Abstract
Computer-aided diagnosis of histopathological images usually requires to examine all cells for accurate diagnosis. Traditional computational methods may have efficiency issues when performing cell-level analysis. In this paper, we propose a robust and scalable solution to enable such analysis in a real-time fashion. Specifically, a robust segmentation method is developed to delineate cells accurately using Gaussian-based hierarchical voting and repulsive balloon model. A large-scale image retrieval approach is also designed to examine and classify each cell of a testing image by comparing it with a massive database, e.g., half-million cells extracted from the training dataset. We evaluate this proposed framework on a challenging and important clinical use case, i.e., differentiation of two types of lung cancers (the adenocarcinoma and squamous carcinoma), using thousands of lung microscopic tissue images extracted from hundreds of patients. Our method has achieved promising accuracy and running time by searching among half-million cells.
Keywords: Histopathological image analysis, cell-level analysis, cell segmentation, large-scale, image retrieval, hashing
1. Introduction
Lung cancer is one of the most common cancers in the world (Siegel et al., 2013), and its diagnosis is an extremely important topic for personalized lung cancer treatment. There are four typical histologic types of lung cancers, including adenocarcinoma, squamous carcinoma, small cell carcinoma, and large cell carcinoma, each of which needs a different treatment Freeman, 2001). Therefore, the accurate staging of lung cancer can help clinicians in determining patient centered treatment, allow for reasonable prognostication, and facilitates comparisons between patient groups in clinical studies. Specifically, current investigations into early detection and adjuvant chemotherapy heavily rely on the proper staging of patients’ cancer type. Not only separating small cell carcinoma (SCC) from non-small cell carcinoma (NSCC) is important, it is also strongly recommended (Travis et al., 2011) to subtype NSCC into more specific types such as adenocarcinoma and squamous cell carcinoma, because 1) adenocarcinomas can be tested for epidermal growth factor receptor (EGFR) mutations as a predictor of response to EGFR tyrosine kinase inhibitors; 2) adenocarcinoma response to pemetrexed therapy is better than squamous; 3) potential life-threatening hemorrhage might occur in patients who have squamous cell carcinoma but misclassified and are given bevacizumab. Bronchial biopsy is one of the most effective diagnosis methods to differentiate them, with the aid of Computer Aided Diagnosis (CAD) systems (Kayser et al., 2002; Thunnissen et al., 1992; Mijović et al., 2008). However, most previous methods have emphasized on the diagnosis of small cell vs. non-small cell (i.e., adenocarcinoma, squamous carcinoma, and large cell carcinoma) types of lung cancers. Few efforts have been put on the differentiation of the adenocarcinoma and squamous carcinoma, both of which belong to NSCC, although this task is clinically significant as their management protocols are different (Edwards et al., 2000).
The main challenge of this task is the need of analyzing all individual cells for accurate diagnosis, since the difference between the adenocarcinoma and squamous carcinoma highly depends on the cell-level information, such as its morphology, shape and appearance. In fact, there are a lot of cellular features used by pathologists to differentiate adenocarcinoma from squamous cell carcinoma. Currently, all of them are estimated in a subjective way without rigorous quantifications. These include, but not limited to: 1) Nucleoli are often more prominent and obvious in adenocarcinoma tumor cells than squamous cell carcinoma; 2) The individual cell borders tend to be sharper in squamous cell carcinoma than Adenocarcinoma; 3) Only squamous cell carcinoma contains intercellular bridges; 4) adenocarcinoma has relatively lower nuclear/cytoplasmic ratios and delicate, vacuolated cytoplasm compared with squamous cell carcinoma. Therefore, rigorously measuring and analyzing each individual cell is important and can assist pathologists for accurate diagnosis. However, a region-of-interest (ROI) image may contain hundreds or thousands of cells. Analyzing each cell can be computationally inefficient using traditional methods. As a result, most previous methods encode the whole image as holistic features by representing the statistics of cell-level information (e.g., architecture features (Doyle et al., 2008) or frequency of local textures (Zhang et al., 2015a)), and may compress high-dimensional features to improve the computational efficiency. Despite the compactness and hence the efficiency, information loss is inevitable in such holistic representation. Therefore, efficiently analyzing each cell is important to investigate. In addition, all the aforementioned cellular features and analysis can only be measured after we complete the accurate cell-level segmentation.
In this paper, we design an automatic framework for the large-scale cell-level analysis of histopathological images, which can examine millions of cells in real-time (preliminary results have been reported in (Zhang et al., 2015b)). Our solution includes two important modules, robust cell segmentation and large-scale cell retrieval. Specifically, segmentation module provides automatic and robust delineation and measurement of cells, enabling effective feature extraction for each cell. The large-scale image retrieval framework can locate similar instances among massive databases of cells, by improving the efficient hashing methods (Datar et al., 2004; Kulis and Grauman, 2009). Given a new image to be diagnosed, our system automatically segments all cells and efficiently discovers the most relevant cells by comparing them with the training database (e.g., millions of cells extracted from thousands of images). The diagnosis is decided by classifying each cell and using the majority logic. We conduct extensive experiments to differentiate lung cancers, i.e., adenocarcinoma and squamous carcinoma, using a large dataset containing thousands of lung microscopic tissue images acquired from hundreds of patients. Our proposed framework achieves 87.3% accuracy in real-time, by searching a massive database of half million cells extracted from this dataset.
The major contribution of this paper is twofold. 1) A comprehensive and real-time framework is designed to analyze histopathological logical images by examining all cells. This framework opens a new avenue for investigating large-scale databases, and is particularly suitable for this challenging use case. 2) In terms of technical contribution, we propose a carefully designed learning method that assigns probabilistic-based importance to different hash values or entries. This scheme alleviates several intrinsic problems of using traditional hashing methods for classification, and significantly improves the accuracy. Furthermore, we also improve the cell segmentation algorithms by handling variations in shape and cell size, which provide robust and accurate delineations of cells.
