Abstract
Schizophrenia research has undergone a recent transformation. By leveraging large sample sizes, genome-wide association studies of common genetic variants have approximately tripled the number of candidate genetic loci. Rare variant studies have identified copy number variants that are schizophrenia risk loci. Among these, the 3q29 microdeletion is now known to be the single largest schizophrenia risk factor. Next-generation sequencing studies are increasingly used for rare variant association testing, and have already facilitated identification of large effect alleles. Collectively, recent findings implicate voltage-gated calcium channel and cytoskeletal pathways in the pathogenesis of schizophrenia. Taken together, these results suggest the possibility of imminent breakthroughs in the molecular understanding of schizophrenia.
Keywords: Schizophrenia Genetics, Psychiatric Genetics, copy number variation, GWAS
Introduction
Schizophrenia (SZ) is a severe psychiatric disorder with a prevalence of approximately 0.5–1% [1]. Symptoms of SZ are classified into positive and negative categories. Positive symptoms include delusions, hallucinations, disorganized speech, and disorganized or catatonic behavior, while alogia (lack of speech), avolition (lowered motivation), and emotional blunting encompass negative symptoms [2]. Schizophrenia is diagnosed based on the observation of at least one core symptom (delusions, hallucinations, or disorganized speech), and at least one additional symptom (a second core symptom, grossly disorganized behavior, or negative symptoms) over a 6-month period in which the individual is disturbed for the majority of a 1-month block. Males have typical onset in their early 20s and females in their late 20s [3].
Established SZ risk factors include paternal age [4], maternal parity [5], obstetric complications [6] including low birth weight for gestational age [5, 6], season of birth [7–10], cannabis use [11], infection by Taxoplasma gondii [12], and urban birth [13]. Evidence of a genetic contribution to risk has accumulated since the 1930's [14]. Initial efforts to establish the relative genetic contribution relied on adoption and fostering study designs, which analyzed phenotypic concordance of parents with offspring that are reared apart. In 1966, Heston reported that children born to affected mothers, when adopted and fostered by different parents, were at increased risk of schizophrenia [15]. Further evidence of a genetic contribution to SZ risk was provided by Kety et al's elegant adoption studies in 1968 and 1976 [16, 17]. Data from twin studies, which analyze the phenotypic concordance between monozygotic and dizygotic twins, suggested that genetic factors account for approximately 80% of the total variance (additive genetic variance: a2= 0.82, C.I=0.71–0.90) [18, 19] although estimates have varied widely [19]. For instance, a 2009 analysis of 9,009,202 individuals across approximately 2 million families concluded that the variance due to additive genetic effects was approximately 64% (C.I. 61.5% – 72.2%) [20]. Thus, it was revealed that genetic factors play a significant role in SZ susceptibility. Furthermore, these findings suggested that genetic studies could help obtain a greater mechanistic understanding of SZ.
While family and twin studies led to the proposition that schizophrenia was under the influence of a “major gene” [14, 21, 22], observations of departures from simple transmission models led to the consideration of polygenic inheritance models [16, 23]. Initial attempts to map SZ loci using linkage analysis had mixed results, and while there were notable successes, including the identification of DISC1 [24] and NRG1 [25], many linkage peaks failed to replicate [26]. These findings led to the conclusion that the genetic architecture of SZ is incredibly heterogeneous. Thus, in order for gene discovery to be a viable strategy for understanding SZ, both larger sample sizes and alternative experimental strategies were needed.
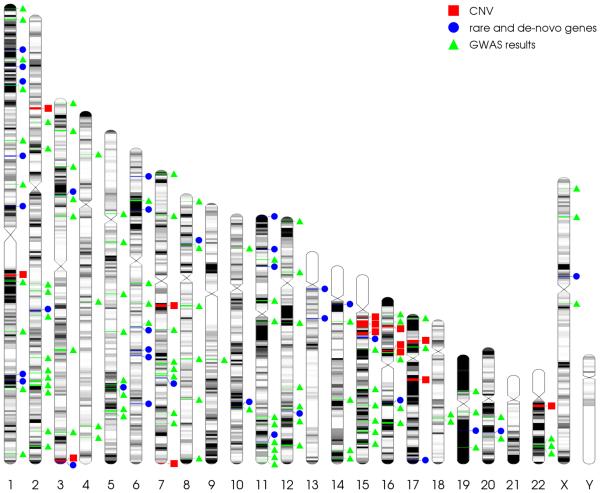
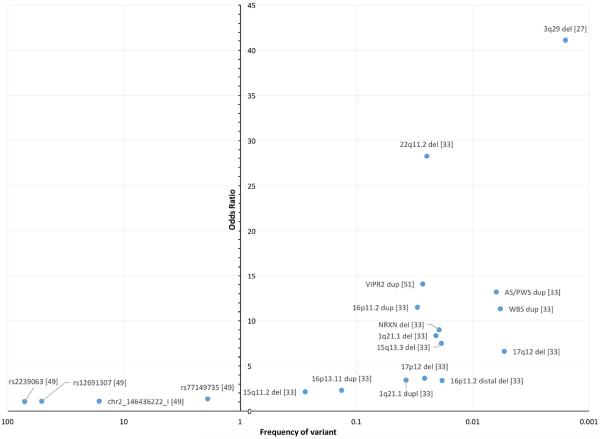
In the last several years, researchers have combined larger sample sizes with methods including whole-exome sequencing (in the analysis of rare variants), copy number variant analysis strategies, and genome wide association studies (GWAS). This work has led to unprecedented success in expanding the understanding of SZ genetic risk factors, and for the first time in the history of SZ genetics, there are now genetic variants that are considered true risk loci. These established alleles include more than 128 common single nucleotide polymorphisms (SNPs), and at least fifteen rare copy number variants (CNVs) (Figure 1). Together, recent studies illuminate several plausible genetic pathways, and support a polygenic model of SZ, in which genetic risk is largely due to either the presence of a very rare, large effect copy number variant (with odds ratios as high as of 41 as seen in the case of the 3q29 deletion [27]), or by the coincident inheritance of many small effect alleles (Figure 2). This review will focus on this very recent history of schizophrenia genetics, including new findings and the biological hypotheses that these findings have inspired.
Figure 1. Locations of SZ-associated alleles.
Chromosomal locations of SZ-associated variants: copy number variants [33, 51] (red squares), recurrent de-novo mutations and rare variants identified by NGS [50, 85] (blue circles), and GWAS loci [49] (green triangles).
Figure 2. A comparison of significant allele odds ratios and population frequencies.
Shown is a comparison of odds ratio (y-axis) and population frequency (log scale; x-axis) for selected GWAS loci (left side of center y-axis) and CNV (right side of center y-axis) associated with SZ. GWAS SNPs from [49] were ranked by odds ratio, and a representative SNP from each quartile was selected for display here.
Copy number variants
In the past decade the success of research focused on the genetics of schizophrenia has advanced remarkably, providing evidence for the association of copy number variants (CNVs) in risk for disease [28, 29]. CNVs, which are defined as gains or losses of genomic material of at least 1 kilobase (kb) in size, can encompass a single exon of a gene, an entire gene, or even multiple genes. Very large CNVs are typically rare in the genome [30]. When comparing SZ cases and unaffected controls, it has been repeatedly demonstrated that case samples have an increased genomic burden of duplications and deletions sized 100kb and larger [31, 32] and this enrichment increases as CNV size increases. Large studies combining case/control cohorts have identified several recurrent loci that are associated with SZ. Replication of the findings across several populations adds credence to the validity of these data and strongly suggests a functional role for these CNVs in the pathophysiology of schizophrenia. There are now at least 15 distinct genomic regions where dosage changes of varying lengths (120kb to 9Mb) are associated with risk for SZ; the increased odds for disease range from 2 to > 40 [27, 33] (Figure 2). Many of these same CNVs have also been implicated in risk for autism, suggesting shared neurological pathways between the psychiatric disorders [34, 35]. In fact, Walsh et al report an enrichment of genes involved in brain development within SZ-associated CNV intervals [32]. Other work has identified abnormalities in signaling complexes, brain structure, and conductivity among carriers of CNV risk loci [36–38]. Further research into the biological pathways disrupted by these CNVs will allow us to better understand the mechanisms responsible for the progression of neuropsychiatric disease such as SZ.
1q21 Deletion and Duplication
The 1.35Mb 1q21.1 deletion was among the first CNV to be associated with SZ, with an odds ratio of 14.83 [39]. It was identified among a combined cohort of 4,718 cases and 41,201 controls from Iceland, United Kingdom, Germany, Finland, Italy, Denmark, Norway, The Netherlands and China. The association at 1q21.1 was supported by a second study published by the International Schizophrenia Consortium (ISC) among a sample comprised of similar ancestry [31]. The reciprocal duplication has also been associated with SZ (odds ratio (OR) = 3.7–4.5) [40], suggesting that any dosage imbalance of these genes confers risk for SZ. Among the many genes within this copy number variable region (approx. 8), the gene for connexin 50, GJA8, has been reported to associate with SZ [41]. Gene ontology and network analysis also supports this association [42].
2p16.3/NRXN1 Deletion
Deletions at 2p16.3 (variable sizes of <1 Mb) that encompass a partial or full-length region of the neurodevelopment and synaptic adhesion gene NXRN1 are associated with SZ [43, 44]. In particular, deletions that span exons of NRXN1 are thought to be responsible for the increase in risk (OR= 8.97, p=0.0027) [45]. The deletion breakpoints are highly variable, and have enabled further investigation into the specific gene regions associated with SZ outcome. Clinical outcome appears to be related to the deletion of specific exons in NRXN1, which have been observed among SZ cases across various studies [46].
3q29 Deletion
The 3q29 deletion (0.84–1.6 kb) has one of the largest effect sizes of any SZ risk factor [27, 47]. This 1.6 Mb CNV contains 22 protein-coding genes, among which PAK2, DLG1 and FBXO45 have been proposed as candidate schizophrenia susceptibility loci [27]. The initial association was detected in 2010, with a 16.98 odds ratio [47]. This finding has subsequently been replicated in two independent studies [40, 48]. As additional cases are ascertained, the strength of association continues to be revised upward. The most recent meta-analysis of 25,904 SZ cases and 62,871 controls found an odds ratio of 41.1 (p= 5.8 × 10−8, 95% C.I 5.6–1953.6) [27].
