Abstract
Irreversible degeneration of the cardiac conduction system is a common disease that can cause activity intolerance, fainting, and death. While electronic pacemakers provide effective treatment, alternative approaches are needed when long-term indwelling hardware is undesirable. Biological pacemakers are composed of electrically active cells that functionally integrate with the heart. Recent findings on cardiac pacemaker cells (PCs) within the sinoatrial node (SAN), along with developments in stem cell technology, have opened a new era in biological pacing. Recent experiments that have derived PC-like cells from non-PCs have brought the field closer than ever before to biological pacemakers that can faithfully recapitulate SAN activity. This review discusses these approaches in the context of SAN biology and addresses the potential for clinical translation.
Cardiac Conduction System Disease and the Need for Biological Pacemakers
Cardiac electrical impulses originate in the sinoatrial node (SAN), a 2–3 centimeter long comma-shaped structure at the junction of the superior vena cava and right atrium.[1] During each heartbeat, the impulse generated in the SAN is transmitted to the neighboring right atrial myocardium.[2] The ensuing wave of depolarization travels throughout the heart and the rest of the cardiac conduction system in a coordinated fashion, triggering sequential contraction of the atria and ventricles. Over an average human lifespan, this sequence is executed over 2 billion times without a major interruption – an extraordinary output that reflects the robustness of cardiac automaticity and impulse transmission. However, under a variety of common pathological conditions, irreversible degeneration or malformation of the cardiac conduction system results in slow heartbeat, activity intolerance, fainting, or even death. At present, there are no drugs appropriate for long-term use that can safely increase heart rate, so the only available treatment is electronic pacemaker implantation for symptomatic or high-risk patients with conduction system disease. In the United States alone, over 200,000 pacemakers are implanted annually, most commonly for degeneration and malfunction of the SAN.[3]
The SAN contains roughly 10,000 specialized pacemaker cells (PCs, see Glossary).[4] Several decades of basic research into the electrophysiological mechanisms involved in PCs automaticity resulted in the identification and cloning of the molecular correlates of critical PC ionic currents [5]. With the ability to introduce exogenous genetic material into human cells in-vitro and in-vivo, there is an ongoing line of research aiming to transform normally quiescent areas of the heart into biological pacemakers. For instance, the introduction of genes encoding ion channels important for PCs, as well as various transplantation approaches of engineered cells represent important advances in the field. These promising studies have been recently reviewed elsewhere and will not be described in this article.[6–9]
A newer line of investigation is premised on the notion that an ideal biological pacemaker would recapitulate the underlying biological properties of genuine PCs in a more comprehensive fashion. In addition to electrophysiological properties, these would include morphology, autonomic responsiveness, source-sink matching, and a dynamic gene expression program unique to PCs. Until fairly recently, the notion of deriving committed PCs from non-PCs would have seemed unfeasible. However, new findings in developmental biology, stem cell biology, and cardiac electrophysiology have now taken PC derivation or reprogramming for cardiac pacing, from the purely theoretical realm, to serious experimental exploration. While recent reviews have explored progress in biological pacing, the aim of this review is to contextualize this work by (1) relating new approaches to biological pacing to recent findings on SAN developmental biology and (2) identifying key basic scientific questions that will need to be addressed so as to move the biological pacing field forward. If existing challenges can be overcome, biological pacemakers created with derived or reprogrammed PCs would constitute a major advance over previous approaches by more faithfully recapitulating the heart’s natural pacing mechanisms.
SAN Structure and Function: Basic Principles
SAN Pacemaker Cells
PCs exhibit automaticity and are responsible for generating the initial electrical impulse that drives each heartbeat (Figure 1).[5] Critical components of the electrical machinery in PCs include the sodium-calcium exchanger [10], voltage-gated calcium channels [11], hyperpolarization-activated cyclic nucleotide gated (hcn) ion channels [12], and spontaneous calcium release from the sarcoplasmic reticulum.[13] Hcn and other channels mediate autonomic responsiveness through their sensitivity to changes in cyclic AMP caused by direct vagal and sympathetic input. Of the Hcn channels, Hcn4 is the most highly expressed in PCs and, when mutated, results in familial sinus bradycardia.[14, 15] Taken together, the electrophysiological output of PCs is the result of a complex interplay of molecules encoded by numerous genes including ion channels, receptors, second messengers and intracellular calcium handling proteins. Moreover, there is a significant debate about the exact mechanisms involved in beat-to-beat regulation of SAN automaticity. Thus, a faithful replication of PC behavior with a biological pacemaker may require reconstitution of the entire PC gene expression program, as opposed to introducing each molecular component individually into quiescent non-PCs.
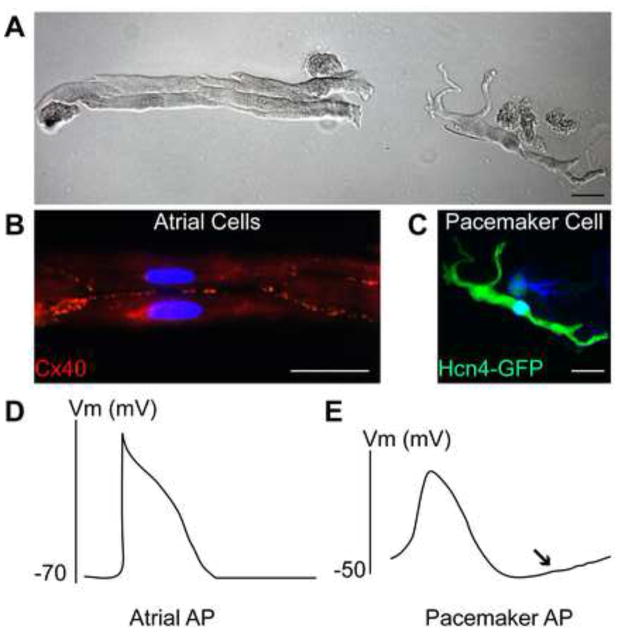
Figure 1. Pacemaker Cells Have Unique Morphology and Electrophysiology [22, 52].
A. Phase-contrast brightfield microscopy image showing isolated mouse cardiomyocytes expressing green fluorescent protein (GFP) under the control of the Hcn4 regulatory region. A cluster of rod-like, striated atrial myocytes is shown on the left. A single pacemaker cell, with striations, but also with several projections and a thinner cell body is shown on the right. B. Fluorescence microscopy image of mouse atrial myocytes as in A. The cluster of GFP-negative atrial myocytes stained with a connexin-40 antibody (red) is indicative of tight cell-cell coupling in atrial tissue. C. Fluorescence microscopy image of a pacemaker cell as in A, positive for Hcn4-GFP and not expressing connexin-40. D. A typical atrial myocyte action potential (AP) has a resting membrane potential of −70 mV or less, a rapid upstroke, and no spontaneous diastolic depolarization. E. A typical pacemaker cell action potential shows a more depolarized maximum diastolic potential, a slow AP upstroke, and a pronounced diastolic depolarization (arrow).
