Abstract
Objective
Suboptimal cytoreductive surgery in advanced epithelial ovarian cancer (EOC) is associated with poor survival but it is unknown if poor outcome is due to the intrinsic biology of unresectable tumors or insufficient surgical effort resulting in residual tumor-sustaining clones. Our objective was to identify the potential molecular pathway(s) and cell type(s) that may be responsible for suboptimal surgical resection.
Methods
By comparing gene expression in optimally and suboptimally cytoreduced patients, we identified a gene network associated with suboptimal cytoreduction and explored the biological processes and cell types associated with this gene network.
Results
We show that primary tumors from suboptimally cytoreduced patients express molecular signatures that are typically present in a distinct molecular subtype of EOC characterized by increased stromal activation and lymphovascular invasion. Similar molecular pathways are present in EOC metastases, suggesting that primary tumors in suboptimally cytoreduced patients are biologically similar to metastatic tumors. We demonstrate that the suboptimal cytoreduction network genes are enriched in reactive tumor stroma cells rather than malignant tumor cells.
Conclusion
Our data suggest that the success of cytoreductive surgery is dictated by tumor biology, such as extensive stromal reaction and increased invasiveness, which may hinder surgical resection and ultimately lead to poor survival.
Keywords: cancer-associated stroma, cytoreductive surgery, debulkin g, desmoplasia, epithelial-mesenchymal transition, invasion, metastasis, ovarian cancer
Introduction
EOC typically presents at an advanced stage with metastatic tumor nodules spread throughout the peritoneal cavity. Standard treatment for EOC is primary surgical cytoreduction followed by adjuvant platinum- and taxane-based chemotherapy. The goal of surgery is to achieve complete cytoreduction as multiple studies have shown that macroscopically visible residual disease is associated with poor progression-free and overall survival (reviewed in [1, 2]). In cases where complete cytoreduction cannot be achieved due to difficulty in resecting tumors that have invaded vital organs, it is preferable to forego primary cytoreduction surgery and use neoadjuvant chemotherapy to reduce the tumor burden and increase the chances of achieving complete cytoreduction through interval cytoreduction surgery. At present, there is no clinically-applicable biomarker that can predict suboptimal cytoreduction [1, 2]. Several preoperative modalities have been evaluated, including computed tomography, serum CA-125, and laproscopic assessment, but did not achieve sufficient specificity and/or sensitivity to be used in clinical decision-making [3–9]. Consequently, many patients are left with a significant amount of residual disease and will not benefit from the aggressive surgical effort yet must endure the potential complications, such as extended recovery time and delayed initiation of chemotherapy.
The crucial question that remains unanswered is what leads to poor survival in suboptimally cytoreduced patients. Two different theories have been proposed. In the first theory (reviewed in [2]), the amount of residual tumor cells dictates the rate of tumor outgrowth and chemotherapeutic accessibility. Specifically, cytoreduction may reduce the number of tumor initiating cells or resistant clones and delay chemoresistance. Additionally, excision of large necrotic masses may improve drug delivery to smaller, less hypoxic tumors with intact vascular systems. The residual microscopic implants may also have a higher fraction of proliferating cells, resulting in better chemosensitivity to standard cytotoxic agents. Finally, removing tumors in specific locations, such as tumors causing bowel obstruction, may improve the patient’s overall health status and immunocompetence. In the second theory (reviewed in [10]), the intrinsic aggressive tumor biology that is responsible for the failure of surgical resection is also responsible for resistance to chemotherapy and a higher rate of growth and invasion. If unresectable tumors are biologically different from resectable tumors, it is expected that they would have different molecular profiles. Two recent studies used expression profile data to identify signatures of suboptimal cytoreduction [11, 12]. Although the two studies used different datasets and parameters of cytoreduction, the resultant gene signatures largely overlap and represent common biological processes, such as extracellular matrix remodeling, invasion and angiogenesis [11, 12]. These processes have been previously associated with EOC progression and metastasis, supporting the idea that the success of surgical cytoreduction is dictated by tumor biology. Here, we analyze molecular pathways associated with suboptimal cytoreduction to identify underlying biological processes that may determine surgical outcome and therapeutic efficacy.
Materials and Methods
Databases and patient eligibility criteria
The three largest gene expression datasets for ovarian cancer that contain information on cytoreduction status, TCGA, GSE26712 [13], and GSE9891 [14], were downloaded from the curatedOvarianData database in R [15]. All datasets in the database had been preprocessed and normalized at the gene level. We restricted our study to primary, late-stage, serous ovarian tumors with information available on cytoreduction status. Samples of low-stage EOC, non-serous EOC, metastases, or other diseases, were excluded from our analysis. There are 468, 182, and 167 patients available with 136, 93, and 66 suboptimally cytoreduced patients in the TCGA, GSE26712, and GSE9891 datasets, respectively. The TCGA and GSE26712 datasets were used to identify the molecular signatures, while the GSE9891 dataset was used for validating the signatures and evaluating their predictive power.
Overall strategy for statistical analysis
Candidate gene signatures were identified based on both differentially expressed genes and differential network structures. First, normalized expression profile data from two datasets (TCGA and GSE26712) were screened for differentially expressed (DE) genes between patients with suboptimally and optimally cytoreduced tumors with the 2-sample t-test for each dataset separately. P < 0.05 was considered to be statistically significant. Common DE genes from both datasets were then merged together and used to build common and differential networks as previously described [16, 17]. Genes on the differential network were then selected as the candidate gene signatures and validated with an independent dataset (GSE9891).
