Abstract
Previously we reported that IL-17+ T cells, primarily IL-17+ γδ cells, are increased in mice lacking the protease inhibitor serpinB1 (serpinb1−/− mice). Here we show that serpinB1-deficient CD4 cells exhibit a cell-autonomous and selective deficiency in suppressing T helper 17 (Th17) cell differentiation. This suggested an opposing role for one or more protease in promoting Th17 differentiation. We found that several SerpinB1-inhibitable cysteine cathepsins are induced in Th17 cells, most prominently cathepsin L (catL); this was verified by peptidase assays, active site labeling and Western blots. Moreover, Th17 differentiation was suppressed by both broad cathepsin inhibitors and catL selective inhibitors. CatL is present in Th17 cells as single chain (SC)- and two-chain (TC)-forms. Inhibiting asparagine endopeptidase (AEP) blocked conversion of SC-catL to TC-catL and increased generation of serpinb1−/− Th17 cells, but not wild-type Th17 cells. These findings suggest that SC-catL is biologically active in promoting Th17 generation and is counter-regulated by serpinB1 and secondarily by AEP. Thus, in addition to regulation by cytokines and transcription factors, differentiation of CD4 cells to Th17 cells is actively regulated by a catL-serpinB1-AEP module. Targeting this protease regulatory module could be an approach to treating Th17 cell-driven autoimmune disorders.
Keywords: Th17 cells, Cathepsins, SerpinB1, Asparagine endopeptidase, Autoimmune disorders
1. Introduction
IL-17 producing T helper cells (Th17 cells) are critically important for protective immunity of mucosal surfaces against fungal infections and extracellular bacteria, but are also implicated in the pathology of severe inflammatory and autoimmune diseases including multiple sclerosis, psoriasis, rheumatoid arthritis, and inflammatory bowel disease. Since the discovery of Th17 cells as the third major T helper lineage distinct from Th1 and Th2 cells [1, 2], much has been learned about the mechanisms that control Th17 cell generation, survival and pathogenesis, in particular about the influence of cytokines and transcription factors. Naive CD4 cells differentiate to Th17 cells in the presence of limiting TGF-β and the proinflammatory cytokine IL-6 [3–5]. During differentiation, the transcription factors BATF and IRF4 establish initial chromatin accessibility and in combination with STAT3 [6] and orphan nuclear receptor RORγt, the master Th17 regulator [7], and the negative regulator cMAF, function to establish a coherent transcriptional program to produce Th17 cells expressing a set of signature cytokines and cytokine receptors, including IL-23R [8] and IL-1R [9]. Overall, the dynamic regulatory program that coordinates Th17 differentiation involves sequential developmental stages coordinated by a network of highly interactive antagonistic interactions of Th17 promoting- and a smaller number of Th17 suppressing-modules [10].
Here we describe an additional layer of regulation whereby the differentiation of Th17 cells is controlled by a protease module that includes cathepsin L (catL), an endosomal/lysosomal cysteine protease, together with asparagine endopeptidase/legumain (AEP), which converts single chain catL (SC-catL) to the two-chain form, and serpinB1, a protease inhibitor previously characterized as a protective anti-inflammatory immune modulator. In prior studies, we correlated the increased inflammation and injury of influenza virus infected serpinb1−/− mice with increased numbers of IL-17-producing T cells, primarily γδ+ T cells [11]. Subsequently, we found that IL17+ γδ T cells, but not IFNγ+ γδ cells, are selectively expanded in naive serpinb1−/− mice [12], indicating a suppressive role for SerpinB1. Here we show, through the use of in vivo immunization and in vitro differentiation, that SerpinB1 deficient CD4 cells exhibit a cell-autonomous and selective deficit of suppression of Th17 cell differentiation. Further experimentation identified the SerpinB1-inhibitable protease that promotes Th17 differentiation as catL and identified AEP as an additional suppressive regulator.
2. Materials and Methods
2.1. Animals
SerpinB1 deficient mice (serpinb1a−/−, hereafter serpinb1−/−) were generated in 129S6/SvEv/Tac (129S6) background [13]. CatH deficient mice (ctsh−/−) in C57BL/6N background [14] were provided by Drs. Thomas Reinheckel (Albert-Ludwig University, Freiburg, Germany) and Johanna Joyce (Memorial Sloan-Kettering Cancer Center, New York). The mice were rederived at Boston Childrens Hospital by mating ctsh−/− males with WT C57BL/6J females and intercrossing the resulting heterozygotes. Ctsh−/− pups from the intercross were selected that carried the C57BL/6J-specific allele at the nicotinamide nucleotide transhydrogenase (Nnt) locus and were backcrossed to C57BL/6J for two additional generations. Ctsh−/− and serpinb1−/− mice were viable and fertile with no gross phenotypes. WT 129S6 mice (Taconic Labs) and WT C57BL/6J (Jackson Labs) were maintained together with serpinb1−/− and ctsh−/− mice in the animal facility of Boston Children’s Hospital. Animal studies were approved by the Institutional Animal Care and Use Committee of Boston Children’s Hospital.
2.2. KLH immunization
WT and serpinb1−/− mice were immunized by injection i.p. of keyhole limpet hemocyanin (KLH) (200 μg, Sigma-Aldrich) in 200 μl of a 1:1 emulsion in Freund’s adjuvant (Sigma-Aldrich). Seven days later, mice were sacrificed and splenocytes were cultured with or without KLH for 2 days with Brefeldin A present during the final 6 h. The cells were collected for flow cytometry and the supernatants for ELISA assay.
2.3. Isolation of naive CD4 cells
Single cell suspensions were prepared from spleens of 4–6 wk old mice. After erythrocyte lysis, pooled splenocytes were depleted of CD11b+, CD8α+ and CD19+ cells using biotinylated primary antibodies (BioLegend) and streptavidin-coated secondary magnetic particles (Stem Cell Technologies). The enriched cells were sorted on the FACS Aria for CD4+CD25negCD44negCD62L+. Purity was >98%.