SAN Architecture
Pacemaker cells within the SAN are surrounded by connective tissue and are heterogeneous in phenotype and morphology. To insure robust source-sink matching, cells at the SAN periphery exhibit tighter electrophysiological coupling to surrounding myocardium than cells in the interior of the SAN, reflecting heterogeneity in gene expression within the SAN. [16] Loss of PCs through chronic injury, fibrosis, or apoptosis causes sinus node dysfunction, as does loss of normal SAN architecture.[17] These findings have important implications for development of biological pacemakers, since cellular material may have to adopt particular architectural features in order to achieve robust pacing.
SAN Development and Gene Expression: Recent Discoveries
Developmental Origins of Pacemaker Cells
In an avian model, PC progenitors were shown to arise from right lateral plate mesoderm just posterior to the heart field shortly after gastrulation in response to Wnt signaling cues.[18] Based on fate mapping experiments, these cells migrated to the right inflow region of the heart at mid-development, where they differentiated into PCs (Figure 2A,B).
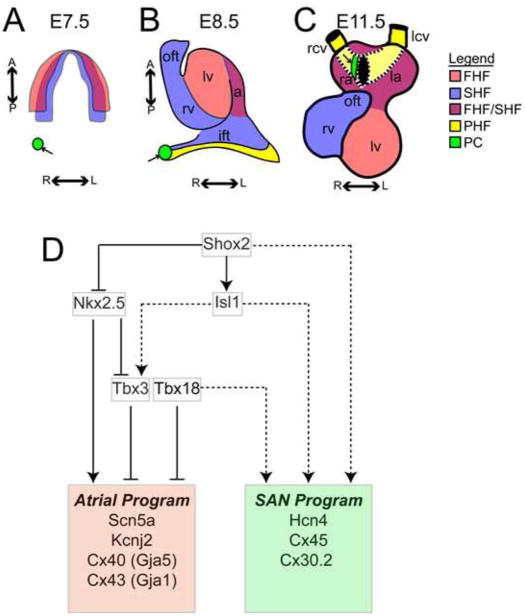
Figure 2. Pacemaker Cell Progenitors Originate Outside the First and Second Heart Fields and Develop a Distinct Transcriptional Network.
A. A chick heart field at stage 8 (corresponding to embryonic day (E) 7.5 in mouse) shows a region forming pacemaker cells (green, arrow), posterior and lateral to the first and second heart fields (FHF/SHF) [18]. B. At chick embryo stage 10 (mouse E8.5), the pacemaker cell progenitors (green, arrow) have migrated to the right inflow tract, a region that is part of the Tbx18+ posterior heart field (PHF) C. The heart is shown at mid-gestation (mouse E11.5) with the anterior atria cut away and the venous inflow to the atria indicated (dashed lines) via the cardinal veins (yellow). The SAN (green, arrow) develops at the junction between the right cardinal vein (yellow, a PHF derived structure) and the right atrium (a mixed FHF/SHF derived structure). D. A contemporary model of the transcriptional network in PCs includes repression of the atrial expression program by Tbx3 and Tbx18[23], activation of the SAN program by Isl1[22, 34] and Shox2, and an antagonistic interaction between Shox2 and Nkx2.5[62]. Solid lines indicate direct transcriptional regulation, while dashed lines indicate transcriptional relationships that may be direct or indirect. Abbreviations: pc, pacemaker cell progenitors; FHF, first heart field; SHF, second heart field; PHF, posterior heart field; oft, outflow tract; rv, right ventricle; lv, left ventricle; ift, inflow tract; ra, right atrium; la, left atrium; rcv, right cardinal vein; lcv, left cardinal vein; R; right, L; left, A; anterior; P; posterior.
Although clonal fate mapping within the mammalian SAN has not yet been reported, the earliest developmental time point at which likely progenitors of PCs can be identified in the mouse is embryonic day (E) 10.5 (corresponding to 4 weeks in human gestation), when a cluster of cells is visible at the junction of the right cardinal vein and posterior right atrium, the location of the future sinoatrial node (Figure 2C).[19, 20] Although the precise moment at which these cells become committed to PC fate in mammals is unknown, explants taken from the mesenchyme posterior to the inflow of the developing heart well before E10.5 can differentiate in-vitro into cells with PC properties, consistent with the findings in avian embryos.[21] Although the earliest pacemaker progenitor cells have not yet been isolated and comprehensively profiled, their rough localization within the embryo and the identification of a few molecular markers have led to novel selection-based approaches to deriving PCs in culture (see below). Because as-yet unidentified local cell autonomous factors may be critical for PC development, these efforts may be greatly facilitated by a deeper understanding of the embryonic microenvironment within which PCs are originally specified.
Transcriptional Regulation in Pacemaker Cells
The transcriptional basis for the PC gene expression program, which exhibits cardiomyocyte as well as neuronal features, is under active investigation. [22] Based on RNA sequencing data, PCs express core cardiac transcription factors Gata4, Mef2C, and Tbx5 at levels similar to working cardiomyocytes, but express Nkx2.5 only at low levels. In addition, a distinct complement of PC-transcription factors is highly expressed, including Tbx18, Tbx3, Shox2, and Isl1 (Figure 2D).
Tbx18 regulates the formation of the entire posterior heart field, the pool of cells from which SAN precursors are believed to emerge. Mice lacking Tbx18 failed to develop a central sinus node and exhibited embryonic lethality with malformation of the sinus venosus.[23] In addition, cultured explants of Tbx18+ mesenchyme from the region of the posterior heart field were able to differentiate into Hcn4+ cells that exhibited spontaneous automaticity and other features of pacemaker cells. Although specific direct transcriptional targets of Tbx18 within pacemaker cells have not been defined, there is evidence that Tbx18 can function as a transcriptional repressor of connexin-43 (Cx43, and other genes) in working cardiomyocytes. [24, 25]
Tbx3 functions as a transcriptional repressor in PCs silencing the gene expression program associated with force-generating cardiomyocytes, which could indirectly promote expression of the PC gene program [26]. In its absence, pacemaker cells lose their distinctive patterns of gene expression, reverting to expression patterns more characteristic of atrial cardiomyocytes.[26] Pacemaker cell function is accordingly compromised in Tbx3 mutants even if Tbx3 loss of function does not prevent PC precursors from differentiating and forming the SAN.[27]
Shox2, like Tbx3, represses the gene expression program of working myocardium, and like Tbx18, is important for the development and morphogenesis of the venous pole of the heart. Mice deficient in Shox2 display embryonic bradycardia, hypoplasia of the venous pole of the heart, and up-regulation of genes normally repressed in PCs.[28, 29] Shox2 also regulates the balance between PC-like properties and working myocyte properties in cells at the specialized interface between the SAN and atrium. This has been shown to occur through Shox2 epistatic interactions with Nkx2.5, promoting Bmp4 expression of.[30, 31] However, its transcriptional role specifically within central SAN PCs has not been fully elucidated.
