Abstract
Background
A subset of atopic dermatitis (AD) is associated with increased susceptibility to eczema herpeticum (ADEH+). We previously reported that common single nucleotide polymorphisms (SNPs) in interferon-gamma (IFNG) and receptor 1 (IFNGR1) were associated with ADEH+ phenotype.
Objective
To interrogate the role of rare variants in IFN-pathway genes for risk of ADEH+.
Methods
We performed targeted sequencing of interferon-pathway genes (IFNG, IFNGR1, IFNAR1 and IL12RB1) in 228 European American (EA) AD patients selected according to their EH status and severity measured by Eczema Area and Severity Index (EASI). Replication genotyping was performed in independent samples of 219 EA and 333 African Americans (AA). Functional investigation of ‘loss-of-function’ variants was conducted using site-directed mutagenesis.
Results
We identified 494 single nucleotide variants (SNVs) encompassing 105kb of sequence, including 145 common, 349 (70.6%) rare (minor allele frequency (MAF) <5%) and 86 (17.4%) novel variants, of which 2.8% were coding-synonymous, 93.3% were non-coding (64.6% intronic), and 3.8% were missense. We identified six rare IFNGR1 missense including three damaging variants (Val14Met (V14M), Val61Ile and Tyr397Cys (Y397C)) conferring a higher risk for ADEH+ (P=0.031). Variants V14M and Y397C were confirmed to be deleterious leading to partial IFNGR1 deficiency. Seven common IFNGR1 SNPs, along with common protective haplotypes (2 to 7-SNPs) conferred a reduced risk of ADEH+ (P=0.015-0.002, P=0.0015-0.0004, respectively), and both SNP and haplotype associations were replicated in an independent AA sample (P=0.004-0.0001 and P=0.001-0.0001, respectively).
Conclusion
Our results provide evidence that both genetic variants in the gene encoding IFNGR1 are implicated in susceptibility to the ADEH+ phenotype.
CAPSULE SUMMARY
We provided the first evidence that rare functional IFNGR1 mutations contribute to a defective systemic IFN-γ immune response that accounts for the propensity of AD patients to disseminated viral skin infections.
Keywords: IFNGR1, genetic variants, atopic dermatitis, eczema herpeticum
INTRODUCTION
Atopic dermatitis (AD) is the most common chronic inflammatory skin disease affecting up to 17% of children in industrialized countries1-6. Eczema herpeticum (EH), a rare but serious complication of AD, occurs in less than 3% of AD patients7. Although it is well known that the primary predisposing factor for a history of EH (ADEH+) is herpes simplex virus-1 (HSV-1) exposure8, previous studies have demonstrated that genetic susceptibility (e.g., null mutation R501X in the FLG gene9, 10) contributes to risk.
Interferons (IFNs) are involved in many host defense functions including antiviral and antimicrobial response, apoptosis, cell cycle control and mediating the action of other cytokines11. There are three classes of IFNs, type I, II and III. IFN gamma (IFN-γ), the only Type-II interferon, is a proinflammatory cytokine that modulates many immune-related genes12, 13. Furthermore, IFN-γ is a T helper type 1 (Th1) cytokine that plays a major role in the host innate and adaptive immune responses by activating macrophages, enhancing natural killer cell activation, promoting T-cell differentiation, as well as regulating B-cell isotype switching, and is implicated in the pathogenesis of allergic diseases14-16. IFN receptors are required for IFNs to exert their biological activity and therefore play a critical role in IFN signaling17, 18.
Previous studies in the NIH/NIAID supported Atopic Dermatitis Research Network (ADRN) found that patients with ADEH+ have markedly reduced levels of IFN-γ compared with AD patients without a history of EH (ADEH−). We also observed that reduced IFN-γ production was associated with IFNG (gene encodes IFN-gamma) and interferon gamma receptor 1 (IFNGR1) SNPs6. Moreover, global gene expression analysis showed that both IFNGR1 and interferon alpha receptor 1 (IFNAR1) were downregulated in vaccinia virus-challenged ADEH+ PBMCs6. Separately, we have observed that PBMCs from patients with ADEH+ respond poorly to interferon alpha and gamma, and some are poor responders to IL-12 stimulation19.
To further interrogate the role of genetic variation in IFN-pathway genes and risk of ADEH+, we performed targeted deep sequencing in a sample of AD patients with (ADEH+) and without EH (ADEH−) to identify common and rare variants associated with ADEH+ susceptibility. Included in the sequencing was IFN-γ (IFNG), interferon receptors (Type-I IFN receptor IFNAR1 and Type-II IFN receptor IFNGR1) and IL12RB1, the gene encoding the b1 subunit of the IL-12 receptor expressed on NK and T cells20. Validation of variants significantly associated with ADEH+ was performed by either genotyping or Sanger sequencing, and exploration of the functional relevance of damaging variants was interrogated by in vitro studies including gamma-IFN activation sequence (GAS) reporter gene assays, testing of IFN-γ responsiveness in IFNgR1 (−/−) cells reconstituted with mutant constructs, and STAT1 phosphorylation upon IFN-γ stimulation.
METHODS
Study subjects and phenotypes
Study subjects for discovery sequencing were recruited through the NIH/NIAID-supported ADRN and included 228 unrelated European American AD patients selected according to their EH status (clinical definition stated in reference 7) and severity measured by Eczema Area and Severity Index (EASI) scores: ADEH+ subjects with the highest Eczema Area and EASI scores (n=121) and AD without a history of EH (ADEH−) subjects with the lowest EASI (n=107, Table 1 and Supplementary Figure 1 in the Online Repository). The total EASI score is calculated by combining information on intensity (redness, thickness, scratching and lichenification) and percentage area affected by eczema within each of the 4 body regions (head and neck, upper limbs, trunk, and lower limbs). To determine whether significantly associated SNVs were specific to risk of ADEH+, we genotyped or sequenced an additional 60 European American ADEH− patients, 159 non-atopic controls, and an independent sample of 169 African American ADEH− patients and 164 non-atopic, healthy controls also participating in ADRN, and for whom DNA samples were available.
Table 1.
Clinical characteristics of 228 European American subjects included in the targeted sequencing phase (discovery cohort).
| Characteristic | Case (ADEH+) | Control (ADEH−) | P-value |
|---|---|---|---|
| N | 121 | 107 | |
| Males; N (%) | 67 (55.4%) | 25 (23.4%) | < 0.0001 |
| Age (years); mean (range) | 22.0 (1.3-80.7) | 38.9 (18.7-73.6) | < 0.0001 |
| IgE (kIU/L); Median (Q1, Q3) | 1836 (300, 4987) | 81.4 (25.2, 253) | < 0.0001 |
| AD severity EASI score; Median (Q1, Q3) | 10.4 (5.4, 16.2) | 1.4 (0.6, 2.8) | < 0.0001 |
IgE calculated in kIU/L. The total EASI score is calculated by combining information on intensity (redness, thickness, scratching and lichenification) and percentage area affected by eczema within each of the 4 body regions (head and neck, upper limbs, trunk, and lower limbs).
AD was diagnosed using the US consensus conference criteria21. ADEH+ subjects were defined as subjects with AD who had a history of at least 1 EH episode. ADEH− subjects were defined as subjects with AD with a negative history of EH. Healthy, non-atopic controls were defined as having no personal history of chronic disease including atopy. All study participants were further evaluated by a detailed history and physical examination as well as a questionnaire to assess history of cutaneous viral infections and concomitant medication use.
