested that pacemaker cells retain properties that are common for the early primitive cardiomyocytes such as poorly organized sarcomere and the expression of Hcn4 that is partially responsible for the high automaticity [55]. Thus it is reasonable to assume that rather than taking a positive acquiring process for pacemaker phenotype, SAN development may undergo a preventive process from maturation towards working myocardial cell fate. Such assumption is well in line with the fact that many machineries required for the maintenance of pacemaker properties are also utilized for sustaining progenitor state of the anterior SHF cells. A good example is that Isl1, which is essential for maintaining the undifferentiated state of the SHF cells, is also crucial for SAN development [10,11]. Moreover, similar to its function in the differentiation of the anterior SHF cells, Nkx2-5 also promotes cardiomyocyte maturation in the venous pole, and Tbx3 was shown to act as a transcription repressor in the SAN for the working myocardial program that is enforced by Nkx2-5 and Tbx5 [28]. However, these transcriptional machineries do not account sufficiently for the pacemaking activities in the SV myocardium and PV myocardium where Tbx3 and Isl1 are not expressed. Given that the derivatives of the SV and PV myocardium are prone to acting as triggers for A-Fib [20], characterization of the molecular mechanism that enforces pacemaker properties in these sites would be important for developing gene based therapeutic approaches for A-fib.
Based on the fact that Shox2 is co-expressed with Hcn4 in the PV and SV myocardium where Isl1 and Tbx3 are not expressed, and that the Shox2+ PV myocardium cells display pacemaker-like properties, Shox2 was believed to be a good candidate responsible for the potential pacemaker properties in the SV and PV myocardium independent of the SAN program genes Tbx3 and Isl1 [9]. Since hypomorphism of Nkx2-5 causes augmentation of Hcn4 expression in the Nkx2-5+/Shox2+ PV myocardium [9], and that the Shox2+/Hcn4+ SV myocardium is negative for Nkx2-5, it was proposed that Nkx2-5 inhibits pacemaker properties that belong to a primitive cell phenotype, and such effect is counter-balanced by Shox2. By genetic approaches, it was established that a Shox2-Nkx2-5 antagonistic mechanism controls pacemaker versus working myocardial cell phenotype in the PV myocardium, SV myocardium, and part of the SAN [9]. Consistent with the observation that hypomorphism of Nkx2-5 results in an “invasion” of SAN phenotype to the surrounding atrial tissue [45], we have recently observed that the tissues being preferentially “invaded” by SAN phenotype are all primed by Shox2 expression [42], suggesting the balance between Shox2 and Nkx2-5 transcription output is critically maintained to restrict the size of the SAN.
3.2. Heterogeneous Development Model of the SAN and the Essential Physiological Function of the Shox2+/Nkx2-5+ SAN Tail/Peripheral Domain
Similar to that in the PV myocardium, Shox2 and Nkx2-5 are also co-expressed in the developing peripheral SAN and function to regulate the development of this SAN domain by their antagonistic action [9]. The development of the SAN tail/peripheral domain appears to be independent of the SAN head, as the deletion of Shox2 in the SAN peripheral domain by Nkx2-5-Cre (Shox2Nkx2-5Cre) led to virtual absence of the SAN tail/peripheral region but an unaffected SAN head [9]. In line with this notion is the fact that null mutation in Tbx18 results in a severely hypoplastic SAN head but leaves the SAN tail/periphery unaffected [8]. The physiological importance of the peripheral region of the SAN was initially implied by Scn5α heterozygous mutants that exhibit sick sinus syndrome associated with SAN exit block [56]. Scn5a encodes a sodium channel protein (Nav1.5) and is expressed in the developing heart including the peripheral region of the SAN but not the SAN head. However, direct evidence supporting an essential role of the peripheral SAN in normal SAN function and S-A conduction was not available, primarily due to the lack of tools or genetic models to inactivate genes specifically in the peripheral SAN domain. In Shox2Nkx2-5Cre model, a severe sick sinus syndrome was observed associated with the absence of the peripheral SAN [9], providing the first line of direct evidence for the functional importance of the peripheral SAN as an integrated part of the