Isl1 is a LIM-homeodomain transcription factor that is expressed transiently in a large portion of cardiac progenitor cells known as the second heart field.[32] Unlike other components of the second heart field, which express Isl1 transiently before they differentiate into cardiomyocytes, PCs express Isl1 throughout development and into adulthood.[33] Conditional deletion of Isl1 in mouse embryos after heart formation results in global dysregulation of gene expression within PCs, as well as bradycardia and SAN malformation.[22, 34] Bioinformatics analysis indicates that Isl1 acts predominantly as a positive transcriptional regulator with PCs, occupying an upstream role in the transcriptional hierarchy.
Novel Approaches to Biological Pacemakers
Recent attempts to create a biological pacemaker have leveraged these new discoveries on SAN molecular and developmental biology (see Table and Figure 3). One approach has been to use molecular markers identified in embryonic PC precursor cells to isolate and expand rare populations of putative PC progenitors from mixed populations of embryonic stem (ES) cell-derived or reprogrammed cardiomyocytes in-vitro. Ultimately, this selected population of cells would be expanded and transplanted into the heart to functionally couple with host myocardium. A second approach has been to drive expression of transcriptional regulators in non-PCs to direct differentiation into PCs, or to effect direct reprogramming from a non-PC to a PC. Theoretically, this approach could be applied to ES cells, induced pluripotent stem cells (iPSC), or another somatic cell type with a view towards cell transplantation. Alternatively, this approach could be used as gene therapy to deliver a reprogramming cocktail directly to the diseased heart in order to convert resident quiescent cells into PCs.
Table 1.
Properties of Induced or Derived Pacemaker-Like Cells
| Ref | Cell Type | Selection Marker |
Programming Factor(s) |
Beat Rate (bpm) |
MDPa (mV) |
Hcn current density |
Autonomic Response |
PC-like Morphology |
Gene Expression | In-Vitro Coupling |
In-Vivo Pacing |
|---|---|---|---|---|---|---|---|---|---|---|---|
| [39] | mouse embryoid body | Shox2 Promoter | None | N/A | N/A | N/A | N/A | No | Tbx3,Shox2,Gata6, Hcn4, Bmp4 | N/A | N/A |
| [38] | mouse embryoid body | Hcn4-GFP | None | N/A | −50 | High | N/A | N/A | Hcn4, Tbx3, Isl1 | N/A | N/A |
| [41] | mouse embryoid body | ALCAM CD166 | None | 360 | −50 | High | Yes | Yes | Tbx3,Tbx18,Isl1, Shox2,Hcn4,Hcn1 | Yes | N/A |
| [52] | mouse fibroblast | Hcn4-GFP | 4F: Gata6, Tbx3, Tbx5, Rxra or Rarg | Inexcitable | N/A | None | N/A | N/A | Hcn4 | N/A | N/A |
| [52] | Mouse fibroblast | Hcn4-GFP | Gata4, Tbx5, Mef2C, Hand2 | 75 | −40 | N/A | N/A | No | Hcn4 | N/A | N/A |
| [45] | Mouse adult atrial myocyte mouse | None | Tbx3 | Inexcitable | −50 | Low | N/A | No | Hcn1, Lbh | N/A | N/A |
| [46] | embryoid body | Myh6 Promoter | Tbx3 | 350 | −55 | High | Yes | Yes | Cx45,Cx30.2,Hcn4 | Yes | N/A |
| [24] | neonatal rat ventricular myocyte | None | Tbx18 | 100 | −47 | Moderate | Yes | Yes | Hcn4 | Yes | Yes-Guine a Pig |
| [48] | porcine ventricular septum | None | Tbx18 | N/A | N/A | N/A | Yes | N/A | N/A | N/A | Yes-Pig |
| [47] | mouse embryoid body | None | Shox2 | 225 | −70 | N/A | Yes | No | Hcn4,Shox2,Cx45 | Yes | Yes-Rat |
| [51] | mouse embryoid body | None | Isl1 | 100 | N/A | N/A | N/A | No | Hcn4,Cx45,Cx30.2, Bmp4,Tbx3 | N/A | N/A |
Abbreviations: a, maximum diastolic potential
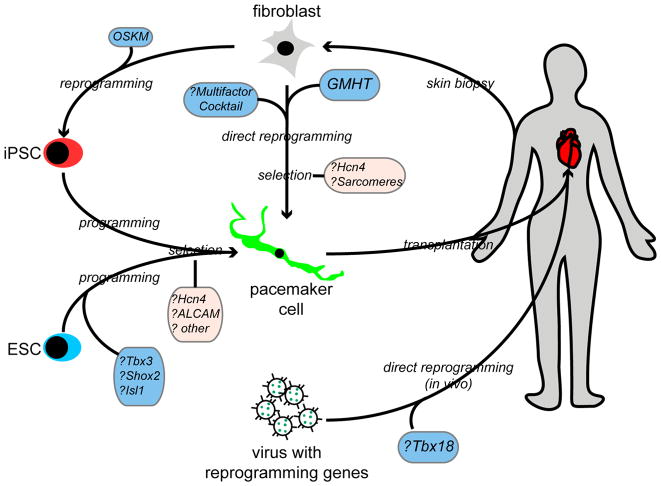
Figure 3. Can Pacemaker-Like Cells be Derived or Programmed from Non-Pacemaker Cells?