2.4. T-helper cell differentiation
Naive CD4 T cells (0.4 × 106) in 24 well plates (Costar) pre-coated with anti-CD3 (154-2C11, 5 μg/ml, BioXcell) and anti-CD28 (37.51, 2 μg/ml, BioXcell) were cultured in RPMI containing 10% FCS and polarizing cytokines. The cytokines were: Th1, mIL-12 (10 ng/ml, Biolegend) and anti-mIL-4 (11B11, 2 μg/ml, BioXcell); Treg, hTGF-β1 (3 ng/ml, Biolegend), mIL-2 (20 ng/ml, Biolegend), anti-mIFN-γ (XMG1.2, 2 μg/ml, BioXcell) and anti-mIL-4; Th17, mIL-6 (10 ng/ml, Biolegend), hTGF-β1 (2 ng/ml), anti-mIFN-γ, and anti-mIL-4. Cells stimulated in ‘neutral’ conditions (anti-mIL-4 plus anti-mIFN-γ without added cytokines) were considered Th0 cells.
Where studied, protease inhibitors, AEBSF (Pefabloc) and E64 (Sigma-Aldrich), E64D (Santa Cruz), z-Phe-Ala-fmk (Enzyme Systems), CA074-OMe (EMD Millipore), Ns-Ile-Trp-CHO (IW-CHO, Enzo Life Sciences), CLIK195 (provided by Guo-Ping Shi), and the AEP inhibitor LI-1 [15], were added at the start of culture. Unless otherwise indicated, differentiated cells were harvested after 3 days for Western blot, peptidase assay or active site labeling or were restimulated for 4h with PMA (50 ng/ml) and ionomycin (750 ng/ml) (Sigma-Aldrich) in the presence or absence of Brefeldin A for flow cytometry or ELISA, respectively.
2.5. Intracellular staining and flow cytometry
Harvested cells were stained with fluorochrome-conjugated antibodies to surface markers (Biolegend). The cells were fixed, permeabilized and stained intracellularly with fluorochrome-conjugated anti-mIL-17A (TC11-18H10) (hereafter IL-17), anti-mIFN-γ (XMG1.2) and anti-FoxP3 (FJK-16s) (all from Biolegend) using FoxP3 fixation/permeabilization reagents and protocols from eBiosciences. Data were acquired on a Canto II cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star).
2.6. ELISA
IL-17A (hereafter IL-17) and IFN-γ were assayed using ELISA kits (eBioscience) according to the manufacturer’s instructions.
2.7. Reverse transcription and qPCR analysis
RNA was isolated using RNeasy kits (Qiagen) and was digested with DNase I (Ambion) and reverse-transcribed using the iScript™ cDNA Synthesis kit (Bio-Rad). The qPCR assays are detailed in Supplemental Materials and Methods.
2.8. Western blot
Differentiated cells were suspended at 12.5 × 106 per ml in PBS with 2 mM AEBSF and lysed with 5X SDS lysis buffer with mercaptoethanol and boiling for 10 min. Alternatively, cell homogenates prepared in NP40-containing buffer (described below) were similarly SDS-solubilized. Samples were resolved on 12% Tris-glycine gels and transferred onto PVDF. Membranes were blocked with 5% milk solids and stained with rabbit antiserum to human SerpinB1 [13], goat antiserum to mouse catL (AF1515, R&D Systems) or sheep antiserum to mouse AEP (AF2058, R&D Systems) followed by HRP-conjugated secondary antibodies (Cell Signaling). Bands were visualized by enhanced chemiluminescence (ECL-Plus, Amersham). Blots were stripped and restained with mouse anti-mouse β-actin antibody (Cell Signaling).
2.9. Enzymes, inhibitor, substrates and peptidase assays
Reagents and peptidase assays (Fig. 4 and 5C) are detailed in Supplemental Materials and Methods.
Fig. 4.
Cathepsin L expression in Th17 cells. Naïve CD4 cells were differentiated as in Fig. 1. A) RNA was extracted at 60 h for qRT-PCR. Normalized results are expressed relative to Th0 cells. B) Peptidase activity. Left: Lysates of Th0, Th1, Treg and Th17 cells assayed for cleavage of z-Phe-Arg-AMC in the presence of the catB inhibitor CA-074. Results are linear rates (units/min). Right: z-Phe-Arg-AMC cleaving activity of Th17 cell lysate in the presence of the indicated inhibitors. C) Cathepsin active site labeling. Left: Differentiated cells incubated with DCG-04 for 30 min are shown as ‘biotin blot’. Arrow indicates the 27 kDa DCG-04 labeled protease. Right: DCG-04 labeling of Th17 cells pre-incubated for 15 min with the indicated inhibitors. D) Western blot for catL. The catL pro-form (38 kDa), single-chain form (SC, 27 kDa) and heavy chain of two-chain form (TC, 20 kDa) are indicated. Right: Densitometric quantitation of the combined mature species. Data are representative findings from (A) 3 experiments (B) 2 experiments, each in duplicate (C) 2–4, and (D) 3 experiments. **P < 0.01, ***P <0.001.
Fig. 5.
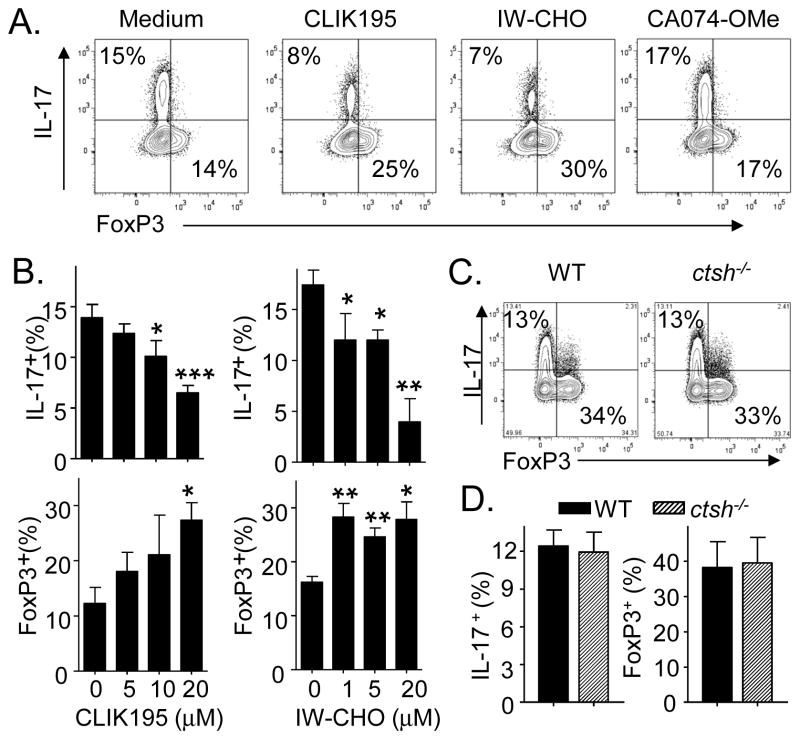
Role of individual cathepsins on Th17 differentiation. A and B) Effect of select cathepsin inhibitors. CD4 cells were differentiated under Th17 polarizing conditions with the indicated inhibitors. A) CLIK195 (20 μM), IW-CHO (20 μM) and CA074-OMe (5 μM). B) Dose-dependent effects of IW-CHO and CLIK195. C) Naïve CD4 T cells from WT and ctsh−/− mice differentiated under Th17 conditions. Data are from (A and B) 3 experiments and (C) 5 mice/group and are shown as (A) representative profiles, (B) means ± SEM and (C) representative profiles (left) and means ± SEM (right). *P < 0.05, **P <0.01.