The study was approved by the Institutional Review Boards at the National Jewish Health, Johns Hopkins University School of Medicine, Oregon Health and Science University, University of California, San Diego, Children’s Hospital of Boston, Ann & Robert H. Lurie Children’s Hospital of Chicago, and University of Rochester. All subjects gave written informed consent before participation.
DNA sequencing
DNA was extracted using standard protocols. Targeted deep sequencing (>40x) of 4 prioritized genes in the interferon pathway was performed on an Illumina HiSeq2000 at the Center for Inherited Disease Research (CIDR) at Johns Hopkins University (see Supplementary methods). Standard Agilent protocols were used to design the target capture for a total of ~105 kb of sequence representing the four interferon pathway genes (the gene itself and 2 kb both up and down-stream), excluding repeat elements. DNA fragmentation was performed on 100ng-1ug of genomic DNA using a Covaris E210 system and ‘libraries’ were prepared; the Agilent Bioanalyzer (HiSensitivity) was used for quality control (fragment size and DNA quality). The high quality next-generation sequencing (NGS) data were processed (e.g., quality control (QC) and variant calling) via an automated pipeline CIDRSeqSuite 2.0, a set of software tools designed to perform secondary and initial tertiary analysis on NGS data from the Illumina HiSeq instrument (see Supplementary methods). The GVS annotation server22, maintained at the Seattle SNPs at the University of Washington, was used for variant annotation. Variants were annotated for 1) position (based on their hg19 position); 2) functional prediction (intronic, coding synonymous, missense, splice site, UTR); 3) novelty (previously reported or not, compared to the HapMap23 (phase 2, release 24) CEU (Utah residents with ancestry from northern and western Europe from the CEPH collection) and YRI (Yoruba in Ibadan, Nigeria) samples and to pilot data from the 1,000 Genomes Project24); and 4) rarity (variants with a minor allele frequency (MAF) <5% were considered as rare). Results were cataloged based on annotation at a gene-based level.
Genotyping
Validation genotyping was performed on both the common and rare variants identified by targeted sequencing in the same discovery cohort (n=228). Replication genotyping was performed to test for association with risk of AD in two independent samples of AD patients (167 European American and 169 African American) and healthy controls (159 European American and 164 African American) who participated in the ADRN. Three out of seven common IFNGR1 variants (rs11914 (coding-synonymous), rs10457655 (intronic) and rs28515059 (C-1179T)), which had not been previously reported for association with ADEH+, were selected for validation (Table 3). Two variants (rs11914 and rs10457655), along with the three rare missense variants (Val14Met, Val61Ile and Tyr397Cys) were validated using TaqMan Allelic Discrimination Assays on the 7900HT Sequence Detection System (Applied Biosystems), as previously described6. Promoter variant rs28515059 (C-1179T) was not compatible with the TaqMan assay and was detected by Sanger sequencing. Briefly, amplicon was designed using Primer3 (http://frodo.wi.mit.edu/primer3/) with default parameters (forward primer sequence: 5′-ACCGTTTAGGCGTCTATGGAG-3′, reverse primer sequence: 5′-CCCATTGCTCTCCTTGGGAAT-3′). PCR amplification was carried out in 25-μL reactions with 1X Taq Gold Buffer, 0.6 μM dNTPs, 0.125 μM forward and reverse primers, 40 ng of DNA, and 1.25 UTaq Gold polymerase (Life Technologies). The reaction was then cycled with the following conditions: initial denaturation at 94°C for 12 min; 40 cycles at 94°C for 20 sec, 54°C for 20 sec, and 72°C for 20 sec; final extension at 72°C for 10 min. PCR products were purified using Exonuclease III and Shrimp Alkaline Phosphatase (Promega, Madison, WI). Automated sequencing was performed by capillary electrophoresis on an ABI3700 (Applied Biosystems) at the Johns Hopkins Genetic Resources Core Facility and all variants were manually called by visual inspection of the electropherogram, using Sequencher software (Gene Codes Corp., Inc.).
Table 3.
Association between common variants in IFNGR1 and protection against ADEH+ among European Americans (P ≤ 0.05)
| Variants | Chr. 6 position (hg38) |
Function | Major allele |
Minor allele |
MAF (ADEH+) |
MAF (ADEH−) |
P value |
|---|---|---|---|---|---|---|---|
| rs11914 | 137198451 | coding- synonymous, +20,980 |
A | C | 0.116 | 0.238 | 0.002 |
| rs17175127 | 137212069 | intron, +7362 | G | A | 0.116 | 0.238 | 0.002 |
| rs1327475* | 137215318 | intron, +4113 | G | A | 0.116 | 0.234 | 0.002 |
| rs10457655* | 137218673 | intron, +758 | G | A | 0.124 | 0.252 | 0.002 |
| rs7749390* | 137219233 | splice site, +198 |
A | G | 0.380 | 0.481 | 0.011 |
| rs2234711* | 137219383 | promoter, −56 | A | G | 0.384 | 0.481 | 0.015 |
| rs28515059 | 137221209 | promoter, −1179 |
C | T | 0.120 | 0.229 | 0.006 |
Denote markers previously reported.
Polymorphic positions are indicated with position 1 refers to the A of the start codon (ATG). Positive number suggests position after start codon, and negative number suggests position before start codon.
Functional testing of IFNGR1 ‘loss-of-function’ variants
Site-directed mutagenesis
Mammalian expression construct of wild type IFNgR1 (IFNgR1WT) in pCMV6-entry vector (RC202761) was purchased from Origene Technologies, Inc. (Rockville, MD). We then used restriction endonuclease Sgl I and Mlu I cut IFNgR1WT cDNA fragment and sub-cloned into pLenti-C-Myc-DDK vector (Origene Technologies, Inc.). We used pLenti-C-Myc-DDK-IFNgR1WT as backbones and QuickChangeII XL Site-directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA) to generate constructs expressing IFNgR1 gene variants. The primers for making IFNgR1Y397C were: forward, 5′-cgctttaaactcgTGTcactccagaaattg-3′; reverse, 5′-caatttctggagtgacacgagtttaaagcg-3′. The primers for making IFNgR1V14M were: forward, 5′-gtcatgcagggtATGagcagggctg-3′; reverse, 5′-cagccctgctcataccctgcatgac-3′. IFNGR1V14MY397C was made by the two sets of primers. All constructs were sequence confirmed and plasmids were purified by Endofree Plasmid Maxi Kit (Qiagen).
GAS reporter gene assay
293 FT cells were maintained in Dulbecco’s Modification of Eagle’s Medium (DMEM) with 4.5g/L glucose, L-glutamine, sodium pyruvate, 10% of fetal bovine serum and penicillin/streptomycin (50 units/ml/50ug/ml) (Life Technologies). The cells were seeded in 24 well dishes the day before transfection. 1 μg plasmids of WT, IFNgR1Y397C, IFNgR1V14M, and IFNGR1V14MY397C with 0.1 μg GAS reporter plasmid and 0.04 ug RSV-Renilla luciferase control vector were co-transfected into each well of 293FT cells using Lipofectamine® 2000 reagent (Life Technologies) as the manufacturer instructed. Luciferase assays were carried out using the Dual-Glo® Luciferase Assay System (Promega) following the manufacture’s guideline. The luciferase intensity was read by a GloMax-Multi+Microplate Multi-mode Reader (Promega).
