Abstract
In plants and fungi, energetics at the plasma membrane is provided by a large protonmotive force (PMF) generated by the family of P-type ATPases specialized for proton transport (commonly called PM H+-ATPases or, in Arabidopsis, AHAs for Arabidopsis H+-ATPases). Studies have demonstrated that this 100-kDa protein is essential for plant growth and development. Posttranslational modifications of the H+-ATPase play critical roles in its regulation. Phosphorylation of several Thr and Ser residues within the carboxy terminal regulatory domain composed of ~ 100 amino acids change in response to environmental stimuli, endogenous hormones, and nutrient conditions. Recently developed mass spectrometric technologies provide a means to carefully quantify these changes in H+-ATPase phosphorylation at the different sites. These chemical modifications can then be genetically tested in planta by complementing the loss-of-function aha mutants with phosphomimetic mutations. Interestingly, recent data suggest that phosphatase-mediated changes in PM H+-ATPase phosphorylation are important in mediating auxin-regulated growth. Thus, as with another hormone (abscisic acid), dephosphorylation by phosphatases, rather than kinase mediated phosphorylation, may be an important focal point for regulation during plant signal transduction. Although interactions with other proteins have also been implicated in ATPase regulation, the very hydrophobic nature and high concentration of this polytopic protein presents special challenges in evaluating the biological significance of these interactions. Only by combining biochemical and genetic experiments can we attempt to meet these challenges to understand the essential molecular details by which this protein functions in planta.
Role of H+-ATPase in maintaining the membrane potential and acidic pH at the plasma membrane surface
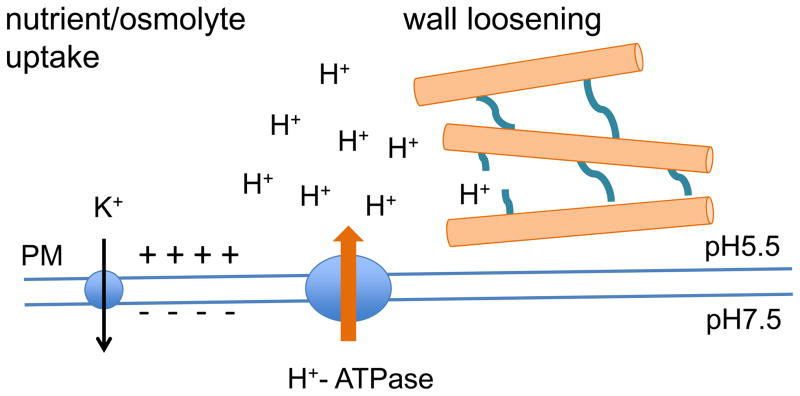
Plant cell growth is highly influenced by the environment, as well as by predetermined developmental programs. Plasma membrane H+-ATPases (H+-pumps) are the primary active transporters that translocate protons to the outside of each cell, providing the electrical and chemical energy that drives solute transport. In plants, this enzyme also provides an acidic environment in the cell wall that is favorable for cell expansion (Figure 1). In animals, a Na+/K+ ATPase provides similar function as the primary active transport system, in creating a membrane potential and a chemical gradient of sodium that drives sodium-coupled transporters and ion channels. In addition to this fundamental difference in the choice of cation used to drive transport, there is also a much higher membrane potential found in plants (e.g., minus 200–250 millivolts in plants versus minus 100–150 millivolts in animals) and this may reflect a need to drive potassium inward to very high concentrations even in soil with low potassium levels, in order to maintain turgor pressure required for plant form and function [1].
Figure 1. Plasma membrane energetics produced by H+-ATPases.
An H+-ATPase creates the H+ electrochemical gradient across the plasma membrane. This enzyme produces both a chemical gradient of protons (delta pH) as well as an outside-positive membrane potential gradient, which together comprise the proton motive force. This transmembrane energy is used by the secondary transporter systems and channels for the transport of osmolytes and nutrients. The cell wall also undergoes apoplastic acidification due to the activation of H+-ATPase, during cell expansion.
An essential role for PM H+-ATPases and the proton motive force was demonstrated in genetic experiments in Arabidopsis with plants containing loss-of-function mutations in the two most highly expressed members, AHA1 and 2, which results in embryonic lethality [2]. The phenotypes of aha2 single mutants also provided clues to the solute transport function of these enzymes during environmental and developmental changes in the vegetative phase [3,4]. For many years it has been recognized that the acidic pH environment created by PM H+-ATPases is an important factor for wall loosening prior to expansion [5–7]. The involvement of PM H+-ATPases in defense-related signaling pathways, such as triggered by the pathogen associated molecular pattern and endogenous peptides, has also recently been addressed [8,9].
