Abstract
Resolution of infection requires the coordinated response of heterogeneous cell types to a range of physiological and pathological signals to regulate their proliferation, migration, differentiation, and effector functions. One mechanism by which immune cells integrate these signals is through modulating metabolic activity. A well-studied regulator of cellular metabolism is the hypoxia-inducible factor (HIF) family, the highly conserved central regulators of adaptation to limiting oxygen tension. HIFs regulation of cellular metabolism and a variety of effector, signaling, and trafficking molecules has made these transcription factors a recent topic of interest in T cell biology. Low oxygen availability, or hypoxia, increases expression and stabilization of HIF in immune cells, activating molecular programs both unique and common among cell types, including glycolytic metabolism. Notably, numerous oxygen-independent signals, many of which are active in T cells, also result in enhanced HIF activity. Here, we discuss both oxygen-dependent and -independent regulation of HIF activity in T cells and the resulting impacts on metabolism, differentiation, function and immunity.
Keywords: Metabolism, T cell, Hypoxia-Inducible Factor, Differentiation, Immunity
1. Introduction
Protective immunity relies on coordinate responses from numerous cell types in order for host tissues to resist incursion by foreign agents. Infection of tissues by pathogens often drives vast changes in the tissue microenvironment as well as the upregulation of systemic signals necessary for marshalling immune cells to the fight. Recently it has become appreciated that antigen, cytokines, chemokines, as well as the balance of nutrients and oxygen can play a role in modulating immune cell metabolism thereby affecting cellular fate and function [1, 2]. Further, T cells must traffic from secondary lymphoid tissues to infected sites, experiencing dramatic shifts in microenvironmental signals unique to tissue milieu, which may play a crucial role in influencing T cell metabolism, altering cellular function and cell fate decisions. How such microenvironmental signals are integrated at the transcriptional level is a topic of considerable importance in understanding T cell immunity.
1.1 T cell metabolism – a modulator of T cell responses
It has been noted for decades that the massive proliferation that occurs following lymphocyte activation is accompanied by marked shifts in cellular metabolism [3]. Quiescent naive T cells primarily rely on oxidative phosophorylation for their energetic demands, but upon activation by cognate peptide and costimulation from antigen presenting cells, T cells rapidly shift towards a reliance on glycolytic metabolism [1, 2]. Glycolytic supply of energy proves to be critical for differentiation and effector function of activated T cells. Following clearance of the pathogen, most effector T cells die, however a small proportion survive and differentiate into long-lived memory populations that exhibit a coordinated return to a reliance on oxidative phosphorylation [1, 2]. Recent work has demonstrated that these metabolic transitions accompany key cell-fate decisions impacting differentiation and effector function of T cell subsets [4–14]. Most intriguingly, data from some of these studies argue that differentiation of memory as well as function of effector cells are dependent on utilization of specific metabolic pathways [6, 8–11, 15]. For example, the transition from predominantly glycolytic metabolism to oxidative phosphorylation, fatty acid metabolism, and generation of spare respiratory capacity by antigen-specific CD8+ T cells may be essential for the formation of long-lived functional memory CD8+ T cells [6]. These data have been recently summarized and are beyond the scope of this review [1, 2]. However, understanding molecular regulators of T cell metabolism has emerged as a topic of intense study, and here we will discuss the master regulators of oxygen homeostasis, the Hypoxia-Inducible Factor transcription factors (HIFs), and their role in regulating T cell metabolism and function.
1.2 HIF – a regulator of cellular metabolism
The importance of cellular metabolism in altering T cell differentiation and function has propelled research into the role of well-established metabolic regulators such as c-Myc, PI3K/AKT, mTOR, Foxo1, and the HIFs in controlling T cell function. HIFs are a family of transcription factors consisting of three alpha subunits, HIF-1α, HIF-2a, and HIF-3α, with differing tissue-specific expression patterns that when stabilized can heterodimerize with HIF-1β, also known as the aryl hydrocarbon receptor nuclear translocator (ARNT), and in the case of HIF-1α and HIF-2α, bind hypoxia response element (HRE) sequences and activate a transcriptional program dedicated to mediating the adaptation of cells to reduced oxygen availability [16–18]. HIF-3α’s lack of a c-terminal activation domain suggests that it is unable to activate transcription despite being capable of heterodimerizing with HIF-1 β and binding HRE sequences. It is currently unclear what role HIF-3α plays in modulating cellular responses to hypoxia [19, 20]. HIFs promote the adaptation of cells to hypoxia primarily by lowering oxygen consumption through the increased expression of two critical glycolytic enzymes, lactate dehydrogenase A (LDHa), which increases the capacity to regenerate NAD+ following reduction of pyruvate produced by glycolysis, and pyruvate dehydrogenase kinase 1 (PDK1), which actively prevents pyruvate from shunting into the TCA cycle [21, 22]. Altering these two metabolic checkpoints along with increased expression of other glycolytic enzymes dramatically shifts cellular metabolism away from oxygen-consuming pathways and drives a reliance on glycolysis for the generation of ATP.
Emerging literature has demonstrated a critical role for HIFs in modulating T cell metabolism via both oxygen-dependent and oxygen-independent pathways and highlighted its regulation of other novel gene targets influencing effector T cell function and differentiation [5, 14, 23–26]. This review will first examine many of the regulators of HIF in T cells, and then what is known about HIF’s impact on T cell metabolism, differentiation, and function. We will conclude by addressing outstanding questions regarding HIF’s role in T cell biology that present attractive areas of future study.
2. Regulators of HIF activity in T cells
Interest in HIF-regulated transcription in T cells stems from HIF’s function as a modulator of metabolism in response to changes in oxygen tension; however, studies in T cells and other immune cell types have revealed that while a primary function of HIFs is the detection and response to low oxygen tensions, a number of other mechanisms regulate HIF activity in an oxygen-independent fashion, allowing T cells to co-opt the HIF pathway for regulation of cellular metabolism and other known transcriptional targets in response to immune stimuli independent of oxygen availability. Here we will cover many of the known regulators of HIF activity in T cells as well as some that remain unexplored in T cell biology, but likely play a role in T cell function and fate given their described function in other cell types.
2.1 Oxygen
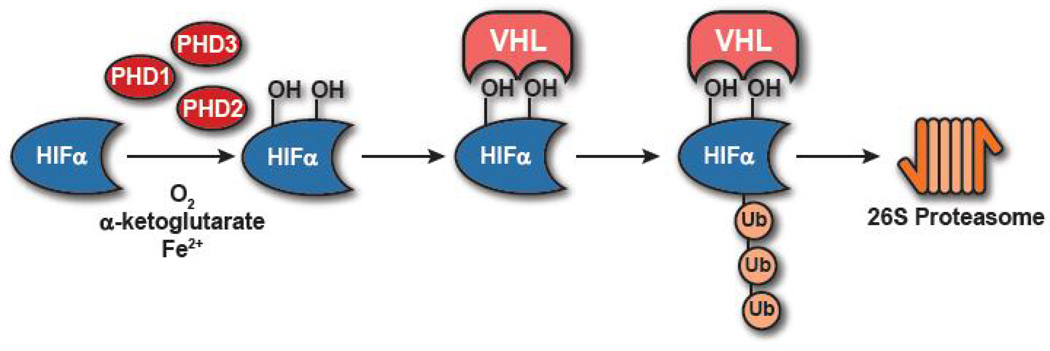
As in other cell types, the HIF pathway serves as the central sensor of oxygen tension in T cells and is regulated in an oxygen-dependent fashion [18, 27]. When T cells are in oxygen-sufficient environments HIFα subunits are rapidly degraded by the proteasome following hydroxylation by prolyl hydroxylase domain proteins (PHDs), primarily PHD2, and ubiquitination by the von Hippel-Lindau tumor suppressor protein complex (pVHL), an E3 ubiquitin ligase that specifically targets hydroxylated HIFα subunits (Figure 1) [28, 29]. Conversely, when cells enter hypoxic environments, (∼1% O2), PHDs, which require oxygen (O2) as a cofactor for their function, along with iron(II) and α-ketoglutarate, are unable to hydroxylate HIF on the appropriate proline residues resulting in a loss of pVHL-dependent ubiquitination and subsequent stabilization of HIFα subunits [28, 29]. HIFα can then dimerize with HIF-1 β upon translocation into the nucleus and activate HIF target genes. While canonical oxygen-dependent regulation of the HIF pathway remains integral to regulating HIFα stability, interestingly, activation by cognate antigen of T cells appears to “unlock” the oxygen-dependent responsiveness of T cells amplifying stabilization of HIFα subunits as naive cells cultured in 1% O2 exhibit mild stabilization of HIFα subunits in comparison to those activated and cultured in 1% O2 (unpublished observation A. Phan and A. Goldrath and [30]).
