Abstract
T cells express specific metabolic programs to promote diverse cellular differentiation states. The activation of naïve T cells upregulates the expression of genes encoding components of the glycolysis, glutaminolysis, and lipid biosynthesis pathways to promote robust proliferation and effector T cell activity. In contrast, memory T cells downregulate these pathways and predominantly rely on catabolic pathways for long-term survival. Dynamic changes in the expression of the genes encoding components of metabolic pathways in part define which metabolic programs are utilized in diverse T cell states. The current data suggest that key transcription factors involved in T cell specialization decisions, including T-bet, Bcl-6, HIF1, IRF4 and Myc, link the selective programming of cellular metabolism with fate decisions. In this review, we will highlight the transcriptional regulatory events that define metabolic pathways involved in effector and memory T cell differentiation.
Keywords: T cell metabolism, transcriptional regulation, epigenetics
Cells of the immune system dynamically regulate metabolic programs to meet their cellular proliferation and functional needs. Similar to cancer cells, robustly proliferating effector cells in the immune system initiate aerobic glycolysis, whereas more quiescent cells, such as the long-lived memory population, utilize catabolic metabolism programs [1, 2]. In recent years, much research has been performed in T cells to define how metabolism is dynamically regulated to promote specialization decisions. These studies have uncovered many mechanistic levels of regulation for cellular metabolic states in T cells. In particular, both transcriptional and post-transcriptional events functionally regulate metabolic programs in T cells [1–4]. In this review, we will focus our discussion on the transcriptional mechanisms that control metabolic programs in effector and memory T cells. The role for metabolism in regulatory T cells has been expertly reviewed recently and will not be covered here [5–7]. Finally, we will discuss how metabolic states might mechanistically contribute to the programming of differentiation decisions and how these concepts might be utilized to develop future treatments for immune disorders.
Transcriptional programming of metabolism in T cells
The genes that encode the enzymes required for metabolic pathways, such as glyceraldehyde-3-phosphate-dehydrogenase (Gapdh), are often referred to as “housekeeping” genes, and as such, are frequently used as experimental controls. This is because it is usually assumed that these genes are similarly expressed in diverse cell populations and that changes in the activity of metabolic enzymes are regulated mostly at the post-transcriptional level. Although accurate in some circumstances, it is now becoming more appreciated that metabolic gene programs are often dynamically regulated at the transcriptional level during development to control the proliferative and differentiation potential of many cell types [8–12]. In the immune system, the precise regulation of metabolic gene expression programs is required for the functional specialization of several immune cell types.
The differentiation of naïve T cells into effector versus memory states represents a cellular setting where metabolism is dynamically regulated at the gene expression level [8, 12–14]. Naïve CD4+ and CD8+ T cells express low levels of the genes that encode enzymes involved in the glycolysis, glutaminolysis, and lipid biosynthesis pathways. When peptide-MHC complexes engage the T cell receptor (TCR), the ensuing TCR-signaling events promote the rapid transcriptional activation of each of these metabolic gene expression pathways. The widespread induction of the genes that encode the transporters and enzymes involved in glycolysis, glutaminolysis and lipid biosynthesis is required for naïve T cells to differentiate into effector cells [8, 10, 13]. In contrast, the glycolysis, glutaminolysis, and lipid biosynthesis pathways are downregulated in memory T cells. Importantly, the transition between the distinct metabolic programs that define effector and memory T cells is also in part regulated at the transcriptional level [14, 15]. In the next few sections, we will discuss the current knowledge surrounding the key transcription factors that control metabolic gene expression programs to promote effector and memory T cell states.
Myc is required for the metabolic programming of effector T cells
At the outset of an infection, there is a rapid burst in the proliferation of antigen specific T cells that initiates the differentiation of the effector T cell program. At this time, a series of molecular events rewire the gene programs that encode fundamental cellular processes, such as metabolism and the cell cycle, to transition the cell from a quiescent resting state to a functionally active proliferative state [16]. For T cells, TCR-signaling initiates this programming change by inducing a number of transcriptional activators that together dramatically upregulate the gene programs for the glycolysis, glutaminolysis, and lipid biosynthesis pathways among others [8, 13]. Elegant studies have shown that the transcription factor Myc plays a non-redundant role in this TCR-driven program [8]. Myc is currently thought to be the first transcription factor that acts downstream of TCR-signaling to initiate the gene program required for the rapid proliferation of effector cells. To accomplish this, Myc serves as a critical transcriptional activator and amplifier of the gene programs for many basic cellular processes that promote cell growth including the glycolysis and glutaminolysis pathways [8, 17]. Not coincidentally, Myc is a well-characterized oncogene because of its role in rewiring cellular metabolism and other programs that support robust proliferation [18]. This means that T cells utilize a common, and potentially pathogenic, functional activity of Myc to initiate the programming required for the proper development of effector T cell responses. Thus, Myc activity needs to be carefully controlled within the context of the effector response.
