Introduction
Mallory-Denk bodies (MDBs) are composed of intracellular aggregations of misfolded proteins in ballooned hepatocytes. They consist of abnormally phosphorylated, ubiquitinated, and cross-linked keratins 8 and 18 (K8/K18) and non-keratin components (Haybaeck et al., 2012). A major player that determines MDB formation is the ballooned hepatocyte. MDB-forming hepatocytes stain positive for numerous markers of preneoplasmic change (French et al., 2011). MDBs form due to the failure of the 26S proteasome protein quality control system which leads to aggresomes composed of cytokeratins (CKs) and undigested proteins such as heat shock proteins (HSPs), Ub, proteasome subunits, tubulin, and the ubiquitin-binding protein p62 (Yuan et al., 1996). It was found that the pathogenesis of MDBs is associated with the downregulation of ufm1-conjugation system (Ufmylation) and FAT10-conjugation system (FATylation) pathways involved in protein quality control (Liu et al., 2014b), The swelling of the balloon cell cytoplasm is due to the osmotic effect of these undigested proteins.
MDBs develop in the liver of DDC re-fed mice. In the DDC fed mouse model where liver cells proliferate, MDBs form and later, after DDC withdrawal (DDC primed hepatocytes), hepatocellular carcinoma (HCC) develops (Li et al., 2008; Oliva et al., 2008).
MicroRNAs (miRNAs) are ~22-nucleotide noncoding RNAs that have key roles in fundamental biological processes including development, cell cycle control, stem-cell differentiation and oncogenesis by regulating the levels of multiple proteins (Ambros, 2004; Miranda et al., 2010). MiRNAs are transcribed from RNA polymerase II or Poly II in the nucleus, and then are transported to the cytoplasm where they are processed into mature miRNAs. They are responsible for the alternation of hundreds of genes by binding to the 3′ or 5′ untranslated (UTR) regions of mRNA (Bartel, 2009). MiRNAs induce mRNA degradation and/or translational inhibition by base-pairing to the target mRNAs (Meijer et al., 2013). Different miRNAs are found to be aberrantly expressed with ethanol exposure, which includes miR-21, miR-155, miR-122, miR-132 and let-7b (McDaniel et al., 2014).
The miR-34 family consists of three members: miR-34a, miR-34b, and miR-34c. MiR-34a is encoded by its own transcript, whereas miR-34b and miR-34c share a common primary transcript. MiR-34a is a key regulator of tumor suppression. It controls the expression of a plethora of target proteins involved in the cell cycle, differentiation and apoptosis, and antagonizes processes that are necessary for basic cancer cell viability as well as cancer stemness, metastasis, and chemoresistance (Hermeking, 2010; Misso et al., 2014). The miR-34 gene promoters contain p53-binding sites that are conserved among humans, implying a p53-dependent regulation of the miR-34 family (He et al., 2007). The complexity of miR-34a expression is reflected in liver disease. MiR-34a is found to be overexpressed in alcoholic liver injury (Dippold et al., 2013; Meng et al., 2012). Recently, HCC tissues with lower miR-34a expression were found to express higher levels of Bcl-2 protein than those with elevated expression of miR-34a (Yang et al., 2014).
MiR-483-3p is another recently reported tumor suppressor exerting anti-tumor properties (Bertero et al., 2013a) and potent anti-proliferative properties in response to cellular injury (Bertero et al., 2011). MiR-483-3p-mediated cell cycle arrest by the direct targeting of the CDC25A phosphatase prevent its association with cyclin D and blocks cells in early G1 phase of the cell cycle (Bertero et al., 2013b), suggesting an important role of miR-483-3p in cell cycle arrest. Despite these reports, the biological significance of miR-34a and miR-483-3p in AH with MDB formation remains unclear.
In this study, significant changes of miR-34a and miR-483-3p are observed by comparing them in AH livers where MDBs had formed with normal livers obtained by RNA sequencing (RNA-Seq) analyses. The altered expression of miR-34a and miR-483-3p was confirmed in the livers of DDC re-fed mice and human liver biopsies from AH livers. P53 is significantly downregulated both in the AH livers and in the livers of DDC re-fed mice, suggesting that the regulation of miR-34a by p53 was reduced during liver MDB formation. The downregulation of miR-483-3p which increases breast cancer susceptibility gene 1 (BRCA1) expression might provide the mechanism to explain how BRCA1 expression was increased in the livers from AH and the DDC re-fed mice.