The rest of the paper is organized as follows. Section 2 reviews relevant work of cell segmentation and content-based image retrieval. Section 3 presents our framework for realtime cell mining. Section 4 shows the experimental results on lung microscopic tissue images. Concluding remarks are given in Section 5.
2. Related Work
2.1. Cell Segmentation
Various approaches of segmentation in pathological image have been investigated. In (Al-Lahham et al., 2012), K-means clustering is used to segment out the cancer cell nuclei at pixel level in a transformed color space. In (Loukas et al., 2003), PCA is applied to learn a color space transform and the cell nuclei are segmented out by globally thresholding the transformed image. In (Markiewicz et al., 2008, 2009), support vector machine (SVM) classifiers are trained to segment background and the cells based on color or morphological features. Because the above approaches mainly rely on color, they do not work well when there exist non-negligible amount of touching cells present in the images.
Watershed transformation and its variants for splitting touching objects have been widely studied (Vincent and Soille, 1991). A RGB color-based segmentation followed by the watershed algorithm is proposed to tackle the touching cells in (Grala et al., 2009), and a 3D watershed algorithm incorporating gradient information and geometric distance of nuclei is represented in (Lin et al., 2003). In order to handle over-segmentation, marker-controlled watershed are investigated in (Grau et al., 2004; Schmitt and Hasse, 2008). In particular, Jung et al. (Jung and Kim, 2010) developed an H-minima transform based marker-controlled watershed algorithm for clustered nucleus segmentation on histopathological images, and an adaptive H-minima transform is reported in (Cheng and Rajapakse, 2009) to generate markers for the watershed algorithm. H-minima transform is relatively robust to noise, but it usually requires a careful choice of the h value. Learning based approaches are also exploited to detect markers for watershed algorithms. Mao et al. (Mao et al., 2006) applied a supervised marker detection based watershed to cell segmentation on bladder inverted papilloma images, where the markers are located by using a classifier with a combination of photometric and shape information. In (Akakin et al., 2012), an SVM classifier is used to automatically detect markers for the watershed algorithm. Compared with unsupervised learning, the supervised marker detection algorithms might provide better performance, but they need sophisticated feature design, which is very challenging due to the complex characteristics of digital pathology images.
Graph-based segmentation methods (Kolmogorov and Zabih, 2004; Boykov and Funka-Lea, 2006) can also be used to automatically segment cells. The nodes of the graph represent pixels or superpixels and each edge corresponds to one pair of neighboring nodes. Image segmentation is achieved by partitioning the graph into several components. Lucchi et al. (Lucchi et al., 2010) exploited a mincut-maxflow algorithm to partition the superpixel based graph, Bernardis and Yu (Bernardis and Yu, 2010) segmented out individual cells based on the normalized cuts (Shi and Malik, 2000), and Zhang et al. (Zhang et al., 2014a) employed a correlation clustering method to achieve superpixel graph partition. Some other graph based methods can be found in (Al-Kofahi et al., 2010; Nath et al., 2006; Faustino et al., 2009; Chen et al., 2008; Wu et al., 2012; Yu et al., 2010; Janowczyk et al., 2012; Lou et al., 2012). Although effcient graph-based segmentation algorithm (Felzenszwalb and Huttenlocher, 2004) is proposed, generally graph partition methods exhibit high time cost, which limits their applications in real cell segmentation.
Deformable models are another popular type of cell seg mentation algorithms in biomedical image analysis. A multireference level set algorithm is used for nucleus segmentation in (Chang et al., 2012), a dynamic watershed scheme is introduced to the level set model with topology dependence for cell segmentation in (Yu et al., 2009), and several repulsive level set approaches are reported in (Yan et al., 2008; Ali et al., 2011; Ali and Madabhushi, 2012; Qi et al., 2012). Xu et al. (Xu et al., 2007) formulated the active contour model into a graph cut framework, which deforms the contour towards a global minimum within the contour neighborhood. In general, these methods are suitable can naturally handle topology changes, but they might create undesired contours with inhomogeneous regions. Therefore, the parametric active contour models are an alternative approach. Li et al. (Li et al., 2007) applied a gradient flow tracking to 3D nuclei segmentation algorithm, and Cai et al. (Cai et al., 2006) developed a repulsive active contour model based on gradient vector flow (GVF) (Xu and Prince, 1998) to segment neuronal axons. However, GVF snake requires clean edge maps to calculate the gradient vector flow, and this might suffer from background clutter in histopathological images.
There exist other types of state-of-the-arts for automatic cell segmentation. Kong et al. (Kong et al., 2011) first separated cellular regions from the background with a supervised pixel-wise classification, and then split touching cells based concave point and radial symmetry. Ozolek et al. (Ozolek et al., 2014) built a statistic model with a set of training nuclei and thereafter performed template matching to segment out individual nuclei. This method can handle touching cases by selecting the best matched model parameters. Another learning based nucleus segmentation is presented in (Kårsnäs et al., 2011), where intensity and label dictionaries are constructed to separate the foreground from the background and then touching nuclei are split by combining region merging with a marker-controlled watershed. Probabilistic models have also attracted research interests. Park et al. (Park et al., 2013) exploited a Gaussian mixture model based on B-splines to achieve cell segmentation, and a generic segmentation framework for pathologic images that employs an EM algorithm with Markov prior is reported in (Monaco et al., 2012). The learning based methods usually require a large number of training data and assume that the training data are sufficient to capture the variations on new testing samples, and the probabilistic models need to be carefully selected such that the used generative models are strong enough to model testing data.
