Abstract
Chronic Obstructive Pulmonary Disease (COPD) is a progressive disease with both environmental and genetic risk factors. Genome-wide association studies (GWAS) have identified multiple genomic regions influencing risk of COPD. To thoroughly investigate the genetic etiology of COPD, however, it is also important to explore the role of copy number variants (CNVs) since the presence of structural variants can alter gene expression and can be causal for some diseases.
Here, we investigated effects of polymorphic CNVs on quantitative measures of pulmonary function and chest CT phenotypes among subjects enrolled in COPDGene, a multi-site study. COPDGene subjects consist of roughly one-third African-American and two-thirds Non-Hispanic white adult smokers (with or without COPD). We estimated CNVs using PennCNV on 9,076 COPDGene subjects using Illumina's Omni-Express genome-wide marker array.
We tested for association between polymorphic CNV components (defined as disjoint intervals of copy number regions) for several quantitative phenotypes associated with COPD within each racial group. Among African-Americans, we identified a polymorphic CNV on chromosome 5q35.2 located between two genes (FAM153B and SIMK1, but also harboring several pseudo-genes) giving genome-wide significance in tests of association with total lung capacity as measured by chest CT scans. This is the first study of genome-wide association tests of polymorphic CNVs and total lung capacity. While the ARIC cohort did not have the phenotype of total lung capacity, we found similar counts of CNV deletions and amplifications among African-American and European subjects in this second cohort.
Keywords: COPD, copy number variant (CNV), total lung capacity (TLCCT), Pulmonary function, Lung hyperinflation, Genome-wide association study (GWAS)
Introduction
Chronic obstructive pulmonary disease (COPD) is the third leading cause of mortality in the US [Heron, 2015]. COPD is characterized by reduced lung function as measured by spirometry (which measures volume and rate of air flow) and includes clinically defined symptoms of chronic bronchitis and anatomically defined emphysema. Chest CT scans reveal finer detail about the internal structure of the lung, and provide quantitative measures of destruction of lung parenchyma, the defining quantitative phenotype of emphysema. Increased total lung capacity (TLCCT) in subjects with moderate-to-severe airflow obstruction (defined as Global Initiative for Obstructive Lung Disease (GOLD) severity classes 2 or higher) typically reflects lung hyperinflation, while reduced TLCCT can also reflect restriction in individuals with interstitial lung disease, chest wall abnormalities, or neuromuscular weakness. Lung hyperinflation, i.e. increased lung gas volume at the end of tidal expiration beyond normal range, is a major co-morbidity in COPD [Gibson, 1996; Vestbo et al., 2013], but details of underlying biological mechanisms remain poorly defined [O'Donnell, 2008].
With the availability of efficient algorithms to call structural variations (including deletions and amplifications of chromosomal segments) from raw intensity level data generated by genome-wide marker panels used for genome-wide association studies (GWAS), useful information about DNA copy number variants (CNVs) can still be extracted from the LRRs and BAFs generated using this array even when these arrays are not specifically designed for CNV studies. We can explore the role of structural variation in the etiology of complex diseases, such as COPD and its related quantitative phenotypes. We undertook a study of polymorphic CNVs and tested for association with several quantitative spirometric measures of pulmonary function along with quantitative measures of density and volume from chest CT scans in African-Americans (AAs) and in non-Hispanic whites (NHWs) adult smokers from the COPDGene study [Regan et al., 2010]. In our study, CNVs were estimated from raw intensity data generated by a genome-wide marker panel using the Hidden Markov Model implemented in PennCNV [Wang et al., 2007] for the entire COPDGene cohort of adult smokers.
Materials and Methods
Subjects
The COPDGene study (NCT00608764, www.copdgene.org) recruited a total of 10,192 adult subjects (age 45 to 80) with at least 10 pack-years of exposure to cigarette smoking [Regan et al., 2010]. This cohort of current and former smokers included individuals ranging across all four GOLD severity classes for COPD, as well as individuals with normal pulmonary function and individuals with abnormal but unclassified spirometric values. All study subjects completed questionnaires covering demographic and behavioral risk factors (including a detailed personal smoking history) [Regan et al., 2010]. Among the entire group of subjects with genotype and CT data passing their respective quality control (QC) steps, about one-third were self-identified African-American (AA; n=2,640) and two-thirds were self-identified NHW (n=5,841) (Supplementary Figure S1).
Measures of pulmonary function
A number of pulmonary function measures were available for COPDGene subjects including forced expiratory volume in the first second (FEV1) and forced vital capacity (FVC). Both pre- and post-bronchodilator spirometry measures (after 180 mcg of albuterol by metered dose inhaler) were available, but here we focused on post-bronchodilator spirometry. These two spirometric quantitative measures and their ratio (FEV1/FVC) define clinical COPD, but there are unclassified spirometric abnormalities, where there is reduced FEV1 but normal FEV1/FVC (also known as preserved ratio with reduced spirometry or PRISm phenotype) [Wan et al., 2014]. CT scan protocols and associated quality measures have been published elsewhere [Regan et al., 2010; Hersh et al., 2013]. The software 3D SLICER was used for image analysis of CT scans (www.slicer.org) [Estepar, 2006; Hersh et al., 2007; Hersh et al., 2013]. The phenotypes considered here included FEV1/FVC, FEV1 percent predicted, percent emphysema, percent gas trapping and total lung capacity. TLCCT (in liters) was measured by volumetric CT scan of the chest while the subject was supine and holding their breath after full inspiration.