STAT1 phosphorylation upon IFN-γ stimulation
50 μg plasmids of IFNgWT, IFNgR1Y397C, IFNgR1V14M, and IFNGR1V14MY397C were transfected into 4×107 human IFNgR1 deficient EBV-line (a kind gift from Dr. Jean-Laurent Casanova, Rockfeller University)25 using an electroporation approach. The cells were suspended in Hepes-buffered saline and transferred into gap 0.4cm cuvettes, then pulsed at voltage 0.300kV and capacitor 960μF using a Gene Pulser Xcell Electroporation Systems (Bio-RAD). After the pulse, cells were recovered in RPMI 1640 with 20% fetal bovine serum and penicillin/streptomycin (50 units/ml/50ug/ml) (Life technologies) for two days. The cells were then stimulated with serial dilution of recombinant human IFN-γ (R&D systems) for 30 minutes at 37°C. After stimulation, the cells were lysed in RIPA buffer supplemented with protease inhibitor and phosphatase inhibitor (Sigma-Aldrich). 100 μg protein from each sample was used for western-blot to detect phosphorylation of STAT1 and wild-type and mutated receptors. Antibodies recognizing phospho-STAT1 and total STAT1 were purchased from Cell Signaling Technology, IFNgR1 antibody (90 Kd) was purchased from Santa Cruz Biotechnology, Inc. (catalog number SC-700). The densitometry analyses of phospho-STAT1 and STAT1 western blots were measured by ImageJ program provided by National Institute of Health (NIH) website.
Statistical analysis
Clinical characteristic summary statistics and comparisons between ADEH+ and ADEH− groups (chi-square test for gender, t-test for age, log10 (total IgE), and log10 (EASI+1)) were calculated using STATA 11.0 (StataCorp. StataCorp LP. College Station, TX).
All variants and subjects passing QC were included in the sequencing data analysis. Additional QC on each variant was performed with PLINK26, including an assessment of deviation from the Hardy-Weinberg equilibrium (HWE). Genetic association analysis for targeted sequencing data was conducted for all variants identified (both common and rare) and haplotype association tests were carried out for the seven common SNPs associated with ADEH+ phenotype. The single-variant tests for IFNG, IFNGR1, IFNAR1 and IL12RB1 and risk of ADEH+ was performed among 121 ADEH+ subjects and 107 ADEH− subjects using logistic regression (for the common variants (MAF≥5%)) and rare variants with MAF<5% were tested under the Fishers exact test framework. Burden tests were also performed collapsing on the rare missense variants in each gene for the combined effects of rare variants. Pairwise linkage disequilibrium (LD) based on the D’ statistic was measured using HaploView (http://www-genome.wi.mit.edu/personal/jcbarret/haplo). LD blocks were defined using the default algorithm which applies the definition of Gabriel et al27.
For additional genotyping in the African American cohort, the Cochran–Armitage trend test was used to test for association between each individual SNP (under an additive model) and risk of ADEH− using PLINK. Tests for association with a P-value of <0.05 were further adjusted by the PLINK permutation test (10,000 permutations), which provided a framework for correction for multiple testing.
RESULTS
Patient characteristics
Targeted sequencing included a total of 121 ADEH+ (case) and 107 ADEH− (control) European American subjects for analysis. Baseline characteristics are presented in Table 1. The ADEH+ group consisted of more male subjects (55.4% vs 23.4%) who were younger (22.0 vs 38.9 years) compared to the ADEH− group. As observed elsewhere7, total serum IgE levels and mean AD severity EASI scores were significantly higher among subjects in the ADEH+ group (1836 vs 81.4 kIU/L and 10.4 vs 1.4, respectively). Detailed information on participants in the ADVN/ADRN has been previously described.6
Sequencing discoveries
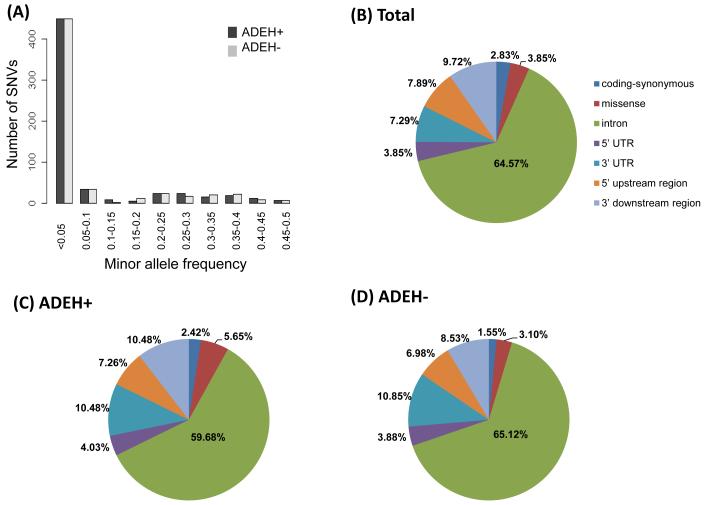
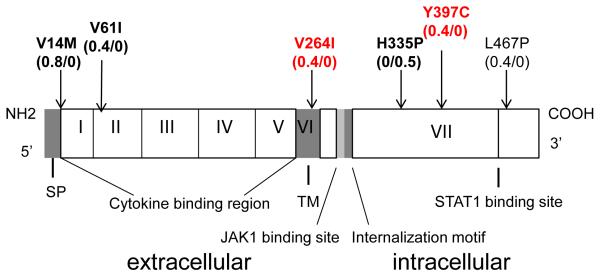
As shown in Table 2, a total of 105kb targeting the four genes and 2 kb both up- and downstream were sequenced; 86,166 base pairs (bp) were successfully returned at a minimum of 40x coverage, after applying the quality control filtering procedure and variant calling. A total of 494 variants were identified across all four genes, including 145 common and 349 (70.6%) rare (minor allele frequency (MAF) <0.05) variants. Of these variants, 124 (25.1%) were private for ADEH+, 129 (26.1%) were private for ADEH−, and 86 (17.4%) were absent in dbSNP Build 141 (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_summary.cgi?view+summary=view+summary&build_id=141). The combined site-frequency spectrum of cases and controls is summarized in Figure 1A, indicating the majority of variants were rare in both the cases and controls (70.6%). Among all the variants detected (Figure 1B), 14 (2.8%) were coding-synonymous, 19 (3.8%) were missense, and 461 (93.3%) were non-coding (introns: 64.6%, 5′ UTR: 3.9%, 3′ UTR: 7.3%, 5′ upstream regions: 7.9%, and 3′ downstream regions: 9.7%). Proportions of variants identified in ADEH+ and ADEH− patients, are summarized according to the major functional categories in Figures 1C and 1D, respectively. All novel rare variants identified by targeted sequencing in 121 ADEH+ (n=37) or 107 ADEH− (n=40) patients are presented in Table E1 (A and B). In addition, six rare missense variants were identified in IFNGR1 (Val14Met, Val61Ile, Val264Ile, His335Pro, Tyr397Cys and Leu467Pro; Figure 2), with 3 (Val14Met, Val61Ile and Tyr397Cys) as possibly damaging variants (with a score of 0.85 or higher on a scale of 0 to 1 (0 is benign)) according to Polymorphism Phenotyping v2 (PolyPhen-2); Table E2)28. Of note, Val14Met, Val61Ile and His335Pro are known IFNGR1 variations (Figure 2) that have been associated with partial or complete IFNGR1 deficiency6, 29. Only one ADEH+ carrier was identified for each of the two variants, Val14Met and Val61Ile, both carriers were positive for HSV-1. In contrast, variant His335Pro was seen in ADEH− patient (Table E2). Two additional rare missense variants Val264Ile and Tyr397Cys were also identified in ADEH+ patients only, but their potential functional role on IFNGR1 deficiency remains unknown. The only carrier identified for the potentially damaging Tyr397Cys variant was positive to HSV-2 and had an EASI score of 40.6. In contrast, the one ADEH+ patient carrying the Val264Ile variant was negative for HSV-1 and HSV-2, and had an EASI score of 6. Thus, we targeted the Tyr397Cys variant in the downstream functional validation. However, no subject was found in our cohort carried a compound heterozygous mutation comprising the IFNGR1 missense variants. As shown in Table 2, missense variants (both common and rare) were also identified in IL12RB1 (n=9) and IFNAR1 (n=3); two of the IL12RB1 rare missense variants (Ser74Arg and Ser320Pro) were possibly damaging and one was novel (Thr517Met) (Table E2). However, no further evidence for association was observed between risk of ADEH+ and missense variants identified in either IL12RB1 or IFNAR1 genes. In contrast, no missense variant was discovered in the IFNG gene (Table 2), for which a region of 8.97kb was sequenced compared to a region of 25.95kb for the IFNGR1 gene.
