Abstract
A common feature of ribonucleic acids (RNAs) is that they can undergo a variety of chemical modifications. As nearly all of these chemical modifications result in an increase in the mass of the canonical nucleoside, mass spectrometry has long been a powerful approach for identifying and characterizing modified RNAs. Over the past several years, significant advances have been made in method development and software for interpreting tandem mass spectra resulting in approaches that can yield qualitative and quantitative information on RNA modifications, often at the level of sequence specificity. We discuss these advances along with instrumentation developments that have increased our ability to extract such information from relatively complex biological samples. With the increasing interest in how these modifications impact the epitranscriptome, mass spectrometry will continue to play an important role in bioanalytical investigations revolving around RNA.
Keywords: modified nucleosides, RNA sequencing, tRNA, rRNA, modified bases, tandem mass spectrometry, LC-MS/MS, MALDI-MS/MS, CARD, epitranscriptome
1. Introduction
Ribonucleic acids (RNA) serve crucial biological roles at the heart of the Central Dogma of molecular biology, where deoxyribonucleic acid (DNA) is transcribed into messenger RNA (mRNA) that is finally translated into proteins. RNAs that do not code for a protein are referred to as non-coding RNAs (ncRNA). A large number of ncRNAs have been discovered and studied including ribosomal (rRNA), transfer (tRNA), long non-coding (lncRNA), small nuclear (snRNA), small nucleolar (snoRNAs), small interfering (siRNAs) and micro (miRNA).
A common feature of nearly all these types of RNAs is that they can undergo a variety of chemical modifications that occur posttranscriptionally.1, 2 These RNA modifications are enzymatically introduced to either the ribose or nucleobase of nucleosides after RNA has been transcribed from the genomic DNA.3–5 Such modifications are extremely diverse, with over 100 now characterized and catalogued.6, 7 Figure 1 contains structures for several posttranscriptionally modified nucleosides that illustrate the diversity of chemical modifications possible. Such modifications can alter canonical and non-canonical RNA-RNA interactions resulting in structures and function beyond those found for the canonical nucleosides adenosine, cytidine, uridine and guanosine.5, 8 The types and levels of modification vary significantly among these various ncRNAs and mRNAs. While most analyses of modified RNAs have focused on the more heavily modified tRNAs and rRNAs,9–13 all RNAs seem to contain at least some chemical modifications and require methods for their analysis. More recently, it has been found that at least some of these modifications are regulated and reversible, leading to the concept of an epitranscriptome, whose modification status can influence gene expression in a manner similar to what is already known about the epigenome.14, 15
Figure 1.
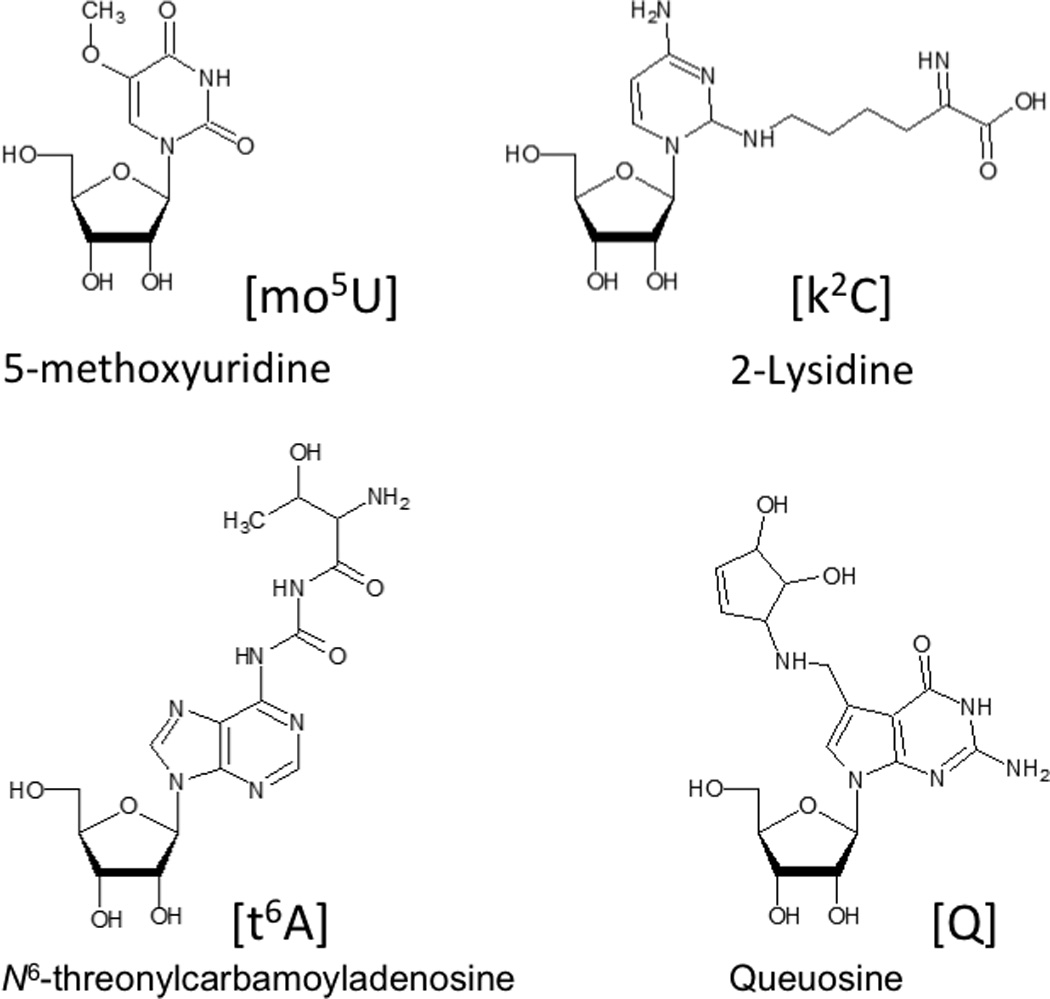
Selected examples of the variety of chemical modifications found in naturally occurring nucleosides.
2. Mass Spectrometric Analysis of Modified Nucleosides
There are a significant number of analytical methods that have been used to detect and identify chemical modifications in RNA. Such methods include reverse transcriptase polymerase chain reaction (RT-PCR), thin layer chromatography (TLC), electrophoresis, microarrays, next-generation sequencing and mass spectrometry.15–23 As shown over 20 years ago by McCloskey,24, 25 the primary utility of using mass spectrometry for such measurements is that this method provides a direct readout on any chemical modifications that change the mass of the canonical nucleoside. Remarkably, with the exception of pseudouridine (ψ) modifications, all other described modified nucleosides result in this desired mass shift that enables direct MS detection.
2.1 Sample Prep for MS Analysis
Modified RNAs are analyzed using matrix-assisted laser desorption/ionization (MALDI) or electrospray ionization (ESI) sources due to the well-documented advantages of these ionization methods. Regardless of the ionization method used, sample preparation remains a key step for successful mass spectrometric analysis of modified oligoribonucleotides and RNAs.