SAN. Furthermore, although previous studies have supported the notion that Nkx2-5 plays an inhibitory role in general on pacemaker program in the venous pole, the conserved expression of Nkx2-5 in the SAN tail/periphery in both mice and humans suggests an underappreciated unique role for Nkx2-5 in SAN development and function. The fact that Nkx2-5 binds directly to a verified enhancer downstream of Scn5α, revealed by Nkx2-5 ChIP-Seq results [9,57,58], suggests that Nkx2-5 regulates Scn5α expression directly in the peripheral SAN domain to confer a unique electrophysiological property to the SAN tail/periphery. This hypothesis is supported by our recent observation that mice bearing the deletion of Nkx2-5 in the SAN peripheral domain also exhibited sick sinus syndrome [42]. We therefore propose a model that the Shox2+/Nkx2-5+/Hcn4+/Scn5α+ SAN peripheral cells possess electrophysiological property intermediate of the Nkx2-5−/Shox2+/Hcn4+/Scn5α− SAN head and the Shox2−/Nkx2-5+/Hcn4−/Scn5α+ atrial cells. Such property enables the Shox2+/Nkx2-5+ SAN tail/peripheral domain to play a unique role in the S-A conduction in addition to its impulse generation capability. Consistent with this model are: (1) Shox2Nkx2-5Cre mice exhibit sick sinus syndrome associated with a virtual absence of the SAN tail/peripheral domain [9]; (2) mice lacking Tbx18 in the heart exhibit normal sinus rhythm regardless the lack of the SAN head [8]; and (3) a computational modeling predicts that the peripheral SAN cells possess higher maximum depolarization slope and higher AP amplitude than the SAN head cells to potentially function as a signal amplifier [59].
4. Genome-Wide Studies on Transcription Regulatory Networks and Chromatin Landscape in the Pro-Pacemaker Cells
It has been established that the SAN and pro-pacemaker cells in the venous pole possess a unique set of cellular properties and genetic features. This notion has been recently elaborated by transcriptome studies that demonstrate the possession of a distinct transcriptome profile by the SAN cells as compared with that in working myocardial cells [10]. It was further demonstrated that such unique SAN cell profile is at least in part maintained by the expression of Isl1, as deletion of Isl1 by Hcn4-CreERT2 led to in the SAN an upregulation of working myocardial genes such as Gja5 and Gja1, and a downregulation of pacemaker genes including Tbx3, Shox2 and Hcn4. As abovementioned, Shox2 and Tbx3 are also responsible for the correct expression level of some of these genes in the SAN including Isl1 itself. Such correlation suggests an epistatic link between Shox2, Tbx3 and Isl1 in SAN development. These genetic interactions, if tested by compounded allelic serial mutations of Shox2, Tbx3, and Isl1, will provide instrumental insights for a better understanding of the genetic cascades governing the unique pacemaker properties for the SAN and other pacemaker cells in the venous pole.
Furthermore, by ChIP-Seq, the molecular mechanisms involved in maintaining the particular properties of the pacemaker cells are being unveiled. Earlier studies of Tbx3 ChIP-Seq on adult heart using a Tbx3 gain-of-function allele revealed a mechanism by which Tbx3 directly competes with Tbx5 to repress the working myocardial cell fate [58]. Similarly, a recent ChIP-Seq study on E12.5 mouse embryonic hearts unraveled an extensive genome-wide co-occupation of Shox2 and Nkx2-5, supporting the Shox2-Nkx2-5 antagonistic mechanism by which Shox2 inhibits the transcription output of Nkx2-5 [9]. These lines of evidence together suggest the importance of transcription repressors in maintaining the pacemaker cell fate in a primitive state. Recent studies have shown that distal acting regulatory elements often reveal crucial information on the lineage specific chromatin landscape and reflect lineage specific transcription events as well as that transcription factor co-occupancy can be used to predict functional enhancers [60]. Our recent in-depth analyses of our published and unpublished ChIP-Seq data further identified co-occupation of Shox2 and Nkx2-5 on distal enhancers of a set of genes. Gene ontology (OG) analysis indicates that A-Fib related biological terms are highly enriched in the Shox2/Nkx2-5 co-occupied sites (Figure 4), suggesting that the co-occupation of Shox2 and Nkx2-5 can serve as a useful criterion for identifying A-Fib related cis-regulatory elements.