2.10. Active site Labeling
Differentiated cells were washed and resuspended at 10 × 106/ml in RPMI without serum and incubated for 30 min with 10 μM DCG-04, a biotin moiety-containing activity-based probe for cysteine cathepsins [16]. The labeled cells were lysed, fractionated by SDS electrophoresis, transfered to PVDF, blocked with 1% BSA and stained with HRP-conjugated avidin. To evaluate the blocking ability of select cathepsin inhibitors, Th17 cells were preincubated with 20 μM E64D, 20 μM CLIK195, 5 nM LHVS (morpholinurea-leucine-homophenylalanine-vinlsulfone phenyl) [17] or 5 μM catK inhibitor-II (Calbiochem).
2.11. Statistical analysis
Data are expressed as means ± SEM and are analyzed by the unpaired Student’s t-test. P < 0.05 was considered significant.
3. Results and Discussion
3.1. SerpinB1 selectively suppresses Th17 differentiation
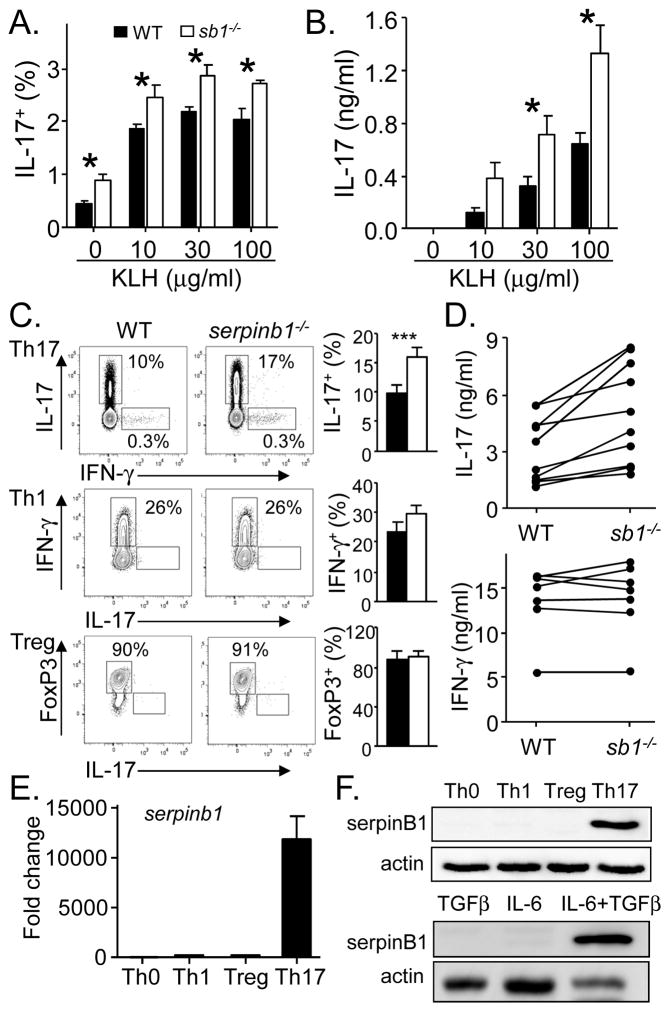
To evaluate the effect of SerpinB1 on Th17 differentiation, we immunized mice with keyhole limpet hemagglutinin (KLH) and evaluated antigen recall responses seven days later. We found that serpinb1−/− mice accumulated increased numbers of antigen-specific Th17 cells compared with WT mice (Fig. 1A), and serpinb1−/− splenocytes produced increased IL-17 (Fig. 1B). To evaluate the selectivity of the serpinB1 effect on Th17 cells, we studied differentiation in vitro. On activation of naive CD4 cells under Th17 polarizing conditions (TGFβ + IL-6), generation of IL-17+ cells was increased for serpinb1−/− cells compared with WT cells, but generation of IFN-γ+ cells under Th1 polarizing conditions and FoxP3+ cells under T regulatory (Treg) polarizing conditions were not different between the genotypes (Fig. 1C). Selective enhancement of Th17 differentiation was seen also at the level of cytokines in that IL-17 production was increased for serpinb1−/− Th17 cells compared with WT cells, but IFN-γ production by Th1 cells was not different between the genotypes (Fig. 1D). These findings are consistent with transcriptional profiling findings that identified SerpinB1 as a Th17 signature gene among the small gene group that suppress the Th17 cell program [10]. Because naive CD4 cells were the only cells in the differentiation studies, the restrictive effect of SerpinB1 on Th17 differentiation is a CD4 cell-intrinsic mechanism.
Fig. 1.