Genotyping
DNA was extracted from whole blood and genotyped on the Illumina Omni-Express chip on 114 plates run in 5 batches. The array contains 728,648 polymorphic probes and included 7,657 non-polymorphic probes among AA subjects and 9,552 non-polymorphic probes among NHW subjects. Initial QC of the genotype data was conducted as part of COPDGene's genome-wide association analysis and has been reported elsewhere [Zhu et al., 2007; Vestbo et al., 2008; Grydeland et al., 2009; Cho et al., 2010; Regan et al., 2010; Cho et al., 2012; Cho et al., 2014; Lee JW, 2014]. This GWAS QC analysis included principal component analysis to confirm the genetic background of all self-reported AA and NHW individuals.
Calling copy numbers
Structural variants that include CNVs in these subjects were delineated using the hidden Markov model-based PennCNV algorithm (HMM) [Wang et al.], which utilizes the log R ratios (LRRs) and B allele frequencies (BAFs) from Illumina's Omni-Express probes to infer deletions and amplifications of chromosomal segments. The LRR is a standardized estimate of the probe intensity, quantifying the total number of allele copies at each marker on the genome-wide panel [Peiffer et al., 2006]. BAF is a standardized estimate for the proportion of B alleles contributing to this total probe intensity, which determines the genotype at each SNP site. BAF is standardized so homozygous genotypes in copy neutral states (i.e., individuals with the expected two copies at each probe) have BAFs of approximately zero or one (representing AA and BB genotypes, respectively), and heterozygous AB genotypes yield BAFs roughly equal to 0.5. ‘Genomic waves’ are a technical artifact, which are highly correlated with GC content and may cause inaccurate CNV calls [Diskin et al., 2008; Leo et al., 2012]. PennCNV addresses genomic waves by incorporating the population GC content at each marker into the HMM. We followed previously established guidelines for QC when calling CNVs [Wang et al., 2007; Scharpf et al., 2012] designed to avoid excessive false positive calls, particularly those due to poor data quality. Specifically, we removed samples with LRR median absolute deviation (MAD) above 0.3 (Figure S2), and dropped all samples on plates showing an excessive number of CNV calls. Genomic regions with poor mappability [Derrien et al., 2012] were also omitted, and only inferred CNVs supported by 10 or more SNP probes were used in this analysis.
After calling CNVs, further QC steps were implemented: 821 samples were judged to be of insufficient quality for calling CNVs, and an additional 73 samples on 4 poorly performing plates (Supplementary Figure S3 and Figure S4) were removed before analysis, leaving 9,076 samples (6,187 NHWs; 2,889 AAs) with valid calls of polymorphic CNVs.
Testing for association
Association tests with a quantitative phenotype (e.g. TLCCT) as the dependent variable were carried out on the estimated CNV components called by PennCNV, i.e. sets of markers of constant copy number state within each subject but showing some variability in copy number states among subjects [Younkin et al., 2014]. Since genomic architecture can differ between ethnic groups, CNV components were derived separately for the AA and NHW groups, and we conducted separate regression analyses on these two groups. Only polymorphic CNV components with DNA alterations in at least 1% of all subjects were considered in this analysis. The primary question of interest was whether polymorphic CNVs influenced a range of quantitative lung phenotypes. Since lung function measurements and chest CT scans were taken at 21 separate study sites, mixed linear models were used to assess the relationship between a quantitative phenotype and polymorphic CNVs adjusting for gender, age, height, current smoking status, and pack-years of cigarette smoking as fixed effects, while study site was treated as a random effect. A log10 transformation was used for the heavily (right) skewed phenotypes (Supplementary Figure S5). We adjusted for different sets of covariates and considered different regression models for each quantitative phenotype (i.e. not every covariate was predictive with each quantitative phenotype). We adjusted for multiple comparisons for the number of polymorphic CNVs with Bonferroni correction, and a test result was considered significant if it was less than the adjusted p-value. Since the number of polymorphic CNVs varied for each phenotype because counts of subjects with missing data varied, the Bonferroni corrected p-value threshold also varied for each phenotype. So we reported Bonferroni corrected p-values for each phenotype separately. Aside from the CNV estimation itself (which was done in PennCNV), all analyses were carried out in the statistical environment R (http://cran.r-project.org/).
Results
Genotype data from 9,970 subjects who passed QC as part of COPDGene's initial GWAS analysis pipeline was drawn from the total 10,192 COPDGene subjects. Further QC yielded 9,076 samples (6,187 NHWs; 2,889 AAs) for analysis of polymorphic CNVs. TLCCT measurements were available for 8,481 of these subjects (2,640 AAs and 5,841 NHWs).