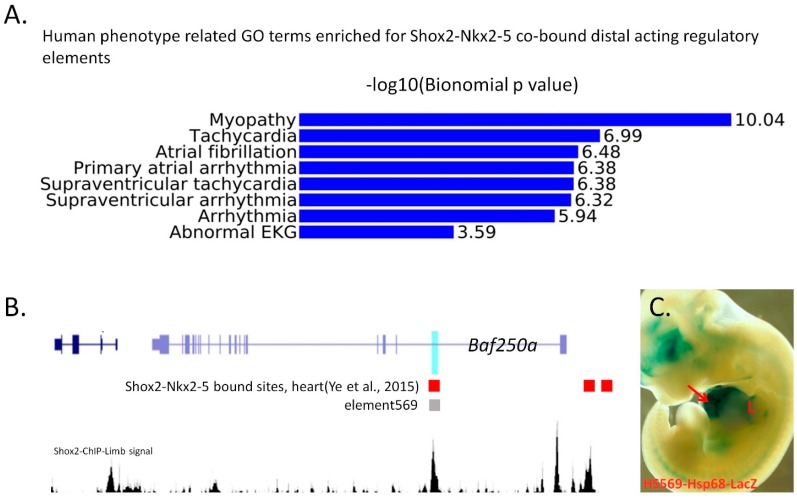
Figure 4.
(A) Gene-ontology analysis reveals highly enriched atrial fibrillation related function in the Shox2-Nkx2-5 co-bound distal acting regulatory elements. (B) Signal track of Shox2-binding in the developing limb at E12.5 in relation to HS569, an enhancer of Baf250a, and Shox2-Nkx2-5 co-bound sites in the embryonic heart. (C) An E11.5 transgenic embryo carrying a reporter construct driven by Shox2-bound distal acting regulatory element (HS569) shows LacZ expression in the heart (arrow) and limb (L).
Interestingly, many of these elements discovered in the embryonic heart are also occupied by Shox2 in other developmental contexts. For example, a co-occupied site by Shox2 and Nkx2-5 in the intron-1 of Arid1a (Baf250a) in the embryonic heart [9] is also bound by Shox2 in the developing limb, suggesting that this site is accessible in both the developing heart and limb. Indeed, the orthologous region of this site in humans (HS569) [61] was characterized to have enhancer activity in the developing heart and limb in mice (Figure 4B,C). Furthermore, integrative interrogation of the distal regulatory elements identified by Islet1 ChIP-Seq in the postnatal SAN and that by Shox2 ChIP-Seq on the developing limb also unveils that a significant proportion of distal regulatory elements (~1/3) occupied by Islet1 is also bound by Shox2, indicating these regulatory elements are utilized in the development of both heart and limb [62]. Notably, a predominant proportion of these chromatin domains is not in an accessible configuration in the Encode heart DNasesHS dataset generated from working myocardium cells [63], suggesting that the pacemaker cells adopt cis-regulatory elements and associated transcription regulatory machinery that are also functioning in other developmental contexts to acquire the cellular phenotype distinct from working myocardial cells. Thus, integrative analysis of transcription factor binding profiles in other developmental context will aid for unraveling the transcriptional machineries operating in the developing SAN and pro-pacemaking cells in the venous pole.
Recent advance and deepened understanding on regulatory chromatin landscape point towards the possibility that based on the accessibility or particular histone modification on tissue/lineage specific cis-regulatory elements, the developmental competence of cells toward the lineage of desire can be predicted. This possibility was nicely exemplified by the studies on human embryonic stem cell derived pancreatic beta cells [64]. It would be important and interesting to explore the features of chromatin landscape in pacemaker cells that are responsible for the specific pacemaker properties including the distinctive sets of gene expression profiles. Such studies on pacemaker cells will similarly benefit the generation of biological pacemakers by providing readouts for the developmental and differentiation competence of precursors toward pacemaker cells. However, technically, it will be very difficult to profile the chromatin landscapes of pacemaker cells directly by DNase-HS, FAIRE-Seq or by ChIP-Seq on various histone modification markers that are predictive of active enhancer/promoter activity. This is due to the relatively scarce SAN cells and that the pro-pacemaker cells in the venous pole are patched with surrounding atrial myocardial cells, precluding accurate isolation of sufficient amount of pacemaker cells in an undisturbed in vivo state. Currently, most of the SAN markers, such as Tbx3 and Hcn4, also label other conduction components, including the AVN. A Shox2 knock-in allele that expresses the Shox2a isoform and DsRed (Shox2HA-DsRed) and allows live imaging of Shox2 expression in the venous pole and the developing SAN [9] would offer a unique tool for FACS isolation of venous pole pacemaker cells. In conjunction with open chromatin sequencing techniques that were designed for rare population of cells such as ATAC-Seq [65], it would be quite possible to characterize the accessible chromatin landscape of pacemaker cells comprehensively. Nevertheless, current ChIP-Seq studies on transcription factors closely related to pacemaker development in the venous pole provide a glimpse into the chromatin landscape of pacemaker cells.