It is yet to be determined which genes in the 3q29 interval are causative. The largest GWAS to date did not find individually associated 3q29 single nucleotide variants [49]. In order to detect association with such infrequent alleles it is useful to consider mutation groups, rather than individual nucleotide associations, and the first study that used this strategy found that 3q29 genes as a group represented the only significant CNV gene set [50]. Although these mutations seemed to be concentrated in the DLG1 gene, with 5 out of 2,536 cases carrying DLG1 variants compared to none among 2,543 controls, this result was not significant after multiple hypothesis correction. Implicating individual genes in the 3q29 interval will require larger samples sizes. In order to understand the mechanisms by which the 3q29 deletion increases SZ risk, biological studies are also necessary. Such investigations are already underway, facilitated by the use of an online registry which has ascertained over 58 carriers (3q29deletion.org).
7q36.3/VIPR2 Duplication
Variably sized microduplications at 7q36.3, spanning a region of 362 kb located upstream and across the coding region of the vasoactive intestinal peptide receptor gene (VIPR2), are associated with SZ [51]. Many of these duplications result in an increase in both VIPR2 transcript and intracellular cAMP within lymphocytes harboring the 7q36.3 variant. VIPR2 encodes for the VPAC2 protein, which is a G-protein coupled receptor that, when activated by its receptor ligand, initiates neuronal signaling cascades [52]. This finding, originally identified using a study cohort of Caucasian SZ cases (OR= 14.1), was replicated among a study cohort of Chinese cases and controls (OR= infinity, 95% CI = 1.327–infinity) [53]. Levinson et al also replicated the association, specifically for the VIPR2 exonic duplications (OR=4.0, p=0.002) [40]. Despite the strong evidence for 7q36.3 duplications in increased susceptibility for SZ, data from a large case/control provided by Rees et al do not support the original findings [33]. Unlike many of the other CNVs associated with SZ, the 7q36.3 duplications are highly variable in breakpoints and consequently, size. Thus replication may depend on which specific duplications are used in association tests.
7q11.23 Duplication
Mulle et al were the first to report an association between a 7q11.23 duplication and SZ [54]. A meta-analysis of 14,387 SZ cases and 28,139 control subjects resulted in a Mantel-Haenszel corrected OR of 10.78. This duplication has also been associated with autism [55, 56]. The reciprocal 1.4–1.5 Mb deletion causes Williams–Beuren Syndrome (WBS) which has been clinically well characterized and results in symptoms that include facial dysmorphisms, short stature, cardiac defects, and cognitive impairments [57]. Individuals with WBS are described as having a “cocktail party personality,” where they are highly social and their verbal abilities are preserved despite cognitive impairments and developmental delay. This highly social phenotype is contrasted to the social withdrawal seen in ASD and SZ [54]. This recent finding awaits replication in another large cohort.
15q11.2 and 15q13.3 Deletions and 15q11-q13 Duplication
Deletions at 15q11.2 (470 kb; OR= 2.73) and 15q13.3 (1.58 Mb; OR= 11.54), like the 1q21.1 deletion, were among the first CNVs identified as SZ risk factors in 2008 [31, 39]. Both these findings have since been replicated and are now well-established SZ susceptibility loci [33, 40, 58–60]. An atypical 129kb duplication at 15q11.2, containing a single gene (UBE3A) involved in synapse development has also been identified in a single SZ case [61, 62].
A maternally derived duplication (> 5 Mb) at 15q11-q13, associated with Prader-Willi syndrome, has been reported with nominal significance (p=0.01; OR = 7.3) in several schizophrenic and schizoaffective individuals [63]. More SZ cases (N=2) harboring the 15q11-q13 duplication were reported in a second study, although no measure of statistical significance was reported [64]. An odds ratio of 13.2 has been predicted for this CNV duplication based on combined data from approximately 15,000 cases [33].
16p11.2 and16p12.1 Deletion, 16p13.1 Deletion and Duplication
SZ associated CNVs along three regions of chromosome 16 have been identified: 16p11.2, 16p12.1, and 16p13.1. Microduplications (but not deletions) at 16p11.2, spanning a region of 600kb and containing 28 genes (29.56–20.11 Mb, hg19) associate with SZ among several thousand cases and controls (OR = 8.4) [65]. At this exact locus, both deletions and duplications are also associated with ASD [66], highlighting the overlapping loci associated with SZ and ASD also observed for at least six other CNVs [34].
A different deletion within the 16p11.2 chromosomal region (28.73 – 28.95 Mb, hg19), located distal to the previously described CNV, has also been identified in two independent cohorts of Ashkenazi Jewish and Bulgarian descent, and has been replicated in a large combined cohort from the US, Europe and Japan (all samples, OR = 6.25) [67]. Rees et al also report a significant finding for this CNV in their large, combined cohort revealing an odds ratio of 3.39 [33].
Among the newest of the SZ associated CNVs, a 480 kb deletion at 16p12.1 was identified in 2014 in a study cohort of approximately 7,000 total SZ cases [68]. This finding was replicated using a separate cohort of nearly 15,000 cases (reported in the same study). This deletion, comprised of 7 genes, is associated with a 2.72-increased risk of developing SZ. Although the data are convincing, this is the only study to date that has reported this finding.
Deletions and duplications within a genomic region at 16p13.1, which had previously been associated with autism and mental retardation [69, 70], were examined for association with SZ among 4,345 affected individuals and 35,079 controls from eight European populations [71]. Several distinct copy number variants exist within this region, and each was tested for association with SZ. Carriers of duplications located in a 1.5 Mb region of chromosome 16 (approximate region = chr16: 15.1–16.6 Mb, hg19) strongly associate with risk for SZ with an OR of 7.27 [71]. This region contains the genes NTAN1 (involved in memory) [72] and NDE1 (involved in neurogenesis) [73]. Notably, for the latter, a rare single nucleotide variant (chr16: 15785118) associates with SZ (p=0.039), strengthening a role for NDE1 in the development of the disease [74]. Support for the association of a 16p13.1 CNV duplication and SZ is provided by two other studies [58, 75]. Most recently, Rees et al report an association with an odds ratio of 2.3 among a total of 12,029 cases and 69,289 [33].
17p12 Deletion, 17q12 Deletion and Duplication
The genomic region 17p12 was first identified in a linkage study of families with SZ affected relatives, but only provided results suggestive of a relationship with SZ [76]. Additional evidence was provided several years later by Kirov et al, where the data showed a 1.3 Mb deletion among several SZ cases; one of which had a parent and three siblings with SZ. This CNV was found to be statistically significant, with an odds ratio of 10, in 5,300 cases and 39,000 controls across two independent cohorts [58]. Another chromosome 17 deletion at 17q12, which is associated with intellectual disability and autism, was observed among schizophrenic study participants from the GAIN and the SGENE Consortium. This 1.4 deletion was not observed among 52,448 healthy controls and has a predicted OR of 4.49 [77]. Rees et al support both deletion findings [33]. A recent study by Szatkiewicz et al also found evidence for an association between SZ and a duplication at 17q12 (OR=4.16, P=0.018) [48].
22q11.2 Deletion
The 22q11.2 deletion, which is an established cause of DiGeorge/VCFS syndrome, was known to be associated with SZ as early as 1994 [78]. The deletion is 3Mb (~65% of carriers) or 1.6 Mb (~35% of carriers) and has one of the largest effect sizes for schizophrenia, with an odds ratio of 28 or greater [33]. The frequency of the deletion in the general population is estimated to be 1 in 4,000, but in SZ it is 1 in 100 [79]. Intriguingly, recent evidence suggests that a duplication within the same general region (22q11.2) provides protection against the development of schizophrenia [80]. It has also been noted that among 22q11.2 deletion carriers, cognitive decline in childhood may signal the beginning of the prodromal period [81].
Rare variant and genome wide association studies
While copy number variant analyses have provided tremendous insight into the genetic architecture of schizophrenia, the role of other classes of genetic variation in SZ risk were also studied. In recent years, two strategies have been used to substantially increase the understanding of the impact that single nucleotide variants have on SZ risk. These strategies differ fundamentally in their approaches to detecting association. Rare variant methods, like Sequence Kernel Association Test (SKAT) [82], associate groups of variants at a genetic locus with a phenotypic outcome. The quality of the underlying sequencing annotation data, generated by tools like PLINK and Seqant [83, 84], may increase the effectiveness of rare variant methods. In contrast, GWAS determine the strength of independent associations between common variants and disease, and is most powerful when common variants have large effect sizes. For heterogeneous disorders like schizophrenia, both study designs require thousands of samples to detect robust genetic signals.
Next generation sequencing for rare variants
In 2014, two complimentary whole-exome sequencing studies analyzed rare variants in SZ cases and controls. Fromer et al looked at the distribution of non-synonymous variants in 623 affected trios (SZ & schizoaffective). In single gene analysis, only the gene TAF13 (encoding transcription initiation factor TFIID subunit 13) contained an exome-wide significant recurrence of loss of function mutations (p= 10–16) [85]. A gene set analysis found significant enrichment in the n-methyl d-aspartate receptor (NMDAR) complex, the actin-mediated cytoskeleton protein (ARC) complex, and Fragile X mental retardation protein (FMRP) target groups [85]. In a second exome sequencing study, Purcell et al reported similar findings among 2,536 SZ case and 2,543 controls. No individual genes were significant under burden or SKAT analysis [50]. However, gene set analysis (conducted by summing individual burden scores) yielded significant enrichments of disruptive rare variants in ARC, postsynaptic density protein 95 (DLG4/PSD-95) complex, NMDAR complex, and voltage gated calcium channel (VGCC) groups [50]. The common findings of rare deleterious variants in both the ARC and NMDAR gene sets between these two studies is highly encouraging. However, these results also suggest that future rare variant analysis will need to use even larger sample sizes to identify individually associated genes.