Network construction
A sparse graphical model [17] was used to simultaneously build the common and the differential networks. The idea is to apply regularized regression for network construction by treating each gene, in turn, as the response variable and the rest of the genes as predictors. In this approach, for each gene xi, the regression model is
where X−i are the expression values of the common DE genes except for gene xi, and y (1/0) indicates suboptimal or optimal cytoreduction. The common and differential networks were formed by collecting all of the ais and bis, respectively. Parameter ai measures the direct dependency between gene xi and the remaining genes, and aij≠ 0 shows there is a partial correlation (edge) between gene xi and xj given the rest of the genes. On the other hand, bi determines y dependent correlation and indicates differential correlation across different clinical conditions, and bij ≠ 0 suggests that there is a differential interaction between gene xi and xj in suboptimally and optimally cytoreduced tumors. The network structures and tuning parameters were determined through stability selection [18]. We first generated 100 sub-samples consisting of 400 patients and then constructed each network for every sub-sample and given tuning parameter. Stability was then defined as the average fraction of disagreements over all edges of the sub-sampled graphs. The network structure and optimal tuning parameter were determined by the most stable sets of edges. The differential network was then visualized and network modules were identified as candidate gene signatures for suboptimal cytoreduction using cytoscape (www.cytoscape.org/) [19].
Multivariate models and validation metrics
To separately validate the candidate gene signatures with an independent dataset (GSE9891), multivariate logistic regression and the two-sided t-test were applied to the expression values of the identified genes in the validation dataset. The receiving operating characteristic (ROC) curve and the area under the ROC curve (AUC) were used to evaluate the prediction power of the identified gene signatures. The statistical toolbox in MATLAB was used for the multivariate logistic regression model and the AUC assessment (Supplemental Materials).
Analysis of public expression profile datasets
R2 Genomics Analysis and Visualization Platform (http://hgserver1.amc.nl/) was used for visual comparison of gene expression in different groups.
Human tissue specimens
Archived human tissue specimens and histochemical staining methods are described in our previous publication [20].
Results
Identification of the suboptimal cytoreduction associated network (SCAN)
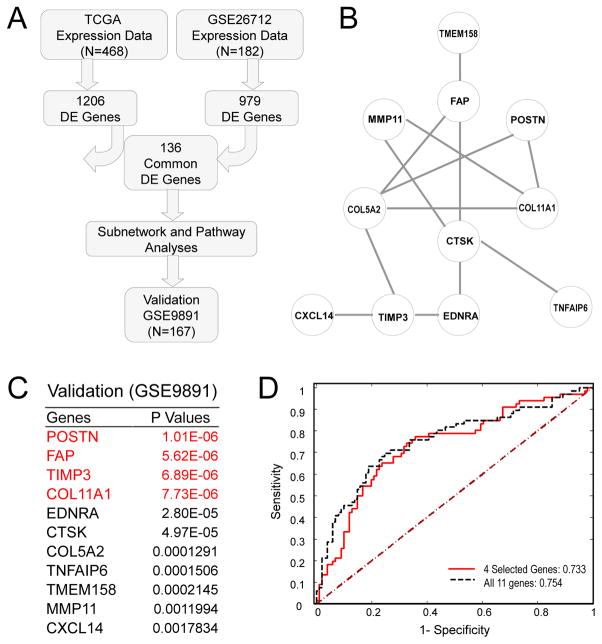
Gene signatures for optimally and suboptimally cytoreduced patients were identified using both differential genes and differential gene-gene interactions. Complex diseases, such as EOC, are not only caused by the mutations of individual genes but also by the dysregulation of molecular networks. Generally, the variations of regulations or interactions between genes under different clinical conditions are associated with the cytoreduction status. Therefore, we adopted a novel approach [16, 17] for gene selection that incorporated both differential expression of genes and differential network structures between patients with or without suboptimal cytoreduction. We used the curatedOvarianData definition of optimal and suboptimal cytoreduction for each dataset [15]. We first compared gene expression levels between ovarian cancer samples from optimally and suboptimally cytoreduced patients in the TCGA and GSE26712 datasets separately using 2-sample t-tests. With a P value of 0.05, 1206 differentially expressed (DE) genes from the TCGA data and 979 DE genes from the GSE26712 data were selected (Fig. 1A). Among the selected DE genes, 136 genes were common to both datasets (Fig. 1A and Table S1). We then merged the two datasets and constructed a common and differential co-expression network using a sparse graphical model [17]. The differential SCAN was created from high-order (partial) correlations conditioning on common (background) correlations. Eleven differentially expressed genes were identified as the candidate gene signature, hereafter referred to as the SCAN genes (Fig. 1B). Based on protein-protein interaction (PPI) network analysis, the SCAN genes likely form a biologically functional network (Fig. S2). The efficiency of the differential network method to identify genes associated with suboptimal cytoreduction was demonstrated by comparing the performance of the 11 SCAN genes identified through the differential network with the top 11 genes identified through the Student’s t-test using an independent validation dataset GSE9891 (Table S1B). All of the 11 SCAN genes had low P values in the validation dataset GSE9891 (Fig. 1C). The four SCAN genes with the lowest P values are highlighted in red (Fig. 1C). The predictive power of the 11 SCAN genes as well as the four SCAN genes with the lowest P values were evaluated separately with logistic regression and predicted Area Under the ROC Curve (AUC) (Fig. 1D). In addition to dividing patients into optimally and suboptimally cytoreduced groups, the TCGA and GSE9891 datasets stratify patients into four groups based on the increasing amount of residual disease. We show that transcript levels of the four SCAN genes with the lowest P values in the validation dataset (POSTN, FAP, TIMP3, and COL11A1) increase proportionally with an increase in the amount of residual disease in both the TCGA and GSE9891 datasets (Fig. S1).