Materials and Methods
Liver biopsy specimens
Human formalin-fixed paraffin-embedded (FFPE) liver biopsies from patients who had alcoholic hepatitis (AH; n=3–5) were obtained from Harbor UCLA hospital archives. In all the cases liver forming MDBs were presented. Normal control livers were used for comparison. The liver biopsies used were also used in previous studies (French et al., 2012; Liu et al., 2015; Liu et al., 2014a; Liu et al., 2014b; Liu et al., 2014c). The liver biopsy sections were 4 μm thick.
Mouse liver
Diethyl 1, 4-dehydro-2, 4, 6-trimethyl-3, 5-pyridine-dicarboxylate (DDC) was used as a model to induce the formation of Mallory-Denk bodies (MDBs) in mice. One-month-old C3H male mice were fed 0.1% DDC added to the control diet and a second group were fed control diet for 10 weeks (Li et al., 2008). The mice were then withdrawn from the drug for 1 month and re-fed DDC for 7 days as previously done (Oliva et al., 2009). Three mice were used in each of two groups as follows: 1) control, 2) DDC. DDC was re-fed for 7 days. Mice livers were placed in isopentane and were then fast frozen with liquid nitrogen and stored at −80°C. The livers used had been used in prior studies (Liu et al., 2014a; Liu et al., 2014b; Oliva et al., 2009). All mice were treated in a humane manner as approved by the Animal Care Committee at Harbor UCLA Laboratory BioMedical Research Institute according to the Guidelines of the National Academy of Science.
Tissue sectioning
Mice liver frozen sections were performed as standard protocol. Mice liver frozen sections were cut 5 μm thick at −20°C and immediately transferred to a micro slide box kept on dry ice and stored at −80°C. A new blade was used for each frozen sample.
RNA isolation
RNA isolation of FFPE sections of human liver biopsies was performed as previously described (Liu et al., 2014b). RNA (5μg) was treated and the quality and yield were assessed by electrophoresis using the Agilent 2100 bioanalyzer (Agilent, Palo Alto, CA, USA).
RNA sequencing (RNA-Seq)
Libraries for RNA-Seq were prepared with Nugen Ovation Human FFPE RNA-Seq Multiplex System. The workflow consists of double-stranded cDNA generation using a mixture of random and poly (T) priming, fragmentation of double stranded cDNA, end repair to generate blunt ends, adaptor ligation, strand selection via nucleotide analog-targeted degradation, InDA-C-mediated adaptor cleavage and PCR amplification to produce the final library. Different adaptors were used for multiplexing samples in one lane. Sequencing was performed on Illumina HiSeq 2500 for a single read 50 run. Data quality check was done on Illumina SAV. Demultiplexing was performed with Illumina CASAVA 1.8.2. The gene expression level was calculated using RSEM software (Li and Dewey, 2011). TPM (transcript per million) was used to normalized the gene expression. Only genes that were significantly modulated (false discovery rate (FDR)-adjusted; p-value <0.05) and more than a 2 fold change were considered differentially expressed in the AH livers compared with normal livers. The pathway and network analysis was performed using Ingenuity Pathway Analysis (IPA). IPA computes a score for each network according to the fit of the set of supplied focus genes. These scores indicate the likelihood of the focus genes to belong to a network versus those obtained by chance. A score >2 indicates a ≤ 99% confidence that a focus gene network was not generated by chance alone. The data presented here are accessible at the UCLA website (http://hpc.ucla.edu/hoffman2/file-transfer/gol.php).