This model figure displays several strategies to create a biological pacemaker through derivation of PCs from fibroblasts, pluripotent cells, or from resident cardiomyocytes, as enumerated in Table 1. First, patient-derived fibroblasts could be cultured and directly reprogrammed to PCs using multifactor cocktails, or via Gata4, Mef2C, Hand2, Tbx5 (GMHT) induction. Subsequently, selection for Hcn4 expression and for contractile apparatus could ensue. Alternatively, pluripotent cells (ES cells or iPSC) could be programmed with PC-specific transcriptional regulators, or differentiated into cardiomyocyte progenitors with subsequent selection for PC-like cells. Finally, viral vectors could be delivered directly into the heart, attempting to reprogram resident cardiomyocytes into PCs in-vivo. Abbreviations: ESC, embryonic stem cell; iPSC, induced pluripotent stem cell; OSKM, Oct4, Sox2, Klf4, Myc.
Selection Methods
Early work with human ES cells showed that beating cardiomyocytes with an embryonic-like phenotype and spontaneous electrical activity could be harvested from human embryoid bodies (EBs). [35] Clusters of such cells could integrate into host myocardium in a large animal model of complete heart block, providing a proof of concept for the idea of using cell therapy to generate a biological pacemaker. [36] The discovery that PCs emerge from the posterior heart field led to the hypothesis that PC progenitors might be recoverable from heterogeneous populations of ES-derived cardiomyocytes; this might be achieved by the use of molecular markers expressed by the mesenchyme near the embryonic inflow tract. An expanded collection of pure PC-like cells could then be transplanted to create a more physiologic biological pacemaker [36].
Hcn4
High-level expression of Hcn4 is a hallmark of PCs and Hcn4 expression is present in a steep gradient from the developing inflow tract to the outflow tract early in heart development. [14, 37] Attempts have been made to select Hcn4+ cells from pools of ES-derived cardiomyocytes or from reprogrammed cells. Using a stable ES cell line expressing green fluorescent protein (GFP) under the control of the Hcn4 promoter, GFP+ cells were isolated during differentiation to cardiomyocytes and were characterized electrophysiologically. These cells exhibited spontaneous beating and expressed Hcn channels. [38] However, extensive characterization relative to PCs was not performed, so it is unclear if these cells could be more similar to embryonic cardiomyocytes or PCs.
Shox2
A Shox2 promoter region capable of driving GFP expression was introduced into mouse ES cells. [39] These cells were then differentiated into EBs, as previous work had identified a role for Shox2 in regulating beating rate. [40] Spontaneously beating Shox2-GFP+ aggregates were observed near beating areas within EBs and individual Shox2-GFP+ cells displayed certain features of PCs, including the expression of Hcn4 and connexin-45 (Cx45), and exhibited spontaneous diastolic depolarization, similar to PCs.
Activated Leukocyte Cell Adhesion Molecule (ALCAM)
ALCAM is expressed in the venous pole of the heart during early mouse heart development, with more extensive expression as the heart matures. Unlike other markers of posterior heart field tissue, ALCAM has been shown to be expressed on the cell surface and ALCAM+ cells can be readily purified using an antibody-based FACS assay. ALCAM+ cells isolated from mouse embryoid bodies were reported to differentiate into cells expressing Hcn4 and to exhibit spontaneous automaticity with nodal-like action potentials. [41] Interestingly, these cells also expressed Shox2, Tbx3, Tbx18, and Isl1, the group of factors specific to PCs, along with low levels of genes associated with working myocardium. Aggregates of these cells could pace ventricular myocytes in culture. The embryonic expression of ALCAM may differ from mouse, so it unknown whether these studies can be translated into the human context. However, this study is the most promising of the pure selection-based methods aimed at isolating and expanding PC-like cells from ES cells.
Programming Methods
Combinations of cardiac transcription factors have been shown to directly reprogram fibroblasts into cardiomyocytes in-vitro and in-vivo (see Box 1). [42–44] These and other findings have led to attempts in programming non-cardiomyocytes (either pluripotent cells or somatic cells) into specific cardiomyocyte subtypes, including PCs.
Box 1. Direct Cardiac Reprogramming.
In a series of seminal papers inspired by the use of direct reprogramming to create iPS cells from fibroblasts[53, 54], three or four cardiac transcription factors (Gata4, Mef2c, Tbx5, +/− Hand2; GMTH) were found to directly reprogram fibroblasts into induced cardiomyocyte-like cells (iCMs). In-vitro, iCMs exhibited cardiomyocyte gene expression programs, and, for a small population of cells, spontaneous electrical activity and contraction[42]. Remarkably, in two of these studies, fibroblasts recruited to the site of myocardial infarction in a mouse model of coronary artery ligation could be reprogrammed into iCMs that integrated within the host myocardium, limiting scar size and reducing the post-infarction decline in heart function.[43, 44] Initial studies on human fibroblasts have been performed[58], and large animal studies are underway to test whether this approach may represent a novel method to regenerate heart tissue. This work has spawned a rapidly advancing cardiac reprogramming field. A current area of intense focus in the field is the development of methods to reprogram cells into specific cardiomyocyte subtypes, including PC-like cells, atrioventricular conduction system cells [59–61], atrial cells, and ventricular myocytes.
Tbx3
In mouse models, Tbx3 overexpression in atrial myocardium outside of the SAN is sufficient to recapitulate certain features of pacemaker cells, including expression of Hcn4, down-regulation of genes associated with force-generating myocytes, and functional ectopic automaticity. [26] In addition, a mouse model with a tamoxifen-induced conditionally expressed Tbx3 allele, resulted in increased Tbx3 levels in mature cardiomyocytes, which could then be recovered from the animal for detailed molecular and physiological studies. [45] While Tbx3 overexpression did not lead to increased expression of genes associated with PC automaticity (e.g., Hcn4), it did result in down-regulation of genes associated with rapid conduction and intercellular connectivity, including Gja1, Gja5, Scn5a, and Kcnj2 [45]. These findings support a putative adjunctive role for Tbx3 in biological pacing, programming nodal myocardium away from a force-generating phenotype which could slow conduction, and thereby potentially improving source sink matching between nodal and extra-nodal tissue.
A promising in-vitro study combined Tbx3 overexpression in ES cells with a promoter-based selection method. [46]ES were cells co-transfected with a Myh6 promoter construct that drove neomycin resistance along with a constitutively expressed human TBX3 and differentiated into cardiomyocytes after selecting with neomycin. A surprisingly large fraction (>80%) of the remaining cells displayed electrophysiological features similar to nodal myocytes, including increased protein levels of Hcn4 and spontaneous action potentials with rates comparable to those of bona fide PCs. Clusters of these cells could pace ventricular slices ex-vivo, and exhibited some characteristic morphological features of PCs [46]. It is noteworthy that aberrant expression of Tbx3 causes such different phenotypes between mature atrial myocardium in-vivo and ES cell-derived cardiomyocytes in-vitro. The more dramatic effects observed in ES cells may reflect a limited developmental window in-vivo during which the factor can specify the fate of a multipotent progenitor.