AP4 and HIF1α sustain Myc-dependent metabolic gene programs
The initial Myc-dependent activation of basic cellular processes in effector T cells is eventually passed on to another series of TCR- and cytokine-inducible transcription factors to sustain this program (Figure 1). These transcription factors play both overlapping and non-redundant roles in maintaining the expression of the glycolysis, glutaminolysis, and cell cycle programs that promote optimal effector T cell activity and expansion. AP4 is one of the transcription factors that play a role downstream of Myc in CD8+ T cells [19]. Myc initially induces AP4 expression, but importantly, AP4 expression is maintained by IL-2-signaling as Myc expression returns to its baseline state when TCR-signaling diminishes. AP4 regulates a substantial portion of the Myc-dependent gene expression program, which includes many of the genes that encode components of the glycolysis pathways [19]. Notably, the proliferation and optimal clonal expansion of effector T cells are impaired in AP4-deficient CD8+ T cells suggesting that AP4 is required to maintain the basic cellular processes needed for these activities after Myc expression declines.
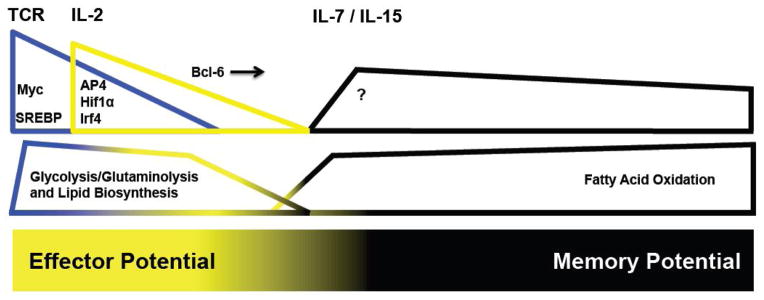
Figure 1. TCR- and cytokine-inducible transcription factors regulate metabolic and differentiation programs in T cells.
In the top panel, TCR-signaling (blue wedge), IL-2-signaling (yellow wedge), and IL-7- and IL-15-signaling (black wedge) are depicted. The key transcription factors that are induced by these signals and are known to influence T cell metabolic programs are shown. The question mark refers to unknown transcriptional regulatory factors. The corresponding metabolic gene programs that are regulated by the signaling events are depicted in the middle panel and a gradient that represents effector and memory potential in T cells is depicted in the bottom panel.
Hypoxia inducible factor 1 alpha (HIF1α) is another transcription factor that is induced by TCR- and IL-2-signaling in T cells to sustain the Myc-dependent program [8, 9]. HIF1α has long been recognized for its role in translating information about oxygen states into specific gene expression programs. One of the well-studied roles for HIF1α relates to its ability to activate the glycolysis gene program in the hypoxic conditions associated with tumor environments [18]. Recent studies in immune cells have now defined an important role for HIF1α in translating diverse environmental conditions into the metabolic gene expression programs that are needed to promote effector cell specialization [9, 12]. In T cells, this means that similar to AP4, HIF1α plays a required role in sustaining the glycolysis and glutaminolysis gene expression programs that are necessary for optimal effector CD8+ T cell expansion and activity [9, 12]. Interestingly, HIF1α also plays a role in CD4+ T cell subtype specialization decisions. HIF1α-dependent gene expression programs, which likely include metabolic programs, contribute to controlling the balance between T helper type 17 (Th17) and regulatory T cells (Treg) [20]. Collectively, the current data indicate that HIF1α plays an important role in immune cell specialization and this is in part due to its role in controlling specific metabolic gene expression programs.
IRF4 fine-tunes metabolic gene expression programs in effector T cells
IRF4 is another transcription factor that acts downstream of Myc to play a role in prolonging the expression of the metabolic gene programs that promote effector CD8+ T cell differentiation [10]. Unlike AP4 and HIF1α, IRF4 expression is highly sensitive to the strength of TCR-signaling [10, 21, 22]. Strong TCR signal strength induces high expression levels of IRF4, whereas less robust TCR signals induce modest IRF4 expression levels. This creates a gradient of IRF4 expression levels within T cell populations that will in part be based upon the intrinsic characteristics of the TCR as well as the antigenic peptide that engages the TCR during a pathogenic encounter. Thus, IRF4 can translate subtle differences in TCR signal strength into fine-tuned gene expression programs, possibly including a metabolic gene program tailored to the antigenic stimulation [10]. This leads to the speculation that after a relatively uniform induction of the metabolic gene program initiated by Myc, there might be subtle differences in the metabolic gene programs that evolve in individual T cell clones based upon TCR-signaling strength.