Quantitative Real-time PCR
Real-time PCR was performed as previously described (Liu et al., 2014b). Primer sequences are as follows: P53 (human; NM_000546) forward primer: 5′-TCTACCTCCCGCCATAAAA-3′, reverse primer: 5′-CTCCTCCCCACAACAAAAC-3′; P53 (mouse; NM_011640) forward primer: 5′-AGCTTT GAGGTTCGTGTTTG-3′, reverse primer: 5′- GGAACATCTCGAAGCGTTTA-3′; P27 (human; NM_004064) forward primer: 5′-TCAAACGTGCGAGTGTCTAA-3′, reverse primer: 5′-CCACTCGTACTTGCCCTCTA-3′; P27 (mouse; NM_009875) forward primer: 5′-AGCGTTTCTTCATTGCCTGT -3′, reverse primer: 5′- CACAAAACATGCCACTTTGG -3′.
MicroRNA assay
Reverse transcription was performed with the above mentioned total RNA (10 ng), and 5×RT primers including U6 snRNA, miR-34a and miR-483-3p using TaqMan MicroRNA Reverse Transcription Kit following the instructions (Applied Biosystems). qPCR amplification was performed using the TaqMan Universal PCR Master Mix plus 20×RT primers including U6 snRNA, miR-34a and miR-483-3p on a StepOnePlus™ Real-time PCR System (Applied Biosystems). Reaction conditions consisted of 50°C for 120 sec, 95°C for 2 min followed by 40 cycles of 95°C for 15 sec, 60 °C for 60 sec. U6 snRNA was used as miRNA control to normalize the target miRNA abundance in each sample. The relative miRNA expression levels were analyzed using the ΔΔCT method. Reaction of each sample was performed in triplicate.
Statistical analysis
Statistical significance was determined using the t-test and One Way ANOVA test with SigmaStat software. P< 0.05 was considered as a statistically significant difference. Regression plots were constructed using SigmaPlot software. All data were presented as the mean ±S.E.M and were representative of at least three-independent experiments done in triplicate.
Results
MiR-34a is upregulated in AH livers and in livers of DDC re-fed mice
The upstream regulators in livers from AH patients with MDBs were analyzed and compared with normal livers. Forty-nine molecules were included in this RNA-Seq database and the complexity of transcriptional expression of these upstream regulators was identified (Unpublished data). The mRNA levels of CCRK (Cdk20), EIF4G1, MYC, TGFβ1 and miR-34a were significantly upregulated, while SATB1, IKK and miR-483-3p were markedly downregulated when compared with normal livers (Fig. 1A). These are mostly factors required for cell cycle progression (CCRK, MYC) (Malumbres, 2014), inflammation (TGFβ1, IKK) and protein-coding genes involved in translation (SATB1, EIF4G1). The expression of miR-34a was first analyzed by real-time PCR in the same AH biopsies used in RNA-Seq. Similar to RNA-Seq observation, miR-34a had an approximately 3-fold upregulation in this test (Fig. 1B).