A key concern with ionization of oligoribonucleotides and RNAs is cation adduction, which can lead to charge neutralization and loss of ion signal. Cations are adducted because oligonucleotides possess a negatively charged phosphodiester backbone at neutral pH. In solution, Coulombic repulsion from adjacent charges is reduced by the presence of solvent. However, once solvent is removed, neutralization of the backbone charges occurs through cation adduction. Cation adducts, except H+ or NH4+, are undesirable as they reduce the sensitivity by spreading the ion species in multiple mass values, cause peak broadening and result in possible spectral interferences making data more difficult to interpret. Thus, a significant and general key to efficiently ionizing oligonucleotides is the need to reduce or eliminate the presence of cation adducts from the sample or solution prior to analysis.26–28
One area of concern is the effect of sample preparation on modification structure.29, 30 A recent example is the work from the Suzuki lab demonstrating that the modified nucleoside N6-threonylcarbamoyladenosine (t6A), which is commonly found in tRNAs, is actually present in vivo as the cyclic form (ct6A) and that sample preparation often leads to hydrolysis and ring opening prior to mass spectrometry detection.31 In addition, it has previously been assumed that modified nucleosides are polar, with few exceptions. However, it was recently found that geranylated RNA, which is lipophilic, can be detected by mass spectrometry provided appropriate sample analysis steps are used.32 These examples illustrate the care that is necessary when preparing and analyzing biological samples by mass spectrometry as an understanding of the types and chemistries of modified RNAs will become ever more important in the future as technology enables more and more sensitive characterization of such samples.
After appropriate sample preparation, modified RNAs can be analyzed at three different regimes: the monomer (i.e., nucleotide/nucleoside) level, as smaller oligonucleotides (e.g., endonuclease digestion products in a bottom-up approach) or as intact RNAs (via a top-down approach). Recent examples of analytical methods for each of these regimes will be discussed below.
2.2 Modified Nucleosides
One of the easiest measurements to conduct on modified RNAs is to enzymatically digest intact RNAs into constituent ribonucleosides, which are amenable to separation and analysis by liquid chromatography tandem mass spectrometry (LC-MS/MS). The key recent development in this regime is the focus on quantitative measurements at both high precision and accuracy. Dedon, Begley and coworkers have created a highly effective targeted LC-MS/MS protocol for the quantitative measurement of modified ribonucleosides.28, 33 They have demonstrated the utility of this protocol for analyzing changes in posttranscriptional modifications in Saccharomyces cerevisiae that result from environmental stress.34, 35
While the Dedon approach typically uses only a single internal standard for quantitative analysis, other researchers have explored options for including a greater number of internal standards. Carell and co-workers have reported the synthesis of multiple modified ribonucleosides, all of which can incorporate one or more isotopic labels.36, 37 An alternative strategy was established by Helm and co-workers, wherein standards for modified ribonucleosides are generated by sample culturing in isotopically labeled medium.38 The added advantage of this latter approach is that the diversity of standards is only limited by the organism’s inherent modification machinery.
Taking advantage of new gas-phase separation technology, Fabris and co-workers have eliminated upfront nucleoside separation by LC and instead used ion mobility coupled to mass spectrometry for the direct analysis of modified nucleosides.39 The potential power of this rapid approach for direct transcriptome-wide detection of modified nucleosides was recently reported by Rose,40 and one can anticipate future developments that allow structural isomers to be more clearly differentiated in these non-chromatographic approaches.
2.3 Bottom-up Modification Mapping
Bottom-up RNA modification mapping uses one or more enzymes, typically an endoribonuclease (RNase), which can digest a larger RNA into smaller oligoribonucleotides that can be conveniently analyzed by MALDI- or LC-MS/MS.41, 42 These analyses take advantage of the advent of genomic sequencing, where ncRNA species can be inferred by analyzing an organism’s genome using motif recognizing software.43 In this manner, the genomic sequence of the RNA of interest is already known, thus the experimental goals are to simply place modified nucleosides into their appropriate sequence location.
We have recently developed several different analytical protocols that are based on bottom-up RNA modification mapping. A multiple enzyme “digest and go” strategy, outlined in Figure 2, was used to create a modification map for the complete set of tRNAs isolated from the bacterial organism Lactococcus lactis.44 The key to this approach is the use of multiple endonucleases to increase mapping coverage as well as high quality MS/MS data. An alternative strategy is to use one or more RNAs of known modification status as a reference for comparison against RNAs of unknown modification status.45, 46 This CARD (comparative analysis of RNA digests) approach is most suitable when comparing wild-type against mutant strains or healthy versus diseased samples.
Figure 2.
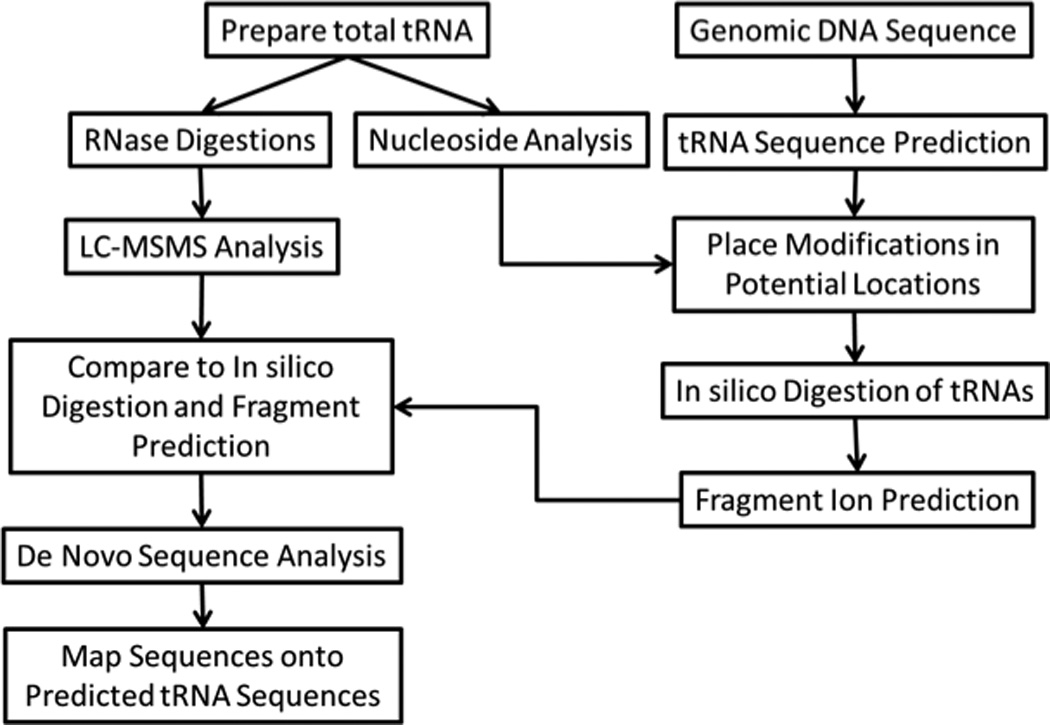
An experimental scheme for mapping modifications on tRNA sequences from total tRNA by a bottom-up strategy is shown. This scheme takes advantage of pre-existing tRNA gene sequences and matches modified nucleosides with MS/MS-based sequence information to map modifications onto the known tRNA sequences.