Role for overlapping and non-redundant metabolic programming mechanisms in effector T cells
An important finding to emerge from the studies defining the transcription factors that regulate the glycolysis and glutaminolysis gene programs in T cells is that there are overlapping, yet non-redundant, roles for several transcription factors that act downstream of TCR- and cytokine-signaling. An interesting biological question to now address is why are several different transcription factors required to coordinate the expression of the glycolysis and glutaminolysis programs in effector T cells? One can speculate that perhaps there is a subtle shift in the quality of these metabolic programs as the effector response proceeds that is necessary for different aspects of cellular proliferation or function. Studies to date have focused on defining the similarities in the genes in the glycolysis and glutaminolysis pathways that are regulated by Myc, AP4, HIF1α and IRF4 [9, 19, 23]. Although there are many common genes that are regulated by each factor, there are also differences. Therefore, future studies that emphasize defining the functional consequences for the differential regulation of the expression of genes that encode specific enzymes or transporters in the glycolysis pathway will be of high importance. One area of particular interest will be to determine how any subtle glycolytic programming differences might affect the functional activity or homing capacity of the effector cell populations. Taken together, there are many intriguing possibilities for how the differential expression of genes in metabolic pathways might uniquely impact differentiation decisions to create diverse T cell populations.
The sterol regulatory element binding (SREBP) proteins and lipid biosynthesis pathway
Much of the focus to date in effector T cells has been on defining the transcription factors that regulate the expression of the genes that encode transporters and enzymes in the glycolysis pathway. This is because of the longstanding knowledge that effector T cells, like cancer cells, switch to aerobic glycolysis [1]. However, there is a comprehensive rewiring of the genes that encode components involved in diverse metabolic pathways to promote the proliferation and functional specialization of effector T cells. One of the additional metabolic pathways that is important for the proliferative and functional capacity of the cell is the sterol or lipid biosynthesis pathway [13, 24]. TCR stimulation, even in the absence of co-stimulation, rapidly, upregulates the expression of SREBP family members [13]. SREBP family members directly induce the expression of genes that encode enzymes in the lipid biosynthesis pathway in several different cell types [25]. Notably, the SREBP-dependent induction of lipids is required for TCR-driven proliferation in effector T cells. This activity is required to support cellular proliferation because SREBP-deficient CD8+ T cells are defective in their proliferative capacity, but the addition of exogenous cholesterol rescues this defect [13]. The SREBP-dependent induction of the lipid biosynthesis pathway might also be important for the activity of signaling complexes in T cells. This relates to the role for the mevalonate pathway in signaling events [26]. Blocking this pathway in CD4+ T cells with the HMG-CoA reductase inhibitor atorvastatin dampens the activity of RAS-dependent signaling events [27]. Taken together, the SREBP-dependent induction of lipids is required for the TCR-driven proliferation and signaling events that influence the expansion and functional activity of effector T cells.
Cytokine-signaling influences metabolic programming in memory T cells
Elegant studies have discovered that metabolic programming changes are also required for the development of memory T cells [15, 28–30]. Coordinating the metabolic gene programming associated with memory T cells is a complex process. It requires both the capacity to dampen the metabolic gene expression programs established during effector T cell differentiation as well as the capacity to promote the expression of genes encoding components of the catabolic pathways that are needed for the quiescent, long-lived phenotype of memory cells. In particular, metabolic pathways such as the fatty acid oxidation (FAO) and lipolysis pathways are upregulated to promote memory cell formation [1, 15, 28, 30]. Similar to the role for IL-2 in enhancing the glycolysis program in effector T cells, cytokines that are known to promote memory T cell formation, such as IL-7 and IL-15, play a role in the metabolic reprogramming of T cells by inducing components of the FAO and lypolysis pathways. For example, in memory CD8+ T cells, IL-15 has been shown to induce the expression of a rate-limiting enzyme for the FAO pathway, carnitine palmitoyl transferase (Cpt1a), as well as the expression of a hydrolase involved in lipolysis, lysosomal acid lipase (Lal) [15, 28]. IL-7 enhances the FAO pathway in part through regulating the expression of the glycerol channel receptor aquaporin 9 (Aqp9) in CD8+ T cells [30]. Importantly, Aqp9-deficient CD8+ T cells are unable to fully induce the expression of numerous components in the FAO pathway and they fail to establish robust long-term memory [30]. Together, these data indicate that IL-7 and IL-15 play a role in initiating the expression of genes involved in the FAO and lipolysis pathways, and this is functionally important for memory CD8+ T cell formation.
Bcl-6 represses the glycolysis gene program
Many transcription factors that are required to establish the metabolic gene expression programs that promote the differentiation of effector T cells have been identified (Figure 1). In contrast, significantly less information is available concerning the transcription factors that regulate the expression of the metabolic gene programs needed for memory T cell development [15, 29]. Insight into one aspect of this topic came from a study addressing the mechanisms that inhibit the expression of the genes involved in the glycolysis pathway in T cells [14]. As discussed, inhibiting glycolysis promotes the transition between effector T cells and memory T cells [31, 32]. In CD4+ T cells, the transcriptional repressor Bcl-6 directly represses the genes that encode transporters and enzymes involved in the glycolysis pathway to effectively dampen the expression of the metabolic gene program associated with effector T cell differentiation [14]. Importantly, Bcl-6 expression is sensitive to environmental IL-2 conditions, which appears to play a role in temporally coordinating the transition between effector and memory T cell responses [14, 33–35]. Bcl-6 expression is low in effector T cells in part because IL-2 is abundant at the onset of the effector immune response and high environmental IL-2 inhibits Bcl-6 expression. In contrast, Bcl-6 expression is induced as IL-2 becomes limiting during the contraction phase of the response, which coincides with the timing of when some effector cells start transitioning into memory potential [16, 36]. Thus, Bcl-6 expression is upregulated in effector T cells as IL-2 becomes limiting, which then allows Bcl-6 to directly repress the genes that encode transporters and enzymes involved in the glycolysis pathway. Together, the current data support the hypothesis that Bcl-6 promotes one of the first mechanistic steps that initiates a metabolic program more compatible with memory T cell formation (Figure 1).