Phosphorylation regulates PM H+-ATPases
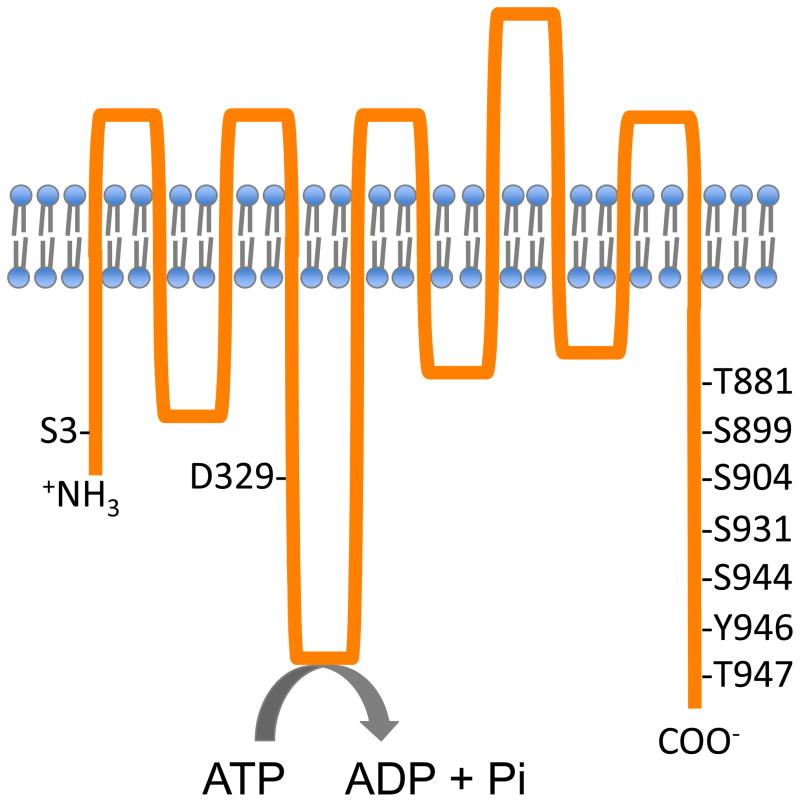
Regulatory phosphorylation sites are mainly clustered in the 100-amino-acid long carboxy terminus, also referred to as the R (regulatory) or auto-inhibitory domain [10]. AHA2 is subject to in vivo phosphorylation on at least eight residues in this C- terminal tail (Figure 2) and changes in AHA2 phosphopeptide abundance has been analyzed via mass spectrometry, either in an untargeted fashion using ion trap based MS/MS or via targeted methods using selected reaction monitoring (SRM) on a triple quadrupole instrument [11,12]. Phosphorylation at the penultimate Thr residue, which can form a complex with a fungal toxin, fusicoccin and a 14-3-3 protein [13,14], appears to be one major stimulatory site involved in transducing the response to auxin and blue light. 14-3-3 protein binding to H+-ATPase has been used as a method to quantify changes in phosphorylation at this pentultimate Thr residue [15]. It also should be noted that activation can occur without H+-ATPase interacting with 14-3-3 protein [16], although the degree by which this occurs in situ remains to be established.
Figure 2. Phosphorylated residues in the AHA2 H+-ATPase.
Structural features of the plasma membrane P-type H+-ATPase, AHA2. H+-ATPase uses ATP energy to translocate H+ during the catalytic cycle, which involve the formation of phosphorylated intermediate of the conserved Asp residue, found at amino acid position, 329. The activity of AHA2 is regulated by the posttranslational phosphorylation of various Ser and Thr residues. Phosphorylation at Thr881 or Thr947 increase AHA2 activity whereas phosphorylation at Ser899 or Ser931 decrease the activity.
Multiple residues respond to various stimuli by different degrees of phosphorylation and the direction of changes at each site can differ. This provides a very flexible but potentially complicated system for regulation of this enzyme’s function (Table 1). For example, in Arabidopsis seedlings, Thr881 phosphorylation of AHA2 is induced by a growth stimulatory peptide, PSY1, whereas Ser899 phosphorylation is induced by a growth inhibitory peptide, Rapid Alkalinization Factor (RALF) [11,17]. In a more complicated situation, flg22 peptide elicits an increase in pSer899 phosphorylation and decreases in pThr881 and pThr947 phosphorylation in cultured cells [8]. Abiotic stresses, such as cold, oxidation, and acid treatments, decreased phosphorylation of the penultimate Thr, as does treatment with ABA [18,19]. Phosphorylation at Tyr946 residue was detected in MS-based sequencing [10]. However, since there can be misinterpretation of the fragment pattern during computer-automated analyses, synthetic standard peptides containing either threonine or tyrosine phosphorylation at this carboxy terminal location can be utilized to carefully examine and compare to the peptides derived from plant extracts. Currently, possible phosphorylation of this tyrosine residue remains to be clarified both chemically and genetically. Although low stoichiometry phosphosites may have critical functions in vivo, it is useful to determine the ratio of phosphorylation over non-phosphorylation for each site. In other words, by knowing what fraction of this protein is phosphorylated at a given time and condition, we will be able to better understand the significance of changes in phosphorylation. For this purpose, the quantitation of phospho- and nonphospho-peptides from the same biological sample is our current goal to reveal the ratio of phosphorylation events at each site [20]. This can be difficult since the methods for enriching phosphopeptides creates a situation where the unphosphorylated peptide may be hard to see without extensive prefractionation. For this purpose, an affinity-tagged H+-ATPase protein provides rapid purification strategies to obtain large quantities of the protein [21]
Table 1.