Figure 1.
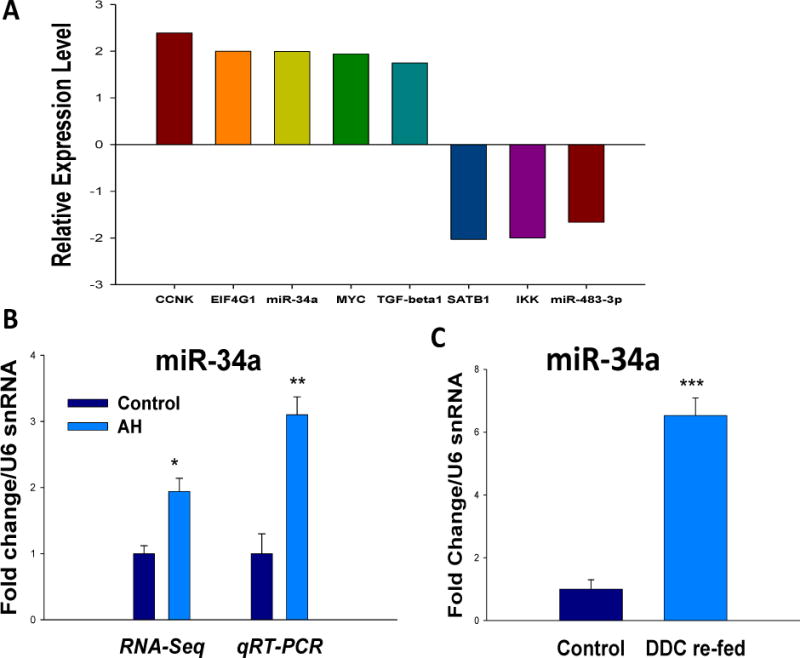
miR-34a is upregulated in AH livers and in the livers of DDC re-fed mice. A Transcription expression of upstream regulators of AH livers by RNA-Seq analyses was shown. B, Increased expression of miR-34a in the livers of alcoholic hepatitis patients was measured by RNA-Seq and qRT-PCR, respectively. C, Induction of miR-34a in the livers of DDC re-fed mice. miRNA expression was quantified by TaqMan miRNA assays (Applied Biosystems). The data were normalized to U6 snRNA (endogenous control) and shown as fold change over the normal livers. Data represent mean values ±S.E.M. *p<0.05, **p<0.01, and ***p<0.001
DDC was used to induce MDB formation in mice. Hepatocyte isolation was then performed from mice fed the control diet and mice re-fed DDC for 7 days as described earlier (Oliva et al., 2009). The miR-34a expression in the livers of DDC re-fed mice was examined using real-time PCR analysis. A 6-fold upregulation of miR-34a was observed in the livers of DDC re-fed mice (Fig. 1C). In conclusion, it is suggested that miR-34a plays an important role in MDB formation.
MiR-34a upregulation is p53-independent during liver MDB formation
The miR-34a gene promoters contain p53-binding sites and one of the strongest inducers of miR-34a is p53 (He et al., 2007; Tarasov et al., 2007). Therefore, the mRNA levels of p53 and its target p27 were analyzed in the AH livers and in the livers of DDC re-fed mice by real-time PCR. Interestingly, p53 was significantly downregulated, both in the livers of DDC re-fed mouse (Fig. 2A) and in AH livers (Fig. 2B). This suggested that miR-34a is upregulated because the downregulation of p53 which failed to inhibit the expression of miR-34a. P27, a cell cycle inhibitor at G1/S growth phase, was shown to be expressed in numerous hepatocytic nuclei in liver biopsies from patients with AH as reported previously (French et al., 2012). P27 was significantly induced in the livers of AH and DDC re-fed mice (Fig. 2C and 2D), with an extreme 200-fold upregulation expression in the livers of AH. This helps explain the mechanism of cell cycle arrest observed in the AJH livers.
Figure 2.
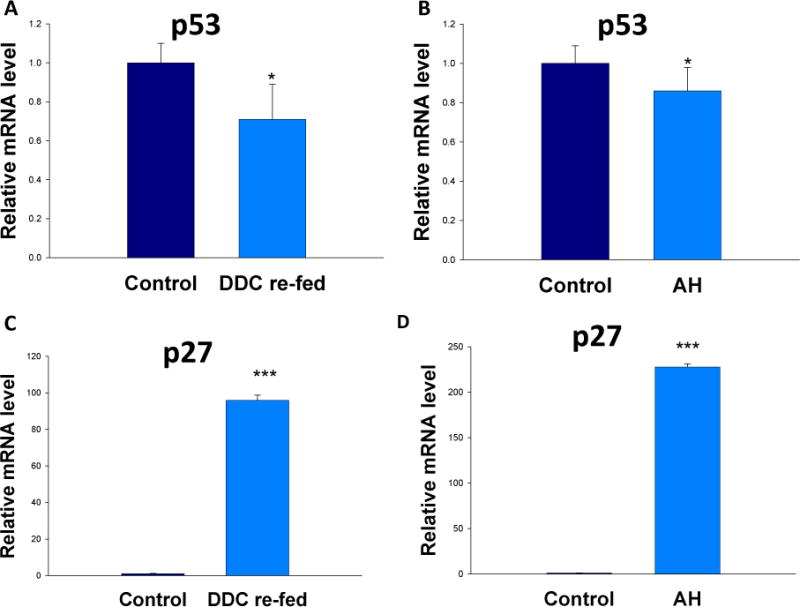
Induction of p53 and p27 in AH livers and in the livers of DDC re-fed mice. Quantification of mRNA was carried out by SYBR real-time PCR assays. * p<0.05 and ***p<0.001 by t-test.