Table 2.
Summary of total number of variants identified by sequencing in four interferon-pathway genes, the number of private variants identified in ADEH+ or ADEH− patients and the break down according to rarity, novelty and major functional categories.
| Gene | Target Region (bp) |
Sequenced Base Pairs* |
Total # of Variants |
Common | Rare | Total # of Private Variants (ADEH+/ ADEH−) |
Novel dbSNP** (Total/ADEH+/ ADEH−) |
Coding synonymous (Total/ADEH +/ADEH−) |
Missense (Total/A DEH+/A DEH−) |
Non- Coding (Total/ADEH+ /ADEH−) |
|---|---|---|---|---|---|---|---|---|---|---|
| IFNG | chr.12:68,5 46,538- 68,555,255 (8,718) |
2,508 | 41 | 12 | 29 | 7/13 | 8/3/5 | 0/0/0 | 0/0/0 | 41/7/13 |
| IFNGR1 | chr.6:137,5 16,664- 137,542,63 3 (25,970) |
18,821 | 120 | 18 | 102 | 47/31 | 23/9/11 | 5/2/0 | 7/5/1 | 108/40/30 |
| IFNAR1 | chr.21:34,6 95,765- 34,734,206 (38,441) |
36,448 | 199 | 59 | 140 | 41/58 | 35/15/15 | 1/0/0 | 3/1/1 | 195/40/57 |
| IL12RB1 | chr.19:18,1 68,352- 18,199,532 (31,180) |
28,389 | 134 | 56 | 78 | 29/27 | 20/11/9 | 8/1/2 | 9/1/2 | 117/27/23 |
| All (%) | ~105kb | 86,166 | 494 | 145 (29.4%) |
349 (70.6% ) |
124/129 (25.1%/26 .1%) |
86/38/40 (17.4%) |
14/3/2 (2.8%/0.6%/0. 4%) |
19/7/4 (3.8%/1. 4%/0.8% ) |
461/114/123 (93.3%/23.1%/ 24.9%) |
The total number of sequenced base pairs that had passed the quality control filtering process.
Variants that were absent in the most recent dbSNP build 141 (the total number of novel variants including those seen in both ADEH+ and ADEH−; variants only seen in ADEH+; and variants only seen in ADEH−).
Fig. 1.
(A) Combined site-frequency spectrum of ADEH+ and ADEH− patients for the 4 interferon-pathway genes. (B) Proportions of all variants identified in the 4 interferon pathway genes according to major functional categories. (C) Proportions of variants identified in ADEH+ and (D) ADEH− patients.
Fig. 2.
Schematic presentation of 6 rare missense variants in IFNGR1. On the top, the variations that identified by targeted sequencing are illustrated with allele frequencies (in ADEH+/ADEH− patients), 3 known functional variations are highlighted in bold and 2 novel variants are in red. On the bottom, the various domain and 7 exons in the 5′ to 3′ direction are indicated. SP=signal peptide, TM=transmembrane domain.
Common IFNGR1 variants and haplotypes associated with protection for ADEH+
Single marker analyses
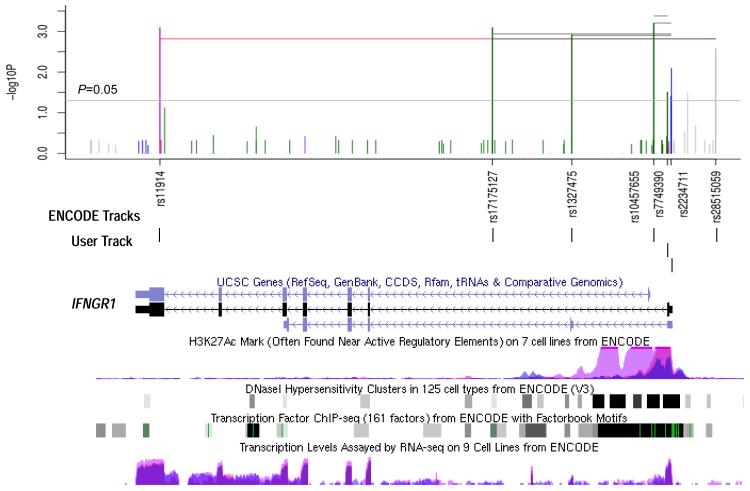
Association tests were performed between variants identified by targeted sequencing and EH status. Seven common variants in IFNGR1 were significantly associated with protection against ADEH+ (P = 0.015-0.002; Table 3 and Figure 3, top panel), four of which were reported previously (rs2234711, rs7749390, rs10457655 and rs1327475)6 and no further validation was attempted. Of the three additional variants identified, 1 was coding-synonymous (rs11914), 1 was intronic (rs17175127) and 1 was in the IFNGR1 promoter (rs28515059, C-1179T). Validation genotyping using TaqMan Assays and Sanger sequencing for these 3 variants described above has confirmed these findings (data not shown).
Fig. 3.
Single marker and haplotype association tests for European Americans and ENCODE regulation tracks on IFNGR1 region (chr6: 137,516,621-137,542,567). SNVs identified by targeted sequencing significantly associated with ADEH+ phenotype (with P values great than 0.01) were illustrated (top panel). The y axis indicates the value of −log10 (P), and the x axis indicates the relative position for each SNV locus in IFNGR1, 3′ to 5′ direction. The vertical lines (green for intron, red for coding-nonsynonymous, purple for coding-synonymous, grey for near-gene-5 and blue for utr-5) represent the position of each SNV on the x axis, and height of the line on the y axis indicates the value of −log10 (P). Similarly, the most significant haplotype results from two to seven-marker windows across the IFNGR1gene were also illustrated by horizontal lines in black (except red color was used for the 7-SNP window) and a P value cutoff of 0.05 was depicted by the grey horizontal line. The bottom panel illustrates the IFNGR1 structure, position of seven SNVs within IFNGR1 gene region (User Track) and regulatory regions with ENCODE regulation tracks including UCSC Gene (IFNGR1), DNase Clusters, Transcription Factor ChIP-seq, Layered H3K27Ac and Transcription Levels on 3 cell lines (NHLF (in pink color), NHEK (in purple color), and K562 (in blue color).