It is interesting to note that IL-2-signaling coordinates the expression of several transcription factors that control metabolic gene expression programs in T cells. In particular, IL-2-signaling sets up an opposing relationship between the expression of the glycolysis-promoting transcription factor HIF1α and the glycolysis-inhibitory transcriptional repressor Bcl-6 in effector versus memory T cells, respectively [9, 14]. Interestingly, experiments examining the forced co-expression of Bcl-6 in the presence of HIF1α suggest that Bcl-6 activity is dominant over HIF1α activity if both factors are present at the same time [14]. This might explain how the balance is shifted away from glycolysis during a transitional state when IL-2 is diminishing, but prior to the loss of the activators involved in effector T cell potential.
T-bet antagonizes the Bcl-6-dependent metabolic gene program
The experimental evidence suggests that the transcription factors discussed above utilize their direct, DNA-binding-dependent potential to regulate metabolic gene expression programs. In contrast, the Th1-lineage-specifying transcription factor T-bet appears to at least in part work by a DNA-binding-independent approach to regulate metabolic gene programming [14]. It has long been established that T-bet is required for the specialization of effector Th1 cells [37, 38]. In this role, T-bet activates Th1-signature genes such as Ifng and Cxcr3 in a DNA-binding-dependent manner [39, 40]. In contrast, new data suggest that T-bet also plays a role in promoting the expression of genes encoding components in the glycolysis pathway in effector Th1 cells in part by utilizing a DNA-binding-independent mechanism [14]. Specifically, T-bet promotes the glycolysis pathway gene program by functionally antagonizing Bcl-6 in CD4+ T cells. This occurs because T-bet-Bcl-6 complex formation requires the DNA-binding zinc fingers of Bcl-6 [14, 33]. Mechanistically, this means that the Bcl-6 DNA-binding domain will not be exposed when T-bet-Bcl-6 complexes form, effectively preventing Bcl-6 from repressing its own target genes. This creates a situation where the relative balance between T-bet and Bcl-6 influences the expression of the glycolysis pathway gene program in CD4+ T cells. Of note, T-bet and Bcl-6 are expressed in CD4+ and CD8+ T cell, innate lymphoid cell, and B cell populations [16, 41–45]. Therefore, it is interesting to speculate that the balance between T-bet and Bcl-6 might functionally regulate genes involved in the glycolysis pathway in a variety of immune cell types. This might play a role in synchronizing the specialization state of immune cells to effectively coordinate the immune response to a specific pathogenic insult.
The realization that T-bet utilizes both DNA-binding-dependent and DNA-binding-independent activities to functionally regulate metabolic gene programming in CD4+ Th1 cells has implications related to how we approach future studies defining the mechanisms that control lineage-specification decisions. For instance, solely assessing the direct DNA-binding-dependent transcriptional programs that are regulated by the factors that control cell fate choices will inevitably miss important biological pathways that are regulated indirectly. Perhaps this is why some of the close connections between cellular metabolism and immune cell specialization programming were initially overlooked. In the example of T-bet and Bcl-6 in T cells, this also sets up an intriguing possibility that the molecular balance between pairs of transcription factors can dynamically integrate environmental signals, such as IL-2 [46]. In some cases, these balances might have the potential to influence metabolic programming to tailor specific subtypes to function in the immediate microenvironment. It is intriguing to speculate that this might influence the breadth of T cell subtypes that are best suited for a specific environmental niche.
Comparing widespread changes in metabolic gene programs in T cells and cancer cells
A somewhat surprising conclusion that has emerged from the current data is that networks of genes encoding the components that make up almost entire metabolic pathways are directly regulated at the transcriptional level by the factors that are important in T cell differentiation decisions [8–10, 13, 14]. That is, each of the transcription factors discussed above does not merely regulate a single gene that encodes one rate-limiting enzyme in a specific metabolic pathway. Rather, each factor regulates a substantial number of genes that encode the transporters and enzymes that control consecutive steps in the metabolic pathway(s). This implies that changes in the expression of metabolic pathways during immune cell differentiation and functional specialization causes more than a mere fine-tuning or a block at an individual step in a predominant metabolic pathway in the cell. Instead, this suggests that the selection of the metabolic program is closely coupled to the differentiation state. This type of reprogramming also occurs in embryonic stem cells where metabolic programming is coupled to differentiation decisions [47, 48].