2.2. Content-Based Image Retrieval
Content-Based Image Retrieval (CBIR) is an effective approach in analyzing medical images. It supports doctors for making clinical decisions by retrieving and visualizing relevant medical images with diagnosis information. To this end, many systems and methods have been developed. For examples, Comaniciu et al. (Comaniciu et al., 1999) designed a CBIR system to support decision making in clinical pathology. In this system, fast color segmenter is used to extract cell features including shape, area, and texture of the nucleus. Its performance was compared with that of a human expert on a database containing 261 digitized specimens. The experimental results demon strated that this system could be used to assist pathologists to improve the analysis. Dy et al. (Dy et al., 2003) described a new hierarchical approach of CBIR based on multiple feature sets and a two-step approach. The query image is classified into different classes with best discriminative features between the classes, and similar images are searched in the predicted class with the features customized to distinguish subclasses. El-Naqa et al. (El-Naqa et al., 2004) proposed a hierarchical learning approach consisting of a cascade of a binary classifier and a regression module to optimize retrieval effectiveness and efficiency. Greenspan et al. (Greenspan and Pinhas, 2007) proposed a CBIR system that consists of a continuous and probabilistic image-representation scheme. It uses Gaussian mixture models (GMM) and information-theoretic image matching via the Kullback-Leibler (KL) measure to match and categorize X-ray images by body regions. Song et al. (Song et al., 2011) designed a hierarchical spatial matching-based image retrieval method using spatial pyramid matching to extract and represent the spatial context of pathological tissues effectively. Recently, Foran et al. (Foran et al., 2011) designed a CBIR system named ImageMiner for comparative analysis of tissue microar-rays by harnessing the benefits of high-performance computing and grid technology.
One of the main limitations of these systems is the scalability. To analyze large-scale datasets, one needs to design efficient CBIR methods. With the goal of comparing CBIR methods on a larger scale, ImageCLEF and VISCERAL provide benchmarks for medical image retrieval tasks (Müller et al., 2005; Langs et al., 2013; Hanbury et al., 2013). In our use case, it is necessary to retrieve among half-million instances in realtime to conduct cell-level analysis in histopathological images. To this end, hashing-based methods have been investigated, which enable fast approximated nearest neighbors (ANN) search to deal with the scalability issue. For examples, the locality sensitive hashing (LSH) (Andoni and Indyk, 2006) uses random projections to map data to binary codes, resulting in highly compact binary codes and enabling efficient comparison within a large database using the Hamming distance. Anchor Graph Hashing (AGH) (Liu et al., 2011) has been proposed to use neighborhood graphs which reveal the underlying manifold of features, leading to a high search accuracy. (Shen et al., 2013) also proposed to leverage manifold information for inductive hashing. Recent work has focused on data-driven hash functions, such as the semi-supervised hashing (SSH) (Wang et al., 2012) incorporating the pairwise semantic similarity and dissimilarity constraints from labeled data. Particularly, supervised hashing methods (Liu et al., 2012; Shen et al., 2015) have also been proposed to leverage annotations into hash function learning. These hashing methods have been employed to solve the dimensionality problem in medical image analysis (Zhang et al., 2015a, 2014b). Specifically, high dimensional features are compressed into 48 bits that are exhaustively compared among thousands of images. However, such high dimensional features only approximately represent cell-level information. It is desired to analyze all cells in our use case, while traditional hashing methods fail to provide accurate results as shown in our experiments.
3. Methodology
3.1. Overview
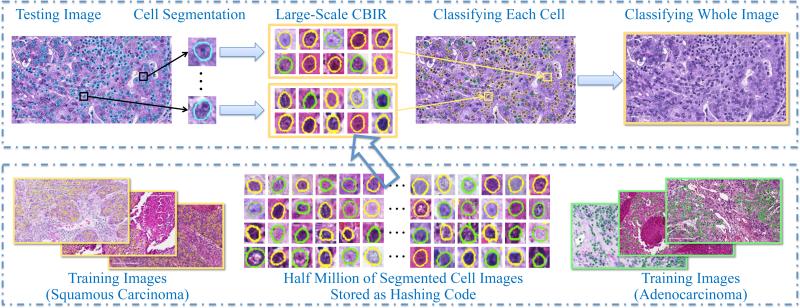
Fig. 1 shows the overview of our proposed framework, which includes offline learning and online classification. During offline learning, our system automatically detects and segments all cells from thousands of images, resulting in half million of cell images. Regarding cell detection and segmentation, we propose to improve the single-pass voting (SPV) scheme (Qi et al., 2012; Xing et al., 2014). Our improvement focuses on handling variations in shape and cell size. After that, texture and appearance features are extracted from these cell images and are compressed as binary codes, i.e., tens of bits. These compressed features are stored in hash table for constant-time access even among millions of images.
Figure 1.
Overview of our proposed framework, based on robust cell segmentation and large-scale cell image retrieval. The top row is the online classification, and the bottom row is the offline learning. Yellow boundaries mean squamous carcinoma, green means adenocarcinoma, and blue means unknown types to be classified.
During online classification, our system segments all cells from a testing image, and same types of features are extracted accordingly and compressed using hashing methods. Then, we perform large-scale cell image retrieval for each segmented cell to classify its category. Finally, the classification result of the testing image is decided by the majority logic, i.e., voting from all cells’ classification. Using this scheme, our system can maximally utilize the cell-level information without sacrificing the computational efficiency, owing to the large-scale retrieval via hashing methods. We also design a content-aware weighting scheme to improve the accuracy of traditional hashing methods, based on the observations and priors in histopathological image analysis. In the following sections, we introduce the details of robust cell segmentation, large-scale cell image retrieval, and weighting techniques.