Fig. 1. Identification and validation of the suboptimal cytoreduction associated network (SCAN).
(A) Statistical analysis workflow chart. (B) Selected biomarkers with both differentially expressed genes and differential networks. (C) External validation of the network genes in the validation dataset (GSE9891). The top four genes with the lowest P values are highlighted in red. (D) Predicted Area Under the ROC Curve (AUC) for the SCAN genes in the validation dataset (GSE9891).
The SCAN genes are enriched in distinct molecular subtypes of ovarian cancer
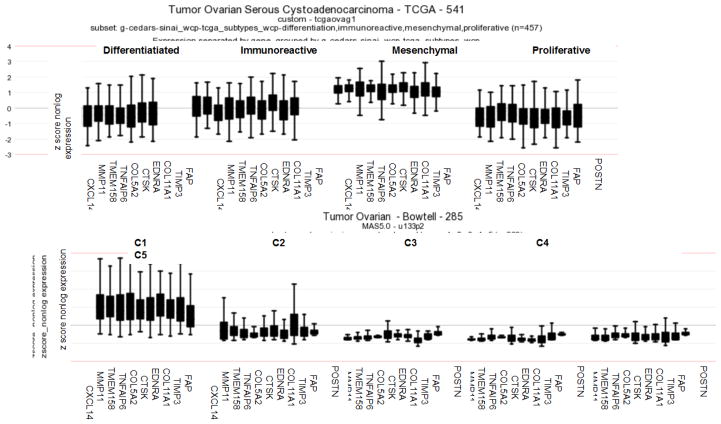
To determine whether the SCAN genes were associated with any previously identified molecular subtypes of EOC, we used expression data from two comprehensive studies that have identified several distinct molecular subtypes of EOC based on expression profiles [14, 21]. In the study by Verhaak et al., 489 high grade serous EOCs from the TCGA dataset clustered into four molecular subtypes (differentiated, immunoreactive, mesenchymal, and proliferative) [21]. In the study by Tothill et al., 251 serous and endometrioid EOC samples clustered into six molecular subtypes, designated C1–C6 [14]. The majority of the serous EOCs were found in the C1–C5 subtypes, while the C6 subtype largely consisted of endometrioid EOCs [14]. In order to identify if the SCAN genes were enriched in any of the identified molecular subtypes of EOC, we plotted the expression levels of the 11 SCAN genes in the differentiated, immunoreactive, mesenchymal, and proliferative molecular subtypes in the TCGA dataset as well as in the C1–C5 subtypes in the Tothill dataset. The analysis revealed that the SCAN genes were most highly expressed in the mesenchymal molecular subtype in the TCGA dataset (Fig. 2A) and in the C1 molecular subtype in the Tothill dataset (Fig. 2B). Notably, the C1 subtype was associated with extensive desmoplasia and the worst survival rate in the Tothill dataset [14] while the mesenchymal molecular subtype was associated with the worst survival rate in the TCGA dataset [22, 23]. Together, these data suggest an association of the SCAN genes with specific molecular subtypes, which are characterized by desmoplasia and/or the presence of a mesenchymal cell state and poor survival.
Fig. 2. The SCAN genes are highly enriched in the C1/mesenchymal molecular subtypes of EOC.
Expression levels of the SCAN genes in (A) differentiated, immunoreactive, mesenchymal, and proliferative molecular subtypes in the TCGA dataset and (B) C1–C5 molecular subtypes in the Tothill dataset.
The SCAN genes are enriched in invasive and metastatic ovarian cancer
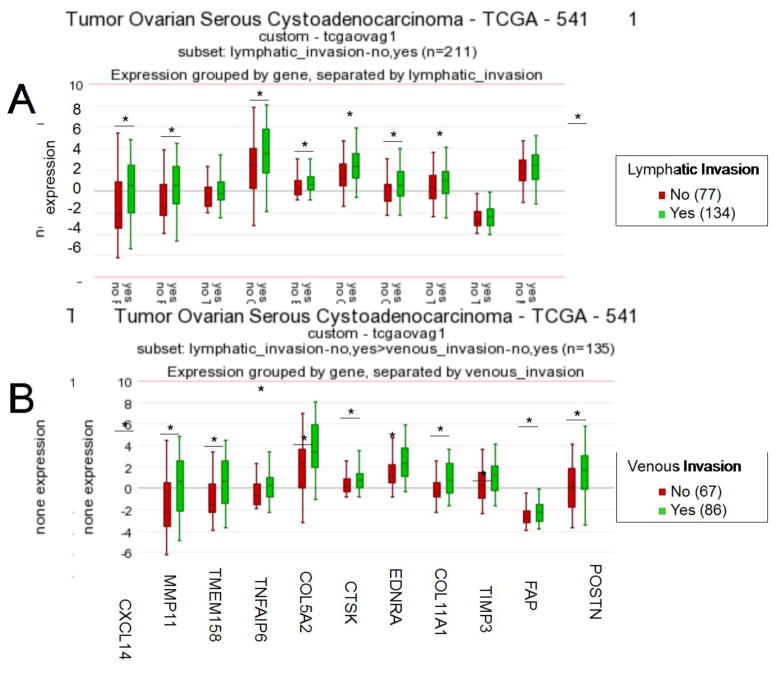
We previously identified three of the four top-scoring SCAN genes (POSTN, TIMP3, and COL11A1) (Fig. 1C) as part of a 10-gene signature of poor survival in EOC and observed their upregulated levels in metastatic EOC in comparison to primary EOC [20, 24]. To identify gene signatures associated with metastasis, we compared the expression profiles of omental EOC metastases to primary EOC using two microarray datasets: Bignotti et al. (17 metastases, 13 primary EOC) and GSE30587 (matched omental metastases and primary EOC from nine patients [25]). The SCAN genes were highly enriched in the signatures of omental metastasis, with five of the 11 SCAN genes (FAP, TIMP3, COL11A1, CTSK, and COL5A2) present in both metastasis signatures (Table S2). The TCGA dataset provides information on the presence or absence of lymphatic and venous invasion in a subset of patients. We show that the expression levels of the SCAN genes are higher in patients with lymphatic and/or venous invasion (Fig. 3). Together, these data suggest that suboptimal cytoreduction may be related to the invasive/metastatic nature of EOC.