Interestingly, this is also reminiscent of the reprogramming that occurs in cancer cells where dramatic alterations in the metabolic program are often controlled by oncogenes at the gene expression level [18]. It has long been recognized that effector cells in the immune system behave like a quasi cancer cell in terms of their proliferative capacity and cellular metabolic program [1]. Thus, it is striking to note that known oncogenes, such as Myc and HIF1α, are key players that establish the metabolic gene programs required for effector T cell proliferation and differentiation [3, 8, 18]. This suggests that the basic cellular processes that effector immune cells are programmed to utilize in their normal lifespan represent the same pathways that are exploited in cancer cells. However, unlike cancerous cells, effector T cells eventually terminate their proliferation program after antigen is cleared. Thus, in a normal setting, a termination program must be in place in effector immune cells that is bypassed in cancer cells.
One component of the termination program will likely relate to active repression mechanisms to downregulate the glycolysis pathway, similar to the role for Bcl-6 in the transition between effector and memory T cell programs [14, 32]. In this case, Bcl-6 serves as a checkpoint to alert the effector T cell to switch off glycolysis to terminate the program associated with robust expansion. Bcl-6 is a member of the BTB-zinc finger (BTB-ZF) transcription factor family [44]. It is interesting to note that another BTB-ZF transcription factor family member, ZBTB7a, acts as a tumor suppressor in part by directly repressing numerous genes in the glycolysis pathway similar to the role for Bcl-6 in T cells [49]. This might suggest that a program to terminate glycolysis is normally in place in cells and this program is bypassed in oncogenic circumstances. It will be interesting in the future to explore whether other BTB-ZF factors play similar roles in regulating this type of metabolic termination program in diverse cellular settings.
Deciphering the roles for metabolites in T cells
It will now be important to understand the biological meaning behind why differentiating immune cells dynamically rewire metabolic pathways at the gene program level. In particular, what are the functional reasons that have selected for the co-evolution of specific metabolic programs with particular immune cell specialization decisions? In some ways, the upregulation of glycolysis in effector immune cells (and cancer cells) does not intuitively make sense when viewed from only the perspective of energy production. Perhaps instead of solely having a centric view on the generation of ATP, another intriguing possibility is that components of the selected metabolic pathways might directly promote the epigenetic programming of specialized immune cells (Figure 2). For instance, some metabolites serve as required co-factors for epigenetic modifying complexes and recent research in the cancer and embryonic stem cell fields are actively exploring the roles that specific metabolites play in modifying the epigenome [47, 50]. For example, histone acetyltransferase complexes (HATs) use acetyl-CoA as a donor to modify histones and create a more permissive chromatin environment [18, 47]. The TCA cycle intermediate alpha-ketoglutarate (αKG) is a required co-factor for both histone and DNA-demethylase complexes, while the methionine cycle intermediate S-adenosylmethionine (SAM) is the donor used in DNA and histone methylation reactions [51–55]. DNA and histone methylation play complex roles in gene expression and are linked to programming developmental cell fate decisions [56, 57]. Thus, it is intriguing to speculate that the close connection between metabolic programs and immune cell specialization states might be related to the role for metabolites in regulating epigenetic programs (Figure 2). This possibility would provide a mechanistic explanation for how changes in cellular metabolism can directly influence the gene expression programs required for effector and memory T cell differentiation [47, 58]. Future studies are needed to explore these exciting areas of research in immune cell populations.
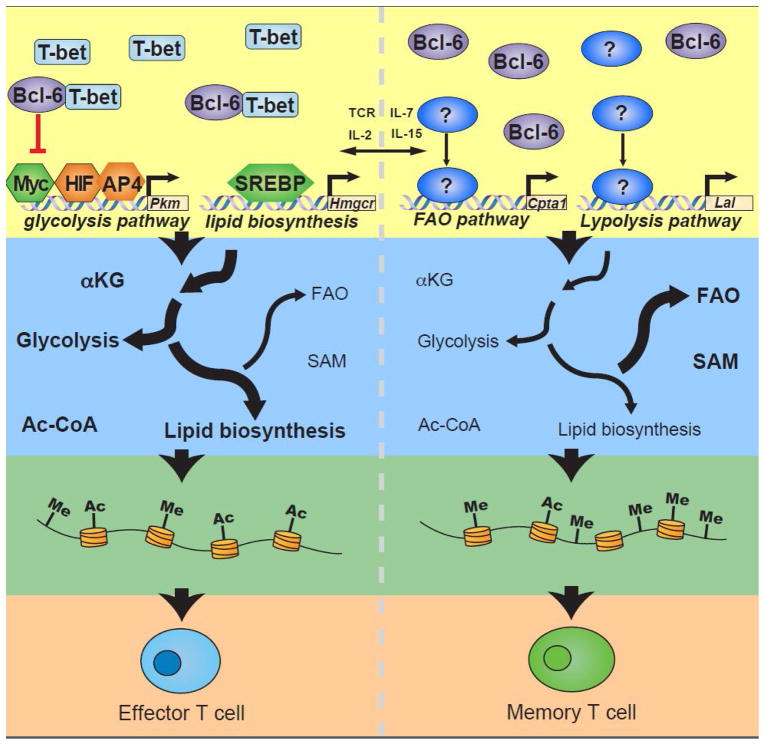
Figure 2. Coordinating metabolic and differentiation programming in T cells.