Fig. 1.
Oxygen-dependent regulation of HIF stability The canonical oxygen-dependent degradation pathway of HIFα requires prolyl-hydroxylation by PHDs and subsequent polyubiquitination by VHL resulting in proteosomal degradation.
2.2 Prolyl hydroxylase domain proteins and Factor Inhibiting HIF-1
Hydroxylation of HIFα subunits is the canonical mechanism for regulating HIF activity [31]. Therefore regulation of PHD expression and activity is critical for regulating HIF function. Three PHD isoforms are present in mice and examination of PHD function shows that PHD2 is the default regulator of HIFα stability in most cell types [32]. Egln1 (PHD2) is constitutively expressed in T cells and hydroxylates HIFα subunits for subsequent degradation at the steady state. Interestingly, a conserved HRE has been identified and shown in mouse embryonic fibroblasts to drive increased PHD2 expression following extended exposure of cells to hypoxia, suggesting a self-regulating circuit designed to prepare cells for re-entry into oxygen rich environments following hypoxia [33, 34]. Egln3, which encodes PHD3, is dynamically expressed in T cells following activation suggesting an importance for hydroxylase activity during T cell responses [35]. PHD3 also appears to be directly regulated by a HRE in a HIF-dependent fashion in in vitro experiments in human cancer cell lines [33, 36]. Further exploration of PHD expression and activity in the context of T cell activation will be informative for defining regulators of HIF activity in the immune response.
In addition to PHDs, another hydroxylase, the Factor Inhibiting HIF-1 (FIH), hydroxylates an asparagine residue in the c-terminal activation domain of both HIF-1α and HIF-2α subunits in normoxia [37, 38]. Asparaginyl-hydroxylation blocks the ability of HIFs to bind transcriptional coactivators CREB-binding protein and p300 [37, 38]. This prevents HIF-mediated transcription, providing an additional layer of post-translational regulation of HIFs that escape degradation by the proteasome.
PHDs and FIH rely on O2, iron(II), and α-ketoglutarate as cofactors. As such, hypoxia or use of competitive inhibitors of α-ketoglutarate or iron chelators have been shown to inhibit prolyl- and asparaginyl- hydroxylase activity and stabilize HIFα subunits [29, 39, 40]. In addition, accumulation of TCA cycle intermediates succinate and fumarate, due to mutations in TCA cycle enzymes, have been shown in renal cell carcinoma cells to competitively inhibit hydroxylase activity by preventing PHD access to α-ketoglutarate thereby promoting HIFα stabilization [41, 42]. This suggests that alterations in T cell metabolism may serve as an additional mechanism regulating HIF stability and activity through modulation of PHD activity.
2.3 T cell receptor
Macrophages have been shown to stabilize HIFα subunits in response to bacterial antigens in an oxygen-independent, TLR-dependent fashion that requires NF-κB activation [43–45]. Much like macrophages, T cells have been shown to stabilize HIFs regardless of oxygen tension in response to activation of antigen receptors [5, 23, 26, 46–49]. T cell receptor (TCR) signaling and costimulation through CD28 results in robust HIFα protein stabilization regardless of oxygen tension which can be further potentiated by hypoxia [26, 49]. Microarray analysis comparing naive and activated CD8+ T cells show increased expression of mRNA for both HIF-1α and HIF-2α following activation in antigen-specific CD8+ T cells responding to viral and bacterial infections, suggesting that TCR signaling regulates both HIF-1α and HIF-2α expression in vivo [35]. Induction of HIF-1a is thought to be mediated by PI3K/mTOR activity downstream of TCR and CD28 signaling which promotes transcription of two splice isoforms of HIF-1α mRNA in human and mouse T cells along with driving increased protein translation [47, 49]. Oxygen-independent stabilization of HIF-2α also occurs at low levels following TCR and CD28 stimulation of CD8+ T cells [26]. However, it is unknown if this occurs through PI3K/mTOR activity similarly to HIF-1α stabilization or if unique molecular pathways drive this stabilization independently. TCR and CD28 signaling have also been shown to activate NF-κB signaling in T cells and given the importance of NF-kB activity in promoting antigen receptor-dependent activation of HIFs in macrophages it stands to reason that NF-kB activity may play a critical role in regulating HIF activity following TCR and CD28 engagement [50, 51]. Additionally, initial studies of TCR-dependent stabilization of HIFα subunits utilized rapamycin, a broad spectrum mTOR inhibitor, to assess mTOR-dependency [49]. However, recent advances in our understanding of the PI3K/mTOR pathway in T cells has revealed additional complexity in the regulation and activity of mTOR (i.e. mTORC1 versus mTORC2, cross-talk with other metabolic pathways) [52]. Further examination of TCR-dependent regulation of HIFα stability in the context of critical T cell activation pathways is necessary to clarify when and where HIF-mediated transcription will influence T cell immunity.
2.4 Cytokines
As interest in the impact of HIFα activity in T cells has increased, the effects of cytokine signaling on HIFα stabilization/activity has begun to be explored. Previous work in human cancer cell lines has demonstrated that TGF-β may drive oxygen-independent regulation of HIF demonstrated by normoxic stabilization of HIFs through Smad-dependent inhibition of PHD2 expression [53]. Intriguingly, in CD4+ T cells, pro-inflammatory IL-6 and anti-inflammatory TGF-β have been implicated in vitro in normoxic stabilization of HIF-1α in a STAT3-dependent manner [23]. However, an additional study demonstrated that HIF-1α stabilization is STAT3 independent suggesting that other cytokines, possibly IL-23, could also play a role in influencing HIF-1α activity in CD4+ T cell differentiation [5]. In macrophages, TH1 and TH2 cytokines could stabilize HIF-1α promoting M1-polarization or HIF-2α driving M2-polarization respectively [54]. Similarly, in vitro activation of CD8+ T cells followed by culture with IL-2 potentiated normoxic stabilization of HIF-1αand little to no stabilization of HIF-2α while culturing with IL-4 promoted normoxic stabilization of both HIF-1α and HIF-2αin expanding CD8s [26]. Culturing activated CD8+ T cells with IL-2 or IL-4 also altered HIF-1α- or HIF-2a–dependence of several differentially expressed effector molecules, transcription factors, and activation-associated receptors. This suggests that cytokine-dependent stabilization of HIFs could be an additional context-specific mechanism for modulating T cell function and fate similar to the bifurcation of M1 and M2 macrophages [26, 54]. Deciphering how individual cytokines affect the different layers of HIFα regulation will provide important insight into the potential impact hypoxia and the inflammatory microenvironment have on T cell differentiation and function.
2.5 Non-coding RNAs
With genome-wide sequencing, it has recently become appreciated that non-coding RNAs (ncRNAs) make up a large proportion of differentially expressed genes following T cell activation. Examination of microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) in T cells has demonstrated important roles for both in the regulation of T cell biology downstream of numerous signals [55–57]. Notably, a number of miRNAs and lncRNAs have been implicated in regulating HIFα accumulation. miR155 and miR210 are hypoxia-regulated which, in mouse intestinal epithelial cells and T cells respectively, provide negative feedback to HIF activity following extended hypoxic signaling [30, 58]. A recent study demonstrated that miR210, but not miR155, is upregulated upon T cell activation in a CD28- and HIF-1α-dependent manner in CD4+ and CD8+ T cells [30]. Surprisingly, computational analysis predicted HIF-1α as a target of miR210 and further study of TH17 CD4+ T cells revealed a negative feedback loop mediated by miR210 for T cells cultured in prolonged hypoxia. Transfer of miR210-deficient CD4+ T cells in a T cell transfer model of colitis resulted in increased severity of disease that correlated with increased stabilization of HIF-1α in CD4+ T cells isolated from the lamina propria of mice. These data suggest that miR210-dependent degradation of HIF-1α mRNA dampens wildtype TH17 responses in chronically hypoxic tissues and may serve as a critical regulator of T cell immunity [30]. Importantly, the impact of miR210 on HIF-1α stability was most striking in conditions of limiting oxygen such as the gut, suggesting that non-canonical regulators of the HIF pathway may provide context-specific tuning of the HIF pathway for regulation of unique cellular functions, such as CD4+ TH subset differentiation.