Downregulation of MiR-483-3p increases expression of BRCA1 in AH livers with MDBs
Another important present was the observation that miR-483-3p was significantly downregulated in AH livers by RNA-Seq analyses. The 3′-UTR of BRCA1 mRNA contains a predicted miR-483-3p target site corresponding perfectly to nucleotides 2–8 of the mature miRNA. The seed target sequence was observed between human and mouse (Fig. 3A). The downregulation expression of miR-483-3p in the RNA-Seq data was further examined by qRT-PCR analysis using AH biopsies (Fig. 3B). The downregulation of miR-483-3p, which increases BRCA1 expression might provide the mechanism of BRCA1 expression was increased in the livers from AH and the DDC re-fed mice.
Figure 3.
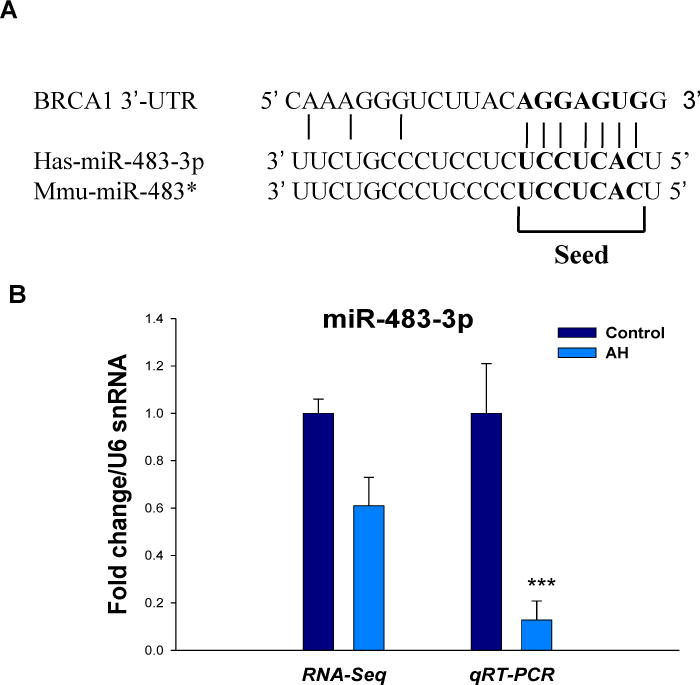
The downregulation of miR-483-3p may regulate BRCA1 expression in AH livers and in the livers of DDC re-fed mice. A, Sequence of putative human and mouse miR-483-3p binding sites in the 3′-UTR of BRCA1 are shown. B, Decreased expression of miR-483-3p in the livers of AH measured by RNA-Seq and qRT-PCR, respectively. miRNA expression was quantified by TaqMan miRNA assays (Applied Biosystems). The data were normalized to U6 snRNA (endogenous control) and shown as fold change over the controls. Data represent mean values ±S.E.M. ***p<0.001.
Discussion
MDBs are prevalent in various hepatic diseases such as AH (Basaranoglu et al., 2011) and can be formed in DDC re-fed mice (French et al., 2010; Li et al., 2008). The function of miRNAs during alcoholic liver disease has recently become of great interest in biological research. Studies have shown that alcoholic liver disease associated miRNAs play a crucial role in the regulation of liver-inflammatory agents such as tumor necrosis factor-alpha (TNFα) (McDaniel et al., 2014). In this study, we specifically discovered an altered regulation of miR-34a and miR-483-3p by a whole-genome expression analysis (RNA-Seq) in AH livers what MDBs had formed. The upregulation of miR-34a and downregulation of miR-483-3p was confirmed by qRT-PCR in the livers of DDC re-fed mice, implying a direct correlation of miR-34a and miR-483-3p with MDB formation. To the best of our knowledge, this is the first study to report the changes in miR-34a and miR-483-3p expression associated with MDB formation in human and mouse livers.