The bottom-up approach has also been enhanced by use of metabolically labeled RNAs that can be used for the identification of modified oligonucleotides47 or for the relative quantification of changes in RNA modification status.48, 49 Figure 3 illustrates the former, wherein the base composition of the RNase digestion product can be determined based on the m/z measurement and then constrained by the number of carbons or nitrogens present, which is determined by the mass shift induced by the metabolic label.47 Alternatively, metabolic labeling to deplete heavy isotopes simplifies detection and identification of larger molecular weight RNase digestion products, and has been used to improve the CARD approach for identification of modified RNAs (Figure 4).50
Figure 3.
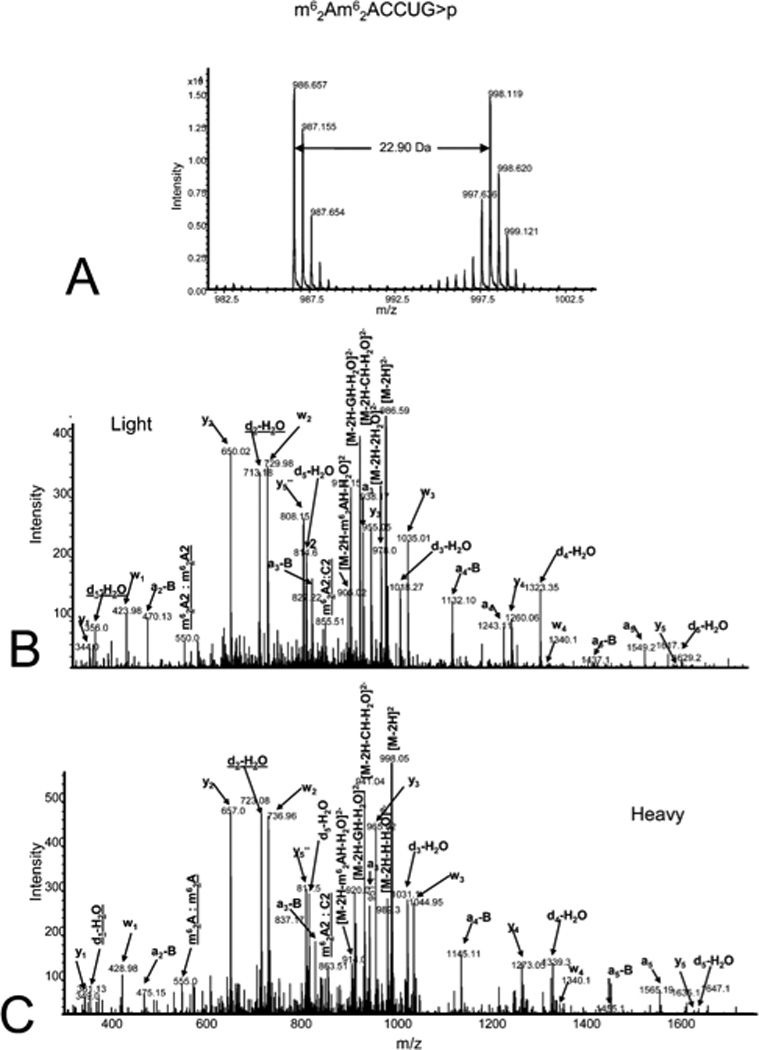
Ultrahigh resolution time-of-flight mass spectrometry analysis of oligoribonucleotide [m62A][m62A]CCUG>p. A. TOF MS spectrum shows both the light (m/z 986.657) and heavy (m/z 998.119) corresponding to monoisotopic masses of 1975.314 and 1998.238 Da, respectively. B. Tandem MS spectra of the light oligoribonucleotide. C. Tandem MS spectra of the heavy oligoribonucleotide. The predominant fragment ions are highlighted. Figure reproduced with permission from S. P. Waghmare; M. J. Dickman. “Characterization and quantification of RNA post-transcriptional modifications using stable isotope labeling of RNA in conjunction with mass spectrometry analysis” Analytical Chemistry 2011, 83, 4894. Copyright 2011 American Chemical Society.
Figure 4.
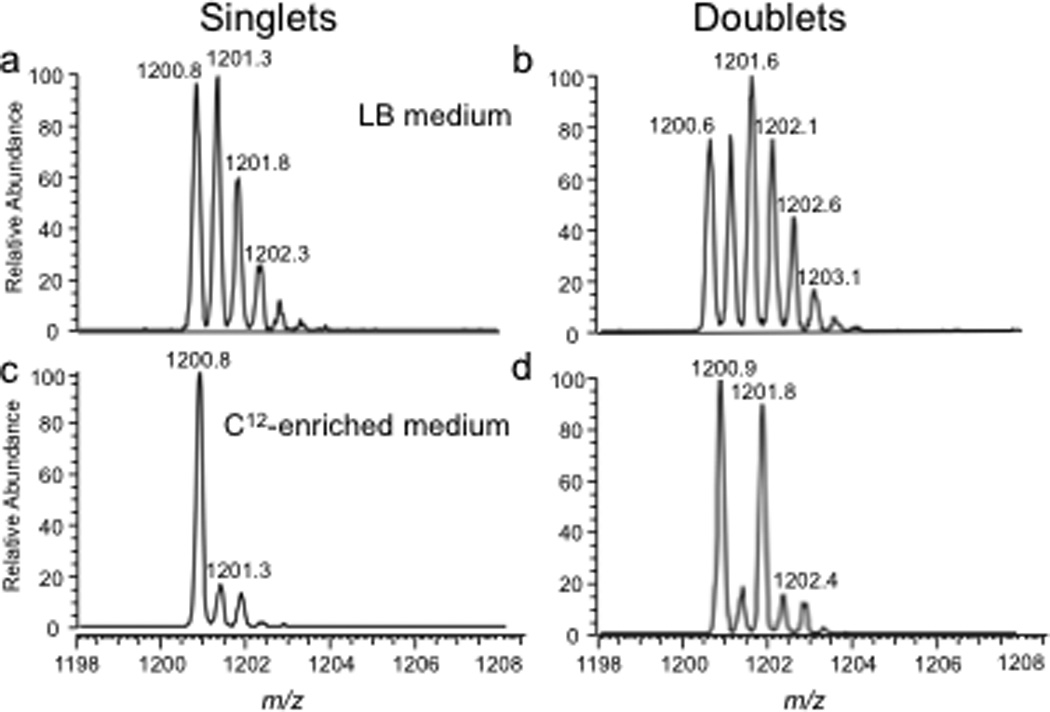
Improvements in singlet and doublet identification using 12C-enriched medium as illustrated with the doubly-charged E. coli total tRNA RNase T1 digestion product A[ms2i6A]AACCGp (MW 2403.4 Da). A. Mass spectrum from sample grown in LB medium and labeled with 16O during RNase T1 digestion. B. Same sample as in (A) except labeled with both 16O and 18O during RNase T1 digestion. C. Mass spectrum obtained when sample grown in 12C-enriched medium and labeled with 16O during RNase T1 digestion. D. Same sample as in (C) except labeled with both 16O and 18O during RNase T1 digestion. Singlet and doublet identifications are simplified in (C) and (D), respectively, by use of 12C-enriched medium. Figure reproduced with permission from C. Wetzel; S. Li; P. Limbach. “Metabolic de-isotoping for improved LC-MS characterization of modified RNAs” Journal of the American Society for Mass Spectrometry 2014, 25, 1114.