In contrast to miRNAs, p21, a lncRNA, has been shown to play a novel role in blocking pVHL-HIFα interactions in transformed and non-transformed human cell lines [59]. In vitro culture of HeLa cells in hypoxia inhibited PHD function and stabilized HIF-1α protein as expected, however, knockdown of p21 lncRNA resulted in an inability to maintain HIF-1α protein levels after 24 hours of hypoxia [59]. Intriguingly, in this study HIF-1α-dependent transcription of egln1 increased PHD2 levels sufficiently to potentiate hypoxic degradation of HIF-1α. Maintenance of HIF-1α stability following increased PHD2 levels required HIF-1α-dependent expression of p21 lncRNA and disruption of pVHL-HIF-1α interactions [59]. Control of HIFα stability by ncRNAs is a recent addition to the HIF pathway’s regulatory roadmap and significant work will need to be done to understand why these loops have evolved and how they impact canonical HIF regulation (i.e. PHD- and VHL-dependent regulation). Also important to note is that the majority of studies on ncRNA regulation of HIFα subunit stability has focused on stabilization of HIF-1α and it remains unclear how other subunits are impacted. Elucidating subunit specific regulation of HIFs will be critical in understanding their individual roles in T cell biology. Due to the emerging importance of ncRNAs in T cell biology investigation into ncRNAs could also provide additional responsive flexibility to the HIF pathway in T cells for the regulation of effector functions.
2.6 Reactive oxygen species (ROS)
T cell activation has been shown to induce mitochondrial activity driving complex III-dependent production of ROS that is essential for activation of nuclear factor of activated T cells (NFAT) and subsequent IL-2 induction [60]. ROS additionally has been implicated in the stabilization of HIFs. However intense study has resulted in two prevailing interpretations of available data regarding the role of mitochondria-derived ROS in regulation of HIF (reviewed [61]). One model argues that mitochondrial ROS is necessary for regulation of HIFα stability. Initial reports examining the role of mitochondria in HIFα stabilization found that loss of mitochondria (Rho cells) or inhibition of electron transport chain (ETC) complexes responsible for ROS production prevented stabilization of HIF-1α in hypoxia (1.5% O2) due to a lack of superoxide (SO) production from the ETC [62, 63]. Additional studies carefully dissecting the contribution of individual ROS species in human cell lines through exogenous addition of ROS or overexpression of ROS-producing or -depleting enzymes revealed that HIFα stabilization was dependent on mitochondrial derived H2O2 rather than SO in hypoxia [64, 65]. Direct mechanistic evidence of how ROS inhibited PHD or VHL function, however, has yet to be shown. These studies are contrasted by experiments which argue that mitochondrial metabolism is essential for HIFα stabilization, but in a ROS-independent fashion. Inhibition of complex III of the ETC of human kidney epithelial cells in the presence of 2,2,4-trimethyl-1,3-pentanediol (TMPD), which allows for continued ETC activity and oxygen consumption without production of SO, resulted in no difference in hypoxic HIFα stabilization arguing that mitochondria-dependent HIFα stabilization in hypoxia is ROS-independent and driven primarily by O2 usage by the ETC which reduces cytosolic O2 availability, thereby reducing PHD activity and culminating in HIFα stabilization [66]. While these two models appear mutually exclusive, a possible explanation for the current data is that a combination of both decreased O2 availability due to mitochondrial metabolism as well as alterations in cellular redox state and iron(II) availability due to ROS production may synergize to promote HIFα stabilization in cells [61]. As mitochondrial metabolism and ROS production are essential for the activation of T cells, this supports a potential role for mitochondrial ROS in regulating stability of HIFα subunits in T cells, similar to observations in other cell types [60, 61]. Experiments demonstrating a direct link between mitochondrial ROS production and HIFα stabilization in T cells could provide mechanistic insight to metabolism-dependent transcriptional programming.
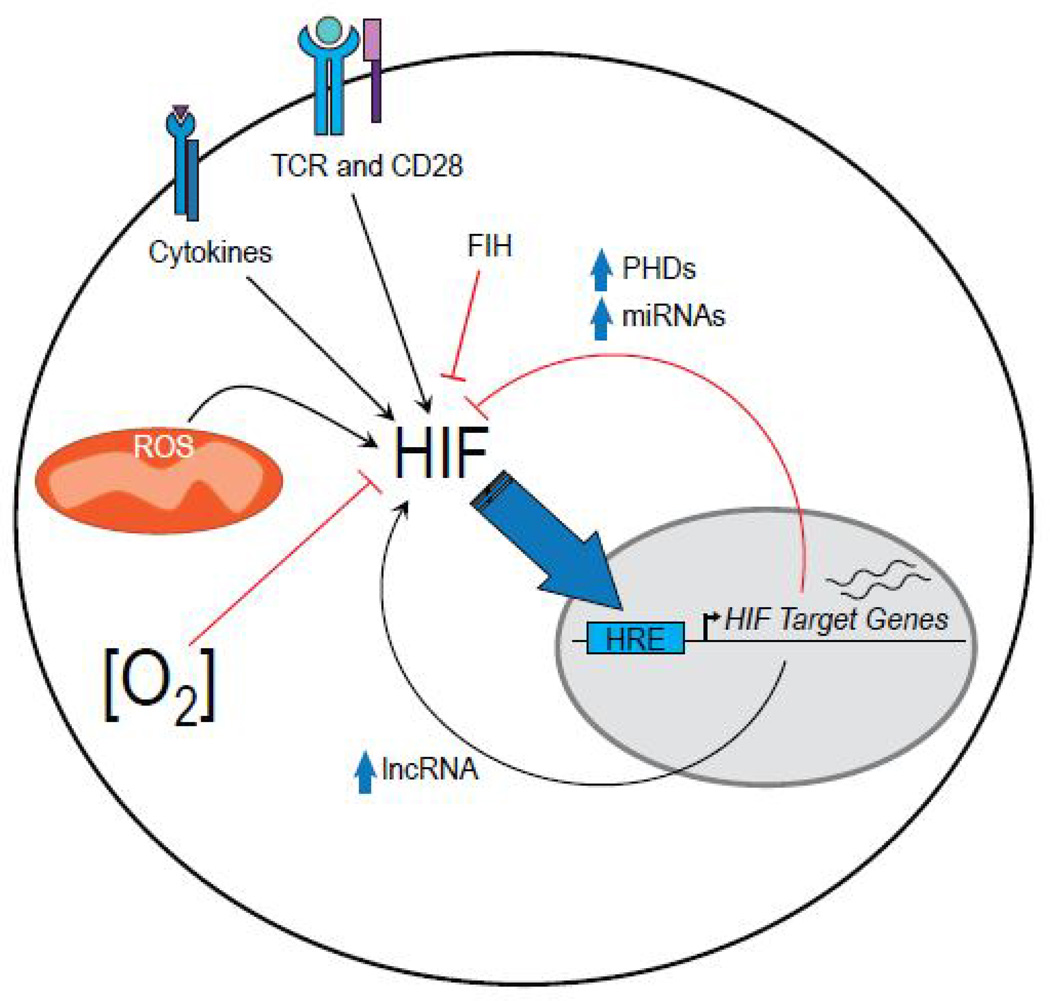
The sheer diversity of signals that have been identified and continue to be implicated in HIF stability and activity reflect the complex role of HIF in the integration of microenvironmental stimuli for the regulation of T cell metabolism, differentiation, and ultimately, function (Figure 2).
Fig. 2.
Regulators of HIF activity in T cells In addition to oxygen, many signals regulate HIF activity in T cells, activating HIF-dependent transcription and potentiating a variety of selfregulatory pathways.
3. Regulation of T cell metabolism, differentiation, and function
T cells undergo rapid metabolic reprogramming within the first 24 hours of activation, and recent work has focused on elucidating key regulators of T cell metabolism in an effort to delineate the direct impact of such changes on T cell immunity [1, 2]. Interestingly, it has been found that HIF-1α activity is unnecessary while c-Myc is essential for metabolic reprogramming of T cells within the first 24 hours of activation [67]. HIF-1α deletion did not impact proliferation, glutaminolysis, glycolysis, or fatty acid oxidation by T cells following activation with anti-CD3 and anti-CD28 in vitro [67]. Induction of LDHa and hexokinase-2 gene expression, two well characterized HIF-1α targets, were moderately downregulated by HIF-1α deletion and mild impairment of glycolytic rate was observed 72 hours after activation. Thus, while unnecessary for the initial transition to glycolytic metabolism, HIF-1α may be involved in sustaining glycolytic throughput during the T cell response [67]. It is important to note these data are from T cells activated in atmospheric oxygen concentrations, leaving open the possibility that in secondary lymphoid tissues, under physiologic oxygen tensions (0.5–4.5% O2), HIFs may play a more significant or different role in regulating early T cell metabolic programming while integrating additional microenvironmental signals to direct trafficking, function, and differentiation [46].