Representation of the possible connections between transcriptional programs, metabolism, epigenetics, and T cell differentiation states. The transcriptional programming of the expression of genes that encode components of metabolic pathways (yellow panel) will cause changes in cellular metabolism programs (blue panel). The changes in the levels of metabolites that serve as donors and co-factors for epigenetic-modifying complexes might then lead to differential epigenetic programming (green panel) that will determine the fate of the effector or memory T cell states (orange panel). The metabolic and epigenetic pathways displayed are an abstract representation of cellular metabolism and epigenetic modifications, respectively, and are not intended to visually represent a specific metabolic pathway or epigenetic program.
Future treatment potentials and conclusions
Another intriguing future direction for the field is to define the consequences for aberrant metabolic programming states in immune cells. In some cases, dysregulated metabolic states might create tumorigenic potential and cause blood cancers. For example, mutations in enzymes such as isocitrate dehydrogenase (IDH) are known to play causative roles in acute myeloid leukemia (AML) [59, 60]. Another possibility is that dysregulated metabolic programming will result in aberrant differentiation capacity and/or cellular functions. This could result in the failure to control an infection, lack of long-term memory, or autoimmune states depending upon the circumstances and the specific metabolic pathway that is dysregulated. In this regard, it is interesting to note that chemotherapeutic agents that target specific metabolic pathways are sometimes repurposed to treat autoimmune diseases. For example, methotrexate, a folate pathway inhibitor that has long been used for cancer treatment, is now used to treat autoimmune diseases such as rheumatoid arthritis, lupus and Crohn’s disease [52]. As a field, we will first need to comprehensively understand the mechanistic reasons that underlie why specific metabolic pathways support immune cell functions to then be able to envision new therapeutic strategies to treat immunological diseases by selectively targeting metabolic pathways. Importantly, repurposing clinically approved metabolic inhibitors that are currently used for cancer treatment has the potential to greatly accelerate translating new mechanistic findings into viable treatment options for immune disorders.
Another area of interest for future studies will be to more thoroughly define the environmental conditions that influence the metabolic programming of immune cells. As discussed in this review, a great deal of effort has been focused on defining the transcriptional mechanisms that control the TCR- and IL-2-inducible changes in metabolic gene programs in effector T cells. It will now be important to define the range of cytokines and receptors that influence metabolic pathways at the gene programming level in T cells and determine the transcription factors that integrate the signals emanating from these events. In this regard, an exciting finding that might impact our understanding of immunotherapy approaches is that signaling through the PD-1 receptor influences metabolic programs in CD8+ T cells [61, 62]. In addition, it will be intriguing to address how the nutrient environment regulates these gene programs because of the role that nutrition plays in the effectiveness of vaccines and immune responses [62, 63]. Especially in the context of therapeutic intervention, it will be critical to define these questions in diverse immune cell types to predict how the overall immune response will be impacted. In summary, there has been immense progress in understanding the relationship between cellular metabolism and immune cell differentiation. Developing our mechanistic knowledge of how the genes in metabolic pathways are functionally regulated, and in turn how metabolism regulates immune cell function, will increase our ability to translate this information into therapeutic potential in the future.
Highlights.
Transcriptional programming of metabolism in effector and memory T cells
Key transcription factors that regulate metabolic programs in T cells
Comparing widespread changes in metabolic gene programs in T cells and cancer cells
Possible role for metabolites in epigenetic programs
Acknowledgments
We would like to thank the National Institutes of Health for funding research in the author’s laboratory (AI061061 and AI113026 to A.S.W.) and the Comprehensive Minority Faculty and Student Development Program for funding to D.A.C. We would also like to thank Weinmann lab members for critical discussions about the concepts presented in this review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342:1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang R, Green DR. Metabolic checkpoints in activated T cells. Nat Immunol. 2012;13:907–915. doi: 10.1038/ni.2386. [DOI] [PubMed] [Google Scholar]