Haplotype analyses
In the sequencing discovery cohort, common protective haplotypes were identified from 2 to 7-SNP windows with an average prevalence of 12% in patients with ADEH+ compared with 23.7% in patients with ADEH− (Table 4 and Figure 3, top). The strongest association was for a common (25% in ADEH+ vs 12% in ADEH−) 2-SNP (rs10457655 and rs7749390) haplotype (AG) that conferred a reduced risk of ADEH+ (P = 0.0004) by over half (odds ratio, 0.41). These two variants are potentially functional: the GG genotype of rs7749390, located in the exon/intron splicing site of IFNGR130, has been associated with increased levels of IFN-γ production and reduced risk of ADEH+6; variant rs10457655 belongs to Category 2b (out of a category system of 1 to 6: 1=likely affect binding and linked to expression of a gene target; 2(a-c)=likely affect binding, subcategories demonstrate direct evidence of binding through ChIP-seq and DNase data, with additional annotations from the most to the least confident; 3=Less likely affect binding; 4-6=Minimal binding evidence) and is likely to directly affect binding of transcription factors according to RegulomeDB31, a database that annotates variants with known and predicted regulatory elements (e.g., DNAase hypersensitivity, binding sites of transcription factors, and promoter regions that have been biochemically characterized to regulate transcription) in non-coding regions32. A 3-SNP haplotype (AGG) also provided strong evidence for association (P = 0.0006) with one additional variant rs2234711 (A-56G or T-56C), which is a functional variant in the promoter associated with several infectious diseases33, 34. It is increasingly clear that a small percentage (7%) of disease-associated SNPs are located in protein-coding regions, and the majority (93%) are located in gene regulatory regions or in intergenic regions35. Because mutations in non-coding portions of a gene can affect how that gene is regulated, we further evaluated regulatory regions for IFNGR1 using the Encyclopedia of DNA Elements (ENCODE) regulatory tracks36. Interestingly, the IFNGR1 haplotype locus (centered on the 3-SNP haplotype) flanks the first exon and overlaps with many regulatory features including DNAaseI hypersensitivity clusters, transcription factors binding sites and enhancer histone H3K27Ac marks (which are often found near active regulatory elements) on 3 out of 7 cell lines: K562 (an immortalized cell line produced from a female patient with chronic myelogenous leukemia), NHEK (normal human epidermal keratinocyte) and NHLF (normal human lung fibroblasts) (Figure 3, bottom).
Table 4.
Common 2 to 7-SNP haplotypes most significantly associated with protection against ADEH+ among EAs in the discovery cohort
| Haplotype | Freq_ADEH+ | Freq_ADEH− | CHIS Q |
OR | P | SNPs |
|---|---|---|---|---|---|---|
| AG | 0.12 | 0.25 | 12.45 | 0.41 | 0.0004 | rs10457655∣rs7749390 |
| AGG | 0.12 | 0.25 | 11.70 | 0.43 | 0.0006 | rs10457655∣rs7749390∣rs2234711 |
| AAGG | 0.12 | 0.23 | 10.41 | 0.44 | 0.0012 | rs1327475∣rs10457655∣rs7749390∣rs2234711 |
| AAAGG | 0.12 | 0.23 | 10.56 | 0.44 | 0.0011 | rs17175127∣rs1327475∣rs10457655∣rs7749390∣rs2234711 |
| AAAGGA | 0.12 | 0.23 | 10.05 | 0.44 | 0.0015 | rs17175127∣rs1327475∣rs10457655∣rs7749390∣rs2234711∣rs28515059 |
| CAAAGG | rs11914∣rs17175127∣rs1327475∣rs10457655∣rs7749390∣rs2234711∣rs2851505 | |||||
| A | 0.12 | 0.23 | 10.05 | 0.44 | 0.0015 | 9 |
Tests for association with susceptibility of AD
To confirm that IFNGR1 variants were specifically associated with risk of ADEH+ and not generic to risk of AD, tests for association between IFNGR1 common and rare variants were performed in two independent samples of AD without EH. Four of the seven IFNGR1 variants were significantly associated with protection against risk of ADEH− (i.e., ADEH− vs non-atopic controls) among the African Americans (P = 0.004-0.0001; Table E3 in the Online Repository). In contrast, no association was found for risk of ADEH− among the European Americans.
Similarly in the African American samples, we observed associations for protection against ADEH− and IFNGR1 haplotypes; a 3-SNP haplotype CAA encompassing the entire IFNGR1 gene provided the strongest evidence (rs11914, rs17175127 and rs28515059; P = 5.9 × 10−4). This strong protective haplotype for AD in the African American sample has not been previously observed6.
Interestingly, a major linkage disequilibrium (LD) block was observed within IFNGR1 for all 7 pairings of contiguous variants spanning 22kb among the African American non-atopic controls using the criteria of Gabriel et al27 (see Supplementary Figure E2 in the Online Repository). No difference was observed for the LD structure among the European American non-atopic controls (Figure E2).
Rare damaging variants associated with risk of ADEH+ and functional validation
We observed the combined evidence of association with rare missense variants in IFNGR1 (P = 0.038, n=6 (Table E2)) when we applied the burden test collapsing on these 6 rare variants, suggesting that rare variants contribute to ADEH+ susceptibility.
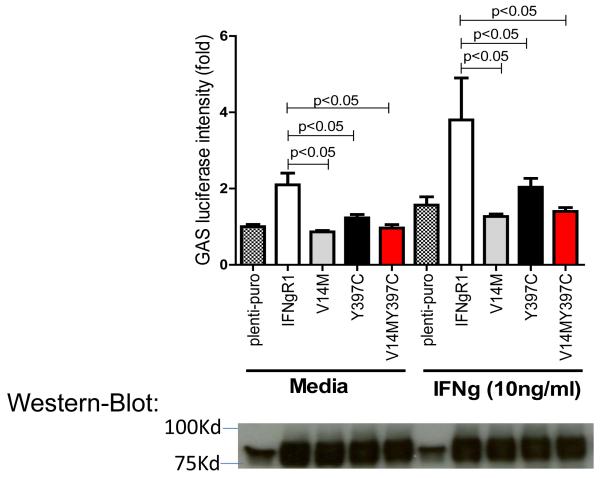
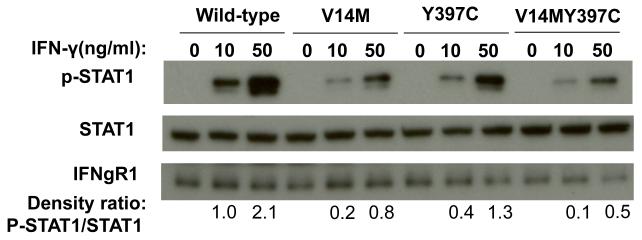
Upon binding to IFN-γ, IFNgR1 and IFNgR2 form as dimers and trigger the activation of JAK1/2. Activated JAK1/2 induces phosphorylation of STAT1, phosphorylated STAT1 then translocates from the cytoplasm into the nucleus to bind to gamma-IFN activation sequence (GAS) in the promoters of IFN-γ targeted genes (Figure 2)37. To confirm whether the two SNVs Val14Met and Tyr397Cys of IFNGR1 alter the receptors’ function, we subcloned these two putative ‘loss-of-function’ variants into mammalian expression plasmids using site-directed mutagenesis methods. We then used two different assays to evaluate the function of the two gene variants compared to the wild-type receptor: GAS report gene assay and western-blot to detect STAT1 phosphorylation upon IFN-γ stimulation. As shown by Figure 4, both V14M and Y397C gene variants had reduced GAS activation as compared to the wild-type receptor. IFNgR1 deficient EBV cells were re-introduced with wildtype IFNgR1, single mutation receptor V14M and Y397C, double mutation of V14M and Y397C. Protein lysates were made from these cells in the presence and absence of IFN-gamma stimulation, the protein expression levels of pSTAT1 and total STAT1 were detected by western blot method. pSTAT1/STAT1 ratio was determined by densitometry measurement. As shown in Figure 5, cells reintroduced with IFNgR1V14M and IFNgR1Y397C have reduced pSTAT1/STAT1 ratio as compared to wildtype receptors; As compared to the single mutated receptors, cells reintroduced with receptors of double mutations of V14M and Y397C have greater reduction of pSTAT1/STAT1 ratio, suggesting that double mutations confer increased damage to the function of the receptor as compared to a single mutation.