Fig. 3. EOCs characterized by lymphatic and venous invasion express higher levels of the SCAN genes.
Relative expression levels of the SCAN genes in patient samples based on the presence or absence of (A) lymphatic invasion and (B) venous invasion. The number of samples in each category is indicated in parentheses.
Tumor stroma, rather than malignant cells, is responsible for increased expression of the SCAN genes
The Gene Set Enrichment Analysis (GSEA) Molecular Signature Database (MSigDB) was used for annotation of the 11 SCAN genes into hallmark genes (H) and GO Gene Sets (C5). The most significant hallmark associated with expression of the SCAN genes was “epithelial-mesenchymal transition (EMT)” in wound healing, fibrosis, and metastasis (p=6.72E-10), while the “extracellular matrix (ECM)” was identified as the most likely site of protein expression (p=6.34E-9) (data not shown).
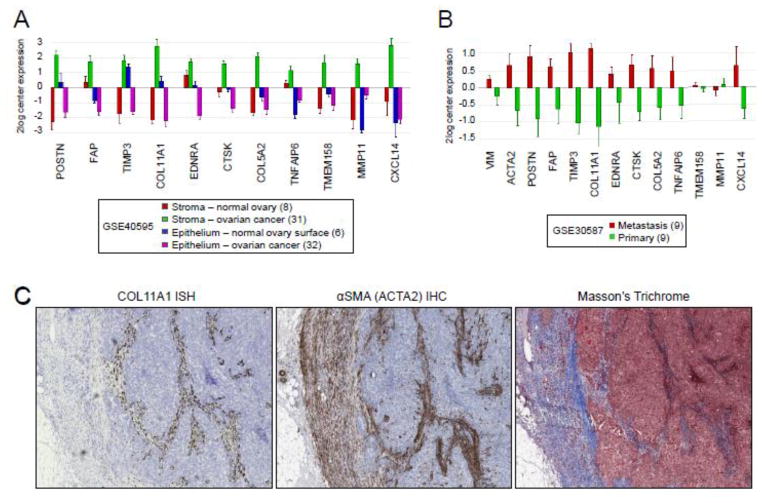
Both malignant epithelial cells and supporting stromal cells secrete ECM in tumors making it difficult to identify the exact source of ECM proteins. To identify which cell type(s) express the SCAN genes, we evaluated the expression levels of the SCAN genes in different ovarian tissue components using the GSE40595 expression profile dataset in which epithelial and stromal cells were laser-microdissected from normal ovaries and ovarian cancers [26]. The results showed that the SCAN genes are most highly expressed in the cancer-associated stroma (Fig. 4A and Fig. S2A).
Fig. 4. The SCAN genes are enriched in the cancer-associated stroma.
(A) Relative expression levels of the SCAN genes in microdissected stromal and epithelial components in the normal ovary and in ovarian cancer. (B) Relative expression levels of the overall stromal marker VIM, reactive stroma marker ACTA2, and the SCAN genes in patient-matched omental metastases and primary tumors. The number of samples in each category is indicated in parentheses. (C) Representative images of sections of a recurrent serous ovarian tumor show COL11A1 expression in a subset of α-SMA-positive fibroblasts. Expression of COL11A1 was determined by in situ hybridization (ISH) with a COL11A1-specific probe. The presence of tumor stroma was determined by immunohistochemical (IHC) staining with α-SMA. The distribution of collagen and smooth muscle connective tissue was determined by Masson’s trichrome staining.
To test whether the SCAN genes are enriched in malignant cancer cells, we ranked 1,036 cancer cell lines in the Cancer Cell Line Encyclopedia (CCLE) by their average expression of the SCAN genes. Ranked at the top were early-passage primary cultures isolated from various human tumors by The Naval Biosciences Laboratory (NBL). Since the cultures were derived from diverse tumor types, including epithelial tumors, gliomas, and sarcomas (Table S3A), it is likely that the SCAN genes represent the stromal cells, which are a common ‘contaminant’ in early-passage primary cultures from tumors. The fibroblast-like morphology characterizing the majority of these cell cultures (Table S3A) is also consistent with the presence of stromal cells. The majority of ovarian cancer cell lines expressed relatively low average levels of the SCAN genes. The highest ranked ovarian cancer cell line, HS571T (rank 55), was an early-passage cell line from the NBL collection (Table S3B). From established ovarian cancer cell lines, the highest ranked were the TOV112D (rank 83) and A2780 (rank 136) cell lines, both of which have been classified as mesenchymal based on functional assays (Table S3B) and [27].