3.2. Robust Cell Segmentation
Accurately delineating cells is critical to the cell-level analysis of histopathological images. It includes cell detection and segmentation. Our detection algorithm is an improved version of single-pass voting (SPV) proposed by Qi et al. (Qi et al., 2012). The improvement focuses on handling variations in shape and cell size. The newly introduced 1) region-based hierarchical voting in a distance transform map handles the shape variation, and 2) Gaussian pyramid based voting suppresses the effect of the scale variation. For an image, a Gaussian pyramid is created. At layer l, an SPV is applied with the distance transform being weighted by a Gaussian kernel. Unlike SPV within which each pixel in the voting area receives uniform vote, this weighted voting enables the pixels that locate more inside the cell to receive more votes. Therefore, this mechanism encourages higher voting scores in the central region of the cells. The final vote value is calculated by summing up all the layers:
| (1) |
where S denotes the set of all voting pixels, Al(m, n) denotes the voting area of pixel (m, n) at layer l and it is defined by a radial range (rmin, rmax) and angular range Δ (Qi et al., 2012). I[·] is an indicator function, and Cl(x, y) represents the distance trans-formation map at layer l. In our experiment, we use Euclidean distance. The g(m, n, μx, μy, σ) is an isotropic Gaussian kernel for pixel (m, n) with mean (μx, μy) = (m+(rmax+rmin)cosθ/2, n+(rmax + rmin)sinθ/2) and scalar σ, where θ represents the angle of the gradient direction with respect to the x axis.
Our segmentation method is based on the active contour (Cohen, 1991) with a newly introduced repulsive term. The repulsive term is used to prevent the evolving contours from crossing and merging with each other. Based on the detection result, a circle is associated with each detected cell as initial contour. The i-th contour vi(s) deforms until it achieves a balance between internal force Fint(vi) and external force Fext(vi) with
| (2) |
| (3) |
| (4) |
where s indexes the points on the contour, and (s) and (s), with their weights α and β, are the second and fourth derivative of vi(s), respectively. ni(s) with its weight γ denotes the internal pressure force and ∇Eext(vi(s)) denotes the edges in the image (∇Eext(vi(s))) = −∇∥T[x(s), y(s)]∥2, T[x(s), y(s)] represents the image). The last term in (4) represents the repulsive force. N is the number of the cells, and dij denotes the Euclidean distance between the points of different contours. λand ω are the weights controlling the edge driven force and repulsive force, respectively. Given initial contours, the contours iteratively deform towards cell boundaries and the cell segmentation is achieved when Eq. 2 is satisfied or the maximum number of iteration is reached.
This method can robustly detect and segment cells from histopathological images, which are used for the cell-level analysis in the next stage. The active contour in Eqs. 2-4 is a parametric model with explicit contour representation, which is different from the level set algorithms (Yan et al., 2008; Ali and Madabhushi, 2012; Qi et al., 2012), which implicitly represent contours. Therefore, given initial contours (based on detection results), our model can take advantage of known topology constraint such that it can prevent contours from splitting or merging; on the other hand, the level set method (Qi et al., 2012) as well as the graph cut based active contour (Xu et al., 2007) allow topology changes such that it might generate undesired small holes inside or outside cells due to intensity heterogeneity, as shown in the experimental section. In addition, our model uses a contour-based repulsive force instead of a region-based term, which is used in (Qi et al., 2012). The in the repulsive term demonstrates that the closer the j-th contour moves to the i-th contour, the more repulsion each contour receives. In this case, the model can effectively handle touching cells by preventing contours from crossing each other.
3.3. Classification via Large-Scale Cell Image Retrieval
Once all cells are segmented from a testing image, our system conducts cell-level classification by exhaustively comparing each cell with all cells in the training database, using hashing-based large-scale image retrieval and majority voting. Hashing has been widely used to compress (high-dimensional) features into binary codes with merely tens of bits (Datar et al., 2004). Therefore, such short binary features allow mapping into a hash table for constant-time retrieval. To improve the accuracy of previous hashing methods, the kernelized scheme (Kulis and Grauman, 2009) is incorporated to handle practical data that is mostly linearly inseparable, which is a common phenomenon of medical images:
| (5) |
where n is the number of training samples, h is the kernelized hashing method mapping a kernel function f(x) with kernel κ to 0 or 1 by taking its sign value, x1, x2, x3, ..., xm are the m random samples selected from the data and aj is the coefficient determining hash functions. The resulting binary codes can be used for indexing and differentiating different categories. Although kernelized scheme well solves the linear inseparability problem of features, it is still not able to provide accurate retrieval or classification, due to the high intra-class variation of histopatho-logical images. Therefore, supervised information (Liu et al., 2012) is also leveraged to design discriminative hash functions that are particularly suitable for analyzing histopathological images:
| (6) |
where r is the number hash bits, S is a matrix encoding the supervised information (e.g., 1 for same category and −1 for different categories) and A is the model parameter to compute hashing code, and is the matrix form of the kernel function, in which is a kernelized vectorial map . The optimization of Q is based on Spectral Relaxation (Weiss et al., 2008) for convexification, which is used as a warm start, and Sigmoid Smoothing for applying standard gradient descent technique, which is used for accurate hashing.
Indexing these compressed features in a hash table, our method can perform image retrieval in constant-time among millions of cells without significantly sacrificing the accuracy. The category of each cell can be decided straightforwardly with the majority logic of retrieved cells, and the whole image is hence classified by accumulating results of its all cells.