2.4 Top-down Modification Mapping
Top-down sequencing methods can also be used to map modifications onto large RNA sequences.51, 52 In this regime, the RNA of interest is first isolated and purified, then subsequently dissociated in the mass spectrometer using one or more activation methods (see below). Unlike bottom-up approaches, a top-down method can provide nearly complete sequence and modification mapping coverage for any particular RNA, although specialized mass spectrometry instrumentation is required. Breuker and co-workers have recently identified optimized ESI conditions that improve MS (and subsequent MS/MS) ion yields for top-down approaches.53 As impressively illustrated by this group, top-down modification mapping offers a powerful alternative to bottom-up approaches and can be used for both synthetically modified as well as biological modified RNAs.
3 Developments in Techniques and Instrumentation
During the past several years, the most significant instrumental advances in the analysis of modified nucleosides have arisen due to improvements in tandem mass spectrometry (MS/MS) and new chromatographic techniques. Moreover, several other technologies including ion mobility and droplet-based sample introduction reveal promising future directions for the field.
3.1 Chromatography
While there are multiple examples of using MALDI-MS for the identification of modified RNAs,41, 54 the more popular approach has been to combine upfront liquid chromatography separation with ESI-MS.55 It was shown very early that ion pair reverse phase liquid chromatography (IP-RP-LC) is most effective for hyphenated LC-MS (and LC-MS/MS) analysis of oligonucleotides.56, 57 The most common mobile-phase contains an ion pairing component (typically triethylamine, TEA) along with an additive, such as hexafluoroisopropanol (HFIP), that improves ionization.58 One challenge of the TEA/HFIP mobile phase for oligoribonucleotides is the carryover effects of TEA, along with the background signal (in negative polarity) from HFIP. These concerns have led to a general consensus in the field that LC-MS of oligonucleotides requires a dedicated system.
To combat this general consensus, multiple labs have investigated alternatives to the TEA/HFIP mobile phase.58–60 One challenge faced by investigators seeking optimal LC conditions for on-line separation and MS analysis is the impact of oligonucleotide sample on the optimization of LC conditions. For example, Gong and McCullagh reported that hexylamine was the best ion pairing reagent for separation of synthetic oligonucleotides (Figure 5).59 In contrast, Erb and Oberacher reported that TEA/HFIP is the best system for quantitative analysis by LC-MS, while cyclohexyldimethylammonium acetate (CycHDMAA) should replace TEA as the ion pairing reagent for qualitative analyses.58
Figure 5.
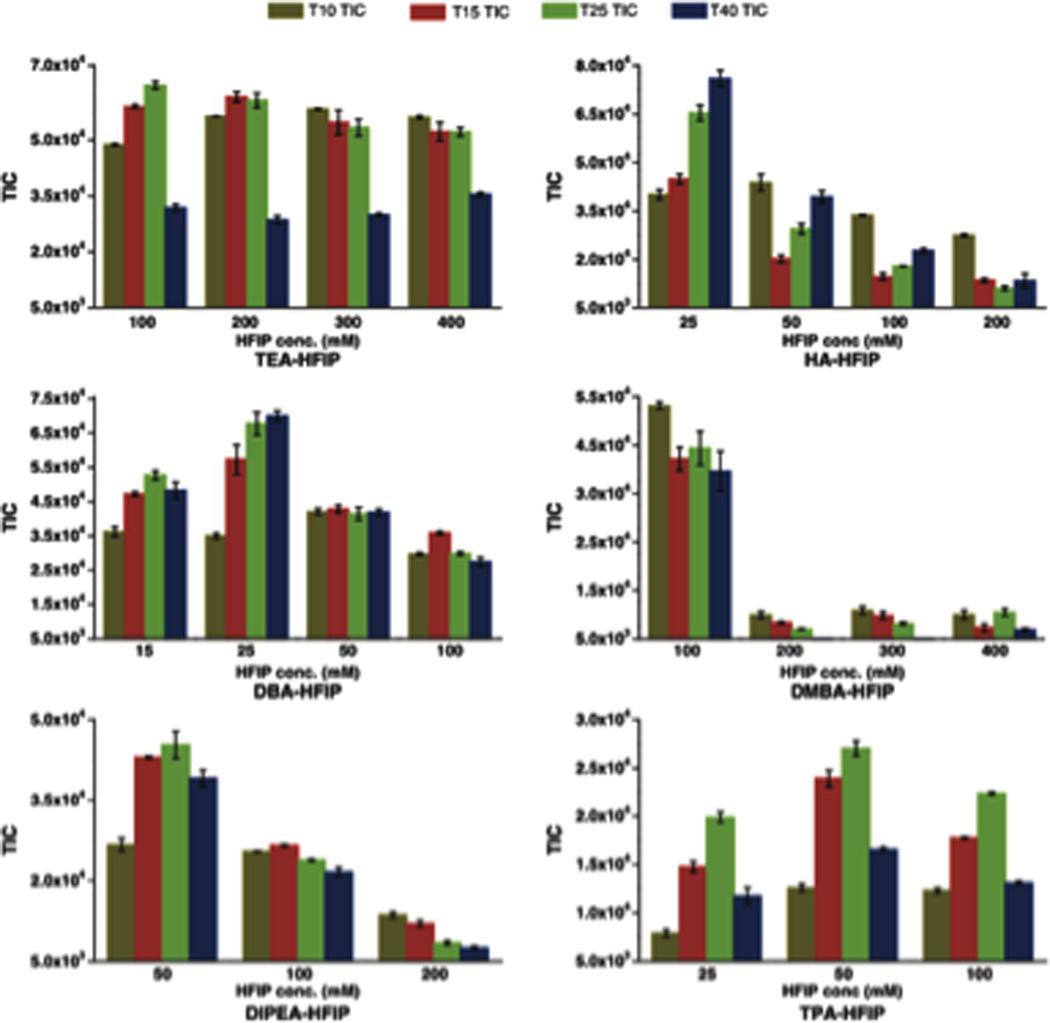
Effect of ion pairing reagent concentration on MS signal sensitivity and the amount of adduct ions formed with oligonucleotides using different IP-HIFP buffers (3 replicates). LC conditions: Mobile phase A (MPA) – IP reagent (5–30 mM) and 100 mM HFIP, Mobile phase B – MPA in 80% MeOH, 4% MeOH in 5 min, 0.2 mL/min, column temperature 45 °C, injected 1 µL except 3 µL for TEA. Figure reproduced with permission from L. Gong; J. S. O. McCullagh. “Comparing ion-pairing reagents and sample dissolution solvents for ion-pairing reversed-phase liquid chromatography/electrospray ionization mass spectrometry analysis of oligonucleotides” Rapid Communications in Mass Spectrometry 2014, 28, 339.
Bartlett and co-workers have conducted one of the most comprehensive studies of alternatives to the TEA/HFIP mobile phase,60 and have reported appropriate experimental conditions for optimal oligoribonucleotide separation and ionization (Figure 6).61 Among their key findings was the development of a model that would predict optimal ion pairing reagents based on sample hydrophobicity. Tripropylamine (TPA), diisopropylamine (DIPA), dimethylbutylamine (DMBA) and diisopropylethylamine (DIEA) were found to be better ion pairing reagents than TEA for samples of moderate (TPA) to high (DMBA, DIEA) hydrophobicity. An added advantage of these findings is that the amount of HFIP counterion can be reduced when using these alternatives.61
Figure 6.