Fig. 4.
Reduced GAS activation by IFNgR1V14M and IFNgR1Y397C variants. Plasmids of empty-vector, IFNgR1WT, IFNgR1V14M, IFNgR1Y397C, and IFNgR1V14MY397C were cotransfected with GAS luciferase reporter plasmids into 293FT cells for overnight. A portion of cells was stimulated with IFN-γ (10ng/ml) for an additional 6 hours. The upper panel shows the luciferase intensity of GAS reporter; the lower panel shows the receptor levels corresponding to the samples shown in the upper panel. This is the representative result of three independent experiments.
Fig. 5.
IFN-γ induced phosphorylation of STAT1 is reduced in IFNgR1(−/−). EBV cells reconstituted with IFNgR1V14M and IFNgR1Y397C variants. Plasmids of IFNgR1WT, IFNgR1V14M, IFNgR1Y397C, and IFNgR1V14MY397C were transfected into IFNgR1 deficient cells. The cells were then stimulated with recombinant human IFN-γ at concentration of 0, 10 ng/ml and 50ng/ml for 30 min. 100μg of protein per lane were loaded for the detection of pSTAT1, STAT1, IFNgR1 wildtype and mutants.
DISCUSSION
Our previous studies have demonstrated that genetic susceptibility plays a pivotal role in the development of ADEH+. A relatively uncommon null mutation in filaggrin (R501X), a major skin barrier protein, was three times more prevalent in patients with ADEH+ than those ADEH− patients9. In addition, we have also previously implicated thymic stromal lymphopoietin (TSLP), an interleukin (IL)-7-like cytokine involved in the pathogenesis of allergic diseases, as a potential causal gene in ADEH+38. Using several different approaches (immunologic, genomic and genetic), we also demonstrated low IFNG, IFNAR1 and IFNGR1 expression in patients with ADEH+ as well as association between IFNG and IFNGR1 tagging SNPs and the ADEH+ phenotype and levels of IFN-γ production6. Subsequently, we reported genetic variants in interferon regulatory factor 2 (IRF2) gene might contribute to risk of ADEH+ and the abnormal immune response to HSV10. These findings suggest genetic variations in the interferon signaling pathway regulate the level of interferon receptor gene expression, and therefore plays a critical role in IFN-γ responsiveness in the development of ADEH+.
It is recognized that a considerable proportion of heritable disease risk in complex traits is in fact associated with rare variants39,40. In this multicenter, case-control study, we conducted targeted deep sequencing with sufficient coverage (including regulatory regions) to discover both the common and rare variants, for four interferon signaling pathway genes with prior evidence for involvement in the immunopathogenesis and genetic risk of ADEH+. It is well known that genetic heterogeneity may limit the identification of risk loci in complex disorders. Therefore, genetic studies of clinically more homogeneous subforms of the disease may strengthen the possibility of identification of genetic susceptibility factors with a special interest for extreme phenotypes. Indeed, data have suggested that the most productive approach for identifying missing heritability in complex traits might be to combine the ‘power of the extreme’ in small, well-phenotyped cohorts, with targeted follow-up in case-control and population cohorts41. We selected 121 ADEH+ patients with the highest EASI score and 107 ADEH− patients with the lowest EASI in the discovery cohort for targeted deep sequencing. This approach represents one of the major strengths of our study, which successfully led us to the discovery of IFNGR1 variants as the most prevalent genetic factors for ADEH+, including those novel deleterious variants previously we were unable to identify.
There are prior reports of the association between IFNGR1 polymorphisms and susceptibility to infectious disease. IFNGR1 promoter polymorphism rs2234711 (A-56G or T-56C) was associated with protection from development of or death from cerebral malaria in the Mandika, the major Gambian ethnic group33. Thye et al.34 reported that allele T-56, along with two other IFNGR1 coding sequence polymorphisms His335Pro and Leu467Pro, was associated with a higher rate of anti-Helicobacter pylori IgG antibodies, susceptibility to allergic disease and the production of high IgE titers42. Findings regarding pulmonary tuberculosis susceptibility and IFNGR1 polymorphisms have also been described43, 44 including recent results on an intronic SNP rs1327474 in African Americans45 and rs7749390, located on the exon/intron splice site, in Han Chinese30. Defects in IFNGR1 are a cause of mendelian susceptibility to mycobacterial disease, also known as familial disseminated atypical mycobacterial infection46. Jouanguy et al. reported a distinct group of exonic and intronic mutations in IFNGR1 including ILE87THR (rs104893973)46 and CYS77TYR (rs104893974)47, along with IL12RB1 deficiency. However, none of them exists in AD patients we sequenced in the current study. Instead, we discovered 6 rare missense variants in IFNGR1 including 3 known functional variants and 1 predicted damaging variant Y397C (Table E2). Moreover, we applied additional molecular biology approaches and confirmed that both V14M (a known functional variant) and Y397C (a predicted functional variant) of IFNGR1 have impaired function in triggering IFN-γ signaling transduction as compared to the wild-type receptor. Thus, both common and rare IFNGR1 variants contribute to susceptibility to ADEH+.
Significant genetic associations were observed in the European American patients by both single marker and haplotype analyses. We have identified common 2 and 3-SNP haplotypes that were protective against ADEH+. These haplotypes are comprised of potentially functional variants. The region harboring the 2 and 3-SNP haplotypes is also highly polymorphic and enriched with histone modification H3K27ac marks (Figure 3). Histone modification is an epigenetic mechanism that influences gene regulation in eukaryotes. In particular, histone modification H3K27ac is known as a promoter mark associated with transcriptional activation (e.g., identifies active enhancers)48, 49. Recent evidence suggests that CpG islands under selective pressure are enriched with H3K27ac marks50. It has long been recognized that innate immunity genes such as IFNGR1 are highly polymorphic or plastic in their promoters, suggesting that these regions of the genome have been under intense evolutionary selective pressure from infectious microorganisms51. Our study revealed the strongest association between genetic variants within the IFNGR1 promoter or surrounding the first exon, where H3K27ac marks are peaked, and the ADEH+ phenotype. These findings suggest IFNGR1 variants may affect gene expression by alternating regulatory elements and cause interferon receptor deficiency and impaired interferon response, which play a key role in the pathogenesis of ADEH+. Furthermore, the difference in significant associations (and frequencies) of the IFNGR1 variants and haplotypes between the two ethnic populations was not entirely unexpected given the difference of population history and natural selection (i.e., exposure to infectious microorganisms) that may shape patterns of genetic variation in the IFNGR1 gene. We recognize the limitation of study that we were not able to replicate our targeted sequencing findings in an independent and similarly powered or larger cohort of ADEH+ patients, in addition to solely validating the findings using alternative platforms for genotyping. ADEH+, however, is an extremely rare phenotype involving <3% of AD, and it is noteworthy that the current study involves the largest cohort of such patients. Given our current limitations of a relatively modestly powered discovery sample, we only discovered ‘loss-of-function’ variants in the IFNGR1 gene. We acknowledge that other genes (e.g., IFNG and IFNAR1) in the same (IFN) or different (e.g., skin barrier) pathways may also harbor genetic defects that could lead to impaired antiviral response against viruses such as HSV and vaccinia and increase the risk of ADEH+, but will require larger samples for discovery.