5. Conclusions
In this review we summarize the previous studies in the perspective of SAN and pro-pacemaking venous pole development (Figure 5). In light of these comprehensive sets of studies, we propose that a delayed maturation towards working myocardium phenotype underpins the pacemaker properties of the Shox2+ pro-pacemaker cells in the developing SV, PV and SAN. Moreover, a converged repressive transcription output of Isl1, Tbx3, Tbx18, and Shox2 for working myocardial program appears essential for the solidification of a morphologically distinct SAN. Additional transcription activators that are expressed in the venous pole, such as Gata4/6 and CoupTFII, may play crucial role to enforce the in vivo developmental program in the SAN.
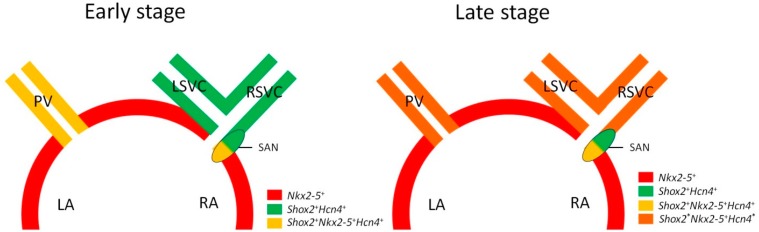
Figure 5.
A schematic illustration summarizing the fate decision of cells in the developing venous pole. In the early developing venous pole, Shox2 is expressed in the SAN head domain as well as the myocardium of the LSVC and RSVC, which are all positive for Hcn4. In addition, the SAN tail/peripheral domain and the PV myocardium are positive for Shox2, Nkx2-5, and Hcn4. At the late developmental stage and adult, the myocardium of the PV, LSVC, and RSVC becomes matured and largely adopts working myocardial phenotype, with potential to reacquire pacemaking activities.
Acknowledgments
The work conducted in the authors’ laboratories and cited in this review article was supported by an American Heart Association Predoctoral Fellowship (13PRE1375003 to Wenduo Ye), and by the NIH as well as by a grant from the National Health and Family Planning Commission of China (No. WKJ-FJ-24) and an International Collaboration Grant from the Science and Technology Department of Fujian Province, China (No. 2015I0011). The HS569 transgenic embryo shown in Figure 4 was kindly provided by the laboratory of L.A. Pennacchio in the Genome Sciences Department at Lawrence Berkeley National Laboratory.
Author Contributions
Y.C. and W.Y. conceived and outlined the manuscript. W.Y. performed most of the experiments described in the manuscript and wrote the manuscript. Y.S. helped in immunohistochemistry experiments. Z.H. and Y.Z. helped in drawing figures for illustration. Y.C. conducted final revision and editing of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Christoffels V.M., Smits G.J., Kispert A., Moorman A.F. Development of the pacemaker tissues of the heart. Circ. Res. 2010;106:240–254. doi: 10.1161/CIRCRESAHA.109.205419. [DOI] [PubMed] [Google Scholar]
- 2.Anderson J.B., Benson D.W. Genetics of sick sinus syndrome. Card. Electrophysiol. Clin. 2010;2:499–507. doi: 10.1016/j.ccep.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keith A., Flack M. The Form and Nature of the Muscular Connections between the Primary Divisions of the Vertebrate Heart. J. Anat. Physiol. 1907;41:172–189. [PMC free article] [PubMed] [Google Scholar]
- 4.Dobrzynski H., Boyett M.R., Anderson R.H. New insights into pacemaker activity: Promoting understanding of sick sinus syndrome. Circulation. 2007;115:1921–1932. doi: 10.1161/CIRCULATIONAHA.106.616011. [DOI] [PubMed] [Google Scholar]
- 5.Herrmann S., Fabritz L., Layh B., Kirchhof P., Ludwig A. Insights into sick sinus syndrome from an inducible mouse model. Cardiovasc. Res. 2011;90:38–48. doi: 10.1093/cvr/cvq390. [DOI] [PubMed] [Google Scholar]
- 6.Morris G.M., Monfredi O., Boyett M.R. Not so fast! Sick sinus syndrome is a complex and incompletely understood disease that might prove hard to model in animals. Cardiovasc. Res. 2011;92:178. doi: 10.1093/cvr/cvr204. [DOI] [PubMed] [Google Scholar]