Effect of SerpinB1 on Th17 cell differentiation. A and B) Immunization studies. WT and serpinb1−/− (sb1−/−) mice were immunized with KLH, and splenocytes harvested on day 7 were restimulated with the KLH as indicated. A) Th17 (IL-17+CD4+) recall cells quantified by flow cytometry. B) IL-17 quantified by ELISA. C and D) In vitro differentiation. Sorted naïve CD4 cells from WT and serpinb1−/− mice were differentiated for 3 days with immobilized α-CD3 plus α-CD28 under Th17 (TGF-β, IL-6, α-IFNγ, α-IL-4) or Th1 (IL-12, α-IL-4), or Treg polarizing conditions (TGF-β, IL-2, α-IFNγ, α-IL-4). C) Differentiated cells evaluated by flow cytometry. D) IL-17 produced by Th17 cells (top); IFNγ produced by Th1 cells (bottom). E and F) SerpinB1 expression in WT cells. Naive CD4 cells were differentiated as in C) except as noted. E) RNA was extracted at 60 h for qRT-PCR, and normalized results are expressed relative to Th0 cells. F) Western blots showing 42 kDa serpinB1. Data are means ± SEM of (A and B) 4 mice/group; (C) 3–5 experiments; (D) each line represents an independent experiment; (E and F) means ± SEM and representative blots of 3–4 experiments. *P < 0.05; ***P < 001.
Under Th17 conditions, Tregs were produced along with Th17 cells as previously described [3] and, in this case, generation of “Tregs” was decreased for serpinb1−/− cells compared with WT cells (Supplemental Fig. 1A). The reciprocal effect of serpinB1 deficiency, increased Th17 cells and decreased Tregs, persisted although its magnitude differed when the ratio of IL-6 to TGF-β was varied (Supplemental Fig. 1B). Total yield of cells in Th17 culture conditions did not differ between WT and serpinb1−/− (1.15 ± 0.28 × 106/ml versus 1.33 ± 0.33 × 106/ml, respectively, n = 4, P=0.4), indicating that the increased percentage of IL-17+ cells and decreased percentage of FoxP3+ serpinb1−/− cells reflect absolute cell numbers. The ability of serpinB1 to reciprocally effect Th17 and Treg generation suggests that serpinB1 acts at or downstream of the common Rorγt+Foxp3+ intermediate cell that serves as precursor to both lineages [18]. Moreover, the effect of serpinB1 to alter Treg production only under Th17 polarizing conditions, suggests a primary effect on Th17 differentiation and a default effect on Tregs. Consistent with this interpretation, serpinb1 mRNA and protein were substantially increased in Th17 cells of WT mice compared with Th0, Th1 and Treg cells (Fig. 1E and F). Also, serpinb1 expression required both TGF-β and IL-6 (Fig. 1F) and occurred relatively late in the Th17 cell program at or downstream of Rorc expression (Supplemental Fig. 1C), findings that are also consistent with a primary action in the Th17 pathway downstream of the branchpoint with Tregs.
3.2. Cysteine proteases are involved in Th17 differentiation
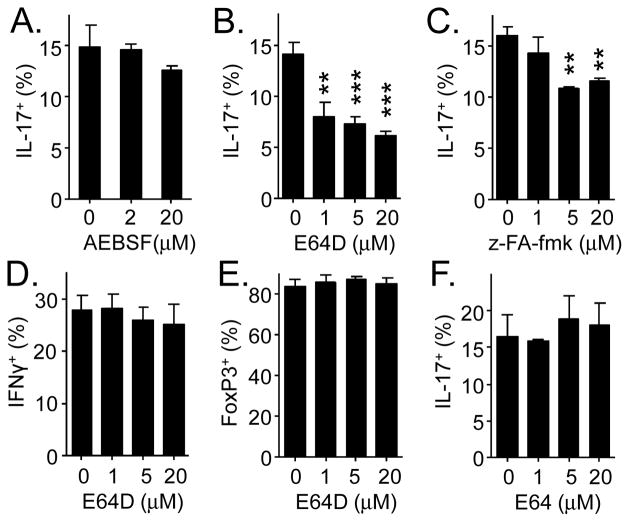
Because inhibition of proteases is SerpinB1’s only known biochemical function, we postulated that the suppressive effect of SerpinB1 on Th17 differentiation involves the intermediacy of a protease. To test this, we performed CD4 cell differentiation in the presence of class-specific protease inhibitors: AEBSF (4-(2-aminoethyl) benzenesulfonyl fluoride), an irreversible inhibitor of serine proteases, and E64D, an irreversible inhibitor of cysteine proteases, especially cysteine cathepsins [19]. Whereas AEBSF did not alter Th17 cell output, E64D caused a dose-dependent decrease, suggesting involvement of a cysteine protease (Fig. 2A and B). This result was surprising because prior studies had characterized SerpinB1 as an inhibitor of serine proteases [20]. Although most serpins inhibit serine proteases (the term originally meant serine protease inhibitor), a sub-group inhibit cysteine proteases (called “cross-class inhibition”) [21]. The E64D effect was verified by finding that z-Phe-Ala-fmk, a chemically unrelated cysteine protease inhibitor, also decreased Th17 cell production (Fig. 2C). In contrast, E64D had no effect on the generation of Th1 cells under Th1 conditions or Tregs under Treg conditions (Fig. 2D and E). Lastly, E64, a cell-impermeable cysteine protease inhibitor, did not alter Th17 cell production (Fig. 2F). The combined findings indicate that one or more CD4 cell intracellular cysteine proteases selectively enhances Th17 cell output, possibly by antagonizing the effect of SerpinB1.
Fig. 2.
Effects of protease inhibitors on Th17 differentiation. Naïve CD4 T cells were differentiated as in Fig. 1 in the presence of proteases inhibitors. A–C) Effect of cell-permeable inhibitors AEBSF, E64D and z-Phe-Ala-fmk on Th17 differentiation. D and E) Effect of E64D on Th1 and Treg differentiation. F) Effect of cell-impermeable inhibitor E64 on Th17 cell differentiation. Data are means ± SEM of 3 experiments. **P < 0.01; ***P < 001.