Genome wide association studies
While rare variant analyses have made important contributions, in recent years the bulk of SZ genetic variant discovery has been driven by genome wide association studies. However, the utility of this design has only become apparent as sample sizes have increased. In 2008, the first GWAS of SZ, conducted across 1,471 samples, reported no genome-wide significant loci [86]. This study was powered to detect 80% of true associations, for single nucleotide polymorphisms (SNPs) of at least 10% minor allele frequency (MAF) and 1.83 effect size (genotype relative risk). Their negative results suggested that common variants in SZ did not have particularly large effect sizes, at least relative to copy number variants (Figure 2). A somewhat larger study in the same year identified a putative association with single nucleotide polymorphism (SNP) rs1344706, located in the zinc finger protein gene ZNF804A [87], which has since been independently replicated [88]. The first major results came in 2009, when the International Schizophrenia Consortium (ISC) combined three cohorts to analyze 8,014 cases and 19,080 controls of European descent. The ISC found robust support for a polygenic model of schizophrenia, suggesting that at least 1/3rd of SZ genetic variance could be explained by common variants of individually small impact [89]. They also found suggestive evidence of an association with VGCC genes, which were previously reported in connection with bipolar disorder (CACNA1C, p = 7.7×10−6) [90]. VGCC genes, such as CACNA1C, are now considered leading candidate SZ risk loci [49, 50]. Two companion studies reported 7 SNPs in the MHC (major histocompatability complex), TCF4 (Transcription Factor 4) and NRGN (Neurogranin) [91], as well as common variants located at 6p22.1[92].
These early successes drove an explosion in SZ GWAS sample sizes. In 2011, a GWAS of 21,856 discovery samples, and 29,839 replication samples were utilized to uncover 5 new loci [93]. Among the genome-wide significant loci were CACNA1C, previously identified by the ISC, and MIR137 (microRNA 137), which is a regulator of neuronal development. A 2013 GWAS conducted across a similar number of samples reported 13 new loci, as well as the replication of several others, including CACNA1C, and MIR137 [94]. Finally, in 2014, a tour-de-force GWAS of 36,989 cases and 113,075 controls reported 128 genome-wide significant SNPs in 108 loci, 83 of which were new [49]. Their polygenic risk analysis suggests that these 128 SNPs explain approximately 3.4% of SZ genetic variance [49]. Many more loci may await discovery in GWAS that interrogate larger numbers of samples. However, by now it is clear that the individual effect sizes of these undiscovered loci are unlikely to approach those seen in copy number analysis.
Biological pathways
Findings from genome wide association studies and exome sequencing studies are converging on genes in the ARC signaling complex, NMDAR complex, VGCC, and FMRP target pathways. Considered a potential “master regulator of synaptic plasticity” [95], ARC is a highly-conserved gene that links neuronal stimulation to synaptic remodeling. ARC is transcribed within neuronal soma in response to synaptic activation [96], and the resulting mRNA is rapidly trafficked to activated dendritic spines, where it undergoes local translation [97]. ARC is critical in the maintenance, but not induction, of long-term potentiation (LTP), which is the persistent strengthening of synapses, long-term depression (LTD), which is the persistent weakening of synapses, and long-term (but not short term) memory consolidation [98]. The stimulus-dependent elevation of ARC expression is in part mediated by the activity of NMDAR and VGCC [99]. At least 28 genes are known to interact with ARC, and form the basis of the “ARC complex” groups tested in the recent exome-sequencing experiments of Fromer et al and Purcell et al [36, 50, 85].
NMDAR are voltage-dependent ligand-binding receptors that mediate excitatory signaling, which plays a crucial role in LTP [100], LTD [101], and both short and long term memory consolidation [102–104]. These receptors allow for the influx of calcium ions in response to binding of glutamate and n-methyl d-aspartate (NMDA), as well as the co-agonist glycine [105]. They are found in a wide range of cell types, including, but not limited to, neurons, astrocytes, and oligodendrocytes [106–109]. In the last 40 years, the hypothesis of NMDAR hypofunction in SZ has gained significant attention. NMDAR hypofunction is seen in the dorsolateral prefrontal cortex and hippocampus of post-mortem brains collected from SZ-diagnosed individuals [110–113]. Unfortunately, drugs targeting NMDAR have shown inconclusive efficacy in clinical trials [114, 115], which may reflect the underlying genetic heterogeneity of the disorder.
Evidence surrounding VGCC involvement in SZ is relatively new: several individual VGCC genes, such as CACNA1C have been replicated in independent GWAS [49, 93]. Furthermore, a VGCC gene set was implicated in a recent whole-exome sequencing experiment [50]. Like NMDAR, VGCC are also critical to cell excitability across a range of cell types, including neurons [116]. They play key roles in LTP and long-term (but not short-term) memory consolidation [104]. Functional studies of rare variants may clarify the role that VGCC plays in SZ occurrence.
Fragile X mental retardation protein (FMRP) is an mRNA-targeting protein, encoded by FMR1 [117]. FMRP impacts LTP and LTD, by regulating translation of a key set of mRNAs. These mRNAs have been identified and make up the gene set of “FMRP targets” [118]. FMRP may also interact directly with some neurologically active proteins. For instance, knockdown experiments have suggested that FMRP may interact with N-type calcium channel Cav2.2 [119] and repress NMDAR subunit translation [120]. Post-mortem ex-vivo studies have reported significantly lower FMRP levels in the cerebella of schizophrenic patients when compared with controls [121]. However, more research is needed to conclusively establish the link.
Overlap with other psychiatric disorders
Targeted sequencing, GWAS, and copy number variant analyses support the possibility that genetic risk factors for schizophrenia overlap with those associated with autism spectrum disorders (ASD) and intellectual disability [122]. Fromer et al, in their 2014 whole exome sequencing experiment, found statistically significant support for concordance with ASD and intellectual disability loss-of-function de-novo gene sets. Genes in the intersection include SCN2A, which encodes a voltage gated sodium channel subunit that is critical in the generation of action potentials (a key step in neuronal signal propagation) and which has been implicated in both ASD and epilepsy [123–126]. Enrichment was also found in NMDAR complex genes, which have been implicated in ASD [127] and ID [128]. In the complimentary case/control whole-exome sequencing study, Purcell et al reported an enrichment of rare mutations in a 738 member FMRP target gene set [50]. Loss of function mutations in FMR1 cause intellectual disability (Fragile X), and are a leading monogenic cause of autism [129]. Furthermore, several FMRP targets overlap with candidate autism risk loci [118]. The VGCC complex gene set was also significantly enriched. Interestingly, both of these rare-variants studies found significant enrichment in ARC complex genes, which interact with NMDAR and VGCC in learning and memory processes, and are implicated in ASD [99, 130].
The largest GWAS of SZ to date reported similar associations. Of note, VGCC genes, including CACNA1C, have now been replicated in several independent GWAS experiments, were enriched in the rare-variant analyses conducted by Purcell et al [49, 93] and represent a leading autism susceptibility pathway [131]. Furthermore, mutations in calcium channel genes have recently been reported in connection with ASD, SZ, attention deficit-hyperactivity disorder (ADD), bipolar disorder (BP), and major depressive disorder (MDD) [132]. Other notable associations include the NMDAR gene, and GRIN2A.
Several SZ-associated copy number variants are also implicated in ASD and ID. Exonic deletions of NRXN1 have been associated with risk for speech delay, mild dysmorphic features, epilepsy and autism spectrum disorders [46]. The 3q29 deletion is associated with mild to moderate intellectual disability and ASD [133]. The 7q11.23 duplication is also associated with ASD [55], while the reciprocal deletion gives rise to Williams-Beuren Syndrome, where the phenotype includes intellectual disability [134]. Several other CNVs have been connected with ASD and/or ID, including deletions in 15q11.2 [135], 15q13.3 [136], 22q11.2 [137], and duplications of 1q21.1 [138]. Future studies, conducted across a larger number of samples, will be needed to delineate the genetic boundaries between SZ, ASD, and intellectual disability.
Future Directions
The estimated heritability of SZ ranges widely, with some estimates as high as 80–88% [18, 19]. At least fifteen rare CNVs, with effect sizes ranging from approximately 2–40, and more than 128 common, small effect-size single nucleotide polymorphisms have been identified as genetic risk factors in schizophrenia (Figure 1). The common variants uncovered by the largest SZ GWAS to date are estimated to explain 3.4% of SZ variance, a fraction of the estimated SZ heritability. Although this may seem like a paucity of the total variance, it is important to note how rapidly the field is advancing: almost all of the common variants now known were discovered in the last four years. This accelerating progress has been driven by the use of larger sample sizes, and suggests that even larger cohorts will lead to a windfall of discovery. Researchers are now well positioned to lead a breakthrough in the understanding of SZ biology, through the combination of genetic variant statistical analysis and functional investigation.
The exciting results discussed here must be translated into biologically meaningful information. This will be accomplished by functional studies of SZ-associated variants, particularly those rare variants with the largest effect sizes. We now have early evidence that many variants affect shared pathways, including voltage gated calcium channel (VGCC), n-methyl d-aspartate receptor complex, activity-regulated cytoskeleton protein (ARC) complex, and FMRP target genes. The study of biological perturbations caused by severe, gene-disrupting mutation may help clarify the means by which SZ presents in the clinic, and in combination with phenotypic characterization of patients carrying specific rare variants, we may be able to transform our understanding of SZ, from a highly heterogeneous disorder, to one that impacts a tractable number of “master systems”. These findings may ultimately be leveraged to offer personalized medicine, refine treatment options, and improve the outcome of individuals with SZ.