In summary, large-scale analyses of IFNGR1 variants in diverse populations are warranted to confirm and quantify the proportion of ADEH+ risk that may be attributed to these variants. Second, functional validation of these variants applying additional molecular genetics approach helped us further understand the molecular mechanisms involved in AD complicated with viral dissemination. A defective systemic IFN-γ immune response that fails to control viral replication plays a key role in the pathogenesis of ADEH+. Finally, genetic variation in these interferon-pathway genes may have broader implications in determining susceptibility to a range of common, chronic human diseases, which have an inflammatory component. In conclusion, our study provides the first evidence that rare functional IFNGR1 mutations contribute to risk of ADEH+. Future studies in larger and diverse populations are warranted to improve our ability to identify patients at greatest risk for ADEH+ and ultimately lead to early intervention to prevent this devastating complication of AD.
Supplementary Material
KEY MESSAGES.
Targeted sequencing identified both common and rare genetic variations in the gene encoding IFNGR1 that are associated with ADEH+ phenotype.
Both Val14Met and Tyr397Cys are ‘loss-of-function’ rare missense variants leading to partial IFNGR1 deficiency and a defective systemic IFN-γ immune response in ADEH+ patients.
Subjects with AD prone to disseminated viral skin infections have defects in their IFN responses.
Acknowledgements
This work was supported by The Atopic Dermatitis Research Network NIH/NIAID contract HHSN272201000020C. K.C.B. was supported in part by the Mary Beryl Patch Turnbull Scholar Program. DYML was supported in part by the Edelstein Family Chair in Pediatric Allergy-Immunology. We thank National Institute of Allergy and Infectious Diseases–DAIT support (Marshall Plaut, MD, and Joy Laurienzo Panza, RN, BSN, CCRC), the DACI Laboratory (Robert Hamilton, PhD), and all of the patients who participated in this study. Special thanks to Gloria David, PhD, Denise C. Babineau, PhD, Brett Jepson, MS, and Jamie Reese, BS, at Rho, Inc, for coordination of the study. The authors thank Monica Campbell, Cassandra Foster, and Pat Oldewurtel for technical assistance. The authors acknowledge Stephanie Boisson-Dupuis, Jacinta Bustamante, Tatiana Kochetkov and Jean-Laurent Casanova for supplying the IFNgR1 deficient cell line. The authors acknowledge Patricia Taylor, Gayle Spears, and The CTRC nurses for their hard work in recruiting human subjects for this study. CTRC is supported in part by the Colorado Clinical and Translational Science Award/Colorado Clinical & Translational Sciences Institute grant UL1 RR025780 from National Center for Research Resources/NIH and UL1 TR000154 from NIH/National Center for Advancing Translational Sciences.
Abbreviations
- IFNGR1
interferon gamma receptor 1
- AD
Atopic dermatitis
- EH
Eczema herpeticum
- ADEH−
Atopic dermatitis without a history of eczema herpeticum
- ADEH+
Atopic dermatitis with a history of eczema herpeticum
- EASI
Eczema Area and Severity Index
- SNP
Single nucleotide polymorphism
- MAF
Minor allele frequency
- LD
Linkage disequilibrium
- GAS
gamma-IFN activation sequence
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Homey B, Steinhoff M, Ruzicka T, Leung DY. Cytokines and chemokines orchestrate atopic skin inflammation. J Allergy Clin Immunol. 2006;118:178–89. doi: 10.1016/j.jaci.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 2.Barnes KC. An update on the genetics of atopic dermatitis: scratching the surface in 2009. J Allergy Clin Immunol. 2010;125:16–29. e1–11. doi: 10.1016/j.jaci.2009.11.008. quiz 30-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boguniewicz M, Leung DY. Recent insights into atopic dermatitis and implications for management of infectious complications. J Allergy Clin Immunol. 2010;125:4–13. doi: 10.1016/j.jaci.2009.11.027. quiz 4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leung DY. Why is eczema herpeticum unexpectedly rare? Antiviral Res. 2013;98:153–7. doi: 10.1016/j.antiviral.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaw TE, Currie GP, Koudelka CW, Simpson EL. Eczema prevalence in the United States: data from the 2003 National Survey of Children’s Health. J Invest Dermatol. 2011;131:67–73. doi: 10.1038/jid.2010.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leung DY, Gao PS, Grigoryev DN, Rafaels NM, Streib JE, Howell MD, et al. Human atopic dermatitis complicated by eczema herpeticum is associated with abnormalities in IFN-gamma response. J Allergy Clin Immunol. 2011;127:965–73. e1–5. doi: 10.1016/j.jaci.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck LA, Boguniewicz M, Hata T, Schneider LC, Hanifin J, Gallo R, et al. Phenotype of atopic dermatitis subjects with a history of eczema herpeticum. J Allergy Clin Immunol. 2009;124:260–9. 9, e1–7. doi: 10.1016/j.jaci.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu F, Sternberg MR, Kottiri BJ, McQuillan GM, Lee FK, Nahmias AJ, et al. Trends in herpes simplex virus type 1 and type 2 seroprevalence in the United States. Jama. 2006;296:964–73. doi: 10.1001/jama.296.8.964. [DOI] [PubMed] [Google Scholar]
- 9.Gao PS, Rafaels NM, Hand T, Murray T, Boguniewicz M, Hata T, et al. Filaggrin mutations that confer risk of atopic dermatitis confer greater risk for eczema herpeticum. J Allergy Clin Immunol. 2009;124:507–13. 13, e1–7. doi: 10.1016/j.jaci.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao PS, Leung DY, Rafaels NM, Boguniewicz M, Hand T, Gao L, et al. Genetic variants in interferon regulatory factor 2 (IRF2) are associated with atopic dermatitis and eczema herpeticum. J Invest Dermatol. 2012;132:650–7. doi: 10.1038/jid.2011.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gough DJ, Levy DE, Johnstone RW, Clarke CJ. IFNgamma signaling-does it mean JAK-STAT? Cytokine Growth Factor Rev. 2008;19:383–94. doi: 10.1016/j.cytogfr.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Platanias LC, Fish EN. Signaling pathways activated by interferons. Exp Hematol. 1999;27:1583–92. doi: 10.1016/s0301-472x(99)00109-5. [DOI] [PubMed] [Google Scholar]
- 13.Biedermann T, Rocken M, Carballido JM. TH1 and TH2 lymphocyte development and regulation of TH cell-mediated immune responses of the skin. J Investig Dermatol Symp Proc. 2004;9:5–14. doi: 10.1111/j.1087-0024.2004.00829.x. [DOI] [PubMed] [Google Scholar]
- 14.Wild JS, Sigounas A, Sur N, Siddiqui MS, Alam R, Kurimoto M, et al. IFN-gamma-inducing factor (IL-18) increases allergic sensitization, serum IgE, Th2 cytokines, and airway eosinophilia in a mouse model of allergic asthma. J Immunol. 2000;164:2701–10. doi: 10.4049/jimmunol.164.5.2701. [DOI] [PubMed] [Google Scholar]