Shox2
The utility of Shox2 to direct differentiation of ES cells into pacemaker cells has been explored in-vitro. [47] Adenoviral transfer of human SHOX2 into mouse ES cells during differentiation resulted in a greater percentage of embryoid bodies with beating cells, faster beating rates, and expression of Hcn4 among other markers. Transplantation of EBs from SHOX2-transduced ES cells into the left ventricles of rat hearts with surgically induced complete heart block resulted in evidence of pacing activity arising from the injection sites, consistent with biological pacemaker activity [47].
Tbx18
In a recent study, neonatal rat ventricular myocytes (NRVMs) were infected with viral expression vectors containing Shox2, Tbx3, Tbx5 or Tbx18 and subsequently assessed for spontaneous automaticity. [25] Of these factors, only Tbx18 resulted in a robust increase in spontaneous beating rate. Tbx18-transduced NRVMs were found to exhibit physiological features of pacemaker cells, including expression of Hcn4, cellular automaticity, and a thinner, elongated cellular morphology. In a porcine model, Tbx18 expressing adenovirus was delivered to the interventricular septa of pigs with complete heart block. Pigs receiving human TBX18 gene transfer experienced accelerated heart rate when compared to controls, and also exhibited automaticity which emanated from the focal site of gene injection. [48] Furthermore, dependence of the pigs on an electronic pacemaker was reduced in the TBX18-transduced pigs relative to controls. Staining of the injection area revealed changes in cellular morphology that could be consistent with induction of pacemaker-like cells, and analysis of mRNA levels showed downregulation of working cardiomyocyte genes (Cx43, Kir2.1, Nkx2.5, and Nav1.5) with upregulation of HCN4. While these results are extremely promising, recovery of TBX18-transduced cells from pigs for single-cell immunohistochemical or electrophysiological analysis remains limited [48]. Because of immunogenicity of the viral vectors that were used to transduce cells with TBX18, the biological pacing effect of TBX18 was transient, so whether this technique could result in robust long-term cellular reprogramming and pacing activity remains to be determined. While these studies represent a pioneering attempt to generate biological pacing through in situ reprogramming, much remains to be discovered about the molecular and genetic mechanisms that underlie these observations.
Isl1
Isl1 was initially identified as a cardiovascular progenitor marker, so Isl1+ cardiomyocyte precursors have long been seen as a potential cell source for cardiac regenerative therapies.[49, 50] Recent discoveries on the role of Isl1 in PC development[22, 34] have prompted a reexamination of its potential to augment differentiation towards the PC fate in culture. Overexpression of Isl1 in ES cells and in Xenopus laevis hearts was shown to result in increased proliferation and differentiation of cardiomyocyte precursors, with a shift in balance of nodal versus working cardiomyocytes. [51] Isl1 overexpression was also reported to result in increased cellular automaticity and increased Hcn4 expression, with reduced expression of genes associated with working myocytes. These results raise the possibility that Isl1 may be able to promote differentiation towards a pacemaker cell fate.
Multifactor Programming
A recent study sought to reprogram fibroblasts specifically to a pacemaker cell fate using a combination of factors deeemed important for pacemaker cell development and function. [52] Fibroblasts from a mouse line expressing GFP under the control of Hcn4 regulatory regions were infected with viral expression vectors bearing 20 candidate transcription factors relevant to PC development. Using an approach pioneered by Shinya Yamanaka and colleagues [53, 54], successively narrowing combinations of the original 20 factors, identified the smallest group of factors that could promote GFP expression. A combination of Tbx5, Tbx3, Gata6, and either Rarg or Rxra induced GFP expression in up to 40% of transfected fibroblasts. Surprisingly, reprogrammed GFP+ cells exhibited neither spontaneous nor provoked electrical activity, and lacked a sarcomeric apparatus, suggesting that while some degree of reprogramming occurred, the cells failed to adopt a cardiac fate [52]. Further work using Hcn4-GFP derived fibroblasts evaluated the propensity of traditional reprogramming factors Gata4, Hand2, Mef2C, and Tbx5 to yield PC-like reprogrammed myocytes. About 32% of reprogrammed fibroblasts exhibited an expression profile consistent with either pacemaker cells or immature non-working cardiomyocytes [52]. These results suggest that while fibroblast reprogramming to a PC lineage may be possible, the optimal combination of reprogramming factors and selection method has yet to be identified.
Limitations of Current Approaches
Thus far, studies have not been consistent in the criteria used to evaluate putative induced, derived, or reprogrammed PC-like cells. Varying combinations of electrophysiological behavior, gene expression, and morphological criteria have been employed, rarely with quantitative comparisons to bona-fide PCs. In this context, it is noteworthy that embryonic cardiomyocytes exhibit robust automaticity due to current through Hcn channels [55], along with the dynamic and combinatorial expression of various cardiac transcription factors. Therefore, it may be difficult to distinguish between an undifferentiated embryonic cardiomyocyte with some unusual features, from a bona fide PC progenitor. This may prove to be particularly challenging in-vitro in the context of different selection methods. The recent publication of a comprehensive expression profile of PCs from developing mouse SAN may contribute in establishing the absolute and relative expression of numerous PC-enriched genes. [22] However, no genome-wide transcriptome data sets have been published for reprogrammed or derived PC-like cells, making detailed genome-wide comparisons with bona fide PCs impossible.
Another potential limitation to current approaches is the lack of a detailed understanding of the earliest committed PC progenitors, which have not yet been isolated or characterized in mammalian embryos. Therefore, there may be important unidentified transcriptional regulators or signaling pathways that are critical to early PC development. Further research on the earliest stages of SAN development may reveal new clues about how to select for and program precursor cells into PCs.
Finally, none of the methods described have successfully translated these new approaches to human cells. Because of considerable interspecies differences between rodent and human cardiovascular systems, translatability of SAN and PC biology from mouse to human is by no means guaranteed. Tbx18-based somatic reprogramming in pigs with complete heart block remains the only demonstration of the potential for reprogramming in a clinically relevant model system.[48] However, even in that study, the cellular and molecular bases for the observed effects remain to be fully elucidated. In the future, it will be essential that better tools and methods be developed in order to carry out mechanistic studies of the reprogramming process in large animals.