Demographic characteristics of subjects with polymorphic CNVs passing all QC steps with TLCCT data available are listed in Table I. The sample size of NHWs with complete data was more than double that of AAs, who tended to be younger and included more current smokers with fewer pack-years of smoking. Among the 2,640 AA subjects, 43% were female compared to 47% female among NHW subjects. AA subjects were an average of 7.6 years younger than NHW subjects. A striking difference was seen in the current smoking status between the two racial groups: 81% of AA subjects currently smoked, whereas only 39% of NHW subjects were current smokers. Despite this, the average pack-years exposure to cigarettes was higher among NHW subjects.
Table I.
Characteristics of study samples.
| African-American (AA) | Non-Hispanic white (NHW) | |
|---|---|---|
| Total (Male : Female) | Total (Male : Female) | |
| Sample size | 2640 (1518 M : 1122 F) | 5841 (3070 M : 2771 F) |
| Mean age at enrollment | 54.5(54.3 M : 54.8 F) | 62.1(62.5 M : 61.7 F) |
| Percent current smokers (%) | 80.7(82.9 M : 77.6 F) | 39.1(40.3 M : 37.9 F) |
| Mean ATS pack-years | 38.1 (38.7 M : 37.2 F) | 47.3(51.5 M : 42.6 F) |
| Average BMI | 29.0(27.7 M : 30.7 F) | 28.6(28.8 M : 28.5 F) |
| Average Height (cm) | 171.4(176.9 M : 164.0 F) | 169.7(176.1 M : 162.7 F) |
| Mean TLCCT (lt) | 4.8 (5.4 M : 4.0 F) | 5.9(6.7 M : 4.9 F) |
Different characteristics of called CNVs between AA and NHW subjects are presented in Supplemental Table S1. The average counts of CNVs differed between AA and NHW subjects, and the mean count was significantly higher among AA subjects. Mean CNV counts estimated here may be lower than those obtained with other genotyping platforms because the OmniExpress SNP array did not include large numbers of monomorphic intensity-only probes [illumina], which can be used to call CNVs in chromosomal segments in the absence of truly polymorphic markers. In both groups, the average count of deletions was higher compared to the average count of amplifications, but the sizes of the called deletions were almost half of the segment size of amplifications in both groups. The quality of the estimated CNVs was slightly better among NHWs (mean MAD=0.128 for LRR, median MAD=0.123) compared to AAs (mean MAD=0.133 for LRR, median MAD =0.128 for LRR), which may result in slightly fewer false positive calls among NHW subjects. Poorly called CNV segments generally result in many short inferred deletions and amplifications, which reflect the “noisy” nature of intensity data, while higher quality raw data generates longer segments of inferred CNVs.
The CNV distribution among current smokers in both racial groups is presented in Supplementary Table S2, which shows while the mean CNV count is significantly different between racial groups, there were only modest differences between current and former smokers. The difference between mean CNV counts among current and former smokers among NHW subjects was not statistically significant, but this difference was just nominally significant among AA subjects (p=0.05). Still, it is highly unlikely that current smoking is the cause for any real difference in CNV counts between groups.
Polymorphic CNV association analysis among AA subjects
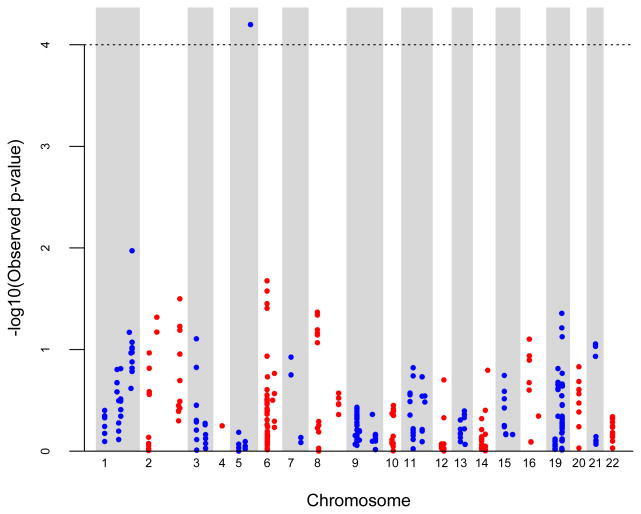
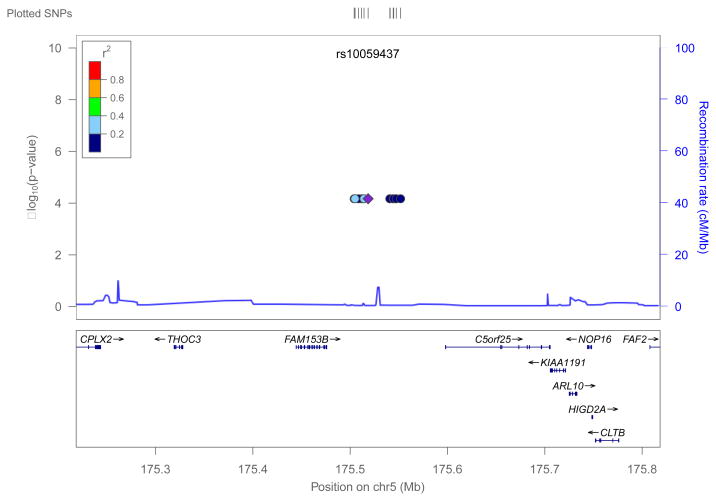