We have previously shown by tumor in situ hybridization that one of the SCAN genes, COL11A1, is primarily expressed in the tumor stroma and that the amount of stromal cells expressing COL11A1 increases during ovarian cancer progression in patient-matched primary, metastatic, and recurrent tumors [20]. Other studies have shown that several of the SCAN genes, including POSTN, TIMP3, and COL11A1, are enriched in the stromal rather than the epithelial tumor component during EOC progression, with the highest levels found in recurrent tumors [28]. The increase in SCAN gene expression in metastatic and recurrent tumors could be a reflection of an increased percentage of stromal cells and decreased percentage of malignant tumor cells. To test this hypothesis, we compared the expression levels of the SCAN genes with the expression levels of the stromal marker vimentin (VIM) in nine patient-matched metastatic and primary tumors in the GSE30587 dataset. Although metastatic tumors in comparison to primary tumors showed an increase in VIM, these changes were modest in comparison to the differential expression of most SCAN genes (Fig. 4B and Fig. S2B). Thus, an increase in the expression of the SCAN genes in metastatic tumors cannot be solely explained by an increased ratio of stromal to epithelial cells. Alternatively, the increased expression of the SCAN genes during cancer progression could be a reflection of a qualitative change in tumor stroma. The progression of epithelial tumors is known to be associated with desmoplasia or the increased presence of ‘reactive stroma’ [29]. Reactive stroma is characterized by increased remodeling of ECM components and production of α-smooth muscle actin (α-SMA) [29]. We show that differential expression of the SCAN genes is more in line with differential expression of the reactive stroma marker ACTA2 (encoding α-SMA) than the overall stroma marker VIM (Fig. 4B and Fig. S2B), indicating that some of the SCAN genes may be associated with the reactive tumor stroma. Consistent with this hypothesis, COL11A1 is expressed in a subset of α-SMA-positive cells in ovarian cancer (Fig. 4C and [20]).
Discussion
Suboptimal primary cytoreductive surgery in advanced epithelial ovarian cancer (EOC) is associated with poor patient survival but it is unknown if this is due to the intrinsic biology of unresectable tumors. Currently, there are no clinically useful predictive models for surgical success, highlighting the need to understand the role of tumor biology in surgical outcome. Recently, two studies have used different statistical approaches to identify gene signatures of suboptimal cytoreduction in the TCGA, GSE26712, and GSE9891 datasets [11, 12]. Six of the 11 SCAN genes (POSTN, FAP, TIMP3, CTSK, TNFAIP6, and CXCL14) are among the top ranked genes in the 200-gene signature of suboptimal debulking identified by Riester [12], and three (FAP, TIMP3, and COL11A1) are present in the 38-gene signature of residual disease identified by Tucker [11]. The prediction accuracy of our SCAN is similar to the prediction accuracy of the signatures identified by Riester [12] and Tucker [11]. While it may be difficult to achieve clinically useful prediction accuracy with molecular signatures, we hypothesized that closer examination of the genes that are consistently identified in association with suboptimal cytoreduction may reveal important information about the biology of unresectable disease.
Our objective was to identify the potential biological pathway(s) and cell type(s) that may be responsible for suboptimal cytoreduction. Significant progress has been made in associating tumor biology with different molecular subtypes of EOC [14, 21]. If tumor biology determines surgical success, it should be possible to link the molecular subtypes of EOC with surgical outcome. Indeed, Tothill et al. observed in their study that the majority of patients with the C1 subtype had extensive residual disease [14]. Our study shows that the molecular pathways associated with suboptimal cytoreduction are highly enriched in the C1/mesenchymal molecular subtype of EOC. Additionally, we show that the SCAN genes are enriched in invasive and metastatic tumors, indicating that primary tumors of the C1/mesenchymal subtype resemble metastatic tumors. Alternatively, the C1/mesenchymal subtype tumors may actually be self-metastases to the ovary rather than primary tumors.
Expression profile data are typically obtained from tumor specimens that contain various types and amounts of stromal cells, making it difficult to discern which cell types contribute to specific gene signatures. Our gene set enrichment analysis identified EMT as the most significant hallmark associated with the SCAN genes. Studies in cancer and fibrosis have demonstrated that epithelial cells can generate tumor stroma through EMT [30, 31]. However, two recent studies in colorectal carcinoma showed that the EMT gene signature in human colorectal cancer is derived from tumor-associated stroma rather than from malignant cells converting to a mesenchymal phenotype [32, 33]. Isella et al. analyzed patient-derived xenograft (PDX) models in which human epithelial tumor cells continued to proliferate when propagated in mice while non-proliferating human stromal cells died out. Human- and mouse-specific RNA sequencing demonstrated that the human mesenchymal signature is decreased in PDXs in comparison to primary tumors, indicating that the EMT signature is derived from stromal cells in human tumors [32]. Callon et al. used FACS to isolate epithelial cells and fibroblasts from primary human tumors and showed that the mesenchymal signature was enriched in tumor fibroblasts [33]. Thus, two studies implementing different techniques came to a similar conclusion that the EMT signature is derived from stromal cells. Consistent with these studies, we show that the SCAN gene signature, which significantly overlaps with the EMT signature in colorectal cancer, is also derived from the tumor stroma in EOC. The tumor microenvironment has been increasingly recognized as a major player in the pathogenesis of EOC [34, 35]. Our data indicate that stromal activation may also impact surgical outcome.