Epigenetic regulation is the main mechanism of liver MDB formation in the mouse model as reported previously (Bardag-Gorce et al., 2008; French et al., 2010; Li et al., 2008). BRCA1 has been shown to trigger miRNA biogenesis and expression of let-7a-1, miR-16-1, miR-145, and miR-34a (Di Martino et al., 2012). By upstream regulator analysis of the RNA-Seq data, a novel miR-483-3p was specifically identified, which was downregulated. This change might be regulated epigenetically by BRCA1 because one of the targets of miRNA-483-3p is BRCA1. Additional miR-483-3p target genes including BBC3/PUMA, SMAD4, CTNNB1 and GDF3 have been identified in other tissues such as Wilm’s tumors, adrenocortical carcinomas, HCT116 colon and HepG2 hepatocellular carcinoma cell lines, and adipose tissue (Ferland-McCollough et al., 2012; Veronese et al., 2011). However, the precise mechanism of regulation of BRCA1 by miR-483-3p is unknown. The downregulation of miR-483-3p could be the mechanism for the upregulation of BRCA1 expression and the upregulation of BRCA1 in turn could be the cause of the cell cycle arrest.
Recent studies showed a significant downregulation of miR-122, a liver-specific miRNA (Girard et al., 2008), which is important for normal lipid metabolism, and the marked upregulation of miR-34a, a critical regulator of apoptosis (Raver-Shapira et al., 2007). Several recent reports have shown that miRNAs miR-122 and miR-34a are two of the most frequently dysregulated miRNAs in steatohepatitis (Li et al., 2009b). Both miR-122 and miR-34a were aberrantly expressed in both alcoholic steatohepatitis and nonalcoholic fatty liver disease (Cheung et al., 2008; Min et al., 2012). We confirmed that miR-34a was significantly upregulated in AH with MDBs. Various transcription factors (TFs) have been shown to regulate miR-34a expression. One of the strongest inducers of miR-34a is p53. The p53 protein regulates multiple cellular pathways and recent studies have shown p53-dependent upregulation of miR-34a in human and mouse cells as a consequence of DNA damage (Bommer et al., 2007). P53 acts through positive feedback loops, which adding robustness to this network have been described, one of them involving sirtuin type 1 (SIRT1) protein (Yamakuchi et al., 2008; Yamakuchi and Lowenstein, 2009). Specific miRNA promotes ethanol-induced fat accumulation in hepatocytes by downregulating SIRT1 (Yin et al., 2012). Once SIRT1 is activated, SIRT1 deacetylates histones and histone methyl-transferases. SIRT1-mediated deacetylation of Lys382 decreases p53-mediated transcriptional activation and reduces the downstream proteins such as p21 and PUMA levels (Li et al., 2009c). Interestingly, significant downregulation of p53 was found both in AH livers and in the livers of DDC re-fed mice, similar to the changes in expression observed in primary fibroblasts and CLL (Baer et al., 2013; Christoffersen et al., 2010). This suggests that miR-34a can be regulated independent of p53 in MDB-forming hepatocytes. The detailed mechanism of this p53-independent regulation in liver MDB formation remains to be determined.
Cancer cells are characterized by hypermethylation of trascription-associated CpG islands of tumor-suppressor genes such as Ufm1 (Liu et al., 2014a), resulting in transcriptional repression and gene inactivation. MiR-34a was shown to be hypermethylated in different types of cancer such as breast, colon and lung cancers (Chim et al., 2010; Li et al., 2009a; Lodygin et al., 2008). Ectopic miR-34a induces a G1 cell cycle arrest, senescence and apoptosis, thereby demonstrating the tumor-suppressive role of miR-34a. Cell cycle arrest is an important mechanism of liver injury since our recent observations imply that the G1/S cell cycle pathway was significantly upregulated in AH livers by RNA-Seq (Unpublished data). Notably, p27 mRNA was found to be significantly upregulated in AH livers and in the livers of DDC re-fed mice. These findings collectively indicate that upregulation of miR-34a is a potent tumor suppressor which regulates the key cell cycle inhibitors causing the liver regeneration failure.
In summary, the upregulation of miR-34a and downregulation of miR-483-3p present in the livers of AH and DDC re-fed mice may provide the explanations for the mechanisms underlying the cause of liver MDB formation and the inhibition of liver cell regeneration.
Acknowledgments
This work was supported by grants from NIH (AAU01021898-03) and P50-11999 Morphology Core.