Indeed in support of HIF-1α maintaining glycolytic metabolism in T cells, a recent study proposes that reduced exposure to IL-2 following T cell activation results in increased expression of Bcl-6 which directly represses glycolytic genes regulated by HIF-1α and c-myc providing a mechanism for dampening glycolytic metabolism and effector function, thereby altering T cell fate decisions in a microenvironment-dependent fashion [68]. These data support a model where HIFs are capable of maintaining glycolytic metabolism in T cells shortly after activation driven by oxygen-independent mechanisms. Identifying additional factors that regulate HIF stability or activity in order to modulate glycolytic maintenance may lend important insights about the molecular “switches” that drive context-dependent metabolic shifts critical for cell fate decisions.
While metabolic reprogramming occurs in all T cells upon activation, recent work has shown that CD4+ and CD8+ T cells show distinct metabolic changes [69]. Following 72 hours of in vitro stimulation with anti-CD3 and anti-CD28 glycolytic and oxidative phosphorylation rates were measured via extracellular flux by CD4+ and CD8+ T cells [69]. Both CD4+ and CD8+ T cells increased total metabolic rates following activation, indicated by increased extracellular acidification rate (ECAR) and oxygen consumption rate (OCR), proxies for glycolysis and oxidative phosphorylation respectively, in comparison to naive cells. However, examination of ECAR/OCR ratios as a measure of reliance on glycolysis or oxidative phosphorylation revealed that while both CD8+ and CD4+ T cells skew towards glycolysis following activation, CD8+ T cells relied more heavily on glycolytic metabolism relative to CD4+ T cells [69]. Interestingly, CD4+ T cell proliferation exhibited greater sensitivity to inhibition of both glycolysis or oxidative phosphorylation despite arguably greater metabolic flexibility relative to CD8+ T cells [69]. These data support a role for both glycolysis and oxidative phosphorylation in promoting T cell proliferation and are supported by studies in both CD4+ and CD8+ T cells where inhibition of glycolytic metabolism reduces but does not abolish proliferation, whereas inhibition of oxidative phosphorylation, surprisingly, produces a more dramatic defect in proliferation by comparison [7, 8, 60]. These data argue that in T cells metabolic reprogramming to a reliance on glycolytic metabolism primarily occurs to promote effector function and likely additional unidentified T cell processes rather than to support proliferation [8]. Importantly, Cao et al also emphasize that context-specific cues will play an essential role in modulating T cell metabolism in vivo. Experiments comparing the effect of fuel availability (glucose versus glutamine), cytokine signaling (IL-2, IL-15, IL-7), titrated levels of TCR stimulation, and addition of co-receptor signaling (41BB or OX40) during activation of T cells found that all microenvironmental signals tuned metabolic pathway skewing, mitochondrial mass, and ROS production in CD4+ and CD8+ T cells [69]. Further studies will need to be done to assess whether these differences in metabolic profiles directly specify lineage and whether HIF regulates these differences. It will be important to determine the differences in energetic demands between CD4+ and CD8+ T cells and how they correlate with unique usage of the HIF pathway in each subset for regulation of metabolism, cellular functions, and fate decisions.
3.1 Regulation of CD4+ T cell metabolism
CD4+ T cells have been shown to differentiate into a wide range of helper subsets following activation. These subsets exhibit distinct effector functions and differentiate following upregulation of key transcription factors. For example in the case of TH1 CD4+ T cells, the transcription factor T-bet is induced upon activation or for TH17 CD4+ T cells the transcription factor RORγt. The wide range of phenotype and function exhibited by these CD4+ helper subsets suggests the possibility for varying metabolic demands, and in line with this hypothesis, culturing activated CD4+ T cells in TH17 conditions drove significantly higher glycolytic rates in comparison to CD4+ T cells cultured in other TH conditions [5]. Notably, HIF-1α protein levels were also high in CD4+ T cells cultured in TH17 conditions [5, 23]. Analysis of gene expression of HIF-1α-deficient CD4+ T cells cultured in TH17 conditions found reduced expression of known HIF-1α targets in the glycolytic pathway in comparison to wildtype cells. Decreased gene expression of genes promoting glycolysis corresponded with greatly reduced glycolytic metabolism, in comparison to HIF-1α-sufficient CD4+ T cells, demonstrating a reliance on HIF-1α in regulating CD4+ T cell metabolism during TH17 differentiation [5]. In support of a role for HIF-1α in regulating CD4+ T cell metabolism, a recent study of Foxp3-regulatory CD4+ T cells (Tr1), that produce IL-10 to control inflammation, demonstrated that HIF-1α also participates in maintaining a shift towards glycolytic metabolism in CD4+ T cells activated and cultured in vitro in the presence of IL-27 [14].
3.2 Impact on CD4+ T cell differentiation and function
The impact of HIF-dependent regulation of cellular metabolism on T cell differentiation and function is less clear than its regulation of glycolytic genes. However, recent work has demonstrated several novel roles for HIF regulation of T cell differentiation and effector function. In vitro experiments with CD4+ T cells demonstrated that inhibiting glycolytic metabolism directly impacted effector function [8]. More specifically, in vitro activation of CD4+ T cells in glucose-free media supplemented with galactose prevented glycolytic metabolism and reduced IFNγ production, dependent on the ability of GAPDH to bind the 3’ UTR of IFNγ mRNA. When the binding site of the 3’ UTR was mutated, inhibition of glycolysis in CD4+ T cells no longer affected the production of IFNγ. These data show that usage of glycolytic metabolism in CD4+ T cells occupies GAPDH, preventing interaction with cytokine mRNA and potentiating effector function [8]. These data provide a potential link between HIF regulation of T cell glycolysis and effector function that warrants further study.
3.2.1 HIF-1 α and the TH17/iTreg balance
Recent reports examining the T cell intrinsic role of HIF-1α on CD4+ T cell differentiation demonstrated that deletion of HIF-1α resulted in reduced capacity for differentiation of TH17 CD4+ T cells in vitro and reduced induction of experimental autoimmune encephalitis, a TH17-driven murine model of autoimmunity in vivo [5, 23]. The reduction in differentiation of TH17 CD4+ T cells in HIF-1α-deficient CD4+ T cells was concomitant with an increase in differentiation of iTreg cells. These initial reports suggested that HIF-1α was critical for expression of IL-17A [5, 23]. However, two distinct mechanisms were proposed to explain how HIF-1α promoted TH17 differentiation and suppressed iTreg formation. One study argued that HIF-1α deficiency primarily perturbed TH17 differentiation through a reduction in glycolytic metabolism and demonstrated in vitro that suppression of glycolysis in wildtype CD4+ T cells by addition of 2-deoxyglucose to TH17 culture conditions inhibited TH17 differentiation and promoted iTreg differentiation [5]. In contrast, Dang et al proposed several novel regulatory functions and targets for HIF-1α[23]. In vitro activation of CD4+ T cells in TH17 polarizing conditions resulted in STAT3 signaling which drove expression of HIF-1α mRNA and HIF-1α protein stabilization. HIF-1α in turn drove expression of RORγt, and in cooperation with RORγt, promoted expression of IL-17A. HIF-1α deficiency resulted in loss of RORγt expression and therefore a loss of IL-17A expression [23]. In addition to driving expression of TH17 signature genes, HIF-1α also inhibited differentiation of iTreg by binding to Foxp3 through a n-terminal domain resulting in proteosomal degradation of Foxp3 in spite of TGF-β signaling present in in vitro TH17 culture conditions [23]. Additional studies by several groups in human and murine models, have both supported and contradicted aspects of these initial reports [70–75]. It is clear that HIF-1α can play a role in the differentiation of TH17 versus iTreg cells. However, the functional effects may be subject to experimental context. Indeed, data from recent reports suggest that HIF-1α may play a role in supporting thymic Treg function in addition to regulating differentiation of TH17/iTreg cells [70, 73, 25].
In addition, as demonstrated in CD8+ T cells, HIFα isoforms may have compensatory regulation that occurs in cytokine-dependent fashion [26]. Therefore, the complicated results seen in various models of TH17 and Treg differentiation may result from differential HIFα expression/stabilization. Careful dissection of the relative contribution of HIFα subunits will be necessary to truly understand the contributions of hypoxia, glycolytic metabolism, and HIFα activity on CD4+ T cell differentiation given that TH differentiation is cytokine dependent.