3.4. Hashing with Content-Aware Weighting
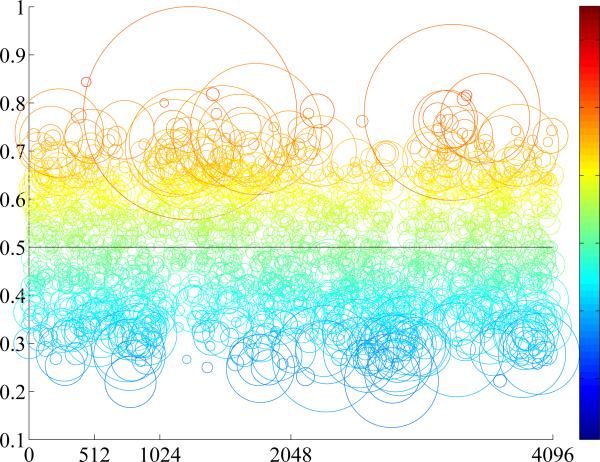
Theoretically, using hashing methods by indexing in a hash table enables constant-time searching, no matter how many training samples are used. However, it also requires that the length of the binary code is sufficiently short, to store in physical memory for fast access. Given limited number of hash bits, an inevitable limitation is that a large number of images may be mapped into the same hash value. In other words, it may result in an unordered set for the same hash value, where exact or near-exact matches may be obscured within a large-scale database due to noisy features, similar instances, or erroneous segmentations. This is particularly true for histopathological image analysis, since the differences of cells are very subtle, and accurate segmentation for all cells is challenging. Consequently, the accuracy of cell classification is adversely affected when choosing the majority of cells mapped into a hash value, and the accuracy of whole image classification is also reduced. Fig. 2 illustrates this inherent limitation of hashing methods in analyzing histopathological images. Half million of cells are mapped into 12 bits, which mean 212 = 4096 hash values. The entries (i.e., hash values) in each hash table are illustrated according to the distribution of cells mapped into them, such as the ratio between two categories (i.e., adenocarcinoma and squamous carcinoma) and the number of cells mapped into that entry. Ideally, each hash value should be discriminative enough, i.e., the number of one type should dominate the other. However, many of them actually contains similar amount of both types of cells, i.e., around 0.5 ratio. In other words, the indecisive hash values are usually around the 0.5 ratio, indicating equal opportunity for either category. Classification based on such hash value is likely inaccurate. The small circles in Fig. 2 are also not reliable, since only few cells are mapped there, which can be easily affected by the image noise or erroneous segmentation. A potential solution is to identify reliable hash values and omit indecisive one, by heuristically select or prune them via feature selection. However, this may involve tuning parameters and lack the consistent measures. Furthermore, there is no guarantee that the hash values from feature selection algorithms are sufficiently discriminative for classification.
Figure 2.
Illustration of the cell distribution in a hash table. X-axis means the hash value using 12 bits, ranging from 0 to 4095, and y-axis means the ratio between two types of cells, ranging from 0 to 1. Each circle means a set of cells mapped to the hash value located in the centroid, its size means the number of cells, and the color map visualizes the ratio of two types of cells, same as the y-axis values.
Therefore, we introduce a probabilistic-based formulation to solve these problems in a principled way, i.e., design a content-aware weighting scheme to re-weight the importance of hash values. Specifically, we aim to assign probability scores to each hash value, based on its ability to differentiate different categories. Such “soft assignment” upon hash values can significantly boost the classification accuracy using hashing-based retrieval. In our framework, kernelized and supervised hashing (KSH) (Liu et al., 2012) is employed as the baseline method to generate initial hash values, because of its efficacy and success in histopathological image analysis (Zhang et al., 2015a). The content-aware weighting scheme can significantly enhance the differentiation ability of hash values generated by this baseline. Intuitively, since cells in certain hash values are not accurate for classification, their weights should be diminished during the process. On the other hand, discriminative hash values should be emphasized, e.g., circles nearby 1 or 0 ratios. In addition, small sizes of circles are not preferred and their weights should be reduced, as they can be easily affected by many factors such as unusual staining color, inaccurate segmentation results and image noise in our use case. Therefore, we designed two metrics to emphasize discriminative hash entries, with generalized notations for multi-class classification:
- Support: Given a specific hash value H, the number of cells mapped into H should be considered. This indicates that such amount of cells are used for the classification of this hash value, each with contribution 1, while all remaining cells are irrelevant, i.e., contribution 0. Therefore, we name this metric as “support”, which is conventionally referred to the set of numbers having non-zero values. Denote SH = {cell : h(cell) = H} as the set of cells mapping into a specific hash value H, where h(cell) is the hash value of the cell. The support WH of the hash value H is defined as:
where |S| is the number of element in set S and r is the number of hash bits, representing 2r hash values.(7) - Certainty: Instead of assigning a certain category label to each hash value, we should consider the confidence of such categorization and assign a probabilistic label to each hash value. Therefore, this “certainty” term defines the probability of a cell belonging to the ith category when its hash value is H:
where l(cell) is the label of a cell image and Li means the ith label or category.(8)
We combine these two weights to advocate the importance of highly discriminative hash values with sufficient support. Specifically, during the training process, WH and P(Li|H) can be computed for all hash values. The category of a whole testing image is decided by:
| (9) |
where Hcell is the hash value of the cell belonging to the query (testing) image.
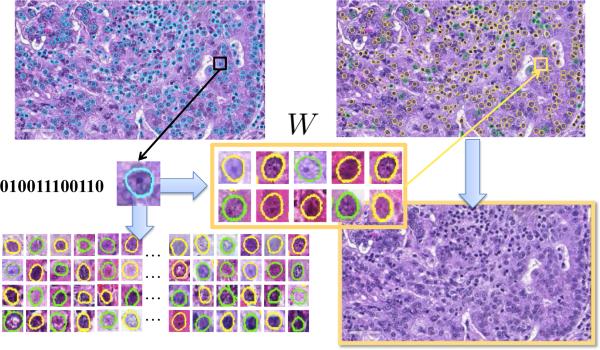
This content-aware weighting scheme effectively solves the issues of using hashing-based retrieval methods for classification. The importance of each cell is decided case-specifically, and accumulating the results of all cells provide accurate classification for the whole image. In addition, this framework is able to accommodate new samples efficiently. The updating scheme can be achieved by storing not only the weights but also the number of cells in each category. Given new samples, we can update the cell number in their mapped hash entries, re-calculate and update the weights based on such information. Regarding the computational complexity, the overhead during the testing stage lies in the weighted combination, which is negligible as demonstrated in the experiments. Therefore, this process is computationally efficient, same as traditional hashing methods. Fig. 3 summarizes the classification procedure using weighted hashing. The whole framework includes cell segmentation, hashing, and retrieval. The probability scores are assigned to each hash entry, and they are aggregated within the whole image for the final classification. Benefited from this thorough analysis of each individual cell, this framework can achieve promising accuracy without sacrificing the efficiency.
Figure 3.