Clinical Translation
If current approaches to biological pacing exhibit long-term success in relevant animal models, significant obstacles will remain before this technology can be widely adopted to treat patients with symptomatic bradycardia. Several of these hurdles, including the development of improved delivery methods of genes and cells to target areas, the possibility of pro-arrhythmic effects of biological pacemakers, and the potential for off-target or oncogenic effects of stem cell and transcription-factor based therapies have been recently reviewed and are not detailed here [6, 7] Moreover, there may be genetic factors that predispose patients – particularly younger ones -- to PC dysfunction. In such cases, reprogramming with the patient’s own cells could result in dysfunctional PCs and suboptimal results.
Perhaps the biggest obstacle facing this promising technology will be to define its clinical niche. Patients with conduction system disease are usually well served by electronic pacemakers, so a biological pacemaker would have to meet a very high standard of performance to supplant existing technology. [56] Nevertheless, some important clinical roles for biological pacing have already been envisioned. For example, patients with active endovascular infections require removal of pacemaker hardware from the body in order to undergo device-free interval antibiotic therapy. Because temporary transvenous pacemakers -- the current standard of care -- are associated with significant morbidity, a technology such as Tbx18-based somatic reprogramming could provide temporary backup pacing during hardware-free intervals.[48] Another possibility would be to implement biological pacing in children with severe bradycardia who face a lifetime with indwelling hardware, battery changes, and lead revisions. [57] A self-sustaining biological pacemaker providing long-term physiological pacing would be transformative for such patients.
More broadly, a full development of this technology could potentially reverse the underlying pathology of conduction system disease. For example, in patients with incipient sinus node dysfunction, would it be possible to deliver reprogramming factors directly to affected areas in the SAN, block the disease-related loss of PCs and restore source-sink matching? Could a diseased SAN be rejuvenated and the need for an electronic pacemaker delayed or prevented? Future research will hopefully address these questions. If the challenges already outlined can be overcome, then this therapy would constitute a true paradigm shift in the management of bradycardia.
Concluding Remarks
The quest for a biological pacemaker has gained momentum from recent discoveries on the molecular biology of the SAN. A variety of stem cell and reprogramming methods have been developed to derive putative PCs from non-PCs. The most promising of these have either used PC-specific markers to select for PC-like progenitors during differentiation of ES cells into cardiomyocytes; or they have used transcriptional regulators that are active in PCs to reprogram non-PCs into PC-like cells. While translation of these studies to human cells has not yet occurred, the earliest large animal proof-of-concept study with direct reprogramming has yielded promising results.[48] Although many obstacles remain, as our understanding of basic SAN biology moves forward (see Outstanding Questions), the field is poised for rapid progress over the next few years. In addition to several technical hurdles, defining the precise clinical roles for biological pacing will be an important challenge as the field matures. If these difficulties can be overcome, a biological pacemaker composed of derived SAN-like cells which circumvents the need for indwelling hardware, would represent a major advance in health care.
Trends Box.
Recent progress in the transcriptional regulation of myocardial sinoatrial node development has guided the derivation of novel biological pacemakers using forward programming of pluripotent cells, or direct somatic reprogramming.
Transduction of embryonic stem (ES) cells with Tbx3 or via selection of ALCAM+ cells from differentiating ES cells has resulted in cell populations with pacemaker cell-like gene expression and automaticity.
Tbx18-based somatic reprogramming has altered differentiated ventricular myocytes into a pacemaker-like phenotype by modifying gene expression, cellular morphology, electrical behavior, and improving heart rhythm in a large animal model of heart block.
Multifactor reprogramming methods simultaneously incorporating several sinoatrial node factors have shown promise but require further development.
Continued progress in understanding early pacemaker cell differentiation and transcriptional regulation at the genome-wide level are poised to accelerate progress in heart programming technology.
Acknowledgments
VV is supported by grants from the NIH/NHLBI (K08-HL101989) and the American Heart Association (14BGIA20490269). The author wishes to thank Dr. Deepak Srivastava, Director of the Gladstone Institute of Cardiovascular Disease, for ongoing mentorship and research support, as well as the Cardiology Division, University of California, San Francisco, for research support.
Glossary
- Automaticity
A cellular electrophysiological behavior whereby an electrically active cell fires rhythmic action potentials spontaneously (without depolarization by another cell). This property is critical for pacemaker cell function and relies on the expression of a particular set of ion channels
- Bradycardia
Slow heart beat, defined as less than 60 beats per minute. Symptomatic bradycardia occurs when heart rate is too low to meet physiological demand, causing activity intolerance, lightheadedness, or fainting. If no reversible causes are present, the only safe and effective long-term treatment is permanent pacemaker implantation
- Biological Pacemaker
A collection of cells engineered for the purpose of electrically pacing a heart with conduction system disease, thereby preserving heart rhythm. Biological pacemakers can be created by engineering cells in-vitro for transplantation in the heart, or by delivery of genes into a specific target area of the heart to create a biological pacemaker in-situ
- Complete Heart Block
A type of cardiac conduction system disease in which electrical transmission is disrupted at the atrioventricular junction or within the His-Purkinje system, so that SAN impulses activate the atria but do not ultimately trigger ventricular activation. To restore heart rhythm in complete heart block, a biological pacemaker would have to be introduced into the ventricular myocardium
- Embryoid Body (EB)
An ES-derived structure consisting of a heterogeneous group of cells in the process of differentiating from ES cells. Embryoid bodies frequently contain collections of ES-derived cardiomyocytes that can be visualized by spontaneous beating. An EB usually contains cell types from all three germ layers in a disorganized syncytium
- Hcn Channels
Hyperpolarization activated, cyclic nucleotide gated ion channels. Unlike other voltage-gated ion channels, members of the Hcn family of channels conduct an inward cation current in response to hyperpolarization, creating a diastolic depolarization and rhythmic firing (automaticity). The current carried by Hcn channels is called “funny current”