Acknowledgements
Thanks to David Cutler for manuscript suggestions. This work was supported by NIH grant MH100917(JGM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Tandon R, Keshavan MS, Nasrallah HA. Schizophrenia, “Just the Facts” What we know in 2008. 2. Epidemiology and etiology, Schizophrenia research. 2008;102:1–18. doi: 10.1016/j.schres.2008.04.011. [DOI] [PubMed] [Google Scholar]
- [2].Tandon R, Gaebel W, Barch DM, Bustillo J, Gur RE, Heckers S, Malaspina D, Owen MJ, Schultz S, Tsuang M, Van Os J, Carpenter W. Definition and description of schizophrenia in the DSM-5. Schizophrenia research. 2013;150:3–10. doi: 10.1016/j.schres.2013.05.028. [DOI] [PubMed] [Google Scholar]
- [3].A. American Psychiatric. A. American Psychiatric. Force DSMT. DOI. 2013. Diagnostic and statistical manual of mental disorders : DSM-5. [Google Scholar]
- [4].Malaspina D, Harlap S, Fennig S, Heiman D, Nahon D, Feldman D, Susser ES. Advancing paternal age and the risk of schizophrenia. Arch Gen Psychiatry. 2001;58:361–367. doi: 10.1001/archpsyc.58.4.361. [DOI] [PubMed] [Google Scholar]
- [5].Hultman CM, Sparen P, Takei N, Murray RM, Cnattingius S. Prenatal and perinatal risk factors for schizophrenia, affective psychosis, and reactive psychosis of early onset: case-control study. BMJ. 1999;318:421–426. doi: 10.1136/bmj.318.7181.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cannon M, Jones PB, Murray RM. Obstetric complications and schizophrenia: historical and meta-analytic review. The American journal of psychiatry. 2002;159:1080–1092. doi: 10.1176/appi.ajp.159.7.1080. [DOI] [PubMed] [Google Scholar]
- [7].Davies G, Welham J, Chant D, Torrey EF, McGrath J. A systematic review and meta-analysis of Northern Hemisphere season of birth studies in schizophrenia. Schizophr Bull. 2003;29:587–593. doi: 10.1093/oxfordjournals.schbul.a007030. [DOI] [PubMed] [Google Scholar]
- [8].Pulver AE, Liang KY, Brown CH, Wolyniec P, McGrath J, Adler L, Tam D, Carpenter WT, Childs B. Risk factors in schizophrenia. Season of birth, gender, and familial risk. Br J Psychiatry. 1992;160:65–71. doi: 10.1192/bjp.160.1.65. [DOI] [PubMed] [Google Scholar]
- [9].Kendell RE, Adams W. Unexplained fluctuations in the risk for schizophrenia by month and year of birth. Br J Psychiatry. 1991;158:758–763. doi: 10.1192/bjp.158.6.758. [DOI] [PubMed] [Google Scholar]
- [10].Torrey EF, Torrey BB, Peterson MR. Seasonality of schizophrenic births in the United States. Arch Gen Psychiatry. 1977;34:1065–1070. doi: 10.1001/archpsyc.1977.01770210079007. [DOI] [PubMed] [Google Scholar]
- [11].Henquet C, Murray R, Linszen D, van Os J. The environment and schizophrenia: the role of cannabis use. Schizophr Bull. 2005;31:608–612. doi: 10.1093/schbul/sbi027. [DOI] [PubMed] [Google Scholar]
- [12].Arias I, Sorlozano A, Villegas E, de Dios Luna J, McKenney K, Cervilla J, Gutierrez B, Gutierrez J. Infectious agents associated with schizophrenia: a meta-analysis. Schizophrenia research. 2012;136:128–136. doi: 10.1016/j.schres.2011.10.026. [DOI] [PubMed] [Google Scholar]
- [13].Krabbendam L, van Os J. Schizophrenia and urbanicity: a major environmental influence--conditional on genetic risk. Schizophr Bull. 2005;31:795–799. doi: 10.1093/schbul/sbi060. [DOI] [PubMed] [Google Scholar]
- [14].Kallmann FJ. DOI. 1938. The genetics of schizophrenia. [Google Scholar]
- [15].Heston LL. Psychiatric disorders in foster home reared children of schizophrenic mothers. Br J Psychiatry. 1966;112:819–825. doi: 10.1192/bjp.112.489.819. [DOI] [PubMed] [Google Scholar]
- [16].Kety SS, Rosenthal D, Wender PH, Schulsinger F. The types and prevalence of mental illness in the biological and adoptive families of adopted schizophrenics. Journal of Psychiatric Research. 1968;6:345–362. [Google Scholar]
- [17].Kety SS, Rosenthal D, Wender PH, Schulsinger F, Jacobsen B. Mental illness in the biological and adoptive families of adopted individuals who have become schizophrenic. Behav Genet. 1976;6:219–225. doi: 10.1007/BF01065721. [DOI] [PubMed] [Google Scholar]
- [18].Cardno AG, Marshall EJ, Coid B, Macdonald AM, Ribchester TR, Davies NJ, Venturi P, Jones LA, Lewis SW, Sham PC, Gottesman II, Farmer AE, McGuffin P, Reveley AM, Murray RM. Heritability estimates for psychotic disorders - The Maudsley Twin psychosis series. Arch Gen Psychiat. 1999;56:162–168. doi: 10.1001/archpsyc.56.2.162. [DOI] [PubMed] [Google Scholar]
- [19].Cannon TD, Kaprio J, Lonnqvist J, Huttunen M, Koskenvuo M. The genetic epidemiology of schizophrenia in a Finnish twin cohort - A population-based modeling study. Arch Gen Psychiat. 1998;55:67–74. doi: 10.1001/archpsyc.55.1.67. [DOI] [PubMed] [Google Scholar]
- [20].Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF, Hultman CM. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kallmann FJ. The genetic theory of schizophrenia: An analysis of 691 schizophrenic twin index families. American Journal of Psychiatry. 1946;103:309–322. doi: 10.1176/ajp.103.3.309. [DOI] [PubMed] [Google Scholar]
- [22].Elston R, Campbell M. Schizophrenia: evidence for the major gene hypothesis. Behavior genetics. 1970;1:3–10. doi: 10.1007/BF01067366. [DOI] [PubMed] [Google Scholar]
- [23].Gottesman II, Shields J. A polygenic theory of schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 1967;58:199. doi: 10.1073/pnas.58.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Blackwood D, Fordyce A, Walker M, Clair DS, Porteous D, Muir W. Schizophrenia and affective disorders—cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. The American Journal of Human Genetics. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, Gunnarsdottir S, Ivarsson O, Chou TT, Hjaltason O, Birgisdottir B, Jonsson H, Gudnadottir VG, Gudmundsdottir E, Bjornsson A, Ingvarsson B, Ingason A, Sigfusson S, Hardardottir H, Harvey RP, Lai D, Zhou M, Brunner D, Mutel V, Gonzalo A, Lemke G, Sainz J, Johannesson G, Andresson T, Gudbjartsson D, Manolescu A, Frigge ML, Gurney ME, Kong A, Gulcher JR, Petursson H, Stefansson K. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. image 45. [DOI] [PubMed] [Google Scholar]
- [27].Mulle JG. DOI. 2015. The 3q29 deletion confers greater than 40-fold increase in risk for schizophrenia, Molecular Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sebat J, Levy DL, McCarthy SE. Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends Genet. 2009;25:528–535. doi: 10.1016/j.tig.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, Krauss RM, Myers RM, Ridker PM, Chasman DI, Mefford H, Ying P, Nickerson DA, Eichler EE. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].C. International Schizophrenia Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- [33].Rees E, Walters JT, Georgieva L, Isles AR, Chambert KD, Richards AL, Mahoney-Davies G, Legge SE, Moran JL, McCarroll SA, O'Donovan MC, Owen MJ, Kirov G. Analysis of copy number variations at 15 schizophrenia-associated loci. Br J Psychiatry. 2014;204:108–114. doi: 10.1192/bjp.bp.113.131052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Crespi BJ, Crofts HJ. Association testing of copy number variants in schizophrenia and autism spectrum disorders. J Neurodev Disord. 2012;4:15. doi: 10.1186/1866-1955-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Guilmatre A, Dubourg C, Mosca AL, Legallic S, Goldenberg A, Drouin-Garraud V, Layet V, Rosier A, Briault S, Bonnet-Brilhault F, Laumonnier F, Odent S, Le Vacon G, Joly-Helas G, David V, Bendavid C, Pinoit JM, Henry C, Impallomeni C, Germano E, Tortorella G, Di Rosa G, Barthelemy C, Andres C, Faivre L, Frebourg T, Saugier Veber P, Campion D. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66:947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, Moran J, Chambert K, Toncheva D, Georgieva L, Grozeva D, Fjodorova M, Wollerton R, Rees E, Nikolov I, van de Lagemaat LN, Bayes A, Fernandez E, Olason PI, Bottcher Y, Komiyama NH, Collins MO, Choudhary J, Stefansson K, Stefansson H, Grant SG, Purcell S, Sklar P, O'Donovan MC, Owen MJ. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Martin AK, Robinson G, Reutens D, Mowry B. Copy number deletion burden is associated with cognitive, structural, and resting-state network differences in patients with schizophrenia. Behav Brain Res. 2014;272:324–334. doi: 10.1016/j.bbr.2014.07.002. [DOI] [PubMed] [Google Scholar]