It is thought that biomarkers of surgical outcome would significantly improve the management of ovarian cancer as patients with a low probability of optimal cytoreduction are more likely to benefit from neoadjuvant chemotherapy. However, it is possible that the same biological features that preclude optimal cytoreduction are responsible for poor response to chemotherapy. For example, the top three ranked SCAN genes in our study (POSTN, FAP, and TIMP3) were also identified as the top three ranked genes associated with therapeutic resistance in EOC [28], suggesting that EOC patients with unresectable disease may be inherently resistant to neoadjuvant chemotherapy. Additionally, five of the 11 SCAN genes (POSTN, FAP, CTSK, COL5A2, and MMP11) have been identified by Farmer et al. in breast cancer as part of the 50-gene signature of resistance to neoadjuvant chemotherapy consisting of 5-fluorouracil, epirubicin and cyclophosphamide [36]. Chemotherapy-resistant breast cancer in the study by Farmer et al. was shown to be associated with increased desmoplasia [36], indicating that the presence of reactive stroma may cause resistance to diverse chemotherapeutic agents or generally restrict chemotherapy access. Thus, it may be necessary to target the reactive tumor stroma before or concurrently with chemotherapy to achieve therapeutic success in suboptimally cytoreduced patients.
The development of agents that target tumor stroma will require a better understanding of the key regulators of stromal activation and the mechanisms by which the reactive stroma contributes to unsuccessful surgical resection, tumor progression, and chemotherapy resistance. A possible treatment strategy may come from outside of the cancer field as stromal activation in cancer has numerous similarities to matrix remodeling in fibrosis, a process that has been extensively studied for targeted therapy. Although there are no Food and Drug Administration (FDA)-approved treatments for organ fibrosis, a large number of compounds have shown promising results in reversing fibrosis in preclinical models and are being tested in human clinical trials for systemic fibrosis conditions [37]. We envision that repurposing these agents for cancer treatment may be effective in reversing stromal activation in cancer and increasing the efficacy of cytoreductive surgery and chemotherapy.
Supplementary Material
Fig. S1. Expression levels of the SCAN genes correlate with the amount of residual disease after cytoreductive surgery. Expression levels of COL11A1, TIMP3, FAP, and POSTN are plotted according to the amount of residual disease in each of the TCGA and GSE9891 datasets. The x axis shows non-transformed expression levels in the TCGA dataset and log2 transformed expression levels in the GSE9891 dataset. The y axis shows samples grouped by the amount of residual disease. The number of samples in each group is indicated in parentheses.
Fig. S2. Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (http://string-db.org/) protein-protein interaction (PPI) network analysis shows that 10 of 11 SCAN genes have physical or functional interactions.
Fig. S3. (A, related to Fig. 4A) Cross-correlations of the SCAN genes between the stroma and cancer cells. In general, the SCAN genes have higher expression in the stroma and lower expression in cancer cells, however, there is no statistically significant (either positive or negative) linear correlation among them. (B, related to Fig. 4B) Correlation coefficients and p values between each of the 11 SCAN genes and ACTA2 (reactive stroma marker) and VIM (general stromal marker). Correlation coefficients > 0.4 are highlighted in yellow. P values < 0.1 are indicated in red.
(A) 136 genes differentially expressed between optimally and suboptimally cytoreduced patients (2-sample t-test; P < 0.05) in both the TCGA and GSE26712 datasets. (B) Comparison of the differential network method and the Student’s t-test method in the identification of statistically significant (P < 0.05) differentially expressed genes between optimal and suboptimal cytoreduction in the validation dataset GSE9891. Genes with P < 0.05 are bolded in red.
The SCAN genes are enriched in omental metastases. Shown are the top 100 upregulated probes/genes that are differentially expressed in each dataset comparing expression profiles of omental metastases to primary EOC. The top 100 upregulated probes/genes were arbitrarily selected as a cut-off point. The SCAN genes are indicated in red.
(A) The Cancer Cell Line Encyclopedia (CCLE; http://www.broadinstitute.org/) cell lines ranked by the average expression levels of the 11 SCAN genes. Relative expression levels of individual genes are indicated numerically and by color (red, high expression; green low expression). The top 55 ranked cell lines are annotated with information provided for individual cell line collections. The SCAN genes are largely enriched in cell lines that represent primary cell cultures containing a mixture of malignant cancer cells and stromal cells. (B) Ovarian cancer CCLE cell lines ranked by the average expression levels of the 11 SCAN genes. Annotation of the ovarian cancer cell lines are based on four published sources that classify cell lines based on their phenotype.
HIGHLIGHTS.
Suboptimal cytoreduction is associated with a specific molecular subtype of ovarian cancer characterized by desmoplastic stromal reaction and poor survival
Tumor biology, including stromal activation, lymphovascular invasion, and metastasis, may preclude optimal cytoreduction despite aggressive surgery
Acknowledgments
Funding
This work was supported by the Office of the Assistant Secretary of Defense for Health Affairs through the Ovarian Cancer Research Program (Award No. W81XWH-14-1-0107) and the National Cancer Institute grant (R21CA194626-01) to SO. BYK is supported by the American Cancer Society Early Detection Professorship Grant (SIOP-06-258-01-COUN). Opinions, interpretations, conclusions and recommendations are those of the authors and are not necessarily endorsed by the funding agencies.
We thank Ann E. Walts for constructive discussions and Kristy J. Daniels for assistance in the preparation of the manuscript.