Changes in the phosphorylation of Arabidopsis plasma membrane H+-ATPases under various genetic and environmental perturbations
| Protein | Condition | Site | Tissue | Detection | References |
|---|---|---|---|---|---|
| AHA1 | flg22 | T881, S899, T948 | cell culture | MS (iTRAQ) | [8] |
| sucrose | T948 | seedling | MS (label free) | [57] | |
| fusicoccin | T948 | seedling | SRM | [12] | |
| phytohormones | T948 | cell culture | MS (label free) | [58] | |
| AHA2 | flg22 | T881, S899, T947 | cell culture | MS (iTRAQ) | [8] |
| flg22 | S944 | cell culture | MS (15N) | [59] | |
| sucrose | T947 | seedling | MS (label free) | [57] | |
| phytohormones | T947 | cell culture | MS (label free) | [58] | |
| RALF | S899 | seedling | MS (15N) | [11] | |
| PSY1 | T881 | seedling | SRM | [17] | |
| aha1, aha2 mutants | T947, T948 | seedling | SRM | [2] | |
| IAA | T947 | hypocotyls | pT947 antibody | [5] | |
| GFP-SAUR19 | T947 | seedling | 14-3-3 overlay | [6] | |
| transgenics | pT947 antibody | ||||
| fusicoccin | T947 | seedling | SRM | [12] | |
| blue light | T947 | seedling | pT947 antibody | [46] | |
| ABA | T947 | seedling | pT947 antibody | [60] | |
| AHA3 | flg22 | T882 | cell culture | MS (iTRAQ) | [8] |
| Other AHAs | blue light | T947 equivalent | guard cell protoplast | pT947 antibody | [61] |
iTRAQ, isobaric tags for relative and absolute quantitation; 15N, nitrogen15 stable-isotope metabolic labeling; SRM, selected reaction monitoring.
Correlation of PM H+-ATPase phosphorylation, catalytic activity, and plant phenotypes
The timing and direction of the changes in H+-ATPase phosphorylation at each of the sites may provide clues on the molecular mechanism of transport function, as well as the roles of the enzyme in plant life. However, since the PMF plays such a fundamental role in many functions (most transporters and channels are coupled to either the delta pH or the membrane potential or both), one needs to carefully interpret a causal direct link between AHA phosphorylation and biological function. For example, an increase in penultimate Thr phosphorylation may indicate activation of H+-ATPase. However, if changes in AHA phosphorylation status are examined in long-term experiments (such as several hours), one may be observing an indirect response of the H+-ATPase to reestablish the proton electrochemical gradient after depletion of proton motive force due to changes in other transporters’ activities. In this context it is clear that to understand the regulation of H+-ATPase by phosphorylation, it is useful to examine the enzyme activity and phenotypic changes associated with phosphorylation changes in short term assays, covering seconds and minutes, rather than hours and days.
With an enriched plasma membrane fraction, in vitro H+-ATPase activity can be quantified by several procedures: 1) quantifying phosphate-molybdate or -malachite complexes to measure released inorganic phosphate [22,23], 2) changes in acridine orange absorbance or pH indicator fluorescence to calculate pH changes in sealed membrane vesicles [24], and 3) NADH-coupled assays to quantify the amount of ATP consumed [24]. Alternatively, in situ measurements of apoplastic pH can be made with glass electrodes [25,26] or pH sensitive dyes [27,28], or the electrical membrane potential can be measured by electrode impalement [29,30]. Ideally, a combination of multiple measurements are useful for corroborating relative changes in H+-ATPase activity since the membrane potential or extracellular pH are likely altered by changes in the activities of other proton coupled transporters or anion transporters (e.g. anions like chloride or nitrate or cations like potassium and calcium). Similarly, some changes in the cytoplasmic pH may be caused by the action of other endomembrane transport systems and those in the apoplastic pH by the secretion of other ions such as NH4+ and these will all alter plasma membrane energetics. Moreover, in vitro measurements of inorganic phosphate release from ATP can be affected by the changes in activity of other ATP-hydrolyzing pumps, which may be co-regulated with the H+-ATPase [8,11]. It is clear that several parameters need to be examined to conclude that the plasma membrane H+-ATPase is directly and specifically altered by an environmental, chemical or genetic perturbation.
Unfortunately, there are no specific inhibitors of the plant PM H+-ATPases analogous to ouabain, which is used in research on the sodium pump in animals. Vanadate is relatively specific for the H+-ATPase in the plasma membrane compared to ATPases in other endomembranes but since vanadate also inhibits protein phosphatases and ABC transporters as well, interpretations using this compound in vitro, and especially in vivo, should be approached with caution. On the other hand, fusicoccin is an activator of PM H+-ATPases, acts at low concentrations and provides a very useful means of ensuring that the response one is measuring is specifically related to the plasma membrane proton pump.
The effects of changes in H+-ATPase activity on plant growth or physiology have also been examined by observing plant phenotypes in response to genetic or environmental perturbations. A change in the cell expansion rate can be studied via root or hypocotyl elongation measurements either under normal or altered growth conditions that place stress on the proton electrochemical gradient. Two independent T-DNA insertion mutants of aha2 show growth retardation compared with the wildtype under high external potassium or alkaline pH conditions that reduce plasma membrane proton motive force. Inversely, the aha2 mutant shows reduced sensitivity to external toxic cations, such as lithium or cesium, whose uptake is coupled with proton motive force. Thus, these growth conditions have been used to examine the phenotypes of mutant plants with altered H+-ATPase activities [3,6]. The effect of H+-ATPase activity changes on phenotype was also visualized by measuring guard cell movements [4,31] and growth rate under salt stress condition [32].