- [38].Maillard AM, Ruef A, Pizzagalli F, Migliavacca E, Hippolyte L, Adaszewski S, Dukart J, Ferrari C, Conus P, Mannik K, Zazhytska M, Siffredi V, Maeder P, Kutalik Z, Kherif F, Hadjikhani N, Beckmann JS, Reymond A, Draganski B, Jacquemont S, C. p11.2 European The 16p11.2 locus modulates brain structures common to autism, schizophrenia and obesity. Mol Psychiatry. 2015;20:140–147. doi: 10.1038/mp.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Moller HJ, Hartmann A, Shianna KV, Ge D, Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, Di Forti M, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Muhleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, Group, Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nothen MM, Peltonen L, Collier DA, St Clair D, Stefansson K. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Levinson DF, Duan J, Oh S, Wang K, Sanders AR, Shi J, Zhang N, Mowry BJ, Olincy A, Amin F, Cloninger CR, Silverman JM, Buccola NG, Byerley WF, Black DW, Kendler KS, Freedman R, Dudbridge F, Pe'er I, Hakonarson H, Bergen SE, Fanous AH, Holmans PA, Gejman PV. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. The American journal of psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ni X, Valente J, Azevedo MH, Pato MT, Pato CN, Kennedy JL. Connexin 50 gene on human chromosome 1q21 is associated with schizophrenia in matched case–control and family-based studies. Journal of Medical Genetics. 2007;44:532–536. doi: 10.1136/jmg.2006.047944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Luo X, Huang L, Han L, Luo Z, Hu F, Tieu R, Gan L. Systematic prioritization and integrative analysis of copy number variations in schizophrenia reveal key schizophrenia susceptibility genes. Schizophr Bull. 2014;40:1285–1299. doi: 10.1093/schbul/sbu045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kirov G, Gumus D, Chen W, Norton N, Georgieva L, Sari M, O'Donovan MC, Erdogan F, Owen MJ, Ropers HH, Ullmann R. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- [44].Kirov G, Rujescu D, Ingason A, Collier DA, O'Donovan MC, Owen MJ. Neurexin 1 (NRXN1) deletions in schizophrenia. Schizophr Bull. 2009;35:851–854. doi: 10.1093/schbul/sbp079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rujescu D, Ingason A, Cichon S, Pietiläinen OPH, Barnes MR, Toulopoulou T, Picchioni M, Vassos E, Ettinger U, Bramon E, Murray R, Ruggeri M, Tosato S, Bonetto C, Steinberg S, Sigurdsson E, Sigmundsson T, Petursson H, Gylfason A, Olason PI, Hardarsson G, Jonsdottir GA, Gustafsson O, Fossdal R, Giegling I, Möller H-J, Hartmann AM, Hoffmann P, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Djurovic S, Melle I, Andreassen OA, Hansen T, Werge T, Kiemeney LA, Franke B, Veltman J, Buizer-Voskamp JE, G. Investigators. Sabatti C, Ophoff RA, Rietschel M, Nöthen MM, Stefansson K, Peltonen L, Clair DS, Stefansson H, Collier DA. Disruption of the neurexin 1 gene is associated with schizophrenia. Human molecular genetics. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dabell MP, Rosenfeld JA, Bader P, Escobar LF, El-Khechen D, Vallee SE, Dinulos MB, Curry C, Fisher J, Tervo R, Hannibal MC, Siefkas K, Wyatt PR, Hughes L, Smith R, Ellingwood S, Lacassie Y, Stroud T, Farrell SA, Sanchez-Lara PA, Randolph LM, Niyazov D, Stevens CA, Schoonveld C, Skidmore D, MacKay S, Miles JH, Moodley M, Huillet A, Neill NJ, Ellison JW, Ballif BC, Shaffer LG. Investigation of NRXN1 deletions: clinical and molecular characterization. Am J Med Genet A. 2013;161A:717–731. doi: 10.1002/ajmg.a.35780. [DOI] [PubMed] [Google Scholar]
- [47].Mulle JG, Dodd AF, McGrath JA, Wolyniec PS, Mitchell AA, Shetty AC, Sobreira NL, Valle D, Rudd MK, Satten G, Cutler DJ, Pulver AE, Warren ST. Microdeletions of 3q29 confer high risk for schizophrenia. Am J Hum Genet. 2010;87:229–236. doi: 10.1016/j.ajhg.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Szatkiewicz JP, O'Dushlaine C, Chen G, Chambert K, Moran JL, Neale BM, Fromer M, Ruderfer D, Akterin S, Bergen SE, Kahler A, Magnusson PK, Kim Y, Crowley JJ, Rees E, Kirov G, O'Donovan MC, Owen MJ, Walters J, Scolnick E, Sklar P, Purcell S, Hultman CM, McCarroll SA, Sullivan PF. Copy number variation in schizophrenia in Sweden. Mol Psychiatry. 2014;19:762–773. doi: 10.1038/mp.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].C. Schizophrenia Working Group of the Psychiatric Genomics Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, O'Dushlaine C, Chambert K, Bergen SE, Kahler A, Duncan L, Stahl E, Genovese G, Fernandez E, Collins MO, Komiyama NH, Choudhary JS, Magnusson PK, Banks E, Shakir K, Garimella K, Fennell T, DePristo M, Grant SG, Haggarty SJ, Gabriel S, Scolnick EM, Lander ES, Hultman CM, Sullivan PF, McCarroll SA, Sklar P. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Vacic V, McCarthy S, Malhotra D, Murray F, Chou HH, Peoples A, Makarov V, Yoon S, Bhandari A, Corominas R, Iakoucheva LM, Krastoshevsky O, Krause V, Larach-Walters V, Welsh DK, Craig D, Kelsoe JR, Gershon ES, Leal SM, Dell Aquila M, Morris DW, Gill M, Corvin A, Insel PA, McClellan J, King MC, Karayiorgou M, Levy DL, DeLisi LE, Sebat J. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature. 2011;471:499–503. doi: 10.1038/nature09884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Piggins HD. Schizophrenia: Zooming in on a gene. Nature. 2011;471:455–456. doi: 10.1038/471455a. [DOI] [PubMed] [Google Scholar]
- [53].Yuan J, Jin C, Sha W, Zhou Z, Zhang F, Wang M, Wang J, Li J, Feng X, Yu S, Wang J. A competitive PCR assay confirms the association of a copy number variation in the VIPR2 gene with schizophrenia in Han Chinese. Schizophrenia research. 2014;156:66–70. doi: 10.1016/j.schres.2014.04.004. [DOI] [PubMed] [Google Scholar]
- [54].Mulle JG, Pulver AE, McGrath JM, Wolyniec P, Dodd AF, Cutler DJ, Sebat J, Malhotra D, Nestadt G, Conrad DF, Hurles M, Barnes CP, Ikeda M, Iwata N, Levinson DF, Gejman PV, Sanders AR, Duan J, Mitchell AA, Peter I, Sklar P, O'Dushlaine CT, Grozeva D, O'Donovan MC, Owen MJ, Hultman CM, Kähler AK, Sullivan PF, Kirov G, Warren ST. Reciprocal duplication of the Williams-Beuren syndrome deletion on chromosome 7q11.23 is associated with schizophrenia. Biological psychiatry. 2014;75:371–377. doi: 10.1016/j.biopsych.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, Mason CE, Bilguvar K, Celestino-Soper PB, Choi M, Crawford EL, Davis L, Wright NR, Dhodapkar RM, DiCola M, DiLullo NM, Fernandez TV, Fielding-Singh V, Fishman DO, Frahm S, Garagaloyan R, Goh GS, Kammela S, Klei L, Lowe JK, Lund SC, McGrew AD, Meyer KA, Moffat WJ, Murdoch JD, O'Roak BJ, Ober GT, Pottenger RS, Raubeson MJ, Song Y, Wang Q, Yaspan BL, Yu TW, Yurkiewicz IR, Beaudet AL, Cantor RM, Curland M, Grice DE, Gunel M, Lifton RP, Mane SM, Martin DM, Shaw CA, Sheldon M, Tischfield JA, Walsh CA, Morrow EM, Ledbetter DH, Fombonne E, Lord C, Martin CL, Brooks AI, Sutcliffe JS, Cook EH, Jr., Geschwind D, Roeder K, Devlin B, State MW. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Van der Aa N, Rooms L, Vandeweyer G, van den Ende J, Reyniers E, Fichera M, Romano C, Delle Chiaie B, Mortier G, Menten B, Destree A, Maystadt I, Mannik K, Kurg A, Reimand T, McMullan D, Oley C, Brueton L, Bongers EM, van Bon BW, Pfund R, Jacquemont S, Ferrarini A, Martinet D, Schrander-Stumpel C, Stegmann AP, Frints SG, de Vries BB, Ceulemans B, Kooy RF. Fourteen new cases contribute to the characterization of the 7q11.23 microduplication syndrome. Eur J Med Genet. 2009;52:94–100. doi: 10.1016/j.ejmg.2009.02.006. [DOI] [PubMed] [Google Scholar]
- [57].Tassabehji M. Williams-Beuren syndrome: a challenge for genotype-phenotype correlations. Hum Mol Genet. 2003;12(Spec No 2):R229–237. doi: 10.1093/hmg/ddg299. [DOI] [PubMed] [Google Scholar]
- [58].Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, C. International Schizophrenia. C. Wellcome Trust Case Control. Craddock N, Owen MJ, O'Donovan MC. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Grozeva D, Conrad DF, Barnes CP, Hurles M, Owen MJ, O'Donovan MC, Craddock N, Kirov G. Wtccc, Independent estimation of the frequency of rare CNVs in the UK population confirms their role in schizophrenia. Schizophrenia research. 2012;135:1–7. doi: 10.1016/j.schres.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Melhem N, Middleton F, McFadden K, Klei L, Faraone SV, Vinogradov S, Tiobech J, Yano V, Kuartei S, Roeder K, Byerley W, Devlin B, Myles-Worsley M. Copy number variants for schizophrenia and related psychotic disorders in Oceanic Palau: risk and transmission in extended pedigrees. Biol Psychiatry. 2011;70:1115–1121. doi: 10.1016/j.biopsych.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, Flavell SW, Kim TK, Griffith EC, Waldon Z, Maehr R, Ploegh HL, Chowdhury S, Worley PF, Steen J, Greenberg ME. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140:704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Noor A, Dupuis L, Mittal K, Lionel AC, Marshall CR, Scherer SW, Stockley T, Vincent JB, Mendoza-Londono R, Stavropoulos DJ. 15q11.2 Duplication Encompassing Only the UBE3A Gene Is Associated with Developmental Delay and Neuropsychiatric Phenotypes. Human Mutation. 2015;36:689–693. doi: 10.1002/humu.22800. [DOI] [PubMed] [Google Scholar]