We performed genome-wide association analysis for AA and NHW groups separately for polymorphic CNV segments. Among AAs, 358 components remained after removing low frequency CNV components (i.e. those with <1% frequency in the total AA group). When testing for association between polymorphic CNV components and the quantitative TLCCT phenotype, we adjusted for covariates listed in Table I (age, gender, height and pack-years of smoking). All four of these covariates were highly significantly associated with quantitative TLCCT levels. We also included recruitment site as a random effects predictor in the model. After adjusting for multiple comparisons, one 47kb polymorphic CNV on chromosome 5q35.2 (spanning hg18 positions 175504185-175551861bp) achieved genome-wide significance (p=6.78e-05) in this analysis of 2,640 AA subjects, as shown in Figure 1. Since we observed significant association only among AA subjects, it is also important to adjust our analysis for admixture. We conducted additional analyses considering average European ancestry in African American subjects. In this analysis, we used the average admixture score for each individual estimated by LAMP [Parker et al., 2014] as a covariate, which was significantly associated with TLCCT, and repeated our analysis. This model yielded the same results as the unadjusted analysis and identified the same region of chromosome 5q35.2 as associated with TLCCT. The –log10(p) in this analysis was somewhat attenuated (-log10(0.0002441299) = 3.61) compared to the previous analysis (-log10(6.344233e-05) = 4.20) and would not exceed the conventional genome-wide critical value (3.85), however, it is clear this 5q35.2 CNV region is still giving a strong signal as being associated with TLCCT. This 47kb segment lies in an intergenic region, between two protein-coding genes and harbors several pseudo-genes. A LocusZoom plot [Pruim et al., 2010] of this region is presented in Figure 2.
Figure 1.
Genome-wide CNV association scan for TLCCT
Figure 2.
Locus Zoom plot of Chromosome 5 (175504185bp to 175551861bp)
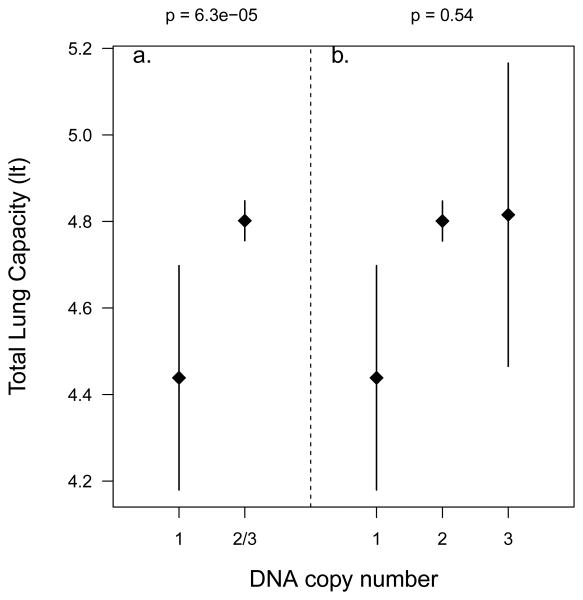
Comparing TLCCT values of AA subjects with and without this genome-wide significant polymorphic deletion revealed TLCCT was lower among the 47 AA subjects with inferred deletions (mean=4.44 liters, 95%CI=4.18-4.70) compared to 2,593 AA subjects without deletions in this region (mean=4.80 liters, 95%CI=4.76-4.85; see Figure 3). These 47 individuals all had hemizygous deletions, and no homozygous deletion carriers were observed in this sample. Among the 2,593 AAs without any deletion in this region, 2,473 were called diploid (normal 2 copies), and 120 individuals had an inferred amplification (3 copies). However, no difference in TLCCT was observed between these latter two groups (p=0.54, Figure 3). These polymorphic CNVs (deletions and amplifications) were observed at the same genomic location (175,504,185 – 175,551,861, hg18) spanning approximately 47 kb, and were clearly visible in the raw intensity data. In Supplementary Figure S7 (a) and (b), we plotted the original BAF and LRR intensities for SNPs on chromosome 5 (175000000-176000000), which shows a clear reduction in intensities among the 47 subjects identified as having a hemizygous deletion in 5q35.2. Similarly, Figure S7 (c) and (d) confirms the called amplifications among the 120 subjects identified as carrying an extra copy in this region. The remaining 2,473 AA subjects were called as normal diploids for probes in this region and plots of a random sample of 50 normal diploids are presented in Figure S7 (e) and (f).
Figure 3.
Mean and Confidence Interval (CI) plot of TLCCT for different polymorphic CNV counts among AA subjects. a. Mean TLCCT and 95%CI in hemizygous deletion carriers vs. normal and those carrying amplifications in chromosome 5q35.2. Mean TLCCT is statistically significantly different (p=6.3e-05) between hemizygous deletion carriers and carriers with 2 or 3 copies in 5q35.2. b. Mean TLCCT and 95%CI plot for hemizygous deletion carriers (i.e. 1 copy), normal diploid individuals (i.e. 2 copies) and carriers of ≥3 copy numbers separately. Mean TLCCT is not statistically significantly different (p=0.54) between carriers with two copies and carriers with 3 copies in 5q35.2.