- 7.Silverman M.E., Hollman A. Discovery of the sinus node by Keith and Flack: On the centennial of their 1907 publication. Heart. 2007;93:1184–1187. doi: 10.1136/hrt.2006.105049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiese C., Grieskamp T., Airik R., Mommersteeg M.T., Gardiwal A., de Gier-de Vries C., Schuster-Gossler K., Moorman A.F., Kispert A., Christoffels V.M. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ Res. 2009;104:388–397. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- 9.Ye W., Wang J., Song Y., Yu D., Sun C., Liu C., Chen F., Zhang Y., Wang F., Harvey R.P., et al. A common Shox2-Nkx2-5 antagonistic mechanism primes the pacemaker cell fate in the pulmonary vein myocardium and sinoatrial node. Development. 2015;142:2521–2532. doi: 10.1242/dev.120220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vedantham V., Galang G., Evangelista M., Deo R.C., Srivastava D. RNA sequencing of mouse sinoatrial node reveals an upstream regulatory role for Islet-1 in cardiac pacemaker cells. Circ. Res. 2015;116:797–803. doi: 10.1161/CIRCRESAHA.116.305913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang X., Zhang Q., Cattaneo P., Zhuang S., Gong X., Spann N.J., Jiang C., Cao X., Zhao X., Zhang X., et al. Transcription factor ISL1 is essential for pacemaker development and function. J. Clin. Investig. 2015;125:3256–3268. doi: 10.1172/JCI68257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rentschler S., Vaidya D.M., Tamaddon H., Degenhardt K., Sassoon D., Morley G.E., Jalife J., Fishman G.I. Visualization and functional characterization of the developing murine cardiac conduction system. Development. 2001;128:1785–1792. doi: 10.1242/dev.128.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blom N.A., Gittenberger-de Groot A.C., DeRuiter M.C., Poelmann R.E., Mentink M.M., Ottenkamp J. Development of the cardiac conduction tissue in human embryos using HNK-1 antigen expression: Possible relevance for understanding of abnormal atrial automaticity. Circulation. 1999;99:800–806. doi: 10.1161/01.CIR.99.6.800. [DOI] [PubMed] [Google Scholar]
- 14.Gittenberger-de Groot A.C., Mahtab E.A., Hahurij N.D., Wisse L.J., Deruiter M.C., Wijffels M.C., Poelmann R.E. Nkx2.5-negative myocardium of the posterior heart field and its correlation with podoplanin expression in cells from the developing cardiac pacemaking and conduction system. Anat. Rec. 2007;290:115–122. doi: 10.1002/ar.20406. [DOI] [PubMed] [Google Scholar]
- 15.Christoffels V.M., Mommersteeg M.T., Trowe M.O., Prall O.W., de Gier-de Vries C., Soufan A.T., Bussen M., Schuster-Gossler K., Harvey R.P., Moorman A.F., et al. Formation of the venous pole of the heart from an Nkx2-5-negative precursor population requires Tbx18. Circ. Res. 2006;98:1555–1563. doi: 10.1161/01.RES.0000227571.84189.65. [DOI] [PubMed] [Google Scholar]
- 16.Xie L., Hoffmann A.D., Burnicka-Turek O., Friedland-Little J.M., Zhang K., Moskowitz I.P. Tbx5-hedgehog molecular networks are essential in the second heart field for atrial septation. Dev. Cell. 2012;23:280–291. doi: 10.1016/j.devcel.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snarr B.S., Wirrig E.E., Phelps A.L., Trusk T.C., Wessels A. A spatiotemporal evaluation of the contribution of the dorsal mesenchymal protrusion to cardiac development. Dev. Dyn. 2007;236:1287–1294. doi: 10.1002/dvdy.21074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ammirabile G., Tessari A., Pignataro V., Szumska D., Sutera Sardo F., Benes J., Jr., Balistreri M., Bhattacharya S., Sedmera D., Campione M. Pitx2 confers left morphological, molecular, and functional identity to the sinus venosus myocardium. Cardiovasc. Res. 2012;93:291–301. doi: 10.1093/cvr/cvr314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun C., Yu D., Ye W., Liu C., Gu S., Sinsheimer N.R., Song Z., Li X., Chen C., Song Y., et al. The short stature homeobox 2 (Shox2)-bone morphogenetic protein (BMP) pathway regulates dorsal mesenchymal protrusion development and its temporary function as a pacemaker during cardiogenesis. J. Biol. Chem. 2015;290:2007–2023. doi: 10.1074/jbc.M114.619007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen B.C., Boukens B.J., Wang T., Moorman A.F., Christoffels V.M. Evolution of the Sinus Venosus from Fish to Human. J. Cardiovasc. Dev. Dis. 2014;1:14–28. doi: 10.3390/jcdd1010014. [DOI] [Google Scholar]