- [63].Ingason A, Kirov G, Giegling I, Hansen T, Isles AR, Jakobsen KD, Kristinsson KT, le Roux L, Gustafsson O, Craddock N, Möller H-J, McQuillin A, Muglia P, Cichon S, Rietschel M, Ophoff RA, Djurovic S, Andreassen OA, Pietiläinen OPH, Peltonen L, Dempster E, Collier DA, St. Clair D, Rasmussen HB, Glenthøj BY, Kiemeney LA, Franke B, Tosato S, Bonetto C, Saemundsen E, Hreidarsson SJ, G. Investigators. Nöthen MM, Gurling H, O'Donovan MC, Owen MJ, Sigurdsson E, Petursson H, Stefansson H, Rujescu D, Stefansson K, Werge T. Maternally Derived Microduplications at 15q11-q13: Implication of Imprinted Genes in Psychotic Illness. The American journal of psychiatry. 2011;168:408–417. doi: 10.1176/appi.ajp.2010.09111660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Stewart LR, Hall AL, Kang SH, Shaw CA, Beaudet AL. High frequency of known copy number abnormalities and maternal duplication 15q11-q13 in patients with combined schizophrenia and epilepsy. BMC Med Genet. 2011;12:154. doi: 10.1186/1471-2350-12-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, Perkins DO, Dickel DE, Kusenda M, Krastoshevsky O, Krause V, Kumar RA, Grozeva D, Malhotra D, Walsh T, Zackai EH, Kaplan P, Ganesh J, Krantz ID, Spinner NB, Roccanova P, Bhandari A, Pavon K, Lakshmi B, Leotta A, Kendall J, Lee YH, Vacic V, Gary S, Iakoucheva LM, Crow TJ, Christian SL, Lieberman JA, Stroup TS, Lehtimaki T, Puura K, Haldeman-Englert C, Pearl J, Goodell M, Willour VL, Derosse P, Steele J, Kassem L, Wolff J, Chitkara N, McMahon FJ, Malhotra AK, Potash JB, Schulze TG, Nothen MM, Cichon S, Rietschel M, Leibenluft E, Kustanovich V, Lajonchere CM, Sutcliffe JS, Skuse D, Gill M, Gallagher L, Mendell NR, C. Wellcome Trust Case Control. Craddock N, Owen MJ, O'Donovan MC, Shaikh TH, Susser E, Delisi LE, Sullivan PF, Deutsch CK, Rapoport J, Levy DL, King MC, Sebat J. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MAR, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu B-L, Daly MJ. Association between Microdeletion and Microduplication at 16p11.2 and Autism. New England Journal of Medicine. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- [67].Guha S, Rees E, Darvasi A, Ivanov D, Ikeda M, Bergen SE, Magnusson PK, Cormican P, Morris D, Gill M, Cichon S, Rosenfeld JA, Lee A, Gregersen PK, Kane JM, Malhotra AK, Rietschel M, Nöthen MM, Degenhardt F, Priebe L, Breuer R, Strohmaier J, Ruderfer DM, Moran JL, Chambert KD, Sanders AR, Shi J, C. Molecular Genetics of Schizophrenia. C. Wellcome Trust Case Control. Kendler K, Riley B, O'Neill T, Walsh D, Malhotra D, Corvin A, Purcell S, Sklar P, Iwata N, Hultman CM, Sullivan PF, Sebat J, McCarthy S, Gejman PV, Levinson DF, Owen MJ, O'Donovan MC, Lencz T, Kirov G. A rare deletion at distal 16p11.2 is implicated in schizophrenia. JAMA psychiatry (Chicago, Ill.) 2013;70:253–260. doi: 10.1001/2013.jamapsychiatry.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Rees E, Walters JTR, Chambert KD, O'Dushlaine C, Szatkiewicz J, Richards AL, Georgieva L, Mahoney-Davies G, Legge SE, Moran JL, Genovese G, Levinson D, Morris DW, Cormican P, Kendler KS, O'Neill FA, Riley B, Gill M, Corvin A, C. Wellcome Trust Case Control. Sklar P, Hultman C, Pato C, Pato M, Sullivan PF, Gejman PV, McCarroll SA, O'Donovan MC, Owen MJ, Kirov G. CNV analysis in a large schizophrenia sample implicates deletions at 16p12.1 and SLC1A1 and duplications at 1p36.33 and CGNL1. Human Molecular Genetics. 2014;23:1669–1676. doi: 10.1093/hmg/ddt540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hannes FD, Sharp AJ, Mefford HC, de Ravel T, Ruivenkamp CA, Breuning MH, Fryns JP, Devriendt K, Van Buggenhout G, Vogels A, Stewart H, Hennekam RC, Cooper GM, Regan R, Knight SJL, Eichler EE, Vermeesch JR. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. Journal of Medical Genetics. 2009;46:223–232. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Ullmann R, Turner G, Kirchhoff M, Chen W, Tonge B, Rosenberg C, Field M, Vianna-Morgante AM, Christie L, Krepischi-Santos AC, Banna L, Brereton AV, Hill A, Bisgaard A-M, Müller I, Hultschig C, Erdogan F, Wieczorek G, Ropers HH. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Human Mutation. 2007;28:674–682. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- [71].Ingason A, Rujescu D, Cichon S, Sigurdsson E, Sigmundsson T, Pietilainen OP, Buizer-Voskamp JE, Strengman E, Francks C, Muglia P, Gylfason A, Gustafsson O, Olason PI, Steinberg S, Hansen T, Jakobsen KD, Rasmussen HB, Giegling I, Moller HJ, Hartmann A, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Bramon E, Kiemeney LA, Franke B, Murray R, Vassos E, Toulopoulou T, Muhleisen TW, Tosato S, Ruggeri M, Djurovic S, Andreassen OA, Zhang Z, Werge T, Ophoff RA, Investigators G, Rietschel M, Nothen MM, Petursson H, Stefansson H, Peltonen L, Collier D, Stefansson K, St Clair DM. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol Psychiatry. 2011;16:17–25. doi: 10.1038/mp.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kwon YT, Balogh SA, Davydov IV, Kashina AS, Yoon JK, Xie Y, Gaur A, Hyde L, Denenberg VH, Varshavsky A. Altered activity, social behavior, and spatial memory in mice lacking the NTAN1p amidase and the asparagine branch of the N-end rule pathway. Mol Cell Biol. 2000;20:4135–4148. doi: 10.1128/mcb.20.11.4135-4148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Pei Z, Lang B, Fragoso YD, Shearer KD, Zhao L, McCaffery PJ, Shen S, Ding YQ, McCaig CD, Collinson JM. The expression and roles of Nde1 and Ndel1 in the adult mammalian central nervous system. Neuroscience. 2014;271:119–136. doi: 10.1016/j.neuroscience.2014.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kimura H, Tsuboi D, Wang C, Kushima I, Koide T, Ikeda M, Iwayama Y, Toyota T, Yamamoto N, Kunimoto S, Nakamura Y, Yoshimi A, Banno M, Xing J, Takasaki Y, Yoshida M, Aleksic B, Uno Y, Okada T, Iidaka T, Inada T, Suzuki M, Ujike H, Kunugi H, Kato T, Yoshikawa T, Iwata N, Kaibuchi K, Ozaki N. Identification of Rare, Single-Nucleotide Mutations in NDE1 and Their Contributions to Schizophrenia Susceptibility. Schizophr Bull. 2015;41:744–753. doi: 10.1093/schbul/sbu147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ikeda M, Aleksic B, Kirov G, Kinoshita Y, Yamanouchi Y, Kitajima T, Kawashima K, Okochi T, Kishi T, Zaharieva I, Owen MJ, O'Donovan MC, Ozaki N, Iwata N. Copy number variation in schizophrenia in the Japanese population. Biol Psychiatry. 2010;67:283–286. doi: 10.1016/j.biopsych.2009.08.034. [DOI] [PubMed] [Google Scholar]
- [76].Park N, Juo SH, Cheng R, Liu J, Loth JE, Lilliston B, Nee J, Grunn A, Kanyas K, Lerer B, Endicott J, Gilliam TC, Baron M. Linkage analysis of psychosis in bipolar pedigrees suggests novel putative loci for bipolar disorder and shared susceptibility with schizophrenia. Mol Psychiatry. 2004;9:1091–1099. doi: 10.1038/sj.mp.4001541. [DOI] [PubMed] [Google Scholar]
- [77].Moreno-De-Luca D, Mulle JG, Kaminsky EB, Sanders SJ, Myers SM, Adam MP, Pakula AT, Eisenhauer NJ, Uhas K, Weik L, Guy L, Care ME, Morel CF, Boni C, Salbert BA, Chandrareddy A, Demmer LA, Chow EW, Surti U, Aradhya S, Pickering DL, Golden DM, Sanger WG, Aston E, Brothman AR, Gliem TJ, Thorland EC, Ackley T, Iyer R, Huang S, Barber JC, Crolla JA, Warren ST, Martin CL, Ledbetter DH. Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am J Hum Genet. 2010;87:618–630. doi: 10.1016/j.ajhg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Pulver AE, Karayiorgou M, Wolyniec PS, Lasseter VK, Kasch L, Nestadt G, Antonarakis S, Housman D, Kazazian HH, Meyers D, et al. Sequential strategy to identify a susceptibility gene for schizophrenia: report of potential linkage on chromosome 22q12-q13.1: Part 1. Am J Med Genet. 1994;54:36–43. doi: 10.1002/ajmg.1320540108. [DOI] [PubMed] [Google Scholar]