The demographic and clinical characteristics of these 47 subjects with hemizygous deletions are presented in Supplementary Table S3. These 47 subjects carrying a hemizygous deletion in 5q35.2 were distributed across 30 different plates during genotyping, making it unlikely any simple batch effects would explain these patterns of called polymorphic CNVs. Among the 47 subjects, 76.6% were male compared to 57.2% male among all remaining 2,593 AA subjects. The mean FEV1/FVC ratio was slightly higher among the 47 deletion carriers (0.74) compared to the other AA subjects (0.72), but this difference was not statistically significant.
Polymorphic CNV association analysis among NHW subjects
Among 5,841 NHW subjects, a total of 257 components remained after removing low frequency CNVs (<1% frequency in the total NHW group). We tested for association between TLCCT and these 257 polymorphic CNV components, while adjusting for the same set of covariates used with AA subjects. There was no evidence of association with quantitative TLCCT measures among the 5,841 NHW subjects (Supplementary Figures S8 and S9). We further looked at the number of subjects with called deletions/amplifications in 5q35.2, the chromosomal region significantly associated with TLCCT among AA subjects. The total counts were much smaller: only 4 subjects were identified has having hemizygous deletions and only 6 subjects had detected amplifications. Thus, there was virtually no statistical power to detect differences associated with these polymorphic CNV in the larger NHW group.
Polymorphic CNV association analysis with other phenotypes
We also performed polymorphic CNV association testing separately for AA and NHW subjects for other quantitative pulmonary and CT imaging measures such as FEV1/FVC, FEV1 percent predicted, percent emphysema and percent gas trapping. None of the polymorphic CNVs identified by PennCNV reached the Bonferroni corrected significance level needed to infer association between these phenotypes and these polymorphic CNV segments. All genome-wide association plots along with their corresponding QQ plots are presented in Supplementary Figures S10 - S13.
Discussion
In this study, we conducted a genome-wide CNV analysis on 9,076 adult smokers (former and current) for quantitative measures of lung function based on spirometry and chest CT scan data. Polymorphic CNVs (including both deletions and amplifications) were identified from intensity data (LRR and BAF) generated on the Illumina OmniExpress chip, and segments of deletions and amplifications across the genome were estimated using the HMM approach implemented in PennCNV [Wang et al., 2007].
We used linear mixed models to test for association with polymorphic CNVs (those with frequency >1%) to identify genomic regions where structural variants were significantly associated with quantitative pulmonary phenotypes in a stratified analysis of NHW and AA groups. Among the five quantitative phenotypes examined, none of the polymorphic CNVs reached genome wide significance for any quantitative phenotypes except TLCCT. For TLCCT, one region of chromosome 5q35.2 showed statistically significant evidence of association among 2,640 AA subjects, where 47 individuals were identified as carrying hemizygous deletions in a 47kb region, with hemizygous deletion carriers showing reduced total lung capacity.
The function of this 47kb long segment is unclear. It contains a lincRNA, RP11-844P9.2 (ENSG00000248596), with unknown function whose expression appears to be primarily in reproductive organs [Consortium, 2013]. This region lies between two protein-coding genes: FAM153B (family with sequence similarity 153, member B) is ∼20kb upstream and SIMC1 (Gene ID: 375484; SUMO-interacting motifs containing 1) is 46kb downstream of the significant deletion region. FAM153B interacts with PLOD3, a membrane-bound enzyme catalyzing hydroxylation of lysine residues in collagen-like peptides [Rual et al., 2005; Franceschini et al., 2013]; although this gene is most highly expressed in brain and reproductive tissue with little expression in lung. SIMC1 (previously known as C5orf25, PLEIAD, and OOMA1) is also expressed predominantly in reproductive tissues but has low levels of expression in many other tissues, including the lung. In the single functional study reported to date, SIMC1 was identified in muscle as an autolysis regulator for CAPN3, a skeletal muscle specific calpain [Ono et al., 2013]. Deletions in the downstream adjacent area of 5q35.2-35.3 cause Sotos syndrome, a childhood overgrowth featuring craniofacial abnormalities, curvature in the spine, heart and kidney defects [Dikow et al., 2013; Ko, 2013; Klaassens et al., 2014]. In the Gene Expression Atlas, this protein also appears to be expressed in bronchial epithelial cells (BEAS-2B) when exposed to the tobacco smoke-borne carcinogen, 2-hydroxy-mino-1-methyl-6-phenylimidazo (1,5) pyridine (N-OH-PhIP) (GSE34635, at http://www.ebi.ac.uk/). The potential biological significance of deletions in this genomic region remain unknown, and further studies will be needed to determine the function of genes or regulatory elements this genomic region, and how they could relate to TLCCT.