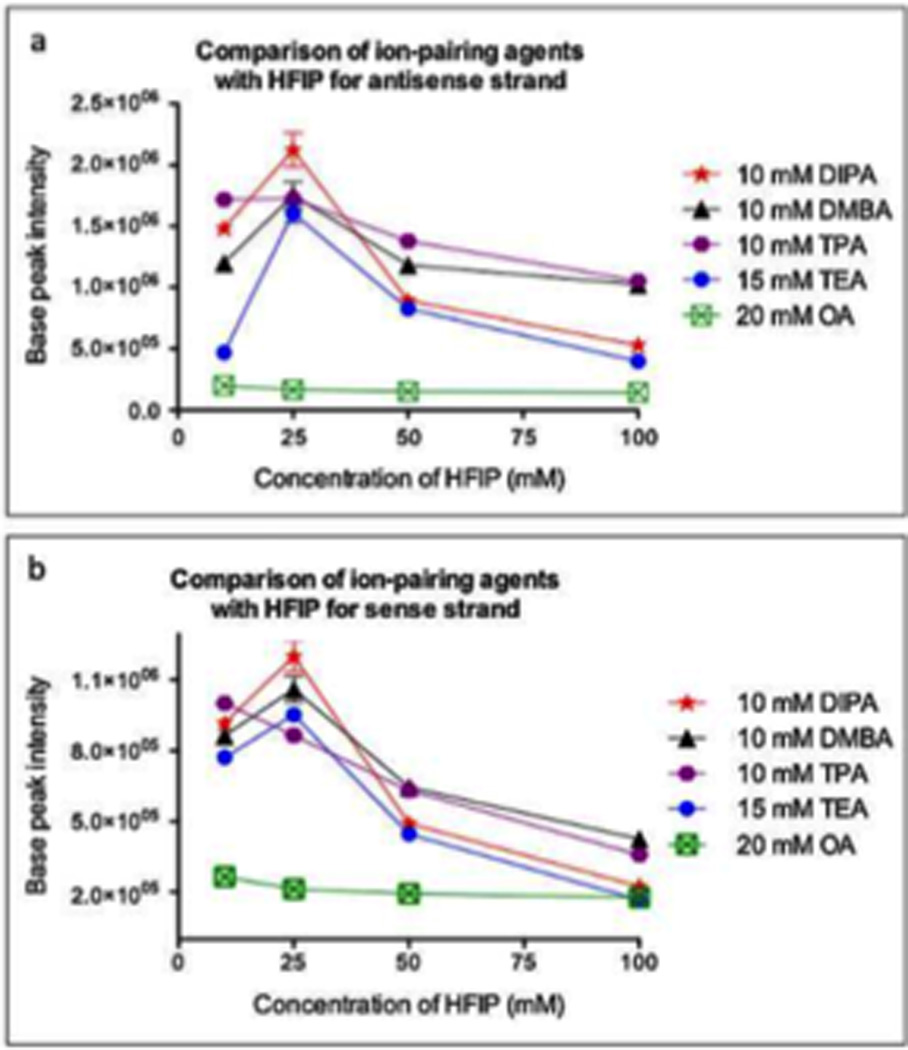
Comparison of the base peak ion intensity of siRNA with alkylamines and various concentrations of HFIP: (a) antisense strand and (b) sense strand. Experiments were performed in triplicate, and data are presented as the mean ± the standard deviation. Figure reproduced with permission from A. C. McGinnis; E. C. Grubb; M. G. Bartlett. “Systematic optimization of ion-pairing agents and hexafluoroisopropanol for enhanced electrospray ionization mass spectrometry of oligonucleotides” Rapid Communications in Mass Spectrometry 2013, 27, 2655.
Another recent development is the use of ammonium acetate-based mobile phases for oligoribonucleotide separations.48 While such mobile phases were among the earliest explored for LC-MS, they were rapidly replaced by the TEA/HFIP method due to the latter’s significant gains in sensitivity. However, for analyses that are not sample limited, these ammonium acetate compositions can reduce any instrumentation-related carry-over, providing practitioners with more flexibility in instrumentation operation.
3.2 Tandem Mass Spectrometry
The analysis of modified RNAs by tandem mass spectrometry has become routine thanks to numerous fundamental studies by several research groups.62–64 Sequence-specific information can be generated by applying energy to intact RNA oligomers in the gas phase, leading to predictable backbone fragmentation, which can locate modifications. More specifically, when activated using collision induced dissociation (CID), oligoribonucleotides typically fragment along the phosphodiester backbone to generate two separate mass ladders. During CID, the most labile fragmentation pathways generate complementary c-type and y-type fragment ions, with w-type and a-base fragment ions also generated although at much lower abundance.62 The presence of CID-generated mass ladders can be exploited to locate modifications within the oligoribonucleotide sequence through the unique mass shift generated by the posttranscriptional modification. Alternatives to CID include higher-energy collisional dissociation (HCD), which yields similar types of fragment ions as CID albeit at slightly greater amounts,65 and electron dissociation detachment (EDD), which predominantly generates d-type and w-type fragment ions.53
As the number of MS/MS investigations of modified RNAs has increased, modification-specific fragmentation patterns have been observed in several cases.63, 66 Labile modified nucleosides can yield strong neutral loss pathways that dominate the MS/MS spectrum (Table 1). Figure 7 illustrates the effect of labile modifications on mass spectral data. Importantly, these non-standard fragmentation pathways can be exploited to generate modification-specific tandem mass spectrometry assays.67–69
Table 1.
Neutral loss occurs when a labile nucleobase fragments instead of the phosphodiester backbone. The neutral losses in this table are those that have been found to dominate MS/MS spectra when present in oligoribonucleotides, often making sequence determination difficult.
| Ion | Neutral Loss (Da) |
|---|---|
| PO3H2 | 80.974 |
| [m7G] | 165.065 |
| [k2C] | 144.090 |
| [Q] | 115.064 |
| [oQ] | 131.058 |
| [t6A] | 162.064 |
Key: m7G – 7-methylguanosine; k2C – lysidine; Q – queuosine; oQ – epoxyqueuosine; t6A – N6-threonylcarbamoyladenosine.
Figure 7.