3.3. Cysteine cathepsins are inhibited by SerpinB1
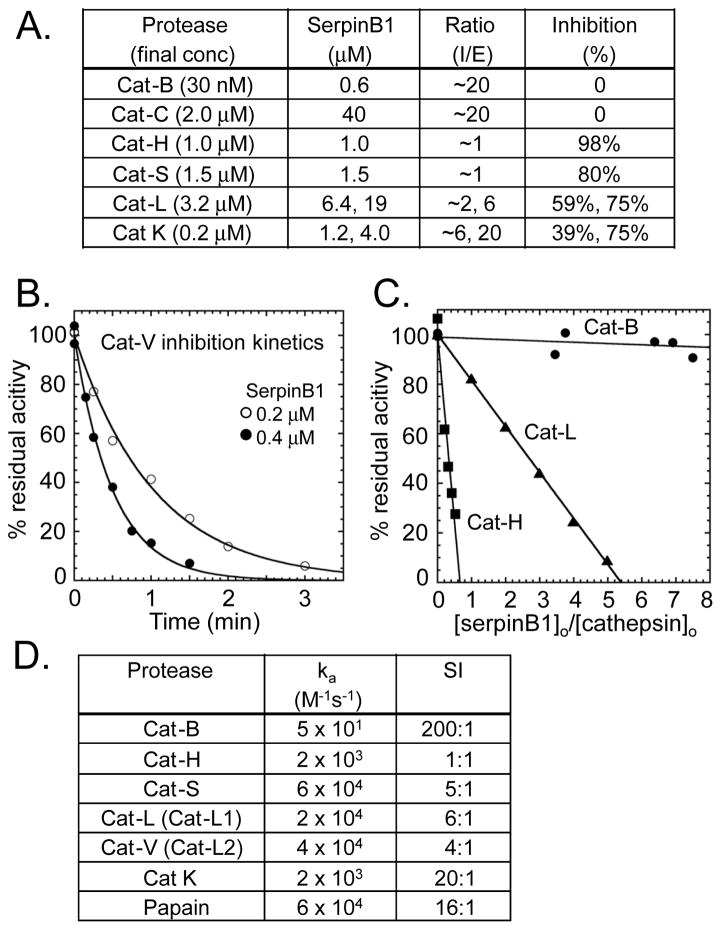
On the premise that SerpinB1 and the E64D-inhibitable protease interact, we performed peptidase inhibition assays to determine whether any cysteine protease can be inhibited by SerpinB1. We focused on cysteine cathepsins of the C1 family of papain-like proteases. These are generally optimally active at acidic pH and have common functions, including terminal protein degradation in lysosomes and also specific functions including antigen presentation (catS and catL), bone resorption (catK) and cytolytic granzyme activation (catC) (reviewed in [22]). With few exceptions (discussed below), CD4 cell cathepsins have received little attention. Because of the different pH optima, we initially screened proteases by brief incubation with excess SerpinB1 at near neutral pH (optimal for SerpinB1) followed by assay of remaining peptidase activity at acidic pH using specific peptides linked to 4-amino-7-methylcoumarin (AMC). Based on this protocol, catH, catS, catL and catK were inhibited by SerpinB1 in that approximate order of efficiency and catB and catC were not inhibited (Fig. 3A).
Fig. 3.
Inhibition of cathepsins by SerpinB1. A) Screening assay. Pure cathepsin and SerpinB1 were co-incubated for 5 min at near neutral pH and residual activity was assayed. I:E (inhibitor: enzyme) ratios are approximate because enzyme concentration was based on suppliers’ information. B–D) Kinetics and stoichiometry of SerpinB1 inhibition of recombinant human cathepsins at pH 6.0. B) Kinetics. Progress curves for inhibition of catV (0.5 nM) by SerpinB1. C) Stoichiometry of inhibition (SI) for catH, catL and catB by the indicated molar ratios of SerpinB1. D) Apparent second-order rate constants and SI for SerpinB1 inhibition of cathepsins. (B and C) Representative data and (A and D) average results of 2–3 experiments.
In the second approach, we maintained pH 6 throughout and measured the kinetics of cathepsin inhibition by SerpinB1 under pseudo first order conditions using active-site titrated enzymes. We included both human catL and catV (also known as catL1 and catL2), which result from a recent gene duplication. Both are orthologous to the single murine catL gene. The inhibition progress curves (e.g. catV in Fig. 3B) allow determination of second order rate constants of inhibition, whereby rate constants >104 M−1s−1 are generally considered physiologically relevant. Stoichiometry of inhibition (SI) values were also determined (Fig. 3C). Typically, serpins form tight complexes with proteases at a stoichiometry of 1, but parallel “substrate reactions” can occur in which the serpin is cleaved, and these are reflected by SI values >1 [21]. The data showed that SerpinB1 inhibits cat-S, L and V with apparent rates >104 M−1s−1, which are comparable to the rate of inhibition of the prototype C1 family protease, papain (Fig. 3D). The SI values varied from 1:1 to 200:1. Although actual physiological SI may differ due to effects of pH, temperature or other interactions [23], the SI values strongly indicate that catH, catS, catL and catV are inhibited with physiologically significant efficiencies, whereas catB and catK are poorly inhibited because they preferentially recognize SerpinB1 as a substrate. The combined results of the two approaches point to cat-S, catL and catV (murine catS and catL) and perhaps catH as candidate SerpinB1-inhibitable E64D-sensitive proteases acting to promote the Th17 differentiation program.