To evaluate our findings of polymorphic CNVs in this area of 5q35.2, we checked estimated CNVs in the Atherosclerosis Risk in Communities (ARIC) study [1989], which included 9,417 European American (EA) subjects and 3,210 African American (AA) subjects. Genotyping was done on the Affymetrix 6.0 chip and PennCNV was used to estimate the CNVs [Scharpf et al., 2014]. The copy number estimates from a mixture model fit to the median LRR from this region among ARIC AA subjects included: 50 homozygous deletions, 3,016 normal diploids, and 144 with a single copy gain. Among EA participants, the CNV frequency was too low to be estimated well under a mixture model. Using the HMM model of PennCNV, CNV estimates were 4 subjects carrying a hemizygous deletion, 19 with a single copy gain, and 9,394 normal diploids. The counts of CNVs from ARIC study in both racial groups are quite comparable with those seen in our COPDGene study, which suggests this polymorphic CNV is more common in AA subjects.
Total lung capacity (TLCCT), the volume of gas in the lungs after a full inspiratory effort, is a key measure of pulmonary function that can be estimated accurately using chest CT scans [Wanger et al., 2005]. TLCCT is known to be associated with age, sex, height, and race in general population samples [Pellegrino et al., 2005]. African Americans have, after adjustment for other known covariates, lower predicted TLCCT values than non-Hispanic White subjects; however, the biological mechanisms for this racial difference have not been determined. It is possible CNVs (including those identified in this study) could contribute to racial differences in TLCCT.
In addition to normal variation in TLCCT values, TLCCT is altered in a range of lung diseases. Emphysema typically causes pathological increases in TLCCT, while interstitial lung diseases (e.g., idiopathic pulmonary fibrosis), chest wall abnormalities (e.g., kyphoscoliosis), and neuromuscular weakness (e.g., amyotrophic lateral sclerosis) cause reductions in TLCCT. Most of the CNV carriers in our study had normal spirometry; thus, these individuals may have developmental differences in their TLCCT values. Other CNV carriers had preserved ratio impaired spirometry (PRISm) [Wan et al., 2011], a poorly understood and heterogeneous group of subjects with normal FEV1/FVC ratio but reduced FEV1 values [Wan et al., 2014] . Further research will be required to determine whether the CNV associated with TLCCT identified here contributes to PRISm.
Lee et al. conducted a conventional genome-wide association study of TLCCT in non-Hispanic whites (NHW) with COPD, testing for association between marker genotypes and TLCCT [Regan et al., 2010; Lee JW, 2014]. They found one genome-wide significant single nucleotide polymorphism (SNP) on chromosome 5p15.2 in a meta-analysis of 4,543 COPD subjects from three different COPD cohorts: COPDGene, the Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE, NCT00292552, www.eclipse-copd.com); and GenKOLS (Bergen, Norway) [Lee et al. 2014]. While this SNP achieved genome-wide significance among NHW subjects in a meta-analysis across these three studies, its minor allele frequency was very low among COPDGene African-Americans, and therefore there was virtually no power to detect association with TLCCT in this group. No previous genome-wide study of polymorphic CNVs has been done for TLCCT.
A genome-wide CNV association study on the Korea Associated Resource (KARE) cohort identified several significant CNVs associated with spirometric measures of pulmonary function (FEV1 and FVC) [Lee et al., 2011], but their genotyping platform and analytical approach were quite different from ours making direct comparisons difficult. Another study of European subjects reported an association between CNVs in the β-defensin gene cluster on chromosome 8p23.1 and clinical COPD [Janssens et al., 2010]. Wain et al. also studied this β-defensin gene cluster and CNVs among European adults and children, but found no evidence of association with clinical COPD, asthma or quantitative measures of lung function [Wain et al., 2014].
The genome-wide marker panel used here was primarily designed to study SNP and was not enriched with monomorphic probes that enhance analysis of CNVs, which may have limited our ability to identify some polymorphic CNVs. Also, because frequencies of polymorphic CNVs differ between AA and NHW subjects, our separate analysis made sample sizes smaller for each group. Clearly, there was reduced power to detect effects of this deleted segment of chromosome 5q35.2 among NHW subjects because there were far fewer hemizygous deletion carriers despite the larger sample size. Better understanding of the role of structural variants on quantitative pulmonary function and CT imaging phenotypes and replication studies will be necessary, although meta-analysis across similar studies could also help. Nonetheless, this study finding provided the first evidence of polymorphic CNVs on quantitative measures of TLCCT
Conclusion
We identified a genome-wide significant 47kb intergenic region on chromosome 5q35.2, which appears to be associated with lower total lung capacity in hemizygous deletion carriers among African-American adult smokers in the COPDGene study. To our knowledge, no previous genome-wide study of polymorphic CNVs has been done for TLCCT. This identified chromosomal region is highly polymorphic for structural variants (both deletions and amplifications) and there were more amplification carriers among African-Americans, but only hemizygous carriers of deleted segments showed phenotypically different TLCCT levels.
Supplementary Material
Acknowledgments
This study was supported by U.S. National Institutes of Health (NIH) grants R01 HL089856 and R01 HL089897. In the past three years, Edwin K. Silverman received honoraria and consulting fees from Merck and grant support and consulting fees from GlaxoSmithKline. We thank Dr. Md. Hafiz Uddin for his thoughtful suggestions. The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
Footnotes
Author contributions: Wrote the paper: All authors. Data analysis: FB, IR, SL. Manuscript draft: FB, IR, TB.