The biological role of phosphorylation was first genetically tested in Arabidopsis by introducing a site-specific mutation changing the penultimate Thr948 of a phloem specific isoform, AHA3, to either a non-phosphorylatable residue, Ala or the phosphomimetic residue, Asp [33]. The Asp mutation was found to complement the pollen lethal phenotype of aha3 knockout mutants, and the quantitative measurement of tissue elongation with these transgenic plants suggested that phosphorylation increased the H+-ATPase activity. It was hypothesized that the Ala mutation at this site acted as a dominant-negative allele since heterozygous aha3 mutants carrying the Thr-to-Ala mutation were male sterile. The role of AHA2 phosphorylation has also been extensively characterized using a powerful heterologous yeast expression system in which the Arabidopsis AHA2 protein rescues the lethality of the yeast’s ATPase gene (pma1), providing a high throughput model for studying AHA2 regulation by phosphorylation [10,34]. This yeast system was also utilized to examine AHA2 regulation by PP2C.D protein phosphatases [6] and the PKS5 kinase and its associated calcium binding protein in co-expression assays [35]. However, the results obtained from the yeast complementation system should be carefully interpreted because plant specific properties, such as hormone or lipid dependent exo-and endo-cytotic mechanisms and/or protein degradation may depend on changes in phosphorylation and the yeast system may not show these effects.
Auxin regulates PM H+-ATPase phosphorylation to promote cell expansion
Based largely on physiological studies correlating auxin-induced elongation growth and apoplastic acidification, auxin has long been hypothesized to activate the plasma membrane H+-ATPases. Indeed, auxin-mediated activation of H+ efflux is a fundamental tenet of the decades-old acid growth theory [7]. Recent work has provided crucial biochemical and genetic support for this hypothesis, as well as the beginnings of mechanistic insight. In a recent study examining the elongation of auxin depleted hypocotyl segments, Takahashi et al. demonstrated that auxin promotes phosphorylation at the penultimate Thr residue, corresponding to Thr947 of Arabidopsis AHA2 [5]. IAA-induced phosphorylation was evident within 10 minutes and peaked by 20 minutes. This effect could be the result of auxin-mediated changes in protein kinase and/or phosphatase activities that target Thr947. Regardless of the mechanism, the kinetics of change in phosphorylation status closely paralleled those of IAA-induced hypocotyl elongation.
More recently, Spartz et al. proposed a mechanism for auxin-mediated PM H+-ATPase activation involving Small Auxin Up-RNA (SAUR) proteins and a family of type 2C protein phosphatases [6]. SAUR genes represent the largest family of auxin-responsive genes, and several are rapidly induced following auxin treatment [36]. Overexpression of SAUR19 [37,38], as well as additional SAUR’s [39–41], promotes hypocotyl elongation and confers several additional cell expansion phenotypes. Interestingly, SAUR19 overexpression also confers several phenotypes indicative of increased PM H+-ATPase activity, including reduced apoplastic pH, and hypersensitivity to lithium and other toxic cations [6]. Indeed, SAUR19 overexpression results in both increased PM H+-ATPase enzymatic activity and Thr947 phosphorylation. These effects appear to be mediated by SAUR regulation of a family of type 2C protein phosphatases (PP2C.Ds) that dephosphorylate Thr947. SAUR19 and other SAURs specifically bind to PP2C.D phosphatases and inhibit their enzymatic activity, suggesting that SAURs and PP2C.D phosphatases act antagonistically. Consistent with this possibility, pp2c.d knockdown plants exhibit phenotypes similar to SAUR19 overexpression lines. PP2C.D1 overexpression, on the other hand, results in severely reduced cell expansion, resistance to lithium, and reductions in both Thr947 phosphorylation and PM H+-ATPase activity. Remarkably, the severe cell expansion defects conferred by PP2C.D1 overexpression were completely suppressed by fusicoccin, strongly suggesting that the growth defects are the direct result of diminished Thr947 phosphorylation of the H+ pump. Furthermore, PP2C.D1 interacts with AHA2 in planta, can dephosphorylate Thr947 in vitro, and negatively regulates AHA2 in a heterologous yeast expression system [6].
Since SAUR genes are rapidly upregulated by auxin, SAUR-mediated inhibition of PP2C.D-mediated Thr947 dephosphorylation, provides an attractive model for how auxin activates PM H+-ATPases. However, important questions remain. Of particular note, auxin-induced Thr947 phosphorylation was unaffected in the tir1 afb2 auxin receptor mutant, or by pre-treatment with the auxin antagonist, PEO-IAA [5], suggesting that auxin-mediated transcription is not required. While this could simply be the result of activity of the remaining AFB receptors and incomplete inhibition by PEO-IAA, prior studies have indeed suggested there are auxin-mediated increases in H+ pump activity that occur too rapidly to be explained by changes in gene expression [42]. Auxin Binding Protein1 (ABP1) has been proposed to mediate this rapid activation, however this possibility needs careful reexamination given the recent finding that abp1 null mutants exhibit no obvious growth phenotypes [43]. Given the difficulty in performing time courses on the short time scale that these experiments require, if the genetic analysis of knockouts indicates that a rapid role of ABP1 in altering cell expansion is unlikely, it is possible that the SAUR mediated phosphatase mechanism induced by rapid changes in SAUR transcription, may be fast enough to explain some, or all, of the early auxin effects on the H+ pump.
Kinases and phosphatases implicated in H+-ATPase regulation
Identifying protein kinases and phosphatases that modify H+-ATPase activity would help understand the regulatory mechanisms of this enzyme during plant growth and adaptation to changes in the environment. In addition to the above-described PP2C.D1 phosphatase involved in SAUR-regulated cell expansion, to date several additional protein kinases and phosphatases have been proposed to directly or indirectly regulate H+-ATPase activity. Consistent with the plasma membrane localization, H+-ATPase directly interacts with and is subject to regulation by receptor kinases for a growth stimulatory peptide, PSY1 and the phytohormone, brassinosteroid [17,44]. The BRassinosteroid Insensitive1, BRI1 receptor activates AHA1 with a kinase function-dependent manner, but without involving phosphorylation of the penultimate Thr. Although the two proteins may exist as a preformed complex, upon treating with brassinolide for 30 min, the plasma membrane was hyperpolarized and cell expansion was observed.