3.3 Regulation of CD8+ T cell metabolism
Multiple lines of evidence also support a role for HIF regulation of metabolism in CD8+ T cells. A recent study prevented formation of functional HIFα transcriptional complexes through conditional deletion of HIF-1 β in T cells. CD8+ T cells deficient in HIF-1 β activated in response to TCR signaling and costimulation were unable to sustain levels of glucose uptake and lactate production following extended culture in vitro [24]. Experiments examining VHL-null CD8+ T cells, which have constitutive stabilization of HIFα subunits, further support a role for HIFα in regulating CD8+ T cell metabolism. Microarray analysis of adoptively transferred P14 TCR transgenic VHL-null CD8+ T cells demonstrated significantly higher expression of glycolysis-associated genes compared to wildtype [26]. Most directly, in vitro activated VHL-null CD8+ T cells have significantly increased glycolytic throughput as well as suppression of oxidative phosphorylation in line with HIF regulation of cellular metabolism seen in other tissues [26].
3.4 Impact on CD8+ T cell differentiation and function
Similar to CD4+ T cells, recent work has established important novel roles for HIFs in the regulation of CD8+ T cell differentiation and function [24, 26]. In addition to preventing the maintenance of glycolytic metabolism in activated CD8+ T cells, conditional deletion of HIF-1 β from T cells resulted in a reduction in gene expression of perforin and some granzymes following in vitro activation and culture with IL-2. Increased stabilization of HIF-1α produced the opposite effect of HIF-1 β deletion, as culture of wildtype cells in 1% O2 drove increased stabilization of HIF-1α and resulted in increased perforin expression [24]. Chromatin immunoprecipitation of RNA Polymerase II (RNAPol II) at the transcription start site or distal exon of perforin showed no difference between wildtype or HIF-1 β-deficient cells in RNAPol II binding suggesting that increases in perforin mRNA and protein are indirectly affected by functional HIF transcriptional complexes [24]. In addition to alterations in cytotoxic molecules, HIF-1 β deletion results in significant increases in mRNA expression for chemokine receptors and trafficking molecules including CCR7, CXCR3, and CD62L [24]. Interestingly, increased gene expression was not restricted to either secondary lymphoid organ homing-receptors or those of inflammatory chemokines, thereby making it difficult to predict whether HIF-1 β-deficient CD8+ T cells may traffic preferentially to certain tissues in vivo following infection. However, in vitro generated wildtype and HIF-1 β-deficient CTLs mixed in equal proportions, labeled, and transferred into naive hosts yielded increased recovery of HIF-1 β-deficient CD8+ T cells from lymph nodes suggesting HIF signaling favored effector CD8+ T cell trafficking to non-lymphoid tissues [24]. In order to assess whether the increased expression of secondary lymphoid homing receptors such as CD62L on HIF-1 β-deficient CTL is a direct transcriptional effect or an indirect result of metabolic perturbation the authors examined CD62L expression on in vitro activated wildtype CD8+ T cells cultured in IL-2 and low levels of glucose and found that limiting levels of glucose prevented downregulation of CD62L following activation, arguing that the increased expression of CD62L in HI F-1 β-deficient cells is the result of reduced glucose uptake due to lower glut1 expression. However, these data do not rule out a direct role for HIF transcriptional activity in regulating T cell trafficking molecules as well [24].
Data regarding loss of HIF-1 β are complimented by studies examining CD8+ T cells following conditional deletion of Vhl, which resulted in constitutive stabilization of HIFα subunits, responding to chronic viral infection [26]. VHL-deficient CTL exhibited increased expression of effector molecules such as GranzymeB as well as resistance to T cell exhaustion in the face of chronic antigen exposure. The resistance of VHL-deficient CTL to exhaustion resulted in reduced viral titers following chronic LCMV clone 13 infection in comparison to wildtype, however, this was accompanied by host death resulting from severe immunopathology [26]. Microarray analysis of VHL-deficient CD8+ T cells responding to either chronic or acute infection demonstrated that constitutive HIF activity drives significant alterations in a variety of effector molecules (GzmB), secreted factors (Ifng), costimulatory receptors (Tnfrsf9), exhaustion/activation associated genes (Lag3), and key CD8+ T cell differentiation transcription factors (Prdm1, Tbx21, Eomes) exhibited significantly altered expression. Many of these changes in gene expression were confirmed at the protein level and analysis of CD8+ T cell subsets during the course of response to viral infections showed a dramatic HIFα-dependent loss of shorter-lived effectors (KLRG1hiCD127lo) in both acute and chronic viral infection [26]. Deletion of both HIF-1α and HIF-2α in addition to VHL rescued host death by eliminating immunopathology and restoring expression of many altered genes to wildtype levels following chronic LCMV infection, demonstrating HIFα dependence of enhanced effector responses to persistent antigen [26]. While it is clear from these data that HIF-1α and HIF-2αplay a role in the regulation of CD8+ T cell exhaustion in the face of persistent antigen, the impact each subunit has on both the metabolic phenotype and the functional phenotype will be important topics to address.
Together these data demonstrate a role for HIFs in the differentiation and function of CD8+ T cells during the response to viral infections. HIFs appear to play a significant role in the modulation of effector molecule expression during the course of an infectious response and likely serve to modulate antigen-dependent responses as constitutive HIFα signaling drives resistance to T cell exhaustion signals. This model is supported by studies of tumor infiltrating CD8+ T cells where HIF-1α increased 4–1BB (Tnfrsf9) expression, a costimulatory receptor, which when ligated has been shown to drive proliferation, IL-2 production, and increased cytolytic activity in response to tumors and chronic viral infections [76, 77]. VHL deletion ultimately impacts both receptor expression and likely signaling events downstream of the receptors as well. Further work exploring how HIFs crosstalk with these signaling pathways will be highly informative and present novel therapeutic approaches.
It is important to note that these studies have focused on effector responses and early CD8+ T cell differentiation events. Whether HIFs regulate the formation of T cell memory remains unknown and experiments exploring HIF’s control of both transcriptional and metabolic targets will be helpful for parsing out the individual contributions of transcription factors and cellular metabolism to the differentiation of protective memory T cells.
4. Questions remaining
The recent interest in the function of HIFs in T cell biology has yielded impressive advances in our understanding of how this family of oxygen-sensing transcription factors impact T cells (Table 1), yet there remain many questions, which could be central to understanding aspects of T cell biology. A primary question is the relevance of tissue oxygen tensions on T cell responses in vivo. Some work has demonstrated that in autoimmune models of colitis and in a variety of infectious contexts that T cells enter hypoxic areas or stabilize HIFs in tissues with localized hypoxia. However, it is unclear how the availability of oxygen does or does not affect T cell function and whether this ultimately impacts clearance of infection or pathology [25, 78]. Determining whether localized hypoxia can impact T cell responses will be critical in understanding the signals that drive T cell fate decisions in vivo.
Table 1.
| Impact of HIF signaling on T cells | Selected References |
|---|---|
 Glycolysis Glycolysis |
Refs: 5, 14, 26 |
 Oxidative Phosphorylation Oxidative Phosphorylation |
|
| Impact on CD4+ T cells | |
 RORγt RORγt |
Refs: 5, 23 |
 Increased IL-17A production Increased IL-17A production |
|
 iTreg differentiation iTreg differentiation |
|
| Impact on CD8+ T cells | |
 Effector molecule expression - perforin and granzymes Effector molecule expression - perforin and granzymes |
Refs: 24, 26 |
 Cytolytic activity Cytolytic activity |
|
 Exhaustion in the face of chronic antigen Exhaustion in the face of chronic antigen |
|
In addition to understanding the in vivo relevance of hypoxia on T cell responses, recent work on HIFs has centered on understanding the role of HIF-1α, leaving the impact of HIF-2α and HIF-3α largely unexplored. Examining the role of HIF-2αand HIF-3α in T cell responses as well as the interactions of HIFα and HIFβ subunits, of which there are a number of ARNT family members, and the interactions of the HIF complex with transcriptional coactivators such as p300 could shed light on HIF regulation of the many novel T cell targets reported and clarify whether regulation of T cell metabolism is HIF-1α-restricted. Further, determining whether independent signals, such as cytokines, may drive differential accumulation of HIFα subunits in vivo and the potential of each subunit to regulate T cell metabolism will be critical for understanding how HIFs respond to complex microenvironmental signals and translate them into T cell function and fate decisions. Clearly defining the targets and interactions regulated by HIFs in T cells may yield unique points for modulation of T cell metabolism and function.