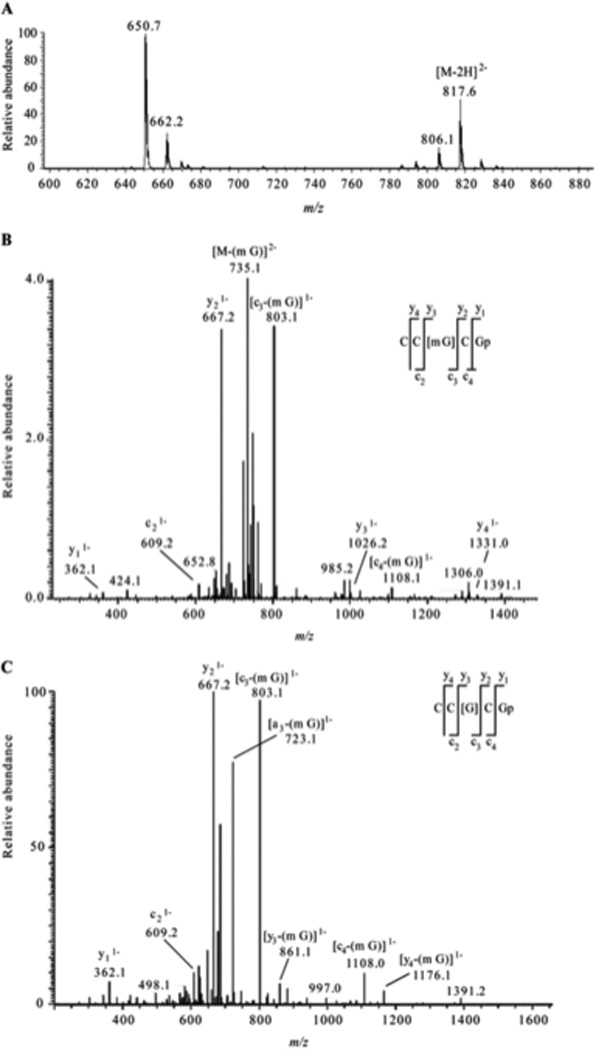
A. Electrospray mass spectrum corresponding to the peak seen in the extracted ion chromatogram for m/z 817.6 of the RNaseT1digest of 5 µg of 16S rRNA from H37Rv M. tuberculosis. The mass spectral data are consistent with the doubly charged ion that would be expected for CC[mG]CG. The other m/z values in this mass spectrum correspond to additional RNase T1 digestion products from 16S rRNA. B. Collision-induced dissociation (CID) mass spectrum of the RNase T1 digestion product at m/z 817.6 shown in panel A. C. CID mass spectrum of the fragment with m/z 735.1 shown in panel B. The observed sequence informative fragments correspond to the expected fragmentation pattern of oligonucleotide CC[mG]CG, which has an mG-base loss. The absence of mG is depicted as [G] in the sequence representation. Sequence-informative fragment ions are labeled following the nomenclature of McLuckey et al.82 Figure reproduced from S. Y. Wong; B. Javid; B. Addepalli; G. Piszczek; M. B. Strader; P. A. Limbach; C. E. Barry, 3rd. “Functional role of methylation of G518 of the 16S rRNA 530 loop by GidB in Mycobacterium tuberculosis” Antimicrob Agents Chemother 2013, 57, 6311.
3.3 Ion Mobility Mass Spectrometry
A promising instrumental advancement, which is underutilized in the mass spectrometry analysis of modified RNAs, is ion mobility spectrometry (IMS), which adds an extra dimension of separation in the gas phase. IMS can separate ions by shape and charge density, referred to as a collisional cross section. Fabris and colleagues have already demonstrated the utility of IMS-MS for the gas-phase separation and analysis of modified nucleosides.39, 40 Further, gas phase mobility mass spectrometry has been used to elucidate structural changes of several intact non-modified microRNAs, RNA complexes, as well as changes in HIV viral RNA assembly.70–72 However, the integration of IMS with MS for the characterization of modified oligoribonucleotides has not yet been reported.
3.4 New Ionization Methods
As mentioned earlier in this review, the most popular ionization methods for modified oligoribonucleotides and RNAs are MALDI and ESI. Recently we investigated droplet-based sample introduction, using induction based fluidics (IBF), as a means of generating ESI-like data from oligonucleotides.73 It appears that droplet-based sample introduction may retain the benefits of ESI, while reducing sampling bias for oligoribonucleotide mixtures. A different alternative has been the use of plasma-based ionization sources for the detection of oligoribonucleotides.74, 75 Although their utility is limited to very specific applications, detection based on the presence of phosphate can introduce selectivity into the analysis. Finally, despite the numerous advances in ambient ionization sources for mass spectrometry,76 there are no significant reports of their use in the analysis of modified RNAs.
3.5 Advances in Software and Bioinformatics
One of the more substantial advances in the area of mass spectrometry of modified RNAs has been in the generation of new software tools and bioinformatics resources. While the number of publicly available RNA programs is significantly smaller than those available in the field of proteomics,77 they are necessary to manage the large datasets, such as those generated by LC-MS/MS approaches.
Schürch and colleagues, building upon their advances in understanding CID fragmentation of modified RNAs, created the OMA-/OPA-suite of software for simplifying interpretation of MS/MS data.78 This software can take inputted RNA sequences, calculate expected MS/MS sequence ladders and then compare the calculated ladders against experimental data to score the strength of the match. A different strategy was used by Sample et al., who created RoboOligo to handle de novo sequence analysis of modified oligoribonucleotides.79 This software program builds up potential oligoribonucleotide sequences with user-selected modified nucleosides based on the input MS/MS data. A useful feature of this software is the ability to map modifications onto an intact RNA in an automated fashion. When these new tools are added to the existing software resources,54, 80 the interested analyst has multiple options available to simplify the interpretation of MS/MS data obtained from modified RNAs.
4. Future Outlook
RNA-based mass spectrometry has advanced significantly over the past several years. The combination of improved analytical methods and protocols combined with advances in software for analysis of MS/MS data greatly simplifies the challenge of characterizing samples that can contain numerous posttranscriptionally modified nucleosides. While RNA-based mass spectrometry still lags the field of proteomics, many of the analytical improvements that first arose within proteomics are now a common feature for characterizing modified RNAs. Yet, despite these advances, there remain areas where improvements would be valued.
As noted earlier in this review, a common challenge for more widespread acceptance of using mass spectrometry for the characterization of modified RNAs is the challenge posed by ion pairing reagents, which can often leave residual contamination within the HPLC system. New developments in HPLC stationary phases or separation modalities that reduce the requirement for an ion pairing reagent would be welcomed. Moreover, additives that can improve ionization efficiency without background effects – especially if combined with less problematic ion pairing reagents such as ammonium acetate – would also benefit this field.
One can anticipate that mass spectrometry will become even more important as additional findings are revealed about the epitranscriptome and the biological importance of modifications becomes better defined.81 While mass spectrometry cannot compete with the throughput available using next-generation sequencing technologies, those technologies cannot match the ability of mass spectrometry to identify nearly all of the modifications present within a single sample. However, should proteomics-like pipelines for RNA modification mapping be developed, then mass spectrometry can provide an important role in validating findings from sequencing approaches at throughputs appropriate for transcriptome-level analysis.
Acknowledgments
Financial support for this work is provided by the National Institutes of Health (GM058843), the National Science Foundation (CHE1507357), the Defense Threat Reduction Agency (HDTRA1-15-1-0033) and the University of Cincinnati.