- 21.Mommersteeg M.T., Christoffels V.M., Anderson R.H., Moorman A.F. Atrial fibrillation: A developmental point of view. Heart Rhythm. 2009;6:1818–1824. doi: 10.1016/j.hrthm.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 22.Mommersteeg M.T., Dominguez J.N., Wiese C., Norden J., de Gier-de Vries C., Burch J.B., Kispert A., Brown N.A., Moorman A.F., Christoffels V.M. The sinus venosus progenitors separate and diversify from the first and second heart fields early in development. Cardiovasc. Res. 2010;87:92–101. doi: 10.1093/cvr/cvq033. [DOI] [PubMed] [Google Scholar]
- 23.Aanhaanen W.T., Mommersteeg M.T., Norden J., Wakker V., de Gier-de Vries C., Anderson R.H., Kispert A., Moorman A.F., Christoffels V.M. Developmental origin, growth, and three-dimensional architecture of the atrioventricular conduction axis of the mouse heart. Circ. Res. 2010;107:728–736. doi: 10.1161/CIRCRESAHA.110.222992. [DOI] [PubMed] [Google Scholar]
- 24.Liang X., Wang G., Lin L., Lowe J., Zhang Q., Bu L., Chen Y., Chen J., Sun Y., Evans S.M. HCN4 dynamically marks the first heart field and conduction system precursors. Circ. Res. 2013;113:399–407. doi: 10.1161/CIRCRESAHA.113.301588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gittenberger-de Groot A.C. The development of the pulmonary vein revisited. Int. J. Cardiol. 2011;147:463–464. doi: 10.1016/j.ijcard.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Douglas Y.L., Jongbloed M.R., Deruiter M.C., Gittenberger-de Groot A.C. Normal and abnormal development of pulmonary veins: State of the art and correlation with clinical entities. Int. J. Cardiol. 2011;147:13–24. doi: 10.1016/j.ijcard.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Moorman A.F.M., Anderson R.H. Development of the pulmonary vein. Int. J. Cardiol. 2011;147:182. doi: 10.1016/j.ijcard.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 28.Hoogaars W.M., Tessari A., Moorman A.F., de Boer P.A., Hagoort J., Soufan A.T., Campione M., Christoffels V.M. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc. Res. 2004;62:489–499. doi: 10.1016/j.cardiores.2004.01.030. [DOI] [PubMed] [Google Scholar]
- 29.Bakker M.L., Boukens B.J., Mommersteeg M.T., Brons J.F., Wakker V., Moorman A.F., Christoffels V.M. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ. Res. 2008;102:1340–1349. doi: 10.1161/CIRCRESAHA.107.169565. [DOI] [PubMed] [Google Scholar]
- 30.Hoogaars W.M., Engel A., Brons J.F., Verkerk A.O., de Lange F.J., Wong L.Y., Bakker M.L., Clout D.E., Wakker V., Barnett P., et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21:1098–1112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frank D.U., Carter K.L., Thomas K.R., Burr R.M., Bakker M.L., Coetzee W.A., Tristani-Firouzi M., Bamshad M.J., Christoffels V.M., Moon A.M. Lethal arrhythmias in Tbx3-deficient mice reveal extreme dosage sensitivity of cardiac conduction system function and homeostasis. Proc. Natl. Acad. Sci. USA. 2012;109:E154–E163. doi: 10.1073/pnas.1115165109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu M., Peng S., Yang J., Tu Z., Cai X., Cai C.L., Wang Z., Zhao Y. Baf250a orchestrates an epigenetic pathway to repress the Nkx2.5-directed contractile cardiomyocyte program in the sinoatrial node. Cell Res. 2014;24:1201–1213. doi: 10.1038/cr.2014.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arnolds D.E., Liu F., Fahrenbach J.P., Kim G.H., Schillinger K.J., Smemo S., McNally E.M., Nobrega M.A., Patel V.V., Moskowitz I.P. TBX5 drives Scn5a expression to regulate cardiac conduction system function. J. Clin. Investig. 2012;122:2509–2518. doi: 10.1172/JCI62617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mommersteeg M.T., Hoogaars W.M., Prall O.W., de Gier-de Vries C., Wiese C., Clout D.E., Papaioannou V.E., Brown N.A., Harvey R.P., Moorman A.F., et al. Molecular pathway for the localized formation of the sinoatrial node. Circ. Res. 2007;100:354–362. doi: 10.1161/01.RES.0000258019.74591.b3. [DOI] [PubMed] [Google Scholar]