Workflow of the weighted hashing-based classification. Starting from an unknown image to be categorized, each segmented cell is classified by searching the most similar instances. Their results are combined via the content-aware weighting scheme, predicting the categorization for the whole image.
4. Experiments
4.1. Data Description
In this section, we conduct extensive experiments to evaluate our weighted hashing with multiple features for cell-level analysis. Our dataset is collected from the Cancer Genome Atlas (TCGA) National Cancer Institute (2013), including 57 adenocarcinoma and 55 squamous carcinoma. 10 patches with 1712×952 resolution, i.e., region-of-interests (ROIs), are cropped from each whole slide scanned pathology specimens, by consulting with certified pathologists. Generally, the ROIs mainly consist of cancer cells. The lymphocytes regions which have different visual patterns than the representative tumor regions are avoided. All the data have been prepared and labeled based on the independent confirmation of the pathologists. In each image, our algorithm detects and segments around 430 cells. In total, 484,136 cells are used to evaluate the segmentation accuracy (195,467 adenocarcinoma cells and 288,669 squamous carcinoma cells). We evaluate the efficacy of our proposed framework in terms of the classification accuracy and computational efficiency. The evaluations are conducted on a 3.40GHz CPU with 4 cores and 16G RAM, in MATLAB and C++ implementation. We empirically set the parameters for cell detection and segmentation algorithms as: σ = 2, δ = 30, rmin = d/8, rmax = 7d/8 (d is the estimated average diameter of all cells in the image) and α = 4.2,β = 0, γ = 0.7, λ = 1.5, ω = 0.5, respectively.
4.2. Evaluation of Cell Segmentation
We demonstrate the performance of the cell detection by comparing it with single-pass voting (SPV) and phase-coded Hough transform (PCHT) (Xie and Ji, 2002). We compute the mean, variance and minimum of the deviation of the detected seeds with respect to their ground truth seeds. Note that only the detected seeds within a 8-pixel circle of its ground truth seed are considered. To evaluate the performance more comprehensively, we define a set of metrics including missing rate (MR), over-detection rate (OR), precision, recall and F1 score. A positive detection is asserted if a detected seed locates within the 8-pixel circle around a ground truth seed, a miss is asserted, otherwise. Over detection is considered as more than one seed are detected in the 12-pixel circle of a ground truth seed. OR is the ratio of the number of such cases over the number of the ground truth seeds. Precision (Prec), recall (Rec) and F1 score are defined as follows: , and , where TP, FP, and FN represent true positive, false positive and false negative, respectively. Note that in our experiment, false positive is defined as the case that a seed is detected out of the 8-pixel circle of a ground truth seed yet within its 12-pixel circle. The performance measurements are shown in Table 1.
Table 1.
Comparative Performance Evaluation of the Detection Accuracy. SPV stands for single-pass voting, and PCHT stands for phase-coded Hough transform. MR stands for the missing rate, and OR stands for the over-detection rate.
| Mean | Variance | Min | MR | OR | |
|---|---|---|---|---|---|
| PCHT | 3.7 | 3.92 | 0.16 | 0.46 | 0.11 |
| SPV | 2.9 | 3.01 | 0.28 | 0.21 | 0.06 |
| Ours | 2.7 | 2.8 | 0.13 | 0.16 | 0.08 |
| FP | TP | Prec | Rec | F 1 | |
| PCHT | 0 | 0.53 | 0.995 | 0.53 | 0.69 |
| SPV | 0.002 | 0.78 | 0.996 | 0.74 | 0.84 |
| Ours | 0.002 | 0.83 | 0.997 | 0.84 | 0.90 |
The performance of the segmentation algorithm is evaluated through comparing our method with four existing methods (mean shift (MS), isoperimetric (ISO) (Grady and Schwartz, 2006), graph-cut and coloring (GCC) (Al-Kofahi et al., 2010), and repulsive level set (RLS) (Qi et al., 2012)), both qualitatively and quantitatively. The segmentation results of a randomly selected patch are shown in Fig. 4. In our quantitative analysis, we define precision and recall where seg represents the segmentation result and gt represents the ground truth. We show the mean, variance and 80 % in Table 2. MS and ISO are general segmentation algorithms which need further postprocessing to achieve satisfied performance, and GCC suffers from over-segmentation. RLS generates undesired small holes inside or outside cells due to topology changes, while the proposed approach address this problem by taking advantage of known topology such that it produces the best segmentation results.
Table 2.
Comparative performance evaluation of the segmentation accuracy for mean shift (MS), ISO (Grady and Schwartz, 2006), GCC (Al-Kofahi et al., 2010) and RLS (Qi et al., 2012). PM and RM represent precision mean and recall mean. PV and RV denote variances of precision and recall. P80% and R80% denote the sorted highest precision and recall, respectively.
| PM | PV | P80% | RM | RV | R80% | |
|---|---|---|---|---|---|---|
| MS | 0.73 | 0.08 | 0.92 | 0.79 | 0.03 | 0.89 |
| ISO | 0.72 | 0.09 | 0.96 | 0.81 | 0.02 | 0.92 |
| GCC | 0.80 | 0.05 | 0.95 | 0.77 | 0.02 | 0.89 |
| RLS | 0.84 | 0.02 | 0.96 | 0.85 | 0.01 | 0.92 |
| Ours | 0.87 | 0.01 | 0.95 | 0.95 | 0.01 | 0.96 |
4.3. Evaluation of Image Classification
In our framework, the image classification (i.e., differentiation of adenocarcinoma and squamous carcinoma) is conducted by examining all cells using hashing-based large-scale image retrieval with content-aware weighting. We compare our hashing-based classification scheme with several effective classifiers employed for histopathological image analysis. Following the convention, k-nearest neighbor (kNN) method is used as the baseline of analyzing histopathological images (Tabesh et al., 2007), owing to its simplicity and efficacy. Dimensionality reduction methods such as principal component analysis (PCA) are effective approaches to improve the computational efficiency and have been employed to analyze histopathological images using high-dimensional features (Sertel et al., 2009). Support Vector Machine (SVM) is a supervised classification method and widely used in grading systems for breast and prostate cancer diagnosis (Doyle et al., 2008). We also compare with the traditional kernelized and supervised hashing (KSH) (Liu et al., 2012). For fair comparison, same features are used for all compared methods, and their parameters and kernel selections are optimized by cross-validation. Specifically, we use an RBF kernel with optimized gamma value for SVM, and k=9 for kNN. Regarding dimensionality reduction, PCA compresses the original features (i.e., 144 dimensional texture feature base on Histogram of Oriented Gradients (Dalal and Triggs, 2005)) into 12 floats, and our hashing method generates 12 bits from each original feature.