References
- The ARIC investigators. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am J Epidemiol. 1989;129(4):687–702. [PubMed] [Google Scholar]
- Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, Hersh CP, DeMeo DL, Hunninghake GM, Litonjua AA, Sparrow D, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42(3):200–2. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MH, Castaldi PJ, Wan ES, Siedlinski M, Hersh CP, Demeo DL, Himes BE, Sylvia JS, Klanderman BJ, Ziniti JP, et al. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet. 2012;21(4):947–57. doi: 10.1093/hmg/ddr524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MH, McDonald ML, Zhou X, Mattheisen M, Castaldi PJ, Hersh CP, Demeo DL, Sylvia JS, Ziniti J, Laird NM, et al. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med. 2014;2(3):214–25. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Estelle J, Marco Sola S, Knowles DG, Raineri E, Guigo R, Ribeca P. Fast computation and applications of genome mappability. PLoS One. 2012;7(1):e30377. doi: 10.1371/journal.pone.0030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikow N, Maas B, Gaspar H, Kreiss-Nachtsheim M, Engels H, Kuechler A, Garbes L, Netzer C, Neuhann TM, Koehler U, et al. The phenotypic spectrum of duplication 5q35.2-q35.3 encompassing NSD1: is it really a reversed Sotos syndrome? Am J Med Genet A. 2013;161A(9):2158–66. doi: 10.1002/ajmg.a.36046. [DOI] [PubMed] [Google Scholar]
- Diskin SJ, Li M, Hou C, Yang S, Glessner J, Hakonarson H, Bucan M, Maris JM, Wang K. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res. 2008;36(19):e126. doi: 10.1093/nar/gkn556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estepar RW, GG, Silverman EK, Reilly JJ, Kikinis R, Westin CF. Accurate airway wall estimation using phase congruency. Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv. 2006;9(Pt 2):125–134. doi: 10.1007/11866763_16. [DOI] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–15. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson GJ. Pulmonary hyperinflation a clinical overview. Eur Respir J. 1996;9(12):2640–9. doi: 10.1183/09031936.96.09122640. [DOI] [PubMed] [Google Scholar]
- Grydeland TB, Dirksen A, Coxson HO, Pillai SG, Sharma S, Eide GE, Gulsvik A, Bakke PS. Quantitative computed tomography: emphysema and airway wall thickness by sex, age and smoking. Eur Respir J. 2009;34(4):858–65. doi: 10.1183/09031936.00167908. [DOI] [PubMed] [Google Scholar]
- Heron M. Deaths: Leading Causes for 2011. Natl Vital Stat Rep. 2015;64(7):1–96. [PubMed] [Google Scholar]
- Hersh CP, Washko GR, Estepar RS, Lutz S, Friedman PJ, Han MK, Hokanson JE, Judy PF, Lynch DA, Make BJ, et al. Paired inspiratory-expiratory chest CT scans to assess for small airways disease in COPD. Respir Res. 2013;14:42. doi: 10.1186/1465-9921-14-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh CP, Washko GR, Jacobson FL, Gill R, Estepar RS, Reilly JJ, Silverman EK. Interobserver variability in the determination of upper lobe-predominant emphysema. Chest. 2007;131(2):424–31. doi: 10.1378/chest.06-1040. [DOI] [PubMed] [Google Scholar]
- illumina. interpreting infinium assay data for whole-genome structural variation. Techical Note: DNA Analysis [Google Scholar]
- Janssens W, Nuytten H, Dupont LJ, Van Eldere J, Vermeire S, Lambrechts D, Nackaerts K, Decramer M, Cassiman JJ, Cuppens H. Genomic copy number determines functional expression of {beta}-defensin 2 in airway epithelial cells and associates with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182(2):163–9. doi: 10.1164/rccm.200905-0767OC. [DOI] [PubMed] [Google Scholar]
- Klaassens M, Morrogh D, Rosser EM, Jaffer F, Vreeburg M, Bok LA, Segboer T, van Belzen M, Quinlivan RM, Kumar A, et al. Malan syndrome: Sotos-like overgrowth with de novo NFIX sequence variants and deletions in six new patients and a review of the literature. Eur J Hum Genet. 2014 doi: 10.1038/ejhg.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JM. Genetic syndromes associated with overgrowth in childhood. Ann Pediatr Endocrinol Metab. 2013;18(3):101–5. doi: 10.6065/apem.2013.18.3.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BY, Cho S, Shin DH, Kim H. Genome-wide association study of copy number variations associated with pulmonary function measures in Korea Associated Resource (KARE) cohorts. Genomics. 2011;97(2):101–5. doi: 10.1016/j.ygeno.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Lee JW, M M, Cho MH, Wan ES, Castaldi PJ, Hunninghake GM, Marchetti N, Lynch DA, Crapo JD, Lomas DA, Coxson HO, Bakke PS, Silverman EK, Hersh CP. DNAH5 is associated with total lung capacity in chronic obstructive pulmonary disease. Respiratory Resch. 2014;15(97) doi: 10.1186/s12931-014-0097-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo A, Walker AM, Lebo MS, Hendrickson B, Scholl T, Akmaev VR. A GC-wave correction algorithm that improves the analytical performance of aCGH. J Mol Diagn. 2012;14(6):550–9. doi: 10.1016/j.jmoldx.2012.06.002. [DOI] [PubMed] [Google Scholar]