Light, one of the most critical factors that affect plant growth and development, is known to influence H+-ATPase activity through the action of the plasma membrane blue-light receptors, phototropins. H+-ATPase and phototropin were reported to be co-purified in a reciprocal manner with pull-down experiments [21,45]; however it is not clear whether H+-ATPase is a direct substrate of phototropin. Hypocotyl bending toward blue light depends on phototropin action and correlates with the change in phosphorylation of the penultimate Thr [46], which is predicted to affect auxin transport and cell expansion rates during phototropism. Additionally, the influence of light on H+-ATPase phosphorylation is evident during the regulation of guard cell movement; Thr947 appears to be phosphorylated by a K252a-insensitive protein kinase and dephosphorylated by an as yet unknown Mg2+-dependent PP2C [47].
Down regulation of H+-ATPase activity is also known to occur after treatment of plants with a growth inhibitory peptide, RALF (Rapid ALkalinization Factor). RALF-induced phosphorylation of AHA2 at Ser899, possibly catalyzed by FERONIA receptor kinase activity, is predicted to inactivate H+-ATPase function leading to the inhibition of cell expansion [11]. Consistent with this model, a feronia mutant secretes protons faster than wildtype and shows a longer root. The FERONIA protein, originally identified as encoded by a gene required for pollen tube arrest [48], is now proposed to be involved in myriad physiological processes including root hair development [49], mechano-sensing [50], early membrane signaling [51], and ethylene production and signaling [52,53]. The broad range of FERONIA functions is consistent with this receptor kinase’s specific effect on modulating plasma membrane energetics, since the PMF is fundamental to so many cellular activities. Phosphorylation of AHA2 at Ser931, observed in an in vitro study, catalyzed by a stress-responsive Ser/Thr protein kinase, PKS5, also down-regulates AHA2 activity [35]. Calcium ions may play a role in the activation of this protein kinase as it also binds a calcium binding protein known as SCaBP1. Earlier studies have also reported that phosphorylation of H+-ATPase occurs in a calcium dependent and kinase-mediated reaction [54,55] but a clear cause and effect has not been demonstrated. Multiple phosphorylated residues are likely substrates of one or more phosphatases and kinases, implicating a complexity of phosphorylation events. It is also clear that one must carefully interpret in vitro experiments in which the H+-ATPase is treated in the absence of the cell since ‘promiscuous’ phosphorylation and dephosphorylation are possible in such experiments that may never occur in vivo. In this context, one needs to identify protein kinases or phosphatases that interact with PM H+-ATPases via approaches such as crosslinking and peptide sequencing and examine the phosphorylation status of the H+-ATPase in plants containing knockout mutatations of such kinases and phosphatases. In addition, one would need to characterize the effect of site-directed mutations of the H+-ATPase on this enzyme’s function, in order to support a causal relationship between phosphorylation changes in the H+-ATPase and phenotypic alterations.
Summary and the future questions
Since the emergence of sophisticated mass spectrometry-based techniques for the identification and quantitation of protein phosphorylation, considerable information has accumulated concerning the chemical basis for H+-ATPase regulation. Genetic mutation and phenotypic assays provide critical insights into the context-specific roles of these chemical changes in vivo. However, the key questions that remain to be solved include: 1) how many protein kinases and phosphatase are required for the regulation of H+-ATPase activity, 2) which sites are modified by those kinases and phosphatases, 3) under what biological conditions are those residues modified, 4) how do the multiple phosphorylation events interact to alter the H+-ATPase catalytic activity, and 5) are there interactions with other proteins in the cytoplasm or apoplast, or within the plane of the membrane, that also regulate the H+-ATPase activity posttranslationally? The three dimensional structure for the minimum catalytic unit of AHA2 (without the C-terminal regulatory sequence) has been solved by X-ray crystallography [56], and yet, it is unknown precisely how the C-terminal domain regulates this enzyme. New advances in mass spectral technologies for studying three dimensional protein structure and in vivo functional characterization using site directed mutagenesis will provide a set of exciting experimental systems to address these questions.
Highlights.
The plasma membrane H+-ATPase 100-kDa protein generates a protonmotive force (PMF).
PMF consists of a membrane potential and a pH gradient, which have separate roles.
H+-ATPase phosphorylation regulates solute transport and cell expansion.
A handful of different phosphosites have opposing effects on pump activity.
With multiple kinases and phosphatases, H+-ATPase regulation is multifaceted and potentially, quite complicated.