Abbreviations
- AH
alcoholic hepatitis
- BRCA1
breast cancer susceptibility gene 1
- DDC
diethyl 1, 4-dehydro-2, 4, 6-trimeth yl-3, 5-pyridine-dicarboxylate
- FFPE
archived formalin-fixed paraffin-embedded
- HCC
hepatocellular carcinoma
- MDB
Mallory-Denk body
- MiRNA
microRNA
- RNA-Seq
RNA sequencing
- SIRT1
sirtuin type 1
- TNFα
tumor necrosis factor-α
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Baer C, et al. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 2013;73:473–7. doi: 10.1158/0008-5472.CAN-12-3731. [DOI] [PubMed] [Google Scholar]
- Bardag-Gorce F, et al. Epigenetic mechanisms regulate Mallory Denk body formation in the livers of drug-primed mice. Exp Mol Pathol. 2008;84:113–21. doi: 10.1016/j.yexmp.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basaranoglu M, et al. Mallory-Denk Bodies in chronic hepatitis. World J Gastroenterol. 2011;17:2172–7. doi: 10.3748/wjg.v17.i17.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertero T, et al. Tumor suppressor function of miR-483-3p on squamous cell carcinomas due to its pro-apoptotic properties. Cell Cycle. 2013a;12:2183–93. doi: 10.4161/cc.25330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertero T, et al. miR-483-3p controls proliferation in wounded epithelial cells. FASEB J. 2011;25:3092–105. doi: 10.1096/fj.10-168401. [DOI] [PubMed] [Google Scholar]
- Bertero T, et al. CDC25A targeting by miR-483-3p decreases CCND-CDK4/6 assembly and contributes to cell cycle arrest. Cell Death Differ. 2013b;20:800–11. doi: 10.1038/cdd.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommer GT, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- Cheung O, et al. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48:1810–20. doi: 10.1002/hep.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim CS, et al. Epigenetic inactivation of the miR-34a in hematological malignancies. Carcinogenesis. 2010;31:745–50. doi: 10.1093/carcin/bgq033. [DOI] [PubMed] [Google Scholar]
- Christoffersen NR, et al. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010;17:236–45. doi: 10.1038/cdd.2009.109. [DOI] [PubMed] [Google Scholar]
- Di Martino MT, et al. Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: in vitro and in vivo evidence. Clin Cancer Res. 2012;18:6260–70. doi: 10.1158/1078-0432.CCR-12-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippold RP, et al. Chronic ethanol feeding alters miRNA expression dynamics during liver regeneration. Alcohol Clin Exp Res. 2013;37(Suppl 1):E59–69. doi: 10.1111/j.1530-0277.2012.01852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferland-McCollough D, et al. Programming of adipose tissue miR-483-3p and GDF-3 expression by maternal diet in type 2 diabetes. Cell Death Differ. 2012;19:1003–12. doi: 10.1038/cdd.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French BA, et al. Mallory-Denk bodies form when EZH2/H3K27me3 fails to methylate DNA in the nuclei of human and mice liver cells. Exp Mol Pathol. 2012;92:318–26. doi: 10.1016/j.yexmp.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SW, et al. The role of innate immunity in the pathogenesis of preneoplasia in drug-induced chronic hepatitis based on a mouse model. Exp Mol Pathol. 2011;91:653–9. doi: 10.1016/j.yexmp.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SW, et al. Mallory-Denk body pathogenesis revisited. World J Hepatol. 2010;2:295–301. doi: 10.4254/wjh.v2.i8.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard M, et al. miR-122, a paradigm for the role of microRNAs in the liver. J Hepatol. 2008;48:648–56. doi: 10.1016/j.jhep.2008.01.019. [DOI] [PubMed] [Google Scholar]
- Haybaeck J, et al. Genetic background effects of keratin 8 and 18 in a DDC-induced hepatotoxicity and Mallory-Denk body formation mouse model. Lab Invest. 2012;92:857–67. doi: 10.1038/labinvest.2012.49. [DOI] [PubMed] [Google Scholar]
- He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–9. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, et al. S-adenosylmethionine prevents Mallory Denk body formation in drug-primed mice by inhibiting the epigenetic memory. Hepatology. 2008;47:613–24. doi: 10.1002/hep.22029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, et al. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009a;275:44–53. doi: 10.1016/j.canlet.2008.09.035. [DOI] [PubMed] [Google Scholar]