Recent work has demonstrated that HIFs play a significant role in the modulation of T cell metabolism, further clarification is required of the mechanistic role HIFs play in T cell function and fate determination. The majority of studies have focused on elucidating the molecular underpinnings that drive T cells towards glycolytic metabolism, however the factors that may either keep cells from adopting highly glycolytic, arguably less memory-like, metabolism or those that “turn off” glycolysis in favor of other metabolic pathways in order to drive long-lived cell fates are currently unknown. Experiments to address the impact of HIF’s maintenance of glycolytic metabolism on differentiation of long-lived memory T cells could shed light on metabolic regulators essential for the transition to memory-promoting metabolism. Furthermore, it is becoming increasingly clear that while a transition to glycolytic metabolism correlates with increased proliferation in T cells, this transition may serve primarily to promote effector function or other unknown processes rather than support rapid expansion of antigen-specific populations [7, 8, 60]. Thus, more detailed analyses of metabolic pathways will be necessary to elucidate the metabolic requirements of T cell function and fate decisions. Careful examination of cellular metabolomics in conjunction with genetic models impacting the HIF pathway could identify important metabolites for the maintenance of T cell function or fate, thereby adding direct evidence for the necessity of metabolic transitions in T cell biology and explain the functional consequences of shifts in metabolic pathway usage.
Finally, it is becoming evident that the regulation of HIFs occurs on multiple levels. While oxygen plays an important role, depending on the context, oxygen availability may be irrelevant to HIF function. Thus, experiments to assess novel regulatory factors of HIF stability and, more importantly, activity will need to be performed. Expanding the model of HIF regulation in T cells will provide a clearer understanding of the scenarios in which HIFs play a role in T cell function and provide mechanistic insight into how T cells translate signals into cell fate decisions.
5. Concluding remarks
T cell responses are initiated by detection of cognate peptide presented by antigen presenting cells in secondary lymphoid tissues and result in migration of T cells to various tissues with disparate local conditions. HIFs are uniquely poised to integrate physiological and disease-derived signals to modulate T cell responses making them an attractive target for further study. Understanding how T cells have taken advantage of the HIF pathway to interpret local conditions and integrate these with the array of mitogenic signals produced during infection is essential for the advancement of our understanding of T cell biology and critical for the development of novel therapeutic approaches.
Highlights.
T cells modulate metabolism in response to environmental signals
Oxygen and immune stimuli stabilize HIF activating a transcriptional program
HIF is a regulator of T cell metabolism, differentiation, and function
HIF integrates numerous signals as well as oxygen to drive unique T cell responses
Many questions remain regarding the HIF -pathway and T cell biology
Acknowledgements
We thank A. Palazon, P. Tyrakis, J. J. Milner, and J. Goulding for critical discussions and review of the manuscript.
Funding
The authors were supported by the UCSD NIH Cell and Molecular Genetics Training Grant (5T32GM007240-36) A.T.P, the NIH (A1096852, A1072117) and the Leukemia and Lymphoma Society for A.W.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Anthony T Phan, Email: atphan@ucsd.edu.
Ananda W Goldrath, Email: agoldrath@ucsd.edu.
References
- 1.Pearce EL, Poffenberger MC, Chang C-H, Jones RG. Fueling Immunity: Insights into Metabolism and Lymphocyte Function. Science. 2013;342:1242454–1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang JT, Wherry EJ, Goldrath AW. Molecular regulation of effector and memory T cell differentiation. Nature Immunology. 2014;15:1104–1115. doi: 10.1038/ni.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang T, Marquardt C, Foker J. Aerobic glycolysis during lymphocyte proliferation. Nature. 1976;261:702–705. doi: 10.1038/261702a0. [DOI] [PubMed] [Google Scholar]
- 4.Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC. Cutting Edge: Distinct Glycolytic and Lipid Oxidative Metabolic Programs Are Essential for Effector and Regulatory CD4+ T Cell Subsets. The Journal of Immunology. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H. HIF1α-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. The Journal of Experimental Medicine. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Windt Gerritje JW, Everts B, Chang C-H, Curtis Jonathan D, Freitas Tori C, Amiel E, Pearce Edward J, Pearce Erika L. Mitochondrial Respiratory Capacity Is a Critical Regulator of CD8+ T Cell Memory Development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kidani Y, Elsaesser H, Hock MB, Vergnes L, Williams KJ, Argus JP, Marbois BN, Komisopoulou E, Wilson EB, Osborne TF, Graeber TG, Reue K, Brooks DG, Bensinger SJ. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nature Immunology. 2013;14:489–499. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang C-H, Curtis Jonathan D, Maggi Leonard B, Faubert B, Villarino Alejandro V, O’Sullivan D, Huang Stanley C-C, van der Windt Gerritje JW, Blagih J, Qiu J, Weber Jason D, Pearce Edward J, Jones Russell G, Pearce Erika L. Posttranscriptional Control of T Cell Effector Function by Aerobic Glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Windt GJW, O’Sullivan D, Everts B, Huang SC-C, Buck MD, Curtis JD, Chang C-H, Smith AM, Ai T, Faubert B, Jones RG, Pearce EJ, Pearce EL. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proceedings of the National Academy of Sciences. 2013;110:14336–14341. doi: 10.1073/pnas.1221740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sukumar M, Liu J, Ji Y, Subramanian M, Crompton JG, Yu Z, Roychoudhuri R, Palmer DC, Muranski P, Karoly ED, Mohney RP, Klebanoff CA, Lal A, Finkel T, Restifo NP, Gattinoni L. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. The Journal of Clinical Investigation. 2013;123:4479–4488. doi: 10.1172/JCI69589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Sullivan D, van der Windt Gerritje JW, Stanley Huang C-C, Curtis Jonathan D, Chang C-H, Buck Michael D, Qiu J, Smith Amber M, Lam Wing Y, DiPlato Lisa M, Hsu F-F, Birnbaum Morris J, Pearce Edward J, Pearce Erika L. Memory CD8+ T Cells Use Cell-Intrinsic Lipolysis to Support the Metabolic Programming Necessary for Development. Immunity. 2014;41:75–88. doi: 10.1016/j.immuni.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin Y, Choi S-C, Xu Z, Perry DJ, Seay H, Croker BP, Sobel ES, Brusko TM, Morel L. Normalization of CD4+ T cell metabolism reverses lupus. Science Translational Medicine. 2015;7:274ra218–274ra218. doi: 10.1126/scitranslmed.aaa0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui G, Staron Matthew M, Gray Simon M, Ho P-C, Amezquita Robert A, Wu J, Kaech Susan M. IL-7-Induced Glycerol Transport and TAG Synthesis Promotes Memory CD8+ T Cell Longevity. Cell. 2015;161:750–761. doi: 10.1016/j.cell.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mascanfroni ID, Takenaka MC, Yeste A, Patel B, Wu Y, Kenison JE, Siddiqui S, Basso AS, Otterbein LE, Pardoll DM, Pan F, Priel A, Clish CB, Robson SC, Quintana FJ. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-α. Nature Medicine, advance online publication. 2015 doi: 10.1038/nm.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gubser PM, Bantug GR, Razik L, Fischer M, Dimeloe S, Hoenger G, Durovic B, Jauch A, Hess C. Rapid effector function of memory CD8+ T cells requires an immediate-early glycolytic switch. Nature Immunology. 2013;14:1064–1072. doi: 10.1038/ni.2687. [DOI] [PubMed] [Google Scholar]