- [79].Bassett AS, Marshall CR, Lionel AC, Chow EW, Scherer SW. Copy number variations and risk for schizophrenia in 22q11.2 deletion syndrome. Hum Mol Genet. 2008;17:4045–4053. doi: 10.1093/hmg/ddn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Rees E, Kirov G, Sanders A, Walters JT, Chambert KD, Shi J, Szatkiewicz J, O'Dushlaine C, Richards AL, Green EK, Jones I, Davies G, Legge SE, Moran JL, Pato C, Pato M, Genovese G, Levinson D, Duan J, Moy W, Goring HH, Morris D, Cormican P, Kendler KS, O'Neill FA, Riley B, Gill M, Corvin A, Craddock N, Sklar P, Hultman C, Sullivan PF, Gejman PV, McCarroll SA, O'Donovan MC, Owen MJ. Evidence that duplications of 22q11.2 protect against schizophrenia. Mol Psychiatry. 2014;19:37–40. doi: 10.1038/mp.2013.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Vorstman JA, Breetvelt EJ, Duijff SN, Eliez S, Schneider M, Jalbrzikowski M, Armando M, Vicari S, Shashi V, Hooper SR, Chow EW, Fung WL, Butcher NJ, Young DA, McDonald-McGinn DM, Vogels A, van Amelsvoort T, Gothelf D, Weinberger R, Weizman A, Klaassen PW, Koops S, Kates WR, Antshel KM, Simon TJ, Ousley OY, Swillen A, Gur RE, Bearden CE, Kahn RS, Bassett AS, B. International Consortium on, S. Behavior in 22q11.2 Deletion Cognitive decline preceding the onset of psychosis in patients with 22q11.2 deletion syndrome. JAMA Psychiatry. 2015;72:377–385. doi: 10.1001/jamapsychiatry.2014.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Shetty AC, Athri P, Mondal K, Horner VL, Steinberg KM, Patel V, Caspary T, Cutler DJ, Zwick ME. SeqAnt: a web service to rapidly identify and annotate DNA sequence variations. BMC Bioinformatics. 2010;11:471. doi: 10.1186/1471-2105-11-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, Carrera N, Humphreys I, Johnson JS, Roussos P, Barker DD, Banks E, Milanova V, Grant SG, Hannon E, Rose SA, Chambert K, Mahajan M, Scolnick EM, Moran JL, Kirov G, Palotie A, McCarroll SA, Holmans P, Sklar P, Owen MJ, Purcell SM, O'Donovan MC. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Sullivan PF, Lin D, Tzeng JY, van den Oord E, Perkins D, Stroup TS, Wagner M, Lee S, Wright FA, Zou F, Liu W, Downing AM, Lieberman J, Close SL. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13:570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CCA, Howie B, Leung H-T, Hartmann AM, Moller H-J, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG, Williams NM, Giegling I, Iwata N, Ikeda M, Darvasi A, Shifman S, He L, Duan J, Sanders AR, Levinson DF, Gejman PV, Cichon S, Nothen MM, Gill M, Corvin A, Rujescu D, Kirov G, Owen MJ. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- [88].Riley B, Thiselton D, Maher BS, Bigdeli T, Wormley B, McMichael GO, Fanous AH, Vladimirov V, O'Neill FA, Walsh D, Kendler KS. Replication of association between schizophrenia and ZNF804A in the Irish Case-Control Study of Schizophrenia sample. Mol Psychiatry. 2010;15:29–37. doi: 10.1038/mp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].C. International Schizophrenia. Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Ferreira MA, O'Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, Fan J, Kirov G, Perlis RH, Green EK, Smoller JW, Grozeva D, Stone J, Nikolov I, Chambert K, Hamshere ML, Nimgaonkar VL, Moskvina V, Thase ME, Caesar S, Sachs GS, Franklin J, Gordon-Smith K, Ardlie KG, Gabriel SB, Fraser C, Blumenstiel B, Defelice M, Breen G, Gill M, Morris DW, Elkin A, Muir WJ, McGhee KA, Williamson R, MacIntyre DJ, MacLean AW, St CD, Robinson M, Van Beck M, Pereira AC, Kandaswamy R, McQuillin A, Collier DA, Bass NJ, Young AH, Lawrence J, Ferrier IN, Anjorin A, Farmer A, Curtis D, Scolnick EM, McGuffin P, Daly MJ, Corvin AP, Holmans PA, Blackwood DH, Gurling HM, Owen MJ, Purcell SM, Sklar P, Craddock N, C. Wellcome Trust Case Control Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, Werge T, Pietiläinen OPH, Mors O, Mortensen PB, Sigurdsson E, Gustafsson O, Nyegaard M, Tuulio-Henriksson A, Ingason A, Hansen T, Suvisaari J, Lonnqvist J, Paunio T, Børglum AD, Hartmann A, Fink-Jensen A, Nordentoft M, Hougaard D, Norgaard-Pedersen B, Böttcher Y, Olesen J, Breuer R, Möller H-J, Giegling I, Rasmussen HB, Timm S, Mattheisen M, Bitter I, Réthelyi JM, Magnusdottir BB, Sigmundsson T, Olason P, Masson G, Gulcher JR, Haraldsson M, Fossdal R, Thorgeirsson TE, Thorsteinsdottir U, Ruggeri M, Tosato S, Franke B, Strengman E, Kiemeney LA, Group, Melle I, Djurovic S, Abramova L, Kaleda V, Sanjuan J, de Frutos R, Bramon E, Vassos E, Fraser G, Ettinger U, Picchioni M, Walker N, Toulopoulou T, Need AC, Ge D, Yoon JL, Shianna KV, Freimer NB, Cantor RM, Murray R, Kong A, Golimbet V, Carracedo A, Arango C, Costas J, Jönsson EG, Terenius L, Agartz I, Petursson H, Nöthen MM, Rietschel M, Matthews PM, Muglia P, Peltonen L, St Clair D, Goldstein DB, Stefansson K, Collier DA. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe'er I, Dudbridge F, Holmans PA, Whittemore AS, Mowry BJ, Olincy A, Amin F, Cloninger CR, Silverman JM, Buccola NG, Byerley WF, Black DW, Crowe RR, Oksenberg JR, Mirel DB, Kendler KS, Freedman R, Gejman PV. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].C. Schizophrenia Psychiatric Genome-Wide Association Study Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, Bergen SE, Collins AL, Crowley JJ, Fromer M, Kim Y, Lee SH, Magnusson PK, Sanchez N, Stahl EA, Williams S, Wray NR, Xia K, Bettella F, Borglum AD, Bulik-Sullivan BK, Cormican P, Craddock N, de Leeuw C, Durmishi N, Gill M, Golimbet V, Hamshere ML, Holmans P, Hougaard DM, Kendler KS, Lin K, Morris DW, Mors O, Mortensen PB, Neale BM, O'Neill FA, Owen MJ, Milovancevic MP, Posthuma D, Powell J, Richards AL, Riley BP, Ruderfer D, Rujescu D, Sigurdsson E, Silagadze T, Smit AB, Stefansson H, Steinberg S, Suvisaari J, Tosato S, Verhage M, Walters JT, Multicenter C, Genetic Studies of Schizophrenia. Levinson DF, Gejman PV, Kendler KS, Laurent C, Mowry BJ, O'Donovan MC, Owen MJ, Pulver AE, Riley BP, Schwab SG, Wildenauer DB, Dudbridge F, Holmans P, Shi J, Albus M, Alexander M, Campion D, Cohen D, Dikeos D, Duan J, Eichhammer P, Godard S, Hansen M, Lerer FB, Liang KY, Maier W, Mallet J, Nertney DA, Nestadt G, Norton N, O'Neill FA, Papadimitriou GN, Ribble R, Sanders AR, Silverman JM, Walsh D, Williams NM, Wormley B, C. Psychosis Endophenotypes International. Arranz MJ, Bakker S, Bender S, Bramon E, Collier D, Crespo-Facorro B, Hall J, Iyegbe C, Jablensky A, Kahn RS, Kalaydjieva L, Lawrie S, Lewis CM, Lin K, Linszen DH, Mata I, McIntosh A, Murray RM, Ophoff RA, Powell J, Rujescu D, Van Os J, Walshe M, Weisbrod M, Wiersma D, C. Wellcome Trust Case Control. Donnelly P, Barroso I, Blackwell JM, Bramon E, Brown MA, Casas JP, Corvin AP, Deloukas P, Duncanson A, Jankowski J, Markus HS, Mathew CG, Palmer CN, Plomin R, Rautanen A, Sawcer SJ, Trembath RC, Viswanathan AC, Wood NW, Spencer CC, Band G, Bellenguez C, Freeman C, Hellenthal G, Giannoulatou E, Pirinen M, Pearson RD, Strange A, Su Z, Vukcevic D, Donnelly P, Langford C, Hunt SE, Edkins S, Gwilliam R, Blackburn H, Bumpstead SJ, Dronov S, Gillman M, Gray E, Hammond N, Jayakumar A, McCann OT, Liddle J, Potter SC, Ravindrarajah R, Ricketts M, Tashakkori-Ghanbaria A, Waller MJ, Weston P, Widaa S, Whittaker P, Barroso I, Deloukas P, Mathew CG, Blackwell JM, Brown MA, Corvin AP, McCarthy MI, Spencer CC, Bramon E, Corvin AP, O'Donovan MC, Stefansson K, Scolnick E, Purcell S, McCarroll SA, Sklar P, Hultman CM, Sullivan PF. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Shepherd JD, Bear MF. New views of Arc, a master regulator of synaptic plasticity. Nat Neurosci. 2011;14:279–284. doi: 10.1038/nn.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Bramham CR, Alme MN, Bittins M, Kuipers SD, Nair RR, Pai B, Panja D, Schubert M, Soule J, Tiron A, Wibrand K. The Arc of synaptic memory. Exp Brain Res. 2010;200:125–140. doi: 10.1007/s00221-009-1959-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- [98].Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, Barnes CA. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J Neurosci. 2000;20:3993–4001. doi: 10.1523/JNEUROSCI.20-11-03993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- [100].Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- [101].Lee HK, Kameyama K, Huganir RL, Bear MF. NMDA induces long-term synaptic depression and dephosphorylation of the GluR1 subunit of AMPA receptors in hippocampus. Neuron. 1998;21:1151–1162. doi: 10.1016/s0896-6273(00)80632-7. [DOI] [PubMed] [Google Scholar]
- [102].Shimizu E, Tang YP, Rampon C, Tsien JZ. NMDA receptor-dependent synaptic reinforcement as a crucial process for memory consolidation. Science. 2000;290:1170–1174. doi: 10.1126/science.290.5494.1170. [DOI] [PubMed] [Google Scholar]
- [103].Lynch MA. Long-term potentiation and memory. Physiol Rev. 2004;84:87–136. doi: 10.1152/physrev.00014.2003. [DOI] [PubMed] [Google Scholar]