Footnotes
Conflict of Interest Statement
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nick AM, Coleman RL, Ramirez PT, Sood AK. A framework for a personalized surgical approach to ovarian cancer. Nat Rev Clin Oncol. 2015;12:239–45. doi: 10.1038/nrclinonc.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schorge JO, McCann C, Del Carmen MG. Surgical debulking of ovarian cancer: what difference does it make? Rev Obstet Gynecol. 2010;3:111–7. [PMC free article] [PubMed] [Google Scholar]
- 3.Bristow RE, Tomacruz RS, Armstrong DK, Trimble EL, Montz FJ. Survival effect of maximal cytoreductive surgery for advanced ovarian carcinoma during the platinum era: a meta-analysis. J Clin Oncol. 2002;20:1248–59. doi: 10.1200/JCO.2002.20.5.1248. [DOI] [PubMed] [Google Scholar]
- 4.Ferrandina G, Sallustio G, Fagotti A, Vizzielli G, Paglia A, Cucci E, et al. Role of CT scan-based and clinical evaluation in the preoperative prediction of optimal cytoreduction in advanced ovarian cancer: a prospective trial. Br J Cancer. 2009;101:1066–73. doi: 10.1038/sj.bjc.6605292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chi DS, Zivanovic O, Palayekar MJ, Eisenhauer EL, Abu-Rustum NR, Sonoda Y, et al. A contemporary analysis of the ability of preoperative serum CA-125 to predict primary cytoreductive outcome in patients with advanced ovarian, tubal and peritoneal carcinoma. Gynecol Oncol. 2009;112:6–10. doi: 10.1016/j.ygyno.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Axtell AE, Lee MH, Bristow RE, Dowdy SC, Cliby WA, Raman S, et al. Multi-institutional reciprocal validation study of computed tomography predictors of suboptimal primary cytoreduction in patients with advanced ovarian cancer. J Clin Oncol. 2007;25:384–9. doi: 10.1200/JCO.2006.07.7800. [DOI] [PubMed] [Google Scholar]
- 7.Ibeanu OA, Bristow RE. Predicting the outcome of cytoreductive surgery for advanced ovarian cancer: a review. Int J Gynecol Cancer. 2010;20 (Suppl 1):S1–11. doi: 10.1111/IGC.0b013e3181cff38b. [DOI] [PubMed] [Google Scholar]
- 8.Rutten MJ, Leeflang MM, Kenter GG, Mol BW, Buist M. Laparoscopy for diagnosing resectability of disease in patients with advanced ovarian cancer. Cochrane Database Syst Rev. 2014;2:CD009786. doi: 10.1002/14651858.CD009786.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomez-Hidalgo NR, Martinez-Cannon BA, Nick AM, Lu KH, Sood AK, Coleman RL, et al. Predictors of optimal cytoreduction in patients with newly diagnosed advanced-stage epithelial ovarian cancer: Time to incorporate laparoscopic assessment into the standard of care. Gynecol Oncol. 2015;137:553–8. doi: 10.1016/j.ygyno.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borley J, Wilhelm-Benartzi C, Brown R, Ghaem-Maghami S. Does tumour biology determine surgical success in the treatment of epithelial ovarian cancer? A systematic literature review. Br J Cancer. 2012;107:1069–74. doi: 10.1038/bjc.2012.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tucker SL, Gharpure K, Herbrich SM, Unruh AK, Nick AM, Crane EK, et al. Molecular biomarkers of residual disease after surgical debulking of high-grade serous ovarian cancer. Clin Cancer Res. 2014;20:3280–8. doi: 10.1158/1078-0432.CCR-14-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riester M, Wei W, Waldron L, Culhane AC, Trippa L, Oliva E, et al. Risk prediction for late-stage ovarian cancer by meta-analysis of 1525 patient samples. J Natl Cancer Inst. 2014;106 doi: 10.1093/jnci/dju048. pii: dju048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonome T, Levine DA, Shih J, Randonovich M, Pise-Masison CA, Bogomolniy F, et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68:5478–86. doi: 10.1158/0008-5472.CAN-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 15.Ganzfried BF, Riester M, Haibe-Kains B, Risch T, Tyekucheva S, Jazic I, et al. curatedOvarianData: clinically annotated data for the ovarian cancer transcriptome. Database-the Journal of Biological Databases and Curation. 2013 doi: 10.1093/database/bat013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, Liu ZP, Zhao XM, Chen L. Identifying disease genes and module biomarkers by differential interactions. J Am Med Inform Assoc. 2012;19:241–8. doi: 10.1136/amiajnl-2011-000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z, Sun F, Braun J, McGovern DP, Piantadosi S. Multilevel regularized regression for simultaneous taxa selection and network construction with metagenomic count data. Bioinformatics. 2015;31:1067–74. doi: 10.1093/bioinformatics/btu778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meinshausen N, Buhlmann P. Stability selection. Journal of the Royal Statistical Society Series B-Statistical Methodology. 2010;72:417–73. [Google Scholar]
- 19.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheon DJ, Tong Y, Sim MS, Dering J, Berel D, Cui X, et al. A collagen-remodeling gene signature regulated by TGF-beta signaling is associated with metastasis and poor survival in serous ovarian cancer. Clin Cancer Res. 2014;20:711–23. doi: 10.1158/1078-0432.CCR-13-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verhaak RG, Tamayo P, Yang JY, Hubbard D, Zhang H, Creighton CJ, et al. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest. 2013;123:517–25. doi: 10.1172/JCI65833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W, Liu Y, Sun N, Wang D, Boyd-Kirkup J, Dou X, et al. Integrating genomic, epigenomic, and transcriptomic features reveals modular signatures underlying poor prognosis in ovarian cancer. Cell Rep. 2013;4:542–53. doi: 10.1016/j.celrep.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 23.Konecny GE, Wang C, Hamidi H, Winterhoff B, Kalli KR, Dering J, et al. Prognostic and therapeutic relevance of molecular subtypes in high-grade serous ovarian cancer. J Natl Cancer Inst. 2014;106 doi: 10.1093/jnci/dju249. pii: dju249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheon DJ, Orsulic S. Ten-gene biomarker panel: a new hope for ovarian cancer? Biomark Med. 2014;8:523–6. doi: 10.2217/bmm.14.