- 3.Palazon A, Goldrath AW, Nizet V, Johnson RS. HIF transcription factors, inflammation, and immunity. Immunity. 2014;41:518–528. doi: 10.1016/j.immuni.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman NM, Chi H. mTOR Links Environmental Signals to T Cell Fate Decisions. Frontiers in immunology. 2014;5:686. doi: 10.3389/fimmu.2014.00686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeng H, Chi H. Metabolic control of regulatory T cell development and function. Trends Immunol. 2014 doi: 10.1016/j.it.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huynh A, Zhang R, Turka LA. Signals and pathways controlling regulatory T cells. Immunol Rev. 2014;258:117–131. doi: 10.1111/imr.12148. [DOI] [PubMed] [Google Scholar]
- 7.Coe DJ, Kishore M, Marelli-Berg F. Metabolic regulation of regulatory T cell development and function. Frontiers in immunology. 2014;5:590. doi: 10.3389/fimmu.2014.00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, Green DR. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finlay DK, Rosenzweig E, Sinclair LV, Feijoo-Carnero C, Hukelmann JL, Rolf J, Panteleyev AA, Okkenhaug K, Cantrell DA. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J Exp Med. 2012;209:2441–2453. doi: 10.1084/jem.20112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Man K, Miasari M, Shi W, Xin A, Henstridge DC, Preston S, Pellegrini M, Belz GT, Smyth GK, Febbraio MA, Nutt SL, Kallies A. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat Immunol. 2013;14:1155–1165. doi: 10.1038/ni.2710. [DOI] [PubMed] [Google Scholar]
- 11.Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doedens AL, Phan AT, Stradner MH, Fujimoto JK, Nguyen JV, Yang E, Johnson RS, Goldrath AW. Hypoxia-inducible factors enhance the effector responses of CD8(+) T cells to persistent antigen. Nat Immunol. 2013;14:1173–1182. doi: 10.1038/ni.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kidani Y, Elsaesser H, Hock MB, Vergnes L, Williams KJ, Argus JP, Marbois BN, Komisopoulou E, Wilson EB, Osborne TF, Graeber TG, Reue K, Brooks DG, Bensinger SJ. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol. 2013;14:489–499. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oestreich KJ, Read KA, Gilbertson SE, Hough KP, McDonald PW, Krishnamoorthy V, Weinmann AS. Bcl-6 directly represses the gene program of the glycolysis pathway. Nat Immunol. 2014;15:957–964. doi: 10.1038/ni.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Windt GJ, Everts B, Chang CH, Curtis JD, Freitas TC, Amiel E, Pearce EJ, Pearce EL. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang JT, Wherry EJ, Goldrath AW. Molecular regulation of effector and memory T cell differentiation. Nat Immunol. 2014;15:1104–1115. doi: 10.1038/ni.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, Zhao K, Levens D. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nature cell biology. 2015;17:351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chou C, Pinto AK, Curtis JD, Persaud SP, Cella M, Lin CC, Edelson BT, Allen PM, Colonna M, Pearce EL, Diamond MS, Egawa T. c-Myc-induced transcription factor AP4 is required for host protection mediated by CD8+ T cells. Nat Immunol. 2014;15:884–893. doi: 10.1038/ni.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nayar R, Schutten E, Bautista B, Daniels K, Prince AL, Enos M, Brehm MA, Swain SL, Welsh RM, Berg LJ. Graded levels of IRF4 regulate CD8+ T cell differentiation and expansion, but not attrition, in response to acute virus infection. J Immunol. 2014;192:5881–5893. doi: 10.4049/jimmunol.1303187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao S, Buzo BF, Pham D, Jiang L, Taparowsky EJ, Kaplan MH, Sun J. Interferon regulatory factor 4 sustains CD8(+) T cell expansion and effector differentiation. Immunity. 2013;39:833–845. doi: 10.1016/j.immuni.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity. 2011;35:337–348. doi: 10.1016/j.immuni.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bensinger SJ, Bradley MN, Joseph SB, Zelcer N, Janssen EM, Hausner MA, Shih R, Parks JS, Edwards PA, Jamieson BD, Tontonoz P. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell. 2008;134:97–111. doi: 10.1016/j.cell.2008.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osborne TF, Espenshade PJ. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it’s been. Genes Dev. 2009;23:2578–2591. doi: 10.1101/gad.1854309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thurnher M, Gruenbacher G. T lymphocyte regulation by mevalonate metabolism. Science signaling. 2015;8:re4. doi: 10.1126/scisignal.2005970. [DOI] [PubMed] [Google Scholar]
- 27.Dunn SE, Youssef S, Goldstein MJ, Prod’homme T, Weber MS, Zamvil SS, Steinman L. Isoprenoids determine Th1/Th2 fate in pathogenic T cells, providing a mechanism of modulation of autoimmunity by atorvastatin. J Exp Med. 2006;203:401–412. doi: 10.1084/jem.20051129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Sullivan D, van der Windt GJ, Huang SC, Curtis JD, Chang CH, Buck MD, Qiu J, Smith AM, Lam WY, DiPlato LM, Hsu FF, Birnbaum MJ, Pearce EJ, Pearce EL. Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity. 2014;41:75–88. doi: 10.1016/j.immuni.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui G, Staron MM, Gray SM, Ho PC, Amezquita RA, Wu J, Kaech SM. IL-7-Induced Glycerol Transport and TAG Synthesis Promotes Memory CD8(+) T Cell Longevity. Cell. 2015;161:750–761. doi: 10.1016/j.cell.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sukumar M, Liu J, Ji Y, Subramanian M, Crompton JG, Yu Z, Roychoudhuri R, Palmer DC, Muranski P, Karoly ED, Mohney RP, Klebanoff CA, Lal A, Finkel T, Restifo NP, Gattinoni L. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J Clin Invest. 2013;123:4479–4488. doi: 10.1172/JCI69589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Man K, Kallies A. Bcl-6 gets T cells off the sugar. Nat Immunol. 2014;15:904–905. doi: 10.1038/ni.2993. [DOI] [PubMed] [Google Scholar]