- Heart Field
A region within the embryonic lateral plate mesoderm that forms the heart. At least three spatially and molecularly distinct subpopulations of cells contribute to different regions of the heart: (1) a first heart field that expresses Tbx5 and Nkx2.5 and forms the left ventricle, part of the atria, and part of the interventricular septum, (2) a second heart field that expresses Isl1 and forms the right ventricle, outflow tract, and large portions of the atria, and (3) a posterior part of the second heart field that expresses Tbx18 and forms the sinus venosus myocardium
- Mesenchyme
Undifferentiated mesodermal tissue. Upon receiving particular cues or signals during development, mesenchymal cells can differentiate into numerous cell types, including specific cardiomyocyte subtypes
- Pacemaker cells
Cardiac pacemaker cells (PCs) are a specialized subtype of cardiomyocyte that exhibit spontaneous electrical activity (automaticity). PCs have distinctive morphological features and a unique set of ion channels and receptors to permit their function. Loss of PCs is an important cause of sinus node dysfunction
- Sinus Node Dysfunction
A common form of cardiac conduction system disease in which a combination of PC loss and fibrosis leads to failure of impulse generation and/or impulse transmission in the SAN
- Source-Sink Matching
A circumstance in which a current source, such as the sinoatrial node (SAN), generates enough current to active a neighboring tissue (the atrium) which is often much larger than the source. Failure of source sink matching will prevent impulse transmission. This leads to the phenomenon of exit block, where SAN generates impulses but they cannot leave the SAN to capture the atrium and no heartbeat is triggered
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Keith A, Flack M. The Form and Nature of the Muscular Connections between the Primary Divisions of the Vertebrate Heart. J Anat Physiol. 1907;41(Pt 3):172–89. [PMC free article] [PubMed] [Google Scholar]
- 2.Fedorov VV, et al. Optical mapping of the isolated coronary-perfused human sinus node. J Am Coll Cardiol. 2010;56(17):1386–94. doi: 10.1016/j.jacc.2010.03.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mond HG, Proclemer A. The 11th world survey of cardiac pacing and implantable cardioverter-defibrillators: calendar year 2009--a World Society of Arrhythmia’s project. Pacing Clin Electrophysiol. 2011;34(8):1013–27. doi: 10.1111/j.1540-8159.2011.03150.x. [DOI] [PubMed] [Google Scholar]
- 4.Bleeker WK, et al. Functional and morphological organization of the rabbit sinus node. Circ Res. 1980;46(1):11–22. doi: 10.1161/01.res.46.1.11. [DOI] [PubMed] [Google Scholar]
- 5.Mangoni ME, Nargeot J. Genesis and regulation of the heart automaticity. Physiol Rev. 2008;88(3):919–82. doi: 10.1152/physrev.00018.2007. [DOI] [PubMed] [Google Scholar]
- 6.Meyers JD, Jay PY, Rentschler S. Reprogramming the conduction system: Onward toward a biological pacemaker. Trends Cardiovasc Med. 2015 doi: 10.1016/j.tcm.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boink GJ, et al. The past, present, and future of pacemaker therapies. Trends Cardiovasc Med. 2015 doi: 10.1016/j.tcm.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Li RA. Gene- and cell-based bio-artificial pacemaker: what basic and translational lessons have we learned? Gene Ther. 2012;19(6):588–95. doi: 10.1038/gt.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosen MR, et al. The road to biological pacing. Nat Rev Cardiol. 2011;8(11):656–66. doi: 10.1038/nrcardio.2011.120. [DOI] [PubMed] [Google Scholar]
- 10.Groenke S, et al. Complete atrial-specific knockout of sodium-calcium exchange eliminates sinoatrial node pacemaker activity. PLoS ONE. 2013;8(11):e81633. doi: 10.1371/journal.pone.0081633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mesirca P, Torrente AG, Mangoni ME. Functional role of voltage gated Ca(2+) channels in heart automaticity. Front Physiol. 2015;6:19. doi: 10.3389/fphys.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baruscotti M, et al. Deep bradycardia and heart block caused by inducible cardiac-specific knockout of the pacemaker channel gene Hcn4. Proc Natl Acad Sci U S A. 2011;108(4):1705–10. doi: 10.1073/pnas.1010122108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lakatta EG, V, Maltsev A, Vinogradova TM. A coupled SYSTEM of intracellular Ca2+ clocks and surface membrane voltage clocks controls the timekeeping mechanism of the heart’s pacemaker. Circ Res. 2010;106(4):659–73. doi: 10.1161/CIRCRESAHA.109.206078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Frigola C, Shi Y, Evans SM. Expression of the hyperpolarization-activated cyclic nucleotide-gated cation channel HCN4 during mouse heart development. Gene Expr Patterns. 2003;3(6):777–83. doi: 10.1016/s1567-133x(03)00125-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milanesi R, et al. Familial sinus bradycardia associated with a mutation in the cardiac pacemaker channel. N Engl J Med. 2006;354(2):151–7. doi: 10.1056/NEJMoa052475. [DOI] [PubMed] [Google Scholar]
- 16.Verheijck EE, et al. Electrophysiological features of the mouse sinoatrial node in relation to connexin distribution. Cardiovasc Res. 2001;52(1):40–50. doi: 10.1016/s0008-6363(01)00364-9. [DOI] [PubMed] [Google Scholar]
- 17.Morris GM, Kalman JM. Fibrosis, electrics and genetics. Perspectives in sinoatrial node disease. Circ J. 2014;78(6):1272–82. doi: 10.1253/circj.cj-14-0419. [DOI] [PubMed] [Google Scholar]
- 18.Bressan M, Liu G, Mikawa T. Early mesodermal cues assign avian cardiac pacemaker fate potential in a tertiary heart field. Science. 2013;340(6133):744–8. doi: 10.1126/science.1232877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mommersteeg MTM, et al. Molecular pathway for the localized formation of the sinoatrial node. Circulation Research. 2007;100(3):354–62. doi: 10.1161/01.RES.0000258019.74591.b3. [DOI] [PubMed] [Google Scholar]
- 20.Viragh S, Challice CE. The development of the conduction system in the mouse embryo heart. Dev Biol. 1980;80(1):28–45. doi: 10.1016/0012-1606(80)90496-0. [DOI] [PubMed] [Google Scholar]
- 21.Mommersteeg MTM, et al. The sinus venosus progenitors separate and diversify from the first and second heart fields early in development. Cardiovascular Research. 2010 doi: 10.1093/cvr/cvq033. [DOI] [PubMed] [Google Scholar]