3.4. Cysteine cathepsins are induced in Th17 cells
A prominent theme of cathepsin biology is the importance of cellular expression patterns to biological function. Examples include MHC class II processing and presentation, which requires catS in B cells and dendritic cells, but catL in thymic epithelial cells, consistent with their restricted gene expression pattern [24]. Similarly, discovery of the abundant and selective expression of catK in osteoclasts predicted its subsequent identification as the principle enzyme in bone resorption [25]. Expression of cathepsins was examined by q-PCR for CD4 cells differentiating under non-polarizing conditions (Th0), as well as Treg and Th17 polarizing conditions. The genes significantly induced in the Th17 lineage compared with Th0 and Treg lineages encode for catB and catC, which are not inhibited by SerpinB1, and the SerpinB1-inhibitable proteases catH and catL (Fig. 4A). The increase of ctsl mRNA in Th17 cells was the most prominent difference (Fig. 4A). Selectively increased catL was demonstrated also by peptidase assays, which revealed increased z-Phe-Arg-AMC cleaving activity in lysates of Th17 cells relative to Th0, Th1 and Treg cells (Fig. 4B). In the lysates prepared from permeabilized Th17 cells, the enzyme responsible for cleaving z-Phe-Arg-AMC was verified as catL by its inhibition by CLIK195, an irreversible catL inhibitor [26], and IW-CHO, a selective reversible catL inhibitor [27, 28], as well as the broad cysteine cathepsin inhibitor E64 (Fig. 4B, right). Selective induction of catL in Th17 cells was further verified by active site labeling with DCG-04, a biotin moiety-containing activity-based probe for cysteine residues of active cathepsins [16]. DCG-04 labeled a single prominent band in Th17 cells at 27 kD, the anticipated molecular weight of mature catL. DCG-04 labeling of the 27 kDa protease was decreased by preincubating Th17 cells with catL inhibitor CLIK195, as well as the broad cysteine cathepsin inhibitor E64D, but not by catS inhibitor LHVS [17] or catK inhibitor II (Fig. 4C, right). Finally, the preferential induction of catL was verified by Western blot, which revealed three catL species in Th17 cells: the inactive proenzyme, and two mature biochemically active species, single chain (SC) catL and the predominant species, two chain (TC) catL (Fig. 4D). Densitometric quantitation of the two mature biochemically active species indicated a four-fold increase of catL in Th17 cells compared with Th0 cells (Fig. 4D, right). Altogether, the findings demonstrate the selective induction of catL during Th17 differentiation.
3.5. Inhibition of cathepsin L suppresses Th17 cell differentiation
To evaluate the putative role of catL in promoting Th17 differentiation, we tested selective cathepsin inhibitors. We did not pursue a gene-deletion approach because ctsl−/− mice are specifically deficient in CD4 cells, the result of failed positive selection by ctsl−/− thymic epithelial cells [24]. The inhibitor studies showed that, similar to E64D, the selective catL inhibitors, CLIK195 and IW-CHO, suppressed Th17 differentiation (Fig. 5A); suppression by both agents was dose-dependent (Fig. 5B, top panels). CA074-OMe (5 μM), a pro-drug form of the catB specific inhibitor CA074, did not alter Th17 cell generation (Fig. 5A) at concentrations that potently inhibited catB activity and was only weakly inhibitory toward catL (Supplemental Fig. 2A). In addition to suppressing Th17 cells, catL inhibitors CLIK195 and IW-CHO (and also E64D, Supplemental Fig. 2B), but not catB inhibitor CA074-OMe increased the generation of Tregs (Fig. 5A, 5B, bottom panels). The reciprocal effects of the catL inhibitors, to decrease Th17 cells and increase Tregs, suggest that catL, like SerpinB1, acts at or downstream of the Rorγt+Foxp3+ precursor common to both lineages. For catH, for which no selective inhibitor was available, we studied cells from gene-deleted mice. On activation of naive CD4 cells under Th17 polarizing conditions, the generation of Th17 cells and Tregs were not different between ctsh−/− and WT CD4 cells (Fig. 5C and D), indicating that catH does not substantially effect Th17 cell differentiation. Together, the cumulative findings point to catL as the protease that promotes Th17 differentiation.
Of note, a genome-wide profiling study of human CD4 cells identified catL1 among the most highly induced genes in human Th17 cells compared with Th0 cells, a finding that was verified by Western blot [29], suggesting that the present findings are relevant for human as well as murine Th17 differentiation. Another study found that catL of human but not murine CD4 cells processes complement component 3 (C3) to active C3b and C3a with the latter required for homeostatic T cell survival and optimal production of Th1 cytokines [28].
3.6. Cathepsin L processing by asparagine endopeptidase (AEP) in Th17 differentiation
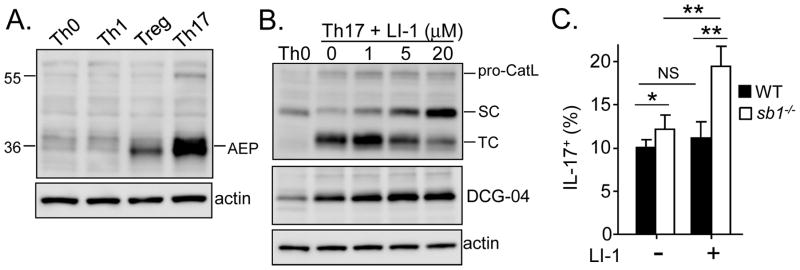
We next questioned whether the SC and TC forms of catL both contribute to Th17 differentiation. CatL is synthesized with a prodomain that protects against premature activation in the endoplasmic reticulum and Golgi and is targeted to the more acidic endosomes where it undergoes autolysis to generate active SC-catL (reviewed in [22]). If present, AEP, an E64-insensitive cysteine protease that hydrolyses substrates C-terminal of asparagine residues, converts SC-catL to disulfide bonded TC-catL; both forms are generally thought to be biochemically active [30, 31]. The presence of both SC- and TC-catL in Th17 cells (Fig. 4D) suggested that AEP is also expressed. Indeed, Western blots confirmed that AEP is strongly induced in Th17 cells, but not in Th0 or Th1 cells and to a lesser extent in Tregs (Fig. 6A). To test for an effect of AEP, naive CD4 cells from WT and serpinb1−/− mice were differentiated under Th17 conditions in the presence of the specific AEP inhibitor LI-1 [15]. Consistent with prior studies, LI-1 dose-dependently decreased TC-catL and increased SC-catL in WT Th17 cells (Fig. 6B) and in serpinb1−/− Th17 cells (Supplemental Fig. 3). The increase of SC-catL was reflected also in an increase of the 27 kDa DCG-04 labeled band (Fig. 6B, middle panel). Of note, although DCG-04 labeling indicates that SC-catL is biochemically active, the absence of DCG-04 labeling of TC-catL has multiple potential explanation such as masking of the protein, inaccessibility due to compartmentation, as well as inhibition by endogenous inhibitors. Whereas LI-1 did not alter Th17 cell differentiation of WT CD4 cells, the inhibitor further increased generation of serpinb1−/− Th17 cells (Fig. 6C). This latter finding points to SC-catL as the biologically active form of catL that promotes Th17 generation and is inhibited in that function in WT cells by the presence of SerpinB1. The combined findings suggest that the Th17-promoting activity of SC-catL is antagonized by the suppressive action of SerpinB1, possibly by direct inhibition (although this has not been tested) and secondarily by degradation by AEP.