Acknowledgments
This study is supported by U.S. Department of Energy (Grant No. DEFG02-88ER13938 to MRS) and the National Science Foundation (Grant No. MCB 0929395 to MRS). Work in WMG’s lab is supported by National Institutes of Health Grant GM067203. The authors thank many other outstanding researchers in the field who contributed towards understanding how the plasma membrane H+-ATPase is regulated. Due to limited space, we were unable to cite many of these excellent H+-ATPase studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
*of special interest
**of outstanding interest
- 1.Hirsch RE, Lewis BD, Spalding EP, Sussman MR. A role for the AKT1 potassium channel in plant nutrition. Science. 1998;280:918–921. doi: 10.1126/science.280.5365.918. [DOI] [PubMed] [Google Scholar]
- 2.Haruta M, Burch HL, Nelson RB, Barrett-Wilt G, Kline KG, Mohsin SB, Young JC, Otegui MS, Sussman MR. Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J Biol Chem. 2010;285:17918–17929. doi: 10.1074/jbc.M110.101733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haruta M, Sussman MR. The effect of a genetically reduced plasma membrane protonmotive force on vegetative growth of Arabidopsis. Plant Physiol. 2012;158:1158–1171. doi: 10.1104/pp.111.189167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Noguchi K, Ono N, Inoue S, Terashima I, Kinoshita T. Overexpression of plasma membrane H+-ATPase in guard cells promotes light-induced stomatal opening and enhances plant growth. Proc Natl Acad Sci U S A. 2014;111:533–538. doi: 10.1073/pnas.1305438111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi K, Hayashi K, Kinoshita T. Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol. 2012;159:632–641. doi: 10.1104/pp.112.196428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6**.Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM. SAUR Inhibition of PP2C-D Phosphatases Activates Plasma Membrane H+-ATPases to Promote Cell Expansion in Arabidopsis. Plant Cell. 2014;26:2129–2142. doi: 10.1105/tpc.114.126037. The molecular mechanism of auxin-induced H+-ATPase activity was shown to be mediated by the inactivation of protein phosphatases that dephosphorylate the penultimate Thr residue of H+-ATPases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rayle DL, Cleland RE. The Acid Growth Theory of auxin-induced cell elongation is alive and well. Plant Physiol. 1992;99:1271–1274. doi: 10.1104/pp.99.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nuhse TS, Bottrill AR, Jones AM, Peck SC. Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 2007;51:931–940. doi: 10.1111/j.1365-313X.2007.03192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huffaker A, Pearce G, Ryan CA. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci U S A. 2006;103:10098–10103. doi: 10.1073/pnas.0603727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rudashevskaya EL, Ye J, Jensen ON, Fuglsang AT, Palmgren MG. Phosphosite mapping of P-type plasma membrane H+-ATPase in homologous and heterologous environments. J Biol Chem. 2012;287:4904–4913. doi: 10.1074/jbc.M111.307264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11**.Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science. 2014;343:408–411. doi: 10.1126/science.1244454. Using phosphoproteomic analyses, this study revealed a peptide-receptor, RALF-FERONIA plasma membrane signaling pathway which leads to the inhibition of cell expansion via the inactivation of H+-ATPases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12**.Stecker KE, Minkoff BB, Sussman MR. Phosphoproteomic Analyses Reveal Early Signaling Events in the Osmotic Stress Response. Plant Physiology. 2014;165:1171–1187. doi: 10.1104/pp.114.238816. With heavy-labeled synthetic peptides, the changes of 58 phosphopeptides abundances were quantitatively analyzed, showing H+-ATPase phosphopeptide abundances changes upon various abiotic- or biotic-stresses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olsson A, Svennelid F, Ek B, Sommarin M, Larsson C. A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14-3-3 binding. Plant Physiol. 1998;118:551–555. doi: 10.1104/pp.118.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jahn T, Fuglsang AT, Olsson A, Bruntrup IM, Collinge DB, Volkmann D, Sommarin M, Palmgren MG, Larsson C. The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell. 1997;9:1805–1814. doi: 10.1105/tpc.9.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svennelid F, Olsson A, Piotrowski M, Rosenquist M, Ottman C, Larsson C, Oecking C, Sommarin M. Phosphorylation of Thr-948 at the C terminus of the plasma membrane H+-ATPase creates a binding site for the regulatory 14-3-3 protein. Plant Cell. 1999;11:2379–2391. doi: 10.1105/tpc.11.12.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piette AS, Derua R, Waelkens E, Boutry M, Duby G. A phosphorylation in the c-terminal auto-inhibitory domain of the plant plasma membrane H+-ATPase activates the enzyme with no requirement for regulatory 14-3-3 proteins. J Biol Chem. 2011;286:18474–18482. doi: 10.1074/jbc.M110.211953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.Fuglsang AT, Kristensen A, Cuin TA, Schulze WX, Persson J, Thuesen KH, Ytting CK, Oehlenschlaeger CB, Mahmood K, Sondergaard TE, et al. Receptor kinase-mediated control of primary active proton pumping at the plasma membrane. Plant J. 2014;80:951–964. doi: 10.1111/tpj.12680. Using yeast two hybrid screen, this study identified a receptor kinase, PSY1R interacts with AHA2, and showed that AHA2 T881 phosphorylation by this receptor kinase increases H+-ATPase activity. [DOI] [PubMed] [Google Scholar]