- 16.Bracken CP, Whitelaw ML, Peet DJ. The hypoxia-inducible factors: key transcriptional regulators of hypoxic responses. Cellular and Molecular Life Sciences CMLS. 2003;60:1376–1393. doi: 10.1007/s00018-003-2370-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartz RS, Eltzschig HK, Carmeliet P. Hypoxia and Inflammation. New England Journal of Medicine. 2011;364:656–665. doi: 10.1056/NEJMra0910283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palazon A, Goldrath Ananda W, Nizet V, Johnson Randall S. HIF Transcription Factors, Inflammation, and Immunity. Immunity. 2014;41:518–528. doi: 10.1016/j.immuni.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makino Y, Kanopka A, Wilson WJ, Tanaka H, Poellinger L. Inhibitory PAS Domain Protein (IPAS) Is a Hypoxia-inducible Splicing Variant of the Hypoxia-inducible Factor-3α Locus. Journal of Biological Chemistry. 2002;277:32405–32408. doi: 10.1074/jbc.C200328200. [DOI] [PubMed] [Google Scholar]
- 20.Maynard MA, Qi H, Chung J, Lee EHL, Kondo Y, Hara S, Conaway RC, Conaway JW, Ohh M. Multiple Splice Variants of the Human HIF-3α Locus Are Targets of the von Hippel-Lindau E3 Ubiquitin Ligase Complex. Journal of Biological Chemistry. 2003;278:11032–11040. doi: 10.1074/jbc.M208681200. [DOI] [PubMed] [Google Scholar]
- 21.Firth JD, Ebert BL, Pugh CW, Ratcliffe PJ. Oxygen-regulated control elements in the phosphoglycerate kinase 1 and lactate dehydrogenase A genes: similarities with the erythropoietin 3’ enhancer. Proceedings of the National Academy of Sciences. 1994;91:6496–6500. doi: 10.1073/pnas.91.14.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J-w, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metabolism. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Dang EV, Barbi J, Yang H-Y, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen H-R, Luo W, Zeller K, Shimoda L, Topalian SL, Semenza GL, Dang CV, Pardoll DM, Pan F. Control of TH17/Treg Balance by Hypoxia-inducible Factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finlay DK, Rosenzweig E, Sinclair LV, Feijoo-Carnero C, Hukelmann JL, Rolf J, Panteleyev AA, Okkenhaug K, Cantrell DA. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. The Journal of Experimental Medicine. 2012;209:2441–2453. doi: 10.1084/jem.20112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clambey ET, McNamee EN, Westrich JA, Glover LE, Campbell EL, Jedlicka P, Zoeten EFd, Cambier JC, Stenmark KR, Colgan SP, Eltzschig HK. Hypoxia-inducible factor-1 alpha-dependent induction of FoxP3 drives regulatory T-cell abundance and function during inflammatory hypoxia of the mucosa. Proceedings of the National Academy of Sciences. 2012;109:E2784–E2793. doi: 10.1073/pnas.1202366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doedens AL, Phan AT, Stradner MH, Fujimoto JK, Nguyen JV, Yang E, Johnson RS, Goldrath AW. Hypoxia-inducible factors enhance the effector responses of CD8+ T cells to persistent antigen. Nature Immunology. 2013;14:1173–1182. doi: 10.1038/ni.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNamee EN, Johnson DK, Homann D, Clambey ET. Hypoxia and hypoxia-inducible factors as regulators of T cell development, differentiation, and function. Immunologic research. 2013;55:58–70. doi: 10.1007/s12026-012-8349-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WSWGK., Jr HIFα Targeted for VHL-Mediated Destruction by Proline Hydroxylation: Implications for O2 Sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 29.Jaakkola P, Mole DR, Tian Y-M, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim Av, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Targeting of HIF-α to the von Hippel-Lindau Ubiquitylation Complex by O2-Regulated Prolyl Hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Flach H, Onizawa M, Wei L, McManus MT, Weiss A. Negative regulation of Hif1a expression and TH17 differentiation by the hypoxia-regulated microRNA miR-210. Nature Immunology. 2014;15:393–401. doi: 10.1038/ni.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Semenza GL. Oxygen Sensing, Hypoxia-Inducible Factors, and Disease Pathophysiology. Annual Review of Pathology: Mechanisms of Disease. 2014;9:47–71. doi: 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- 32.Appelhoff RJ, Tian Y-M, Raval RR, Turley H, Harris AL, Pugh CW, Ratcliffe PJ, Gleadle JM. Differential Function of the Prolyl Hydroxylases PHD1, PHD2, and PHD3 in the Regulation of Hypoxia-inducible Factor. Journal of Biological Chemistry. 2004;279:38458–38465. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- 33.Marxsen Jan H, Stengel P, Doege K, Heikkinen P, Jokilehto T, Wagner T, Jelkmann W, Jaakkola P, Metzen E. Hypoxia-inducible factor-1 (HIF-1) promotes its degradation by induction of HIF-α-prolyl-4-hydroxylases. Biochemical Journal. 2004;381:761–767. doi: 10.1042/BJ20040620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metzen E, Stiehl Daniel P, Doege K, Marxsen Jan H, Hellwig-Bürgel T, Jelkmann W. Regulation of the prolyl hydroxylase domain protein 2 (phd2/egln-1) gene: identification of a functional hypoxia-responsive element. Biochemical Journal. 2005;387:711–717. doi: 10.1042/BJ20041736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heng TSP, Painter MW, Elpek K, Lukacs-Kornek V, Mauermann N, Turley SJ, Koller D, Kim FS, Wagers AJ, Asinovski N, Davis S, Fassett M, Feuerer M, Gray DHD, Haxhinasto S, Hill JA, Hyatt G, Laplace C, Leatherbee K, Mathis D, Benoist C, Jianu R, Laidlaw DH, Best JA, Knell J, Goldrath AW, Jarjoura J, Sun JC, Zhu Y, Lanier LL, Ergun A, Li Z, Collins JJ, Shinton SA, Hardy RR, Friedline R, Sylvia K, Kang J. The Immunological Genome Project: networks of gene expression in immune cells. Nature Immunology. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 36.Pescador N, Cuevas Y, Naranjo S, Alcaide M, Villar D, Landázuri Manuel O, del Peso L. Identification of a functional hypoxia-responsive element that regulates the expression of the egl nine homologue 3 (egln3/phd3) gene. Biochemical Journal. 2005;390:189–197. doi: 10.1042/BJ20042121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1 α and VHL to mediate repression of HIF-1 transcriptional activity. Genes & Development. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine Hydroxylation of the HIF Transactivation Domain: A Hypoxic Switch. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 39.Epstein ACR, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian Y-M, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ. C. elegans EGL-9 and Mammalian Homologs Define a Family of Dioxygenases that Regulate HIF by Prolyl Hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 40.Bruick RK, McKnight SL. A Conserved Family of Prolyl-4-Hydroxylases That Modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 41.Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 42.Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung Y-L, Merino M, Trepel J, Zbar B, Toro J, Ratcliffe PJ, Linehan WM, Neckers L. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: Novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8:143–153. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 43.Blouin CC, Pagé EL, Soucy GM, Richard DE. Hypoxic gene activation by lipopolysaccharide in macrophages: implication of hypoxia-inducible factor 1α. Blood. 2004;103:1124–1130. doi: 10.1182/blood-2003-07-2427. [DOI] [PubMed] [Google Scholar]
- 44.Peyssonnaux C, Datta V, Cramer T, Doedens A, Theodorakis EA, Gallo RL, Hurtado-Ziola N, Nizet V, Johnson RS. HIF-1α expression regulates the bactericidal capacity of phagocytes. Journal of Clinical Investigation. 2005;115:1806–1815. doi: 10.1172/JCI23865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rius J, Guma M, Schachtrup C, Akassoglou K, Zinkernagel AS, Nizet V, Johnson RS, Haddad GG, Karin M. NF-κB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1α. Nature. 2008;453:807–811. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caldwell CC, Kojima H, Lukashev D, Armstrong J, Farber M, Apasov SG, Sitkovsky MV. Differential Effects of Physiologically Relevant Hypoxic Conditions on T Lymphocyte Development and Effector Functions. The Journal of Immunology. 2001;167:6140–6149. doi: 10.4049/jimmunol.167.11.6140. [DOI] [PubMed] [Google Scholar]
- 47.Lukashev D, Caldwell C, Ohta A, Chen P, Sitkovsky M. Differential Regulation of Two Alternatively Spliced Isoforms of Hypoxia-inducible Factor-1α in Activated T Lymphocytes. Journal of Biological Chemistry. 2001;276:48754–48763. doi: 10.1074/jbc.M104782200. [DOI] [PubMed] [Google Scholar]