Fig. 6.
Effect of cathepsin L processing by AEP on Th17 differentiation. A) Western blot for AEP in differentiated T cells. Mature AEP (42 kDa) is indicated. B and C) Effect of the AEP inhibitor LI-1 on Th17 differentiation. Naive CD4 cells from (B) WT or (C) WT and serpinb1−/− mice were differentiated under Th17 conditions in the presence of the indicated concentration of LI-1. B) Effect on catL evaluated by (top) catL blot, (middle) DCG 04-labeling and (bottom) actin blot. C) Effect on Th17 cell generation. Data from 3 experiments are shown as (A and B) representative Western blots and (C) flow cytometric profiles and means ± SEM. *P < 0.05; **P< 01.
4. Conclusions
The overall findings strongly suggest that catL, serpinB1 and AEP form an intracellular regulatory module that contributes to the control of Th17 differentiation. Cumulative evidence suggests that the module functions relatively late in Th17 differentiation and has a reciprocal effect on Treg generation. All three molecules are selectively induced in Th17 cells as shown previously for human catL [29] and shown here for murine catL (Fig. 4), serpinB1 (Fig. 1E) and AEP (Fig. 6A). Based on confocal microscopy of kidney cells, which revealed AEP localized to endosomes separate from the lysosomes containing the bulk of cathepsins [30], we speculate that the two forms of catL localize in Th17 cells to different compartments: SC-catL in endosomes and TC-catL in lysosomes. AEP may function by regulating the effective lifetime of biologically active endosomal SC-catL. Collectively, the current study implicates a novel protease module as contributor to adaptive Th17 cell differentiation. Future studies will be needed to identify the target(s) of catL in differentiating Th17 cells and learn how the catL cleavage event impinges on known and not-yet-discovered elements of the complex Th17 regulatory network. Targeting the catL-serpinB1-AEP module, once it is better defined, might offer a strategy to treat Th17 cell-driven inflammation and autoimmune diseases.
Supplementary Material
Fig. S1. Relates to Fig. 1. (A,B) Effect of serpinB1 on Treg differentiation under Th17 conditions. Naive CD4 cells from WT and serpinb1−/− mice were differentiated (A) as in Fig. 1C (2 ng/ml TGF-β, 10 ng/ml IL-6) or (B) with the indicated TGF-β and IL-6 concentrations. (C) Time course of expression of Serpinb1 and Rorc during Th17 differentiation of CD4 cells. Representative findings and means ± SEM of (A) 5 experiments from Fig. 1C and (B,C) each 2 experiments. **P < 0.01.
Fig. S2A. Relates to Fig. 5A. Effect of CA074-OMe treatment of cells on catB and catL activities. CD4 cells were differentiated under Th17 polarizing conditions in the absence (medium) or presence of CA074-OMe (5 μM). CatL activity in cell lysates was assayed as in Fig. 4B; catB activity was assayed at pH 6 with z-Gly-Arg-AMC. Results are of 2 experiments.
Fig. S2B. Relates representative to Fig. 2B and 5B. Effect of E64D on generation of FoxP3+ cells under Th17 conditions. Naive CD4 cells from WT and serpinb1−/− mice were differentiated with varying dose E64D. Means ± SEM of 3 experiments from Fig. 2B **P < 0.01; ***P < 001.
Fig. S3. Relates to Fig. 6B. Effect of the AEP inhibitor LI-1 on catL of serpinb1−/− Th17 cells. Naive CD4 cells from serpinb1−/− mice were differentiated under Th17 conditions in the presence of the indicated concentration of LI-1. Western blot for catL representative of 2 experiments.
Cathepsin L, serpinB1 and asparagine endopeptidase (AEP) are induced in Th17 cells.
SerpinB1 inhibits Cathepsin L and restricts Th17 differentiation.
Cathepsin L inhibition by pharmacological agents suppresses Th17 differentiation.
Inhibition of AEP prevents Cathepsin L degradation.
Th17 differentiation is further enhanced in serpinB1−/− cells on inhibition of AEP.
Acknowledgments
We thank Thomas Reinheckel and Johanna Joyce for providing ctsh−/− mice, Dieter Bromme for cathepsin V, Phillip Bird for initial discussions and material and Norma Gerard for use of CFX96 Real-Time System. This work was supported by NIH RO1 HL066548, R21 AI103407 and an RRRC award from Boston Children’s Hospital (to E.R.O) and NIH R37 HL39888 (to S.T.O.).