- 15.Gariboldi S, Palazzo M, Zanobbio L, Dusio GF, Mauro V, Solimene U, et al. Low dose oral administration of cytokines for treatment of allergic asthma. Pulm Pharmacol Ther. 2009;22:497–510. doi: 10.1016/j.pupt.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Herberth G, Heinrich J, Roder S, Figl A, Weiss M, Diez U, et al. Reduced IFN-gamma- and enhanced IL-4-producing CD4+ cord blood T cells are associated with a higher risk for atopic dermatitis during the first 2 yr of life. Pediatr Allergy Immunol. 2010;21:5–13. doi: 10.1111/j.1399-3038.2009.00890.x. [DOI] [PubMed] [Google Scholar]
- 17.Farrar MA, Schreiber RD. The molecular cell biology of interferon-gamma and its receptor. Annu Rev Immunol. 1993;11:571–611. doi: 10.1146/annurev.iy.11.040193.003035. [DOI] [PubMed] [Google Scholar]
- 18.Goodbourn S, Didcock L, Randall RE. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J Gen Virol. 2000;81:2341–64. doi: 10.1099/0022-1317-81-10-2341. [DOI] [PubMed] [Google Scholar]
- 19.Bin L, Edwards MG, Heiser R, Streib JE, Richers B, Hall CF, et al. Identification of novel gene signatures in patients with atopic dermatitis complicated by eczema herpeticum. J Allergy Clin Immunol. 2014;134:848–55. doi: 10.1016/j.jaci.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altare F, Durandy A, Lammas D, Emile JF, Lamhamedi S, Le Deist F, et al. Impairment of mycobacterial immunity in human interleukin-12 receptor deficiency. Science. 1998;280:1432–5. doi: 10.1126/science.280.5368.1432. [DOI] [PubMed] [Google Scholar]
- 21.Eichenfield LF, Hanifin JM, Beck LA, Lemanske RF, Jr., Sampson HA, Weiss ST, et al. Atopic dermatitis and asthma: parallels in the evolution of treatment. Pediatrics. 2003;111:608–16. doi: 10.1542/peds.111.3.608. [DOI] [PubMed] [Google Scholar]
- 22.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–6. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The International HapMap Project Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 24.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jouanguy E, Altare F, Lamhamedi S, Revy P, Emile JF, Newport M, et al. Interferon-gamma-receptor deficiency in an infant with fatal bacille Calmette-Guerin infection. N Engl J Med. 1996;335:1956–61. doi: 10.1056/NEJM199612263352604. [DOI] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 28.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van de Wetering D, de Paus RA, van Dissel JT, van de Vosse E. Functional analysis of naturally occurring amino acid substitutions in human IFN-gammaR1. Mol Immunol. 2010;47:1023–30. doi: 10.1016/j.molimm.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 30.He J, Wang J, Lei D, Ding S. Analysis of functional SNP in ifng/ifngr1 in Chinese Han population with tuberculosis. Scand J Immunol. 2010;71:452–8. doi: 10.1111/j.1365-3083.2010.02393.x. [DOI] [PubMed] [Google Scholar]
- 31.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koch O, Awomoyi A, Usen S, Jallow M, Richardson A, Hull J, et al. IFNGR1 gene promoter polymorphisms and susceptibility to cerebral malaria. J Infect Dis. 2002;185:1684–7. doi: 10.1086/340516. [DOI] [PubMed] [Google Scholar]
- 34.Thye T, Burchard GD, Nilius M, Muller-Myhsok B, Horstmann RD. Genomewide linkage analysis identifies polymorphism in the human interferon-gamma receptor affecting Helicobacter pylori infection. Am J Hum Genet. 2003;72:448–53. doi: 10.1086/367714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar V, Westra HJ, Karjalainen J, Zhernakova DV, Esko T, Hrdlickova B, et al. Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet. 2013;9:e1003201. doi: 10.1371/journal.pgen.1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–86. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 38.Gao PS, Rafaels NM, Mu D, Hand T, Murray T, Boguniewicz M, et al. Genetic variants in thymic stromal lymphopoietin are associated with atopic dermatitis and eczema herpeticum. J Allergy Clin Immunol. 2010;125:1403–7. e4. doi: 10.1016/j.jaci.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–9. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lohmueller KE, Sparso T, Li Q, Andersson E, Korneliussen T, Albrechtsen A, et al. Whole-exome sequencing of 2,000 Danish individuals and the role of rare coding variants in type 2 diabetes. American Journal of Human Genetics. 2014;93:1072–86. doi: 10.1016/j.ajhg.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walters RG, Jacquemont S, Valsesia A, de Smith AJ, Martinet D, Andersson J, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–5. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aoki M, Matsui E, Kaneko H, Inoue R, Fukao T, Watanabe M, et al. A novel single-nucleotide substitution, Leu 467 Pro, in the interferon-gamma receptor 1 gene associated with allergic diseases. Int J Mol Med. 2003;12:185–91. [PubMed] [Google Scholar]
- 43.Fraser DA, Bulat-Kardum L, Knezevic J, Babarovic P, Matakovic-Mileusnic N, Dellacasagrande J, et al. Interferon-gamma receptor-1 gene polymorphism in tuberculosis patients from Croatia. Scand J Immunol. 2003;57:480–4. doi: 10.1046/j.1365-3083.2003.01253.x. [DOI] [PubMed] [Google Scholar]
- 44.Newport MJ, Awomoyi AA, Blackwell JM. Polymorphism in the interferon-gamma receptor-1 gene and susceptibility to pulmonary tuberculosis in The Gambia. Scand J Immunol. 2003;58:383–5. doi: 10.1046/j.1365-3083.2003.01328.x. [DOI] [PubMed] [Google Scholar]
- 45.Velez DR, Hulme WF, Myers JL, Weinberg JB, Levesque MC, Stryjewski ME, et al. NOS2A, TLR4, and IFNGR1 interactions influence pulmonary tuberculosis susceptibility in African-Americans. Hum Genet. 2009;126:643–53. doi: 10.1007/s00439-009-0713-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jouanguy E, Lamhamedi-Cherradi S, Altare F, Fondaneche MC, Tuerlinckx D, Blanche S, et al. Partial interferon-gamma receptor 1 deficiency in a child with tuberculoid bacillus Calmette-Guerin infection and a sibling with clinical tuberculosis. J Clin Invest. 1997;100:2658–64. doi: 10.1172/JCI119810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jouanguy E, Dupuis S, Pallier A, Doffinger R, Fondaneche MC, Fieschi C, et al. In a novel form of IFN-gamma receptor 1 deficiency, cell surface receptors fail to bind IFN-gamma. J Clin Invest. 2000;105:1429–36. doi: 10.1172/JCI9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–26. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akhtar MM, Scala G, Cocozza S, Miele G, Monticelli A. CpG islands under selective pressure are enriched with H3K4me3, H3K27ac and H3K36me3 histone modifications. BMC Evol Biol. 2013;13:145. doi: 10.1186/1471-2148-13-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lazarus R, Vercelli D, Palmer LJ, Klimecki WJ, Silverman EK, Richter B, et al. Single nucleotide polymorphisms in innate immunity genes: abundant variation and potential role in complex human disease. Immunol Rev. 2002;190:9–25. doi: 10.1034/j.1600-065x.2002.19002.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.