- 48.Lukashev D, Klebanov B, Kojima H, Grinberg A, Ohta A, Berenfeld L, Wenger RH, Ohta A, Sitkovsky M. Cutting Edge: Hypoxia-Inducible Factor 1α and Its Activation-Inducible Short Isoform I.1 Negatively Regulate Functions of CD4+ and CD8+ T Lymphocytes. The Journal of Immunology. 2006;177:4962–4965. doi: 10.4049/jimmunol.177.8.4962. [DOI] [PubMed] [Google Scholar]
- 49.Nakamura H, Makino Y, Okamoto K, Poellinger L, Ohnuma K, Morimoto C, Tanaka H. TCR Engagement Increases Hypoxia-Inducible Factor-1α Protein Synthesis via Rapamycin-Sensitive Pathway under Hypoxic Conditions in Human Peripheral T Cells. The Journal of Immunology. 2005;174:7592–7599. doi: 10.4049/jimmunol.174.12.7592. [DOI] [PubMed] [Google Scholar]
- 50.Cheng J, Montecalvo A, Kane LP. Regulation of NF-κB induction by TCR/CD28. Immunologic research. 2011;50:113–117. doi: 10.1007/s12026-011-8216-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paul S, Schaefer BC. A new look at TCR signaling to NF-κB. Trends in immunology. 2013;34:269–281. doi: 10.1016/j.it.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pollizzi KN, Powell JD. Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nature reviews. Immunology. 2014;14:435–446. doi: 10.1038/nri3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McMahon S, Charbonneau M, Grandmont S, Richard DE, Dubois CM. Transforming Growth Factor β1 Induces Hypoxia-inducible Factor-1 Stabilization through Selective Inhibition of PHD2 Expression. Journal of Biological Chemistry. 2006;281:24171–24181. doi: 10.1074/jbc.M604507200. [DOI] [PubMed] [Google Scholar]
- 54.Takeda N, O’Dea EL, Doedens A, Kim J-w, Weidemann A, Stockmann C, Asagiri M, Simon MC, Hoffmann A, Johnson RS. Differential activation and antagonistic function of HIF-α isoforms in macrophages are essential for NO homeostasis. Genes & Development. 2010;24:491–501. doi: 10.1101/gad.1881410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, Chen W, Mattick JS. Genome-Wide Identification of Long Noncoding RNAs in CD8+ T Cells. The Journal of Immunology. 2009;182:7738–7748. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- 56.Pagani M, Rossetti G, Panzeri I, de Candia P, Bonnal RJP, Rossi RL, Geginat J, Abrignani S. Role of microRNAs and long-non-coding RNAs in CD4+ T-cell differentiation. Immunological Reviews. 2013;253:82–96. doi: 10.1111/imr.12055. [DOI] [PubMed] [Google Scholar]
- 57.Jeker LT, Bluestone JA. microRNA regulation of T-cell differentiation and function. Immunological reviews. 2013;253:65–81. doi: 10.1111/imr.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bruning U, Cerone L, Neufeld Z, Fitzpatrick SF, Cheong A, Scholz CC, Simpson DA, Leonard MO, Tambuwala MM, Cummins EP, Taylor CT. MicroRNA-155 Promotes Resolution of Hypoxia-Inducible Factor 1α Activity during Prolonged Hypoxia. Molecular and Cellular Biology. 2011;31:4087–4096. doi: 10.1128/MCB.01276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang F, Zhang H, Mei Y, Wu M. Reciprocal Regulation of HIF-1α and LincRNA-p21 Modulates the Warburg Effect. Molecular Cell. 2014;53:88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 60.Sena Laura A, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman David A, Wang C-R, Schumacker Paul T, Licht Jonathan D, Perlman H, Bryce Paul J, Chandel Navdeep S. Mitochondria Are Required for Antigen-Specific T Cell Activation through Reactive Oxygen Species Signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor Cormac T. Mitochondria and cellular oxygen sensing in the HIF pathway. Biochemical Journal. 2008;409:19. doi: 10.1042/BJ20071249. [DOI] [PubMed] [Google Scholar]
- 62.Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proceedings of the National Academy of Sciences. 1998;95:11715–11720. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chandel NS, McClintock DS, Feliciano CE, Wood TM, Melendez JA, Rodriguez AM, Schumacker PT. Reactive Oxygen Species Generated at Mitochondrial Complex III Stabilize Hypoxia-inducible Factor-1α during Hypoxia A MECHANISM OF O2 SENSING. Journal of Biological Chemistry. 2000;275:25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- 64.Brunelle JK, Bell EL, Quesada NM, Vercauteren K, Tiranti V, Zeviani M, Scarpulla RC, Chandel NS. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metabolism. 2005;1:409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 65.Guzy RD, Hoyos B, Robin E, Chen H, Liu L, Mansfield KD, Simon MC, Hammerling U, Schumacker PT. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metabolism. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 66.Chua YL, Dufour E, Dassa EP, Rustin P, Jacobs HT, Taylor CT, Hagen T. Stabilization of Hypoxia-inducible Factor-1α Protein in Hypoxia Occurs Independently of Mitochondrial Reactive Oxygen Species Production. The Journal of Biological Chemistry. 2010;285:31277–31284. doi: 10.1074/jbc.M110.158485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, Green DR. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oestreich KJ, Read KA, Gilbertson SE, Hough KP, McDonald PW, Krishnamoorthy V, Weinmann AS. Bcl-6 directly represses the gene program of the glycolysis pathway. Nature Immunology. 2014;15:957–964. doi: 10.1038/ni.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cao Y, Rathmell JC, Macintyre AN. Metabolic Reprogramming towards Aerobic Glycolysis Correlates with Greater Proliferative Ability and Resistance to Metabolic Inhibition in CD8 versus CD4 T Cells. PLoS ONE. 2014;9:e104104. doi: 10.1371/journal.pone.0104104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ben-Shoshan J, Maysel-Auslender S, Mor A, Keren G, George J. Hypoxia controls CD4+CD25+ regulatory T-cell homeostasis via hypoxia-inducible factor-1α. European Journal of Immunology. 2008;38:2412–2418. doi: 10.1002/eji.200838318. [DOI] [PubMed] [Google Scholar]
- 71.Ikejiri A, Nagai S, Goda N, Kurebayashi Y, Osada-Oka M, Takubo K, Suda T, Koyasu S. Dynamic regulation of Th17 differentiation by oxygen concentrations. International Immunology. 2012;24:137–146. doi: 10.1093/intimm/dxr111. [DOI] [PubMed] [Google Scholar]
- 72.Darce J, Rudra D, Li L, Nishio J, Cipolletta D, Rudensky AY, Mathis D, Benoist C. An N-terminal mutation of the Foxp3 transcription factor alleviates arthritis but exacerbate diabetes. Immunity. 2012;36:731–741. doi: 10.1016/j.immuni.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Higashiyama M, Hokari R, Hozumi H, Kurihara C, Ueda T, Watanabe C, Tomita K, Nakamura M, Komoto S, Okada Y, Kawaguchi A, Nagao S, Suematsu M, Goda N, Miura S. HIF-1 in T cells ameliorated dextran sodium sulfate-induced murine colitis. Journal of Leukocyte Biology. 2012;91:901–909. doi: 10.1189/jlb.1011518. [DOI] [PubMed] [Google Scholar]
- 74.Bollinger T, Gies S, Naujoks J, Feldhoff L, Bollinger A, Solbach W, Rupp J. HIF-1α-and hypoxia-dependent immune responses in human CD4+CD25high T cells and T helper 17 cells. Journal of Leukocyte Biology. 2014;96:305–312. doi: 10.1189/jlb.3A0813-426RR. [DOI] [PubMed] [Google Scholar]
- 75.Hsiao H-W, Hsu T-S, Liu W-H, Hsieh W-C, Chou T-F, Wu Y-J, Jiang S-T, Lai M-Z. Deltex1 antagonizes HIF-1α and sustains the stability of regulatory T cells in vivo. Nature Communications. 2015;6 doi: 10.1038/ncomms7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Palazón A, Martínez-Forero I, Teijeira A, Morales-Kastresana A, Alfaro C, Sanmamed MF, Perez-Gracia JL, Peñuelas I, Hervás-Stubbs S, Rouzaut A, Landázuri MOd, Jure-Kunkel M, Aragonés J, Melero I. The HIF-1α Hypoxia Response in Tumor-Infiltrating T Lymphocytes Induces Functional CD137 (4-1BB) for Immunotherapy. Cancer Discovery. 2012;2:608–623. doi: 10.1158/2159-8290.CD-11-0314. [DOI] [PubMed] [Google Scholar]
- 77.Vezys V, Penaloza-MacMaster P, Barber DL, Ha S-J, Konieczny B, Freeman GJ, Mittler RS, Ahmed R. 4–1 BB Signaling Synergizes with Programmed Death Ligand 1 Blockade To Augment CD8 T Cell Responses during Chronic Viral Infection. The Journal of Immunology. 2011;187:1634–1642. doi: 10.4049/jimmunol.1100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Werth N, Beerlage C, Rosenberger C, Yazdi AS, Edelmann M, Amr A, Bernhardt W, von Eiff C, Becker K, Schäfer A, Peschel A, Kempf VAJ. Activation of Hypoxia Inducible Factor 1 Is a General Phenomenon in Infections with Human Pathogens. PLoS ONE. 2010;5:e11576. doi: 10.1371/journal.pone.0011576. [DOI] [PMC free article] [PubMed] [Google Scholar]