To conduct the comparison, we randomly select 20% patients as testing data (around 230 images, or 96, 000 cells), and use the images from remaining patients as training. This procedure is repeated for 30 times to obtain the mean and standard deviation. Table 3 shows the quantitative results of the classification accuracy. Despite the efficacy of kNN in many applications, it fails to produce reasonable results in this challenging problem, due to the large variance of cell images, noise in such large-scale database and unbalanced number of two classes. PCA reduces the feature dimensions, which could be redundancy information or noise. The classification accuracy is significantly improved, while still only around 70%. SVM incorporates supervised information, i.e., labels of adenocarcinoma and squamous carcinoma. Not surprisingly, it largely outperforms unsupervised methods, with an accuracy of 81.6%. KSH has the same merit of using supervised information, and hence achieves comparable accuracy as SVM. Our proposed hashing method not only utilizes kernels and supervision, but also is equipped with the content-award weighting scheme to solve the inherent problems of hashing methods. Therefore, it outperforms all other methods, with an accuracy of 87.3%. In addition, the standard deviation of our algorithm is also relatively small, indicating the stableness of our algorithm. Table 3 also shows the individual accuracy of adenocarcinoma and squamous carcinoma. Besides the superior accuracy, our method also achieves the most balanced results for both cases, which is important to this clinical problem as both cases should be recognized and sacrificing the accuracy of one case is not acceptable.
Table 3.
Quantitative comparisons of the classification accuracy (the mean value and standard deviation) and running time. Compared methods include kNN (Tabesh et al., 2007), PCA (Sertel et al., 2009), SVM (Doyle et al., 2008), KSH (Liu et al., 2012) and ours.
| Adeno | Squam | Average | Time(s) | |
|---|---|---|---|---|
| kNN | 0.309 ± 0.058 | 0.710 ± 0.072 | 0.514 | 2605.80 |
| PCA | 0.458 ± 0.084 | 0.954 ± 0.057 | 0.711 | 460.20 |
| SVM | 0.929 ± 0.085 | 0.704 ± 0.092 | 0.816 | 46.82 |
| KSH | 0.861 ± 0.076 | 0.763 ± 0.084 | 0.812 | 1.22 |
| Ours | 0.887 ± 0.069 | 0.854 ± 0.062 | 0.873 | 1.68 |
Table 3 also compares the computational efficiency of these methods, i.e., the testing time for classification. Our hashing method compresses each feature into merely 12 bits, resulting in a hash table with 4096 values, which allow instant access to images mapped into any hash value. Therefore, KSH and our method is real-time, i.e., around 1-2 seconds. Our method uses content-aware weighting and is slightly slower than KSH, due to a small overhead for computing the weighted average. Such computational overhead (i.e., 0.4s) is negligible in practice. Other methods are all significantly slower, ranging from 46 to 2600 seconds. This is the main factor preventing previous methods from being used for cell-level analysis. Note that the detection and segmentation takes around tens of seconds for each image, and feature extraction takes half second, both of which are the same for all compared methods. The overall speed is quite efficient for practical use.
4.4. Discussions
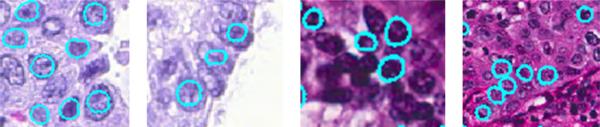
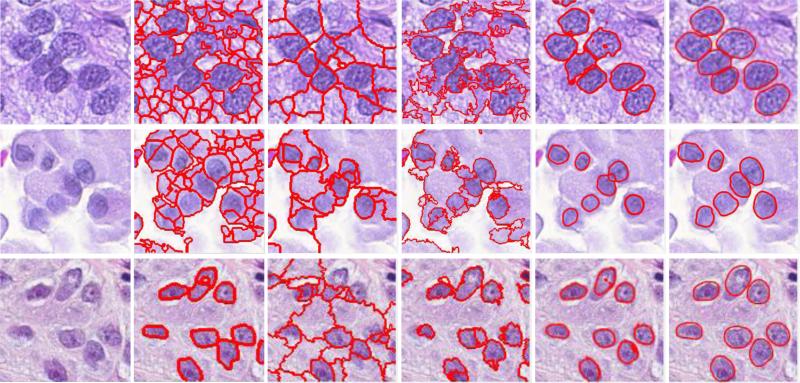
In this section, we discuss the parameters, implementation issues and some limitations of our system, and their potential solutions. Fig. 5 shows several failure cases of our cell segmentation algorithm. The first two cases have under-segmentation problem. This issue is caused by the following reasons: 1) weak boundaries of cell images, 2) and the significantly strong edges within the cells that can mislead the evolution process of the active contours. Note that although our algorithm fails to accurately delineate the cell boundaries, the results still implicitly preserve structure of the cell images. A possible improvement is to incorporate the output of a learning based edge detector into Eq. 4. The other two cases in Fig. 5 fail to detect several cells. This is possibly caused by the largely overlapped cells and/or high similarity with the background, which may introduce uncertainty for cell detection. Note that these are challenging cases to segment. For those densely clustered cells with missing cell boundaries, the cell detection and segmentation algorithms may fail in some cases. Particularly, the current model can effectively handle touching cells, but not largely occluded or overlapped cells. One potential solution to tackle the occlusion is to incorporate shape prior modeling. In fact, our segmentation framework can accurately segment the majority of images, demonstrated in Table 2.