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brodsky AS, Fischer A, Miller DH, Vang S, MacLaughlan S, Wu HT, et al. Expression profiling of primary and metastatic ovarian tumors reveals differences indicative of aggressive disease. PLoS One. 2014;9:e94476. doi: 10.1371/journal.pone.0094476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeung TL, Leung CS, Wong KK, Samimi G, Thompson MS, Liu J, et al. TGF-beta modulates ovarian cancer invasion by upregulating CAF-derived versican in the tumor microenvironment. Cancer Res. 2013;73:5016–28. doi: 10.1158/0008-5472.CAN-13-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang RY, Wong MK, Tan TZ, Kuay KT, Ng AH, Chung VY, et al. An EMT spectrum defines an anoikis-resistant and spheroidogenic intermediate mesenchymal state that is sensitive to e-cadherin restoration by a src-kinase inhibitor, saracatinib (AZD0530) Cell Death Dis. 2013;4:e915. doi: 10.1038/cddis.2013.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryner L, Guan Y, Firestein R, Xiao Y, Choi Y, Rabe C, et al. Upregulation of Periostin and Reactive Stroma Is Associated with Primary Chemoresistance and Predicts Clinical Outcomes in Epithelial Ovarian Cancer. Clin Cancer Res. 2015;21:2941–51. doi: 10.1158/1078-0432.CCR-14-3111. [DOI] [PubMed] [Google Scholar]
- 29.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 30.Iwano M, Plieth D, Danoff TM, Xue C, Okada H, Neilson EG. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin Invest. 2002;110:341–50. doi: 10.1172/JCI15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petersen OW, Nielsen HL, Gudjonsson T, Villadsen R, Rank F, Niebuhr E, et al. Epithelial to mesenchymal transition in human breast cancer can provide a nonmalignant stroma. Am J Pathol. 2003;162:391–402. doi: 10.1016/S0002-9440(10)63834-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isella C, Terrasi A, Bellomo SE, Petti C, Galatola G, Muratore A, et al. Stromal contribution to the colorectal cancer transcriptome. Nat Genet. 2015;47:312–9. doi: 10.1038/ng.3224. [DOI] [PubMed] [Google Scholar]
- 33.Calon A, Lonardo E, Berenguer-Llergo A, Espinet E, Hernando-Momblona X, Iglesias M, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet. 2015;47:320–9. doi: 10.1038/ng.3225. [DOI] [PubMed] [Google Scholar]
- 34.Schauer IG, Sood AK, Mok S, Liu J. Cancer-associated fibroblasts and their putative role in potentiating the initiation and development of epithelial ovarian cancer. Neoplasia. 2011;13:393–405. doi: 10.1593/neo.101720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saad AF, Hu W, Sood AK. Microenvironment and pathogenesis of epithelial ovarian cancer. Horm Cancer. 2010;1:277–90. doi: 10.1007/s12672-010-0054-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farmer P, Bonnefoi H, Anderle P, Cameron D, Wirapati P, Becette V, et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009;15:68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
- 37.Wynn TA, Ramalingam TR. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med. 2012;18:1028–40. doi: 10.1038/nm.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Expression levels of the SCAN genes correlate with the amount of residual disease after cytoreductive surgery. Expression levels of COL11A1, TIMP3, FAP, and POSTN are plotted according to the amount of residual disease in each of the TCGA and GSE9891 datasets. The x axis shows non-transformed expression levels in the TCGA dataset and log2 transformed expression levels in the GSE9891 dataset. The y axis shows samples grouped by the amount of residual disease. The number of samples in each group is indicated in parentheses.
Fig. S2. Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (http://string-db.org/) protein-protein interaction (PPI) network analysis shows that 10 of 11 SCAN genes have physical or functional interactions.
Fig. S3. (A, related to Fig. 4A) Cross-correlations of the SCAN genes between the stroma and cancer cells. In general, the SCAN genes have higher expression in the stroma and lower expression in cancer cells, however, there is no statistically significant (either positive or negative) linear correlation among them. (B, related to Fig. 4B) Correlation coefficients and p values between each of the 11 SCAN genes and ACTA2 (reactive stroma marker) and VIM (general stromal marker). Correlation coefficients > 0.4 are highlighted in yellow. P values < 0.1 are indicated in red.
(A) 136 genes differentially expressed between optimally and suboptimally cytoreduced patients (2-sample t-test; P < 0.05) in both the TCGA and GSE26712 datasets. (B) Comparison of the differential network method and the Student’s t-test method in the identification of statistically significant (P < 0.05) differentially expressed genes between optimal and suboptimal cytoreduction in the validation dataset GSE9891. Genes with P < 0.05 are bolded in red.
The SCAN genes are enriched in omental metastases. Shown are the top 100 upregulated probes/genes that are differentially expressed in each dataset comparing expression profiles of omental metastases to primary EOC. The top 100 upregulated probes/genes were arbitrarily selected as a cut-off point. The SCAN genes are indicated in red.
(A) The Cancer Cell Line Encyclopedia (CCLE; http://www.broadinstitute.org/) cell lines ranked by the average expression levels of the 11 SCAN genes. Relative expression levels of individual genes are indicated numerically and by color (red, high expression; green low expression). The top 55 ranked cell lines are annotated with information provided for individual cell line collections. The SCAN genes are largely enriched in cell lines that represent primary cell cultures containing a mixture of malignant cancer cells and stromal cells. (B) Ovarian cancer CCLE cell lines ranked by the average expression levels of the 11 SCAN genes. Annotation of the ovarian cancer cell lines are based on four published sources that classify cell lines based on their phenotype.