- 35.Weinberger F., Mehrkens D., Friedrich F.W., Stubbendorff M., Hua X., Muller J.C., Schrepfer S., Evans S.M., Carrier L., Eschenhagen T. Localization of Islet-1-positive cells in the healthy and infarcted adult murine heart. Circ. Res. 2012;110:1303–1310. doi: 10.1161/CIRCRESAHA.111.259630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tessadori F., van Weerd J.H., Burkhard S.B., Verkerk A.O., de Pater E., Boukens B.J., Vink A., Christoffels V.M., Bakkers J. Identification and functional characterization of cardiac pacemaker cells in zebrafish. PLoS ONE. 2012;7:e47644. doi: 10.1371/journal.pone.0047644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kapoor N., Liang W., Marban E., Cho H.C. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat. Biotechnol. 2013;31:54–62. doi: 10.1038/nbt.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munshi N.V., Olson E.N. Translational medicine. Improving cardiac rhythm with a biological pacemaker. Science. 2014;345:268–269. doi: 10.1126/science.1257976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christoffels V.M., Grieskamp T., Norden J., Mommersteeg M.T., Rudat C., Kispert A. Tbx18 and the fate of epicardial progenitors. Nature. 2009;458:E8–E9. doi: 10.1038/nature07916. [DOI] [PubMed] [Google Scholar]
- 40.Blaschke R.J., Hahurij N.D., Kuijper S., Just S., Wisse L.J., Deissler K., Maxelon T., Anastassiadis K., Spitzer J., Hardt S.E., et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- 41.Espinoza-Lewis R.A., Yu L., He F., Liu H., Tang R., Shi J., Sun X., Martin J.F., Wang D., Yang J., Chen Y. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev. Biol. 2009;327:376–385. doi: 10.1016/j.ydbio.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye W., Chen Y. Revealing the default pacemakers in the venous pole. 2016. to be submitted for publication.
- 43.Puskaric S., Schmitteckert S., Mori A.D., Glaser A., Schneider K.U., Bruneau B.G., Blaschke R.J., Steinbeisser H., Rappold G. Shox2 mediates Tbx5 activity by regulating Bmp4 in the pacemaker region of the developing heart. Hum. Mol. Genet. 2010;19:4625–4633. doi: 10.1093/hmg/ddq393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye W., Chen Y. (Department of Cell and Molecular Biology, Tulane University, New Orleans, LA, USA). 2015. Unpublished Observation.
- 45.Mommersteeg M.T., Brown N.A., Prall O.W., de Gier-de Vries C., Harvey R.P., Moorman A.F., Christoffels V.M. Pitx2c and Nkx2-5 are required for the formation and identity of the pulmonary myocardium. Circ. Res. 2007;101:902–909. doi: 10.1161/CIRCRESAHA.107.161182. [DOI] [PubMed] [Google Scholar]
- 46.Prall O.W., Menon M.K., Solloway M.J., Watanabe Y., Zaffran S., Bajolle F., Biben C., McBride J.J., Robertson B.R., Chaulet H., et al. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128:947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakashima Y., Yanez D.A., Touma M., Nakano H., Jaroszewicz A., Jordan M.C., Pellegrini M., Roos K.P., Nakano A. Nkx2-5 suppresses the proliferation of atrial myocytes and conduction system. Circ. Res. 2014;114:1103–1113. doi: 10.1161/CIRCRESAHA.114.303219. [DOI] [PubMed] [Google Scholar]
- 48.Ai D., Liu W., Ma L., Dong F., Lu M.F., Wang D., Verzi M.P., Cai C., Gage P.J., Evans S., et al. Pitx2 regulates cardiac left-right asymmetry by patterning second cardiac lineage-derived myocardium. Dev. Biol. 2006;296:437–449. doi: 10.1016/j.ydbio.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Franco D., Campione M. The role of Pitx2 during cardiac development. Linking left-right signaling and congenital heart diseases. Trends Cardiovasc. Med. 2003;13:157–163. doi: 10.1016/S1050-1738(03)00039-2. [DOI] [PubMed] [Google Scholar]