References
- 1.Carell T, Brandmayr C, Hienzsch A, Müller M, Pearson D, Reiter V, Thoma I, Thumbs P, Wagner M. Angewandte Chemie (International ed in English) 2012;51:7110–7131. doi: 10.1002/anie.201201193. [DOI] [PubMed] [Google Scholar]
- 2.Helm M, Alfonzo JD. Chem Biol. 2014;21:174–185. doi: 10.1016/j.chembiol.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moukadiri I, Garzon MJ, Bjork GR, Armengod ME. Nucleic Acids Res. 2013;42:2602–2623. doi: 10.1093/nar/gkt1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiaville PC, Iwata-Reuyl D, de Crécy-Lagard V. RNA Biol. 2014;11:1529–1538. doi: 10.4161/15476286.2014.992277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giegé RJL. In: DNA and RNA modification enzymes: comparative structure, mechanism, functions, cellular interactions and evolution. Grosjean H, editor. Austin, TX: Landes Bioscience; 2009. p. 18. [Google Scholar]
- 6.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FAP, Fabris D, Agris PF. Nucleic Acids Res. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Machnicka M, Milanowska K, Osman O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother K, Helm M, Bujnicki J, Grosjean H. Nucleic Acids Res. 2012:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doudna JA. Nature Struct. Biol. 2000;7:954–956. doi: 10.1038/80729. [DOI] [PubMed] [Google Scholar]
- 9.Akashi H. Curr Opin Genet Dev. 2001;11:660–666. doi: 10.1016/s0959-437x(00)00250-1. [DOI] [PubMed] [Google Scholar]
- 10.Fujishima K, Kanai A. Frontiers in Genetics. 2014;5:142. doi: 10.3389/fgene.2014.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guymon R, Pomerantz SC, Crain PF, McCloskey JA. Biochemistry. 2006;45:4888–4899. doi: 10.1021/bi052579p. [DOI] [PubMed] [Google Scholar]
- 12.Ermolaeva MD. Curr Issues Mol Biol. 2001;3:91–97. [PubMed] [Google Scholar]
- 13.Dittmar KA, Mobley EM, Radek AJ, Pan T. J Mol Biol. 2004;337:31–47. doi: 10.1016/j.jmb.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 14.Fu L, Guerrero C, Zhong N, Amato N, Liu Y, Liu S, Cai Q, Ji D, Jin S, Niedernhofer L, Pfeifer G, Xu G, Wang Y. J Amer Chem Soc. 2014;136:3. doi: 10.1021/ja505305z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Y, Dominissini D, Rechavi G, He C. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 16.Yokogawa T, Kumazawa Y, Miura K, Watanabe K. Nucleic Acids Res. 1989;17:2623–2638. doi: 10.1093/nar/17.7.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanduc D. Prep. Biochem. 1994;24:167–174. doi: 10.1080/10826069408010090. [DOI] [PubMed] [Google Scholar]
- 18.Kumazawa Y, Yokogawa T, Hasegawa E, Miura K, Watanabe K. J Biol Chem. 1989;264:13005–13011. [PubMed] [Google Scholar]
- 19.Kumazawa Y, Yokogawa T, Tsurui H, Miura K, Watanabe K. Nucleic Acids Res. 1992;20:2223–2232. doi: 10.1093/nar/20.9.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaefer M, Pollex T, Hanna K, Lyko F. Nucleic Acids Res. 2009;37:e12. doi: 10.1093/nar/gkn954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cattenoz PB, Taft RJ, Westhof E, Mattick JS. RNA. 2013;19:257–270. doi: 10.1261/rna.036202.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song C-X, Yi C, He C. Nature Biotechnology. 2012;30:1107–1116. doi: 10.1038/nbt.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birkedal U, Christensen-Dalsgaard M, Krogh N, Sabarinathan R, Gorodkin J, Nielsen H. Angewandte Chemie International Edition. 2015;54:451–455. doi: 10.1002/anie.201408362. [DOI] [PubMed] [Google Scholar]
- 24.McCloskey JA, Nishimura S. Acc Chem Res. 1977;10:403–409. [Google Scholar]
- 25.Kowalak JA, Pomerantz SC, Crain PF, McCloskey JA. Nucleic Acids Res. 1993;21:4577–4584. doi: 10.1093/nar/21.19.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castleberry CM, Rodicio LP, Limbach PA. Curr Protoc Nucleic Acid Chem. 2008;Chapter 10(Unit 10 12) doi: 10.1002/0471142700.nc1002s35. [DOI] [PubMed] [Google Scholar]
- 27.Van Dongen WD, Niessen WMA. Bioanalysis. 2011;3:541–564. doi: 10.4155/bio.11.8. [DOI] [PubMed] [Google Scholar]
- 28.Cai WM, Chionh YH, Hia F, Gu C, Kellner S, McBee ME, Ng CS, Pang YLJ, Prestwich EG, Lim KS, Ramesh Babu I, Begley TJ, Dedon PC. In: Methods Enzymol. Chuan H, editor. Vol. 560. Academic Press; 2015. pp. 29–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasai H, Murao K, Nishimura S, Liehr JG, Crain PF, McCloskey JA. European Journal of Biochemistry. 1976;69:435–444. [Google Scholar]
- 30.Crain PF. Methods Enzymol. Vol. 193. Academic Press; 1990. pp. 782–790. [DOI] [PubMed] [Google Scholar]
- 31.Miyauchi K, Kimura S, Suzuki T. Nature Chem Biol. 2013;9:105–111. doi: 10.1038/nchembio.1137. [DOI] [PubMed] [Google Scholar]
- 32.Dumelin C, Chen Y, Leconte A, Chen Y, Liu D. Nat Chem Biol. 2012;8:913–919. doi: 10.1038/nchembio.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su D, Chan CTY, Gu C, Lim KS, Chionh YH, McBee ME, Russell BS, Babu IR, Begley TJ, Dedon PC. Nat. Protocols. 2014;9:828–841. doi: 10.1038/nprot.2014.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan CT, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ. PLoS Genet. 2010;6:e1001247. doi: 10.1371/journal.pgen.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dedon PC, Begley TJ. Chem Res Toxicol. 2014;27:330–336. doi: 10.1021/tx400438d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brandmayr C, Wagner M, Brückl T, Globisch D, Pearson D, Kneuttinger AC, Reiter V, Hienzsch A, Koch S, Thoma I, Thumbs P, Michalakis S, Müller M, Biel M, Carell T. Angewandte Chemie (International ed in English) 2012;51:11162–11165. doi: 10.1002/anie.201203769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Globisch D, Pearson D, Hienzsch A, Brückl T, Wagner M, Thoma I, Thumbs P, Reiter V, Kneuttinger AC, Müller M, Sieber SA, Carell T. Angewandte Chemie (International ed in English) 2011;50:9739–9742. doi: 10.1002/anie.201103229. [DOI] [PubMed] [Google Scholar]
- 38.Kellner S, Neumann J, Rosenkranz D, Lebedeva S, Ketting RF, Zischler H, Schneider D, Helm M. Chemical communications. 2014;50:3516–3518. doi: 10.1039/c3cc49114e. [DOI] [PubMed] [Google Scholar]