- 33.Oestreich KJ, Mohn SE, Weinmann AS. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol. 2012;13:405–411. doi: 10.1038/ni.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ballesteros-Tato A, Leon B, Graf BA, Moquin A, Adams PS, Lund FE, Randall TD. Interleukin-2 Inhibits Germinal Center Formation by Limiting T Follicular Helper Cell Differentiation. Immunity. 2012 doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pipkin ME, Sacks JA, Cruz-Guilloty F, Lichtenheld MG, Bevan MJ, Rao A. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32:79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 38.Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 39.Beima KM, Miazgowicz MM, Lewis MD, Yan PS, Huang TH, Weinmann AS. T-bet binding to newly identified target gene promoters is cell type-independent but results in variable context-dependent functional effects. J Biol Chem. 2006;281:11992–12000. doi: 10.1074/jbc.M513613200. [DOI] [PubMed] [Google Scholar]
- 40.Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 2008;22:2980–2993. doi: 10.1101/gad.1689708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lazarevic V, Glimcher LH, Lord GM. T-bet: a bridge between innate and adaptive immunity. Nat Rev Immunol. 2013;13:777–789. doi: 10.1038/nri3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weinmann AS. Roles for helper T cell lineage-specifying transcription factors in cellular specialization. Adv Immunol. 2014;124:171–206. doi: 10.1016/B978-0-12-800147-9.00006-6. [DOI] [PubMed] [Google Scholar]
- 43.Chang PP, Barral P, Fitch J, Pratama A, Ma CS, Kallies A, Hogan JJ, Cerundolo V, Tangye SG, Bittman R, Nutt SL, Brink R, Godfrey DI, Batista FD, Vinuesa CG. Identification of Bcl-6-dependent follicular helper NKT cells that provide cognate help for B cell responses. Nat Immunol. 2012;13:35–43. doi: 10.1038/ni.2166. [DOI] [PubMed] [Google Scholar]
- 44.Beaulieu AM, Sant’Angelo DB. The BTB-ZF family of transcription factors: key regulators of lineage commitment and effector function development in the immune system. J Immunol. 2011;187:2841–2847. doi: 10.4049/jimmunol.1004006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Basso K, Dalla-Favera R. Roles of BCL6 in normal and transformed germinal center B cells. Immunol Rev. 2012;247:172–183. doi: 10.1111/j.1600-065X.2012.01112.x. [DOI] [PubMed] [Google Scholar]
- 46.Oestreich KJ, Weinmann AS. Master regulators or lineage-specifying? Changing views on CD4(+) T cell transcription factors. Nat Rev Immunol. 2012;12:799–804. doi: 10.1038/nri3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaelin WG, Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teslaa T, Teitell MA. Pluripotent stem cell energy metabolism: an update. The EMBO journal. 2015;34:138–153. doi: 10.15252/embj.201490446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu XS, Haines JE, Mehanna EK, Genet MD, Ben-Sahra I, Asara JM, Manning BD, Yuan ZM. ZBTB7A acts as a tumor suppressor through the transcriptional repression of glycolysis. Genes Dev. 2014;28:1917–1928. doi: 10.1101/gad.245910.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, Asara JM, Daley GQ, Cantley LC. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ko M, An J, Pastor WA, Koralov SB, Rajewsky K, Rao A. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol Rev. 2015;263:6–21. doi: 10.1111/imr.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 55.Shen L, Song CX, He C, Zhang Y. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu Rev Biochem. 2014;83:585–614. doi: 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nottke A, Colaiacovo MP, Shi Y. Developmental roles of the histone lysine demethylases. Development. 2009;136:879–889. doi: 10.1242/dev.020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, Lahesmaa R, Orkin SH, Rodig SJ, Daley GQ, Rao A. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell stem cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell metabolism. 2012;16:9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang F, Travins J, DeLaBarre B, Penard-Lacronique V, Schalm S, Hansen E, Straley K, Kernytsky A, Liu W, Gliser C, Yang H, Gross S, Artin E, Saada V, Mylonas E, Quivoron C, Popovici-Muller J, Saunders JO, Salituro FG, Yan S, Murray S, Wei W, Gao Y, Dang L, Dorsch M, Agresta S, Schenkein DP, Biller SA, Su SM, de Botton S, Yen KE. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340:622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 61.Patsoukis N, Bardhan K, Chatterjee P, Sari D, Liu B, Bell LN, Karoly ED, Freeman GJ, Petkova V, Seth P, Li L, Boussiotis VA. PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nature communications. 2015;6:6692. doi: 10.1038/ncomms7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Siska PJ, Rathmell JC. T cell metabolic fitness in antitumor immunity. Trends Immunol. 2015;36:257–264. doi: 10.1016/j.it.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]