- 18.Bobik K, Duby G, Nizet Y, Vandermeeren C, Stiernet P, Kanczewska J, Boutry M. Two widely expressed plasma membrane H+-ATPase isoforms of Nicotiana tabacum are differentially regulated by phosphorylation of their penultimate threonine. Plant J. 2010;62:291–301. doi: 10.1111/j.1365-313X.2010.04147.x. [DOI] [PubMed] [Google Scholar]
- 19.Bobik K, Boutry M, Duby G. Activation of the plasma membrane H+ -ATPase by acid stress: antibodies as a tool to follow the phosphorylation status of the penultimate activating Thr. Plant Signal Behav. 2010;5:681–683. doi: 10.4161/psb.5.6.11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kline-Jonakin KG, Barrett-Wilt GA, Sussman MR. Quantitative plant phosphoproteomics. Curr Opin Plant Biol. 2011;14:507–511. doi: 10.1016/j.pbi.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21**.Rodrigues RB, Sabat G, Minkoff BB, Burch HL, Nguyen TT, Sussman MR. Expression of a translationally fused TAP-tagged plasma membrane proton pump in Arabidopsis thaliana. Biochemistry. 2014;53:566–578. doi: 10.1021/bi401096m. By expressing a tagged protein of H+-ATPase, the lethality caused by aha1/aha2 double mutation was rescued and a subset of potential interactors of H+-ATPase were identified via mass spectrometry-based sequencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Surowy TK, Sussman MR. Immunological cross-reactivity and inhibitor sensitivities of the plasma membrane H+-ATPase from plants and fungi. Biochimica et Biophysica Acta (BBA) -Bioenergetics. 1986;848:24–34. [Google Scholar]
- 23.Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 24.Palmgren MG. An H+-ATPase Assay: Proton Pumping and ATPase Activity Determined Simultaneously in the Same Sample. Plant Physiol. 1990;94:882–886. doi: 10.1104/pp.94.3.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 26.Pearce G, Moura DS, Stratmann J, Ryan CA. Production of multiple plant hormones from a single polyprotein precursor. Nature. 2001;411:817–820. doi: 10.1038/35081107. [DOI] [PubMed] [Google Scholar]
- 27.Monshausen GB, Bibikova TN, Messerli MA, Shi C, Gilroy S. Oscillations in extracellular pH and reactive oxygen species modulate tip growth of Arabidopsis root hairs. Proc Natl Acad Sci U S A. 2007;104:20996–21001. doi: 10.1073/pnas.0708586104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santi S, Schmidt W. Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 2009;183:1072–1084. doi: 10.1111/j.1469-8137.2009.02908.x. [DOI] [PubMed] [Google Scholar]
- 29.Spalding EP, Cosgrove DJ. Mechanism of blue-light-induced plasma-membrane depolarization in etiolated cucumber hypocotyls. Planta. 1992;188:199–205. doi: 10.1007/BF00216814. [DOI] [PubMed] [Google Scholar]
- 30.Lohse G, Hedrich R. Characterization of the plasma-membrane H+-ATPase from Vicia faba guard cells : Modulation by extracellular factors and seasonal changes. Planta. 1992;188:206–214. doi: 10.1007/BF00216815. [DOI] [PubMed] [Google Scholar]
- 31.Merlot S, Leonhardt N, Fenzi F, Valon C, Costa M, Piette L, Vavasseur A, Genty B, Boivin K, Muller A, et al. Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J. 2007;26:3216–3226. doi: 10.1038/sj.emboj.7601750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gevaudant F, Duby G, von Stedingk E, Zhao R, Morsomme P, Boutry M. Expression of a constitutively activated plasma membrane H+-ATPase alters plant development and increases salt tolerance. Plant Physiol. 2007;144:1763–1776. doi: 10.1104/pp.107.103762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robertson WR, Clark K, Young JC, Sussman MR. An Arabidopsis thaliana plasma membrane proton pump is essential for pollen development. Genetics. 2004;168:1677–1687. doi: 10.1534/genetics.104.032326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Axelsen KB, Venema K, Jahn T, Baunsgaard L, Palmgren MG. Molecular dissection of the C-terminal regulatory domain of the plant plasma membrane H+-ATPase AHA2: mapping of residues that when altered give rise to an activated enzyme. Biochemistry. 1999;38:7227–7234. doi: 10.1021/bi982482l. [DOI] [PubMed] [Google Scholar]
- 35.Fuglsang AT, Guo Y, Cuin TA, Qiu Q, Song C, Kristiansen KA, Bych K, Schulz A, Shabala S, Schumaker KS, et al. Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+ -ATPase by preventing interaction with 14-3-3 protein. Plant Cell. 2007;19:1617–1634. doi: 10.1105/tpc.105.035626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren H, Gray WM. SAUR Proteins as Effectors of Hormonal and Environmental Signals in Plant Growth. Mol Plant. 2015 doi: 10.1016/j.molp.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inze D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012;70:978–990. doi: 10.1111/j.1365-313X.2012.04946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci U S A. 2011;108:20231–20235. doi: 10.1073/pnas.1110682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chae K, Isaacs CG, Reeves PH, Maloney GS, Muday GK, Nagpal P, Reed JW. Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J. 2012;71:684–697. doi: 10.1111/j.1365-313X.2012.05024.x. [DOI] [PubMed] [Google Scholar]