- [104].Bauer EP, Schafe GE, LeDoux JE. NMDA receptors and L-type voltage-gated calcium channels contribute to long-term potentiation and different components of fear memory formation in the lateral amygdala. J Neurosci. 2002;22:5239–5249. doi: 10.1523/JNEUROSCI.22-12-05239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg PH. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256:1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- [106].Verkhratsky A, Kirchhoff F. NMDA Receptors in glia. Neuroscientist. 2007;13:28–37. doi: 10.1177/1073858406294270. [DOI] [PubMed] [Google Scholar]
- [107].Anaparti V, Ilarraza R, Orihara K, Stelmack GL, Ojo OO, Mahood TH, Unruh H, Halayko AJ, Moqbel R. NMDA receptors mediate contractile responses in human airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2015;308:L1253–1264. doi: 10.1152/ajplung.00402.2014. [DOI] [PubMed] [Google Scholar]
- [108].Lee KH, Park JY, Kim K. NMDA receptor-mediated calcium influx plays an essential role in myoblast fusion. FEBS Lett. 2004;578:47–52. doi: 10.1016/j.febslet.2004.10.076. [DOI] [PubMed] [Google Scholar]
- [109].Said SI, Berisha HI, Pakbaz H. Excitotoxicity in the lung: N-methyl-D-aspartate-induced, nitric oxide-dependent, pulmonary edema is attenuated by vasoactive intestinal peptide and by inhibitors of poly(ADP-ribose) polymerase. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:4688–4692. doi: 10.1073/pnas.93.10.4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Vrajova M, Stastny F, Horacek J, Lochman J, Sery O, Pekova S, Klaschka J, Hoschl C. Expression of the hippocampal NMDA receptor GluN1 subunit and its splicing isoforms in schizophrenia: postmortem study. Neurochem Res. 2010;35:994–1002. doi: 10.1007/s11064-010-0145-z. [DOI] [PubMed] [Google Scholar]
- [111].Hahn CG, Wang HY, Cho DS, Talbot K, Gur RE, Berrettini WH, Bakshi K, Kamins J, Borgmann-Winter KE, Siegel SJ, Gallop RJ, Arnold SE. Altered neuregulin 1-erbB4 signaling contributes to NMDA receptor hypofunction in schizophrenia. Nat Med. 2006;12:824–828. doi: 10.1038/nm1418. [DOI] [PubMed] [Google Scholar]
- [112].Kristiansen LV, Patel SA, Haroutunian V, Meador-Woodruff JH. Expression of the NR2B-NMDA receptor subunit and its Tbr-1/CINAP regulatory proteins in postmortem brain suggest altered receptor processing in schizophrenia. Synapse. 2010;64:495–502. doi: 10.1002/syn.20754. [DOI] [PubMed] [Google Scholar]
- [113].Weickert CS, Fung SJ, Catts VS, Schofield PR, Allen KM, Moore LT, Newell KA, Pellen D, Huang XF, Catts SV, Weickert TW. Molecular evidence of N-methyl-D-aspartate receptor hypofunction in schizophrenia. Mol Psychiatry. 2013;18:1185–1192. doi: 10.1038/mp.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Javitt DC, Zukin SR, Heresco-Levy U, Umbricht D. Has an angel shown the way? Etiological and therapeutic implications of the PCP/NMDA model of schizophrenia. Schizophr Bull. 2012;38:958–966. doi: 10.1093/schbul/sbs069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Iwata Y, Nakajima S, Suzuki T, Keefe RS, Plitman E, Chung JK, Caravaggio F, Mimura M, Graff-Guerrero A, Uchida H. Effects of glutamate positive modulators on cognitive deficits in schizophrenia: a systematic review and meta-analysis of double-blind randomized controlled trials. Mol Psychiatry. doi: 10.1038/mp.2015.68. DOI 10.1038/mp.2015.68(2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Catterall WA. Voltage-gated calcium channels. Cold Spring Harb Perspect Biol. 2011;3:a003947. doi: 10.1101/cshperspect.a003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- [118].Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Ferron L, Nieto-Rostro M, Cassidy JS, Dolphin AC. Fragile X mental retardation protein controls synaptic vesicle exocytosis by modulating N-type calcium channel density. Nat Commun. 2014;5:3628. doi: 10.1038/ncomms4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Fatemi SH, Kneeland RE, Liesch SB, Folsom TD. Fragile X mental retardation protein levels are decreased in major psychiatric disorders. Schizophrenia research. 2010;124:246–247. doi: 10.1016/j.schres.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, Walker MF, Ober GT, Teran NA, Song Y, El-Fishawy P, Murtha RC, Choi M, Overton JD, Bjornson RD, Carriero NJ, Meyer KA, Bilguvar K, Mane SM, Sestan N, Lifton RP, Gunel M, Roeder K, Geschwind DH, Devlin B, State MW. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Weiss LA, Escayg A, Kearney JA, Trudeau M, MacDonald BT, Mori M, Reichert J, Buxbaum JD, Meisler MH. Sodium channels SCN1A, SCN2A and SCN3A in familial autism. Mol Psychiatry. 2003;8:186–194. doi: 10.1038/sj.mp.4001241. [DOI] [PubMed] [Google Scholar]
- [125].Ogiwara I, Ito K, Sawaishi Y, Osaka H, Mazaki E, Inoue I, Montal M, Hashikawa T, Shike T, Fujiwara T, Inoue Y, Kaneda M, Yamakawa K. De novo mutations of voltage-gated sodium channel alphaII gene SCN2A in intractable epilepsies. Neurology. 2009;73:1046–1053. doi: 10.1212/WNL.0b013e3181b9cebc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Kamiya K, Kaneda M, Sugawara T, Mazaki E, Okamura N, Montal M, Makita N, Tanaka M, Fukushima K, Fujiwara T, Inoue Y, Yamakawa K. A nonsense mutation of the sodium channel gene SCN2A in a patient with intractable epilepsy and mental decline. J Neurosci. 2004;24:2690–2698. doi: 10.1523/JNEUROSCI.3089-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Purcell AE, Jeon OH, Zimmerman AW, Blue ME, Pevsner J. Postmortem brain abnormalities of the glutamate neurotransmitter system in autism. Neurology. 2001;57:1618–1628. doi: 10.1212/wnl.57.9.1618. [DOI] [PubMed] [Google Scholar]
- [128].Hamdan FF, Gauthier J, Araki Y, Lin DT, Yoshizawa Y, Higashi K, Park AR, Spiegelman D, Dobrzeniecka S, Piton A, Tomitori H, Daoud H, Massicotte C, Henrion E, Diallo O, Group SD, Shekarabi M, Marineau C, Shevell M, Maranda B, Mitchell G, Nadeau A, D'Anjou G, Vanasse M, Srour M, Lafreniere RG, Drapeau P, Lacaille JC, Kim E, Lee JR, Igarashi K, Huganir RL, Rouleau GA, Michaud JL. Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet. 2011;88:306–316. doi: 10.1016/j.ajhg.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- [130].Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, Napolitano C, Schwartz PJ, Joseph RM, Condouris K, Tager-Flusberg H, Priori SG, Sanguinetti MC, Keating MT. CaV1.2 Calcium Channel Dysfunction Causes a Multisystem Disorder Including Arrhythmia and Autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- [132].C. Cross-Disorder Group of the Psychiatric Genomics Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Cox DM, Butler MG. A clinical case report and literature review of the 3q29 microdeletion syndrome. Clin Dysmorphol. 2015;24:89–94. doi: 10.1097/MCD.0000000000000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Ramocki MB, Bartnik M, Szafranski P, Kolodziejska KE, Xia Z, Bravo J, Miller GS, Rodriguez DL, Williams CA, Bader PI, Szczepanik E, Mazurczak T, Antczak-Marach D, Coldwell JG, Akman CI, McAlmon K, Cohen MP, McGrath J, Roeder E, Mueller J, Kang SH, Bacino CA, Patel A, Bocian E, Shaw CA, Cheung SW, Mazurczak T, Stankiewicz P. Recurrent distal 7q11.23 deletion including HIP1 and YWHAG identified in patients with intellectual disabilities, epilepsy, and neurobehavioral problems. Am J Hum Genet. 2010;87:857–865. doi: 10.1016/j.ajhg.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Cox D, Butler M. The 15q11.2 BP1–BP2 Microdeletion Syndrome: A Review. International Journal of Molecular Sciences. 2015;16:4068. doi: 10.3390/ijms16024068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Ben-Shachar S, Lanpher B, German JR, Qasaymeh M, Potocki L, Nagamani SC, Franco LM, Malphrus A, Bottenfield GW, Spence JE, Amato S, Rousseau JA, Moghaddam B, Skinner C, Skinner SA, Bernes S, Armstrong N, Shinawi M, Stankiewicz P, Patel A, Cheung SW, Lupski JR, Beaudet AL, Sahoo T. Microdeletion 15q13.3: a locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J Med Genet. 2009;46:382–388. doi: 10.1136/jmg.2008.064378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Niklasson L, Rasmussen P, Oskarsdottir S, Gillberg C. Autism, ADHD, mental retardation and behavior problems in 100 individuals with 22q11 deletion syndrome. Res Dev Disabil. 2009;30:763–773. doi: 10.1016/j.ridd.2008.10.007. [DOI] [PubMed] [Google Scholar]
- [138].Girirajan S, Dennis Megan Y., Baker C, Malig M, Coe Bradley P., Campbell Catarina D., Mark K, Vu Tiffany H., Alkan C, Cheng Z, Biesecker Leslie G., Bernier R, Eichler Evan E. Refinement and Discovery of New Hotspots of Copy-Number Variation Associated with Autism Spectrum Disorder. American Journal of Human Genetics. 2013;92:221–237. doi: 10.1016/j.ajhg.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]