Footnotes
Conflict of interest disclosure
The authors declare no conflicts of interest related to this study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nature immunology. 2005;6:1123–32. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 2.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nature immunology. 2005;6:1133–41. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–8. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 4.Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–4. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 5.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nature immunology. 2007;8:967–74. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 6.Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–15. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–33. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 8.McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, Blumenschein WM, et al. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nature immunology. 2009;10:314–24. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496:461–8. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong D, Farley K, White M, Hartshorn KL, Benarafa C, Remold-O’Donnell E. Critical role of serpinB1 in regulating inflammatory responses in pulmonary influenza infection. J Infect Dis. 2011;204:592–600. doi: 10.1093/infdis/jir352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao P, Hou L, Farley K, Sundrud MS, Remold-O’Donnell E. SerpinB1 regulates homeostatic expansion of IL-17+ gammadelta and CD4+ Th17 cells. J Leukoc Biol. 2014;95:521–30. doi: 10.1189/jlb.0613331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benarafa C, Priebe GP, Remold-O’Donnell E. The neutrophil serine protease inhibitor serpinb1 preserves lung defense functions in Pseudomonas aeruginosa infection. J Exp Med. 2007;204:1901–9. doi: 10.1084/jem.20070494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buhling F, Kouadio M, Chwieralski CE, Kern U, Hohlfeld JM, Klemm N, et al. Gene targeting of the cysteine peptidase cathepsin H impairs lung surfactant in mice. PloS one. 2011;6:e26247. doi: 10.1371/journal.pone.0026247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, Bogyo M. Synthesis and evaluation of aza-peptidyl inhibitors of the lysosomal asparaginyl endopeptidase, legumain. Bioorganic & medicinal chemistry letters. 2012;22:1340–3. doi: 10.1016/j.bmcl.2011.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenbaum D, Medzihradszky KF, Burlingame A, Bogyo M. Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chemistry & biology. 2000;7:569–81. doi: 10.1016/s1074-5521(00)00014-4. [DOI] [PubMed] [Google Scholar]
- 17.Shi GP, Bryant RA, Riese R, Verhelst S, Driessen C, Li Z, et al. Role for cathepsin F in invariant chain processing and major histocompatibility complex class II peptide loading by macrophages. J Exp Med. 2000;191:1177–86. doi: 10.1084/jem.191.7.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, Victora GD, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–40. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett AJ, Kembhavi AA, Brown MA, Kirschke H, Knight CG, Tamai M, et al. L-trans-Epoxysuccinyl-leucylamido(4-guanidino)butane (E-64) and its analogues as inhibitors of cysteine proteinases including cathepsins B, H and L. The Biochemical journal. 1982;201:189–98. doi: 10.1042/bj2010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooley J, Takayama TK, Shapiro SD, Schechter NM, Remold-O’Donnell E. The serpin MNEI inhibits elastase-like and chymotrypsin-like serine proteases through efficient reactions at two active sites. Biochemistry. 2001;40:15762–70. doi: 10.1021/bi0113925. [DOI] [PubMed] [Google Scholar]
- 21.Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. The Journal of biological chemistry. 2001;276:33293–6. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- 22.Reiser J, Adair B, Reinheckel T. Specialized roles for cysteine cathepsins in health and disease. The Journal of clinical investigation. 2010;120:3421–31. doi: 10.1172/JCI42918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gettins PG. Serpin structure, mechanism, and function. Chemical reviews. 2002;102:4751–804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 24.Nakagawa TY, Rudensky AY. The role of lysosomal proteinases in MHC class II-mediated antigen processing and presentation. Immunological reviews. 1999;172:121–9. doi: 10.1111/j.1600-065x.1999.tb01361.x. [DOI] [PubMed] [Google Scholar]
- 25.Drake FH, Dodds RA, James IE, Connor JR, Debouck C, Richardson S, et al. Cathepsin K, but not cathepsins B, L, or S, is abundantly expressed in human osteoclasts. The Journal of biological chemistry. 1996;271:12511–6. doi: 10.1074/jbc.271.21.12511. [DOI] [PubMed] [Google Scholar]
- 26.Katunuma N, Murata E, Kakegawa H, Matsui A, Tsuzuki H, Tsuge H, et al. Structure based development of novel specific inhibitors for cathepsin L and cathepsin S in vitro and in vivo. FEBS letters. 1999;458:6–10. doi: 10.1016/s0014-5793(99)01107-2. [DOI] [PubMed] [Google Scholar]
- 27.Yasuma T, Oi S, Choh N, Nomura T, Furuyama N, Nishimura A, et al. Synthesis of peptide aldehyde derivatives as selective inhibitors of human cathepsin L and their inhibitory effect on bone resorption. Journal of medicinal chemistry. 1998;41:4301–8. doi: 10.1021/jm9803065. [DOI] [PubMed] [Google Scholar]
- 28.Liszewski MK, Kolev M, Le Friec G, Leung M, Bertram PG, Fara AF, et al. Intracellular complement activation sustains T cell homeostasis and mediates effector differentiation. Immunity. 2013;39:1143–57. doi: 10.1016/j.immuni.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tuomela S, Salo V, Tripathi SK, Chen Z, Laurila K, Gupta B, et al. Identification of early gene expression changes during human Th17 cell differentiation. Blood. 2012;119:e151–60. doi: 10.1182/blood-2012-01-407528. [DOI] [PubMed] [Google Scholar]
- 30.Shirahama-Noda K, Yamamoto A, Sugihara K, Hashimoto N, Asano M, Nishimura M, et al. Biosynthetic processing of cathepsins and lysosomal degradation are abolished in asparaginyl endopeptidase-deficient mice. The Journal of biological chemistry. 2003;278:33194–9. doi: 10.1074/jbc.M302742200. [DOI] [PubMed] [Google Scholar]
- 31.Maehr R, Hang HC, Mintern JD, Kim YM, Cuvillier A, Nishimura M, et al. Asparagine endopeptidase is not essential for class II MHC antigen presentation but is required for processing of cathepsin L in mice. J Immunol. 2005;174:7066–74. doi: 10.4049/jimmunol.174.11.7066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Relates to Fig. 1. (A,B) Effect of serpinB1 on Treg differentiation under Th17 conditions. Naive CD4 cells from WT and serpinb1−/− mice were differentiated (A) as in Fig. 1C (2 ng/ml TGF-β, 10 ng/ml IL-6) or (B) with the indicated TGF-β and IL-6 concentrations. (C) Time course of expression of Serpinb1 and Rorc during Th17 differentiation of CD4 cells. Representative findings and means ± SEM of (A) 5 experiments from Fig. 1C and (B,C) each 2 experiments. **P < 0.01.
Fig. S2A. Relates to Fig. 5A. Effect of CA074-OMe treatment of cells on catB and catL activities. CD4 cells were differentiated under Th17 polarizing conditions in the absence (medium) or presence of CA074-OMe (5 μM). CatL activity in cell lysates was assayed as in Fig. 4B; catB activity was assayed at pH 6 with z-Gly-Arg-AMC. Results are of 2 experiments.
Fig. S2B. Relates representative to Fig. 2B and 5B. Effect of E64D on generation of FoxP3+ cells under Th17 conditions. Naive CD4 cells from WT and serpinb1−/− mice were differentiated with varying dose E64D. Means ± SEM of 3 experiments from Fig. 2B **P < 0.01; ***P < 001.
Fig. S3. Relates to Fig. 6B. Effect of the AEP inhibitor LI-1 on catL of serpinb1−/− Th17 cells. Naive CD4 cells from serpinb1−/− mice were differentiated under Th17 conditions in the presence of the indicated concentration of LI-1. Western blot for catL representative of 2 experiments.