- 39.Quinn R, Basanta-Sanchez M, Rose RE, Fabris D. J Mass Spectrom. 2013;48:703–712. doi: 10.1002/jms.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rose RE, Quinn R, Sayre JL, Fabris D. RNA. 2015;21:1361–1374. doi: 10.1261/rna.049429.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Douthwaite S, Kirpekar F. Methods Enzymol. 2007;425:1–20. doi: 10.1016/S0076-6879(07)25001-3. [DOI] [PubMed] [Google Scholar]
- 42.Gaston KW, Limbach PA. RNA Biol. 2014;11:1568–1585. doi: 10.4161/15476286.2014.992280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowe TM, Eddy SR. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puri P, Wetzel C, Saffert P, Gaston KW, Russell SP, Cordero Varela JA, van der Vlies P, Zhang G, Limbach PA, Ignatova Z, Poolman B. Mol Microbiol. 2014;93:944–956. doi: 10.1111/mmi.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li S, Limbach P. Anal Chem. 2012;84:8607–8613. doi: 10.1021/ac301638c. [DOI] [PubMed] [Google Scholar]
- 46.Li S, Limbach P. Analyst. 2013;138:1386–1394. doi: 10.1039/c2an36515d. [DOI] [PubMed] [Google Scholar]
- 47.Waghmare SP, Dickman MJ. Anal Chem. 2011;83:4894–4901. doi: 10.1021/ac200547y. [DOI] [PubMed] [Google Scholar]
- 48.Popova AM, Williamson JR. JACS. 2014;136:2058–2069. doi: 10.1021/ja412084b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taoka M, Nobe Y, Hori M, Takeuchi A, Masaki S, Yamauchi Y, Nakayama H, Takahashi N, Isobe T. Nucleic Acids Research. 2015;43:e115. doi: 10.1093/nar/gkv560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wetzel C, Li S, Limbach P. J Am Soc Mass Spectrom. 2014;25:1114–1123. doi: 10.1007/s13361-014-0889-9. [DOI] [PubMed] [Google Scholar]
- 51.Taucher M, Ganisl B, Breuker K. International Journal of Mass Spectrometry. 2011;304:91–97. doi: 10.1016/j.ijms.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang TY, Liu J, McLuckey SA. J Am Soc Mass Spectrom. 2010;21:890–898. doi: 10.1016/j.jasms.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 53.Taucher M, Breuker K. Angewandte Chemie (International ed in English) 2012;51:11289–11292. doi: 10.1002/anie.201206232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matthiesen R, Kirpekar F. Nucleic Acids Res. 2009;37:e48. doi: 10.1093/nar/gkp139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGinnis AC, Chen B, Bartlett MG. Journal of Chromatography B. Analytical Technologies in the Biomedical and Life Sciences. 2012;883–884:76–94. doi: 10.1016/j.jchromb.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 56.Fountain KJ, Gilar M, Gebler JC. Rapid Commun Mass Spectrom. 2003;17:646–653. doi: 10.1002/rcm.959. [DOI] [PubMed] [Google Scholar]
- 57.Wetzel C, Limbach P. J Proteomics. 2011;75:3450–3464. doi: 10.1016/j.jprot.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 58.Erb R, Oberacher H. Electrophoresis. 2014;35:1226–1235. doi: 10.1002/elps.201300269. [DOI] [PubMed] [Google Scholar]
- 59.Gong L, McCullagh JSO. Rapid Commun Mass Spectrom. 2014;28:339–350. doi: 10.1002/rcm.6773. [DOI] [PubMed] [Google Scholar]
- 60.Chen B, Mason SF, Bartlett MG. J Am Soc Mass Spectrom. 2013;24:257–264. doi: 10.1007/s13361-012-0509-5. [DOI] [PubMed] [Google Scholar]
- 61.McGinnis AC, Grubb EC, Bartlett MG. Rapid Commun Mass Spectrom. 2013;27:2655–2664. doi: 10.1002/rcm.6733. [DOI] [PubMed] [Google Scholar]
- 62.Huang T, Kharlamova A, Liu J, McLuckey S. J Am Soc Mass Spectrom. 2008;19:1832–1840. doi: 10.1016/j.jasms.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 63.Nyakas A, Stucki S, Schürch S. J Am Soc Mass Spectrom. 2011;22:875–886. doi: 10.1007/s13361-011-0106-z. [DOI] [PubMed] [Google Scholar]
- 64.Nyakas A, Eberle RP, Stucki SR, Schürch S. J Am Soc Mass Spectrom. 2014;25:1155–1166. doi: 10.1007/s13361-014-0873-4. [DOI] [PubMed] [Google Scholar]
- 65.Kullolli M, Knouf E, Arampatzidou M, Tewari M, Pitteri SJ. J Am Soc Mass Spectrom. 2014;25:80–87. doi: 10.1007/s13361-013-0759-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.N A, Hari Y, Stucki SR, Schürch S. CHIMIA. 2014;68:164–167. doi: 10.2533/chimia.2014.164. [DOI] [PubMed] [Google Scholar]
- 67.Addepalli B, Limbach PA. J Am Soc Mass Spectrom. 2011;22:1363–1372. doi: 10.1007/s13361-011-0137-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wetzel C, Limbach P. Analyst. 2013;138:6063–6072. doi: 10.1039/c3an01224g. [DOI] [PubMed] [Google Scholar]
- 69.Wong SY, Javid B, Addepalli B, Piszczek G, Strader MB, Limbach PA, Barry CE., 3rd Antimicrob Agents Chemother. 2013;57:6311–6318. doi: 10.1128/AAC.00905-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takebayashi K, Hirose K, Izumi Y, Bamba T, Fukusaki E. J Biosci and Bioeng. 2013;115:332–338. doi: 10.1016/j.jbiosc.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 71.Fabris D. Biopolymers. 2009;91 [Google Scholar]
- 72.B C, Legiewicz M, Turner KB, Fabris D, Hamm TE, Rekosh D, Hammarskjöld ML, Le Grice SF. Proc Natl Acad Sci U S A. 2008;105:5. doi: 10.1073/pnas.0804461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ross RL, Sauter AD, Jr, Limbach PA. J Mass Spectrom. 2015;50:1175–1179. doi: 10.1002/jms.3636. [DOI] [PubMed] [Google Scholar]
- 74.Easter RN, Kroning KK, Caruso JA, Limbach PA. Analyst. 2010;135:2560–2565. doi: 10.1039/c0an00399a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Catron B, Caruso JA, Limbach PA. J Am Soc Mass Spectrom. 2012;23:1053–1061. doi: 10.1007/s13361-012-0366-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hsu CC, Dorrestein PC. Curr Op in Biotechnol. 2015;31:24–34. doi: 10.1016/j.copbio.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakayama H, Takahashi N, Isobe T. Mass Spectrom Rev. 2011;30:1000–1012. doi: 10.1002/mas.20325. [DOI] [PubMed] [Google Scholar]
- 78.Nyakas A, Blum LC, Stucki SR, Reymond JL, Schürch S. J Am Soc Mass Spectrom. 2013;24:249–256. doi: 10.1007/s13361-012-0529-1. [DOI] [PubMed] [Google Scholar]
- 79.Sample PJ, Gaston KW, Alfonzo JD, Limbach PA. Nucleic Acids Res. 2015;43:e64–e64. doi: 10.1093/nar/gkv145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nakayama H, Akiyama M, Taoka M, Yamauchi Y, Nobe Y, Ishikawa H, Takahashi N, Isobe T. Nucleic Acids Res. 2009;37:e47. doi: 10.1093/nar/gkp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meyer KD, Jaffrey SR. Nat Rev Mol Cell Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McLuckey SA, Van Berker GJ, Glish GL. J Am Soc Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]