- Li S, et al. Differential expression of microRNAs in mouse liver under aberrant energy metabolic status. J Lipid Res. 2009b;50:1756–65. doi: 10.1194/jlr.M800509-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, et al. Inhibition of SUV39H1 methyltransferase activity by DBC1. J Biol Chem. 2009c;284:10361–6. doi: 10.1074/jbc.M900956200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, et al. IL-8 signaling is up-regulated in alcoholic hepatitis and DDC fed mice with Mallory Denk Bodies (MDBs) present. Exp Mol Pathol. 2015;99:320–325. doi: 10.1016/j.yexmp.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, et al. Mallory-Denk Body (MDB) formation modulates ufmylation expression epigenetically in alcoholic hepatitis (AH) and non-alcoholic steatohepatitis (NASH) Exp Mol Pathol. 2014a;97:477–83. doi: 10.1016/j.yexmp.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, et al. Ufmylation and FATylation pathways are downregulated in human alcoholic and nonalcoholic steatohepatitis, and mice fed DDC, where Mallory-Denk bodies (MDBs) form. Exp Mol Pathol. 2014b;97:81–88. doi: 10.1016/j.yexmp.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, et al. TLR3/4 signaling is mediated via the NFkappaB-CXCR4/7 pathway in human alcoholic hepatitis and non-alcoholic steatohepatitis which formed Mallory-Denk bodies. Exp Mol Pathol. 2014c;97:234–40. doi: 10.1016/j.yexmp.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodygin D, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- Malumbres M. Cyclin-dependent kinases. Genome Biol. 2014;15:122. doi: 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel K, et al. The functional role of microRNAs in alcoholic liver injury. J Cell Mol Med. 2014;18:197–207. doi: 10.1111/jcmm.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HA, et al. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science. 2013;340:82–5. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- Meng F, et al. Epigenetic regulation of miR-34a expression in alcoholic liver injury. Am J Pathol. 2012;181:804–17. doi: 10.1016/j.ajpath.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min HK, et al. Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab. 2012;15:665–74. doi: 10.1016/j.cmet.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda RC, et al. MicroRNAs: master regulators of ethanol abuse and toxicity? Alcohol Clin Exp Res. 2010;34:575–87. doi: 10.1111/j.1530-0277.2009.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misso G, et al. Mir-34: a new weapon against cancer? Mol Ther Nucleic Acids. 2014;3:e194. doi: 10.1038/mtna.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva J, et al. Fat10 is an epigenetic marker for liver preneoplasia in a drug-primed mouse model of tumorigenesis. Exp Mol Pathol. 2008;84:102–12. doi: 10.1016/j.yexmp.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva J, et al. Betaine prevents Mallory-Denk body formation in drug-primed mice by epigenetic mechanisms. Exp Mol Pathol. 2009;86:77–86. doi: 10.1016/j.yexmp.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raver-Shapira N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–43. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Tarasov V, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–93. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- Veronese A, et al. Mutated beta-catenin evades a microRNA-dependent regulatory loop. Proc Natl Acad Sci U S A. 2011;108:4840–5. doi: 10.1073/pnas.1101734108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M, et al. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105:13421–6. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle. 2009;8:712–5. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- Yang F, et al. MicroRNA-34a targets Bcl-2 and sensitizes human hepatocellular carcinoma cells to sorafenib treatment. Technol Cancer Res Treat. 2014;13:77–86. doi: 10.7785/tcrt.2012.500364. [DOI] [PubMed] [Google Scholar]
- Yin H, et al. MicroRNA-217 promotes ethanol-induced fat accumulation in hepatocytes by down-regulating SIRT1. J Biol Chem. 2012;287:9817–26. doi: 10.1074/jbc.M111.333534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan QX, et al. Mallory body induction in drug-primed mouse liver. Hepatology. 1996;24:603–12. doi: 10.1002/hep.510240324. [DOI] [PubMed] [Google Scholar]