- 22.Vedantham V, et al. RNA Sequencing of Mouse Sinoatrial Node Reveals an Upstream Regulatory Role for Islet-1 in Cardiac Pacemaker Cells. Circ Res. 2015 doi: 10.1161/CIRCRESAHA.116.305913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiese C, et al. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circulation Research. 2009;104(3):388–97. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- 24.Kapoor N, et al. Transcriptional suppression of connexin43 by TBX18 undermines cell-cell electrical coupling in postnatal cardiomyocytes. J Biol Chem. 2011;286(16):14073–9. doi: 10.1074/jbc.M110.185298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapoor N, et al. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat Biotechnol. 2013;31(1):54–62. doi: 10.1038/nbt.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoogaars WM, et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21(9):1098–112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frank DU, et al. Lethal arrhythmias in Tbx3-deficient mice reveal extreme dosage sensitivity of cardiac conduction system function and homeostasis. Proc Natl Acad Sci U S A. 2012;109(3):E154–63. doi: 10.1073/pnas.1115165109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Espinoza-Lewis RA, et al. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2–5. Dev Biol. 2009;327(2):376–85. doi: 10.1016/j.ydbio.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blaschke RJ, et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115(14):1830–8. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- 30.Ye W, et al. A common Shox2-Nkx2–5 antagonistic mechanism primes the pacemaking cell fate in the pulmonary vein myocardium and sinoatrial node. Development. 2015 doi: 10.1242/dev.120220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun C, et al. The short stature homeobox 2 (Shox2)-bone morphogenetic protein (BMP) pathway regulates dorsal mesenchymal protrusion development and its temporary function as a pacemaker during cardiogenesis. J Biol Chem. 2015;290(4):2007–23. doi: 10.1074/jbc.M114.619007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai CL, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5(6):877–89. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinberger F, et al. Localization of Islet-1-positive cells in the healthy and infarcted adult murine heart. Circ Res. 2012;110(10):1303–10. doi: 10.1161/CIRCRESAHA.111.259630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang X, et al. Transcription factor ISL1 is essential for pacemaker development and function. J Clin Invest. 2015 doi: 10.1172/JCI68257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Satin J, et al. Mechanism of spontaneous excitability in human embryonic stem cell derived cardiomyocytes. J Physiol. 2004;559(Pt 2):479–96. doi: 10.1113/jphysiol.2004.068213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kehat I, et al. Electromechanical integration of cardiomyocytes derived from human embryonic stem cells. Nat Biotechnol. 2004;22(10):1282–9. doi: 10.1038/nbt1014. [DOI] [PubMed] [Google Scholar]
- 37.Vedantham V, et al. Spatiotemporal regulation of an Hcn4 enhancer defines a role for Mef2c and HDACs in cardiac electrical patterning. Dev Biol. 2013;373(1):149–62. doi: 10.1016/j.ydbio.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morikawa K, et al. Identification, isolation and characterization of HCN4-positive pacemaking cells derived from murine embryonic stem cells during cardiac differentiation. Pacing Clin Electrophysiol. 2010;33(3):290–303. doi: 10.1111/j.1540-8159.2009.02614.x. [DOI] [PubMed] [Google Scholar]
- 39.Hashem SI, Claycomb WC. Genetic isolation of stem cell-derived pacemaker-nodal cardiac myocytes. Mol Cell Biochem. 2013;383(1–2):161–71. doi: 10.1007/s11010-013-1764-x. [DOI] [PubMed] [Google Scholar]
- 40.Hashem SI, et al. Shox2 regulates the pacemaker gene program in embryoid bodies. Stem Cells Dev. 2013;22(21):2915–26. doi: 10.1089/scd.2013.0123. [DOI] [PubMed] [Google Scholar]
- 41.Scavone A, et al. Embryonic stem cell-derived CD166+ precursors develop into fully functional sinoatrial-like cells. Circ Res. 2013;113(4):389–98. doi: 10.1161/CIRCRESAHA.113.301283. [DOI] [PubMed] [Google Scholar]
- 42.Ieda M, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142(3):375–86. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qian L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485(7400):593–8. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song K, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485(7400):599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bakker ML, et al. T-box transcription factor TBX3 reprogrammes mature cardiac myocytes into pacemaker-like cells. Cardiovasc Res. 2012;94(3):439–49. doi: 10.1093/cvr/cvs120. [DOI] [PubMed] [Google Scholar]
- 46.Jung JJ, et al. Programming and isolation of highly pure physiologically and pharmacologically functional sinus-nodal bodies from pluripotent stem cells. Stem Cell Reports. 2014;2(5):592–605. doi: 10.1016/j.stemcr.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ionta V, et al. SHOX2 overexpression favors differentiation of embryonic stem cells into cardiac pacemaker cells, improving biological pacing ability. Stem Cell Reports. 2015;4(1):129–42. doi: 10.1016/j.stemcr.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu YF, et al. Biological pacemaker created by minimally invasive somatic reprogramming in pigs with complete heart block. Sci Transl Med. 2014;6(245):245ra94. doi: 10.1126/scitranslmed.3008681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laugwitz KL, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433(7026):647–53. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moretti A, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127(6):1151–65. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 51.Dorn T, et al. Direct nkx2–5 transcriptional repression of isl1 controls cardiomyocyte subtype identity. Stem Cells. 2015;33(4):1113–29. doi: 10.1002/stem.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nam YJ, et al. Induction of diverse cardiac cell types by reprogramming fibroblasts with cardiac transcription factors. Development. 2014;141(22):4267–78. doi: 10.1242/dev.114025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 55.Yasui K, et al. I(f) current and spontaneous activity in mouse embryonic ventricular myocytes. Circ Res. 2001;88(5):536–42. doi: 10.1161/01.res.88.5.536. [DOI] [PubMed] [Google Scholar]
- 56.Udo EO, et al. Long term quality-of-life in patients with bradycardia pacemaker implantation. Int J Cardiol. 2013;168(3):2159–63. doi: 10.1016/j.ijcard.2013.01.253. [DOI] [PubMed] [Google Scholar]
- 57.Wilhelm BJ, et al. Complications and Risk Assessment of 25 Years in Pediatric Pacing. Ann Thorac Surg. 2015;100(1):147–53. doi: 10.1016/j.athoracsur.2014.12.098. [DOI] [PubMed] [Google Scholar]
- 58.Fu JD, et al. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Reports. 2013;1(3):235–47. doi: 10.1016/j.stemcr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rentschler S, et al. Myocardial Notch signaling reprograms cardiomyocytes to a conduction-like phenotype. Circulation. 2012;126(9):1058–66. doi: 10.1161/CIRCULATIONAHA.112.103390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsai SY, et al. Efficient Generation of Cardiac Purkinje Cells from ESCs by Activating cAMP Signaling. Stem Cell Reports. 2015;4(6):1089–102. doi: 10.1016/j.stemcr.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maass K, et al. Isolation and characterization of embryonic stem cell-derived cardiac Purkinje cells. Stem Cells. 2015;33(4):1102–12. doi: 10.1002/stem.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ye W, et al. A common Shox2-Nkx2–5 antagonistic mechanism primes the pacemaker cell fate in the pulmonary vein myocardium and sinoatrial node. Development. 2015;142(14):2521–32. doi: 10.1242/dev.120220. [DOI] [PMC free article] [PubMed] [Google Scholar]