- 50.St Amand T.R., Ra J., Zhang Y., Hu Y., Baber S.I., Qiu M., Chen Y. Cloning and expression pattern of chicken Pitx2: A new component in the SHH signaling pathway controlling embryonic heart looping. Biochem. Biophys. Res. Commun. 1998;247:100–105. doi: 10.1006/bbrc.1998.8740. [DOI] [PubMed] [Google Scholar]
- 51.Campione M., Ros M.A., Icardo J.M., Piedra E., Christoffels V.M., Schweickert A., Blum M., Franco D., Moorman A.F. Pitx2 expression defines a left cardiac lineage of cells: Evidence for atrial and ventricular molecular isomerism in the iv/iv mice. Dev. Biol. 2001;231:252–264. doi: 10.1006/dbio.2000.0133. [DOI] [PubMed] [Google Scholar]
- 52.Franco D., Campione M., Kelly R., Zammit P.S., Buckingham M., Lamers W.H., Moorman A.F. Multiple transcriptional domains, with distinct left and right components, in the atrial chambers of the developing heart. Circ. Res. 2000;87:984–991. doi: 10.1161/01.RES.87.11.984. [DOI] [PubMed] [Google Scholar]
- 53.Wang J., Bai Y., Li N., Ye W., Zhang M., Stephanie B.G., Tao Y., Chen Y., Wehrens X., Martin J. Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. Proc. Natl. Acad. Sci. USA. 2014;111:9181–9186. doi: 10.1073/pnas.1405411111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J., Klysik E., Sood S., Johnson R.L., Wehrens X.H., Martin J.F. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc. Natl. Acad. Sci. USA. 2010;107:9753–9758. doi: 10.1073/pnas.0912585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bakker M.L., Christoffels V.M., Moorman A.F. The cardiac pacemaker and conduction system develops from embryonic myocardium that retains its primitive phenotype. J. Cardiovasc. Pharmacol. 2010;56:6–15. doi: 10.1097/FJC.0b013e3181e775d3. [DOI] [PubMed] [Google Scholar]
- 56.Butters T.D., Aslanidi O.V., Inada S., Boyett M.R., Hancox J.C., Lei M., Zhang H. Mechanistic links between Na+ channel (SCN5A) mutations and impaired cardiac pacemaking in sick sinus syndrome. Circ. Res. 2010;107:126–137. doi: 10.1161/CIRCRESAHA.110.219949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He A., Kong S.W., Ma Q., Pu W.T. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc. Natl. Acad. Sci. USA. 2011;108:5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van den Boogaard M., Wong L.Y., Tessadori F., Bakker M.L., Dreizehnter L.K., Wakker V., Bezzina C.R., Hoen P.A., Bakkers J., Barnett P., et al. Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer. J. Clin. Investig. 2012;122:2519–2530. doi: 10.1172/JCI62613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lei M., Zhang H., Grace A.A., Huang C.L. SCN5A and sinoatrial node pacemaker function. Cardiovasc. Res. 2007;74:356–365. doi: 10.1016/j.cardiores.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 60.Attanasio C., Nord A.S., Zhu Y., Blow M.J., Li Z., Liberton D.K., Morrison H., Plajzer-Frick I., Holt A., Hosseini R., et al. Fine tuning of craniofacial morphology by distant-acting enhancers. Science. 2013;342 doi: 10.1126/science.1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Visel A., Minovitsky S., Dubchak I., Pennacchio L.A. VISTA Enhancer Browser—A database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ye W., Chen Y. Shox2 functions as a patterning factor in limb development. 2016. to be submitted for publication.
- 63.Yue F., Cheng Y., Breschi A., Vierstra J., Wu W., Ryba T., Sandstrom R., Ma Z., Davis C., Pope B.D., et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang A., Yue F., Li Y., Xie R., Harper T., Patel N.A., Muth K., Palmer J., Qiu Y., Wang J., et al. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell. 2015;16:386–399. doi: 10.1016/j.stem.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pott S., Lieb J.D. Single-cell ATAC-seq: Strength in numbers. Genome Biol. 2015;16:172. doi: 10.1186/s13059-015-0737-7. [DOI] [PMC free article] [PubMed] [Google Scholar]