- 40.Kong Y, Zhu Y, Gao C, She W, Lin W, Chen Y, Han N, Bian H, Zhu M, Wang J. Tissue-specific expression of SMALL AUXIN UP RNA41 differentially regulates cell expansion and root meristem patterning in Arabidopsis. Plant Cell Physiol. 2013;54:609–621. doi: 10.1093/pcp/pct028. [DOI] [PubMed] [Google Scholar]
- 41.Stamm P, Kumar PP. Auxin and gibberellin responsive Arabidopsis SMALL AUXIN UP RNA36 regulates hypocotyl elongation in the light. Plant Cell Rep. 2013;32:759–769. doi: 10.1007/s00299-013-1406-5. [DOI] [PubMed] [Google Scholar]
- 42.Rück A, Palme K, Venis MA, Napier RM, Felle HH. Patch-clamp analysis establishes a role for an auxin binding protein in the auxin stimulation of plasma membrane current in Zea mays protoplasts. The Plant Journal. 1993;4:41–46. [Google Scholar]
- 43.Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci U S A. 2015;112:2275–2280. doi: 10.1073/pnas.1500365112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caesar K, Elgass K, Chen Z, Huppenberger P, Witthoft J, Schleifenbaum F, Blatt MR, Oecking C, Harter K. A fast brassinolide-regulated response pathway in the plasma membrane of Arabidopsis thaliana. Plant J. 2011;66:528–540. doi: 10.1111/j.1365-313X.2011.04510.x. [DOI] [PubMed] [Google Scholar]
- 45.Kaiserli E, Sullivan S, Jones MA, Feeney KA, Christie JM. Domain swapping to assess the mechanistic basis of Arabidopsis phototropin 1 receptor kinase activation and endocytosis by blue light. Plant Cell. 2009;21:3226–3244. doi: 10.1105/tpc.109.067876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hohm T, Demarsy E, Quan C, Allenbach Petrolati L, Preuten T, Vernoux T, Bergmann S, Fankhauser C. Plasma membrane H+ -ATPase regulation is required for auxin gradient formation preceding phototropic growth. Mol Syst Biol. 2014;10:751. doi: 10.15252/msb.20145247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayashi Y, Nakamura S, Takemiya A, Takahashi Y, Shimazaki K, Kinoshita T. Biochemical characterization of in vitro phosphorylation and dephosphorylation of the plasma membrane H+-ATPase. Plant Cell Physiol. 2010;51:1186–1196. doi: 10.1093/pcp/pcq078. [DOI] [PubMed] [Google Scholar]
- 48.Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, Grossniklaus U. The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science. 2007;317:656–660. doi: 10.1126/science.1143562. [DOI] [PubMed] [Google Scholar]
- 49.Duan Q, Kita D, Li C, Cheung AY, Wu HM. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci U S A. 2010;107:17821–17826. doi: 10.1073/pnas.1005366107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shih HW, Miller ND, Dai C, Spalding EP, Monshausen GB. The receptor-like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Curr Biol. 2014;24:1887–1892. doi: 10.1016/j.cub.2014.06.064. [DOI] [PubMed] [Google Scholar]
- 51.Li C, Yeh FL, Cheung AY, Duan Q, Kita D, Liu MC, Maman J, Luu EJ, Wu BW, Gates L, et al. Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. Elife. 2015:4. doi: 10.7554/eLife.06587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mao D, Yu F, Li JB, VDP, Tan D, Li J, Liu Y, Li X, Dong M, Chen L, et al. FERONIA Receptor Kinase Interacts with S-adenosylmethionine Synthetase and Suppresses S-adenosylmethionine Production and Ethylene Biosynthesis in Arabidopsis. Plant Cell Environ. 2015 doi: 10.1111/pce.12570. [DOI] [PubMed] [Google Scholar]
- 53.Deslauriers SD, Larsen PB. FERONIA is a key modulator of brassinosteroid and ethylene responsiveness in Arabidopsis hypocotyls. Mol Plant. 2010;3:626–640. doi: 10.1093/mp/ssq015. [DOI] [PubMed] [Google Scholar]
- 54.Lino B, Carrillo-Rayas MT, Chagolla A, Gonzalez de la Vara LE. Purification and characterization of a calcium-dependent protein kinase from beetroot plasma membranes. Planta. 2006;225:255–268. doi: 10.1007/s00425-006-0343-8. [DOI] [PubMed] [Google Scholar]
- 55.Schaller GE, Sussman MR. Phosphorylation of the plasma-membrane H+-ATPase of oat roots by a calcium-stimulated protein kinase. Planta. 1988;173:509–518. doi: 10.1007/BF00958964. [DOI] [PubMed] [Google Scholar]
- 56.Pedersen BP, Buch-Pedersen MJ, Morth JP, Palmgren MG, Nissen P. Crystal structure of the plasma membrane proton pump. Nature. 2007;450:1111–1114. doi: 10.1038/nature06417. [DOI] [PubMed] [Google Scholar]
- 57.Niittyla T, Fuglsang AT, Palmgren MG, Frommer WB, Schulze WX. Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Mol Cell Proteomics. 2007;6:1711–1726. doi: 10.1074/mcp.M700164-MCP200. [DOI] [PubMed] [Google Scholar]
- 58.Chen Y, Hoehenwarter W, Weckwerth W. Comparative analysis of phytohormone-responsive phosphoproteins in Arabidopsis thaliana using TiO2-phosphopeptide enrichment and mass accuracy precursor alignment. Plant J. 2010;63:1–17. doi: 10.1111/j.1365-313X.2010.04218.x. [DOI] [PubMed] [Google Scholar]
- 59.Benschop JJ, Mohammed S, O’Flaherty M, Heck AJ, Slijper M, Menke FL. Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteomics. 2007;6:1198–1214. doi: 10.1074/mcp.M600429-MCP200. [DOI] [PubMed] [Google Scholar]
- 60.Hayashi Y, Takahashi K, Inoue S, Kinoshita T. Abscisic acid suppresses hypocotyl elongation by dephosphorylating plasma membrane H+-ATPase in Arabidopsis thaliana. Plant Cell Physiol. 2014;55:845–853. doi: 10.1093/pcp/pcu028. [DOI] [PubMed] [Google Scholar]
- 61.Ueno K, Kinoshita T, Inoue S, Emi T, Shimazaki K. Biochemical characterization of plasma membrane H+-ATPase activation in guard cell protoplasts of Arabidopsis thaliana in response to blue light. Plant Cell Physiol. 2005;46:955–963. doi: 10.1093/pcp/pci104. [DOI] [PubMed] [Google Scholar]