- O'Donnell D. [Dynamic lung hyperinflation and its clinical implication in COPD] Rev Mal Respir. 2008;25(10):1305–18. doi: 10.1016/s0761-8425(08)75094-0. [DOI] [PubMed] [Google Scholar]
- Ono Y, Iemura S, Novak SM, Doi N, Kitamura F, Natsume T, Gregorio CC, Sorimachi H. PLEIAD/SIMC1/C5orf25, a novel autolysis regulator for a skeletal-muscle-specific calpain, CAPN3, scaffolds a CAPN3 substrate, CTBP1. J Mol Biol. 2013;425(16):2955–72. doi: 10.1016/j.jmb.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MM, Foreman MG, Abel HJ, Mathias RA, Hetmanski JB, Crapo JD, Silverman EK, Beaty TH, Investigators CO. Admixture mapping identifies a quantitative trait locus associated with FEV1/FVC in the COPDGene Study. Genet Epidemiol. 2014;38(7):652–9. doi: 10.1002/gepi.21847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer DA, Le JM, Steemers FJ, Chang W, Jenniges T, Garcia F, Haden K, Li J, Shaw CA, Belmont J, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16(9):1136–48. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino R, Viegi G, Brusasco V, Crapo RO, Burgos F, Casaburi R, Coates A, van der Grinten CP, Gustafsson P, Hankinson J, et al. Interpretative strategies for lung function tests. Eur Respir J. 2005;26(5):948–68. doi: 10.1183/09031936.05.00035205. [DOI] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, Crapo JD. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7(1):32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437(7062):1173–8. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Scharpf RB, Beaty TH, Schwender H, Younkin SG, Scott AF, Ruczinski I. Fast detection of de novo copy number variants from SNP arrays for case-parent trios. BMC Bioinformatics. 2012;13:330. doi: 10.1186/1471-2105-13-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharpf RB, Mireles L, Yang Q, Kottgen A, Ruczinski I, Susztak K, Halper-Stromberg E, Tin A, Cristiano S, Chakravarti A, et al. Copy number polymorphisms near SLC2A9 are associated with serum uric acid concentrations. BMC Genet. 2014;15:81. doi: 10.1186/1471-2156-15-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestbo J, Anderson W, Coxson HO, Crim C, Dawber F, Edwards L, Hagan G, Knobil K, Lomas DA, MacNee W, et al. Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE) Eur Respir J. 2008;31(4):869–73. doi: 10.1183/09031936.00111707. [DOI] [PubMed] [Google Scholar]
- Vestbo J, Hurd SS, Agusti AG, Jones PW, Vogelmeier C, Anzueto A, Barnes PJ, Fabbri LM, Martinez FJ, Nishimura M, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2013;187(4):347–65. doi: 10.1164/rccm.201204-0596PP. [DOI] [PubMed] [Google Scholar]
- Wain LV, Odenthal-Hesse L, Abujaber R, Sayers I, Beardsmore C, Gaillard EA, Chappell S, Dogaru CM, McKeever T, Guetta-Baranes T, et al. Copy number variation of the beta-defensin genes in europeans: no supporting evidence for association with lung function, chronic obstructive pulmonary disease or asthma. PLoS One. 2014;9(1):e84192. doi: 10.1371/journal.pone.0084192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan ES, Castaldi PJ, Cho MH, Hokanson JE, Regan EA, Make BJ, Beaty TH, Han MK, Curtis JL, Curran-Everett D, et al. Epidemiology, genetics, and subtyping of preserved ratio impaired spirometry (PRISm) in COPDGene. Respir Res. 2014;15(1):89. doi: 10.1186/s12931-014-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan ES, Hokanson JE, Murphy JR, Regan EA, Make BJ, Lynch DA, Crapo JD, Silverman EK, Investigators CO. Clinical and radiographic predictors of GOLD-unclassified smokers in the COPDGene study. Am J Respir Crit Care Med. 2011;184(1):57–63. doi: 10.1164/rccm.201101-0021OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome research. 2007;17(11):1665–74. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanger J, Clausen JL, Coates A, Pedersen OF, Brusasco V, Burgos F, Casaburi R, Crapo R, Enright P, van der Grinten CP, et al. Standardisation of the measurement of lung volumes. Eur Respir J. 2005;26(3):511–22. doi: 10.1183/09031936.05.00035005. [DOI] [PubMed] [Google Scholar]
- Younkin SG, Scharpf RB, Schwender H, Parker MM, Scott AF, Marazita ML, Beaty TH, Ruczinski I. A genome-wide study of de novo deletions identifies a candidate locus for non-syndromic isolated cleft lip/palate risk. BMC Genet. 2014;15:24. doi: 10.1186/1471-2156-15-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Warren L, Aponte J, Gulsvik A, Bakke P, Anderson WH, Lomas DA, Silverman EK, Pillai SG, International CGNI The SERPINE2 gene is associated with chronic obstructive pulmonary disease in two large populations. Am J Respir Crit Care Med. 2007;176(2):167–73. doi: 10.1164/rccm.200611-1723OC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.