Figure 5.
Some failure cases of our cell segmentation algorithms, including under-segmentation and misdetection.
Since the image classification relies on the features extracted from the segmented cells, inaccurate segmentation may adversely affect the classification accuracy. Nonetheless, our system still generates accurate classification results, because of two reasons: 1) Most segmented cells are correct, which is reflected by the high precision and recall. 2) More importantly, the weighting scheme reduces the importance of unreliable features, most of which are extracted from inaccurate segmentations. Particularly, this weighting scheme ensures the robustness of the classification module, making it less sensitive to the segmentation precision. Therefore, our content-aware hashing method not only benefits the classification accuracy, but also is compatible with the paradigm of cell-level analysis, given the fact that most existing cell segmentation methods are still not perfect.
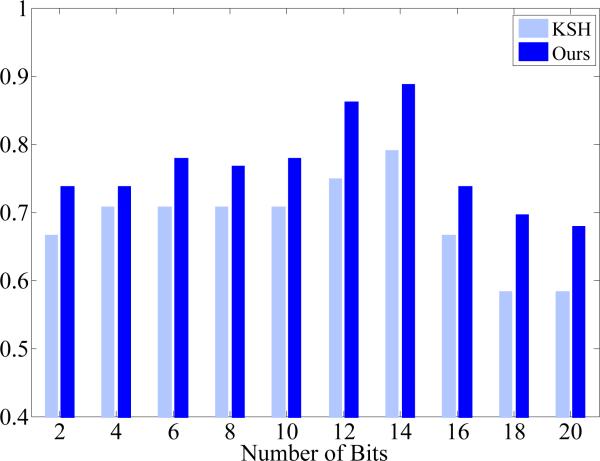
Our hashing-based classification has few parameters that are easy to choose and not sensitive. This is critical to an automatic framework for histopathological image analysis, since tuning sensitive parameters is infeasible when conducting this large-scale and cell-level analysis. Particularly, our hashing-based classification only has one parameter, i.e., the number of hash bits. In our experiments, we have used 12 bits for classification, indicating 4096 hash values. Theoretically, using one bit is already sufficient for binary classification purpose, i.e., diffrentiation of two types of cells. However, as shown in Fig. 2, some hash values may not be reliable and have to be pruned, due to image noise and several inaccurate segmentations. Therefore, it is necessary to use many hash values, which also enable multi-label classification. On the other hand, it is also desired to have enough samples mapped into each hash value, so the support weight can be effective and benefit the classification accuracy. Therefore, the number of hash bits should not be very large either. In fact, using 20 hash bits can result in one million different hash values, sufficiently representing half million cells in our dataset. In addition, using a large number of hash bits (e.g., 64 bits) may reduce the computational and memory efficiency, since the hash table is no longer an option owing to the memory constraint. Therefore, we have chosen 12 bits for this task, mapping half million cells to 4096 hash values and hence ensuring sound accuracy of classification without sacrificing the computational and memory efficiency. This is also demonstrated by our experiments shown in Fig. 6. Note that our model is able to generate accurate results within a certain range of parameter values, i.e., not that sensitive to parameters, making it suitable for the large-scale analysis. Furthermore, Fig. 6 also shows that our content-aware weighting scheme consistently improves the hashing method for classification accuracy, when using different number of hash bits.
Figure 6.
Classification accuracy of our content-aware hashing and KSH (Liu et al., 2012), using different number of hashing bits (2 to 20).
Currently, we have validated our framework on around one thousand images with half million cells. We expect to apply it on much larger databases (e.g., hundreds of millions cells) or whole slide images in the future. In this case, parallel computing may be necessary to ensure the computational efficiency. Our framework for cell-level analysis can be straightforwardly parallelled. For example, the whole slide image can be divided as multiple patches, and each patch can be processed by one node of the cluster for cell segmentation and classification independently. Note that if holistic features are used, e.g., architecture features, such parallel computing can only be applied on the cell detection and segmentation, but not the feature extraction, which needs to analyze the whole image simultaneously. In general, the computational efficiency of our framework is very promising and has the potential to handle large-scale databases.
5. Conclusions
In this paper, we proposed a robust and efficient framework to analyze histopathological images at cell-level. This is achieved by segmenting all cells and discovering the most relevant instances for each cell among a massive database. The main contribution of this proposed framework is to enable real-time and cell-level analysis of histopathological images, benefited from our weighted hashing-based classification. This weighting scheme alleviates the intrinsic problems of traditional hashing methods. It significantly improves the diagnosis accuracy of a challenging clinical problem, i.e., differentiating two types of lung cancers as the adenocarcinoma and squamous carcinoma using histopathological images. We envision that it can provide usable tools to assist clinicians’ diagnoses of cellular images and support efficient data management. In the future, we plan to investigate various types of features, such as geometry features and cell shapes, and fuse them in the supervised hashing framework to boost the accuracy. Although this weighting scheme is specifically designed for cell-level analysis of histopathological images, resulting in promising performance in this challenging application, it may also benefit the classification accuracy of other applications such as natural image categorization. We plan to investigate this in the future as well.
Highlights.
A comprehensive and real-time framework is designed to perform cell-level analysis for histopathological images, by leveraging the robust cell segmentation and hashing-based large-scale image retrieval.
For large-scale image retrieval, we propose a content-aware hashing method that adaptively decides the importance of each hash value. This scheme alleviates several intrinsic problems of traditional hashing methods, and significantly improves the classification accuracy.
We also improve the cell segmentation algorithms by handling variations in shape and cell size, to provide robust and accurate delineations of cells.
Our framework will potentially provide useable tools to assist clinicians’ diagnoses and support efficient medical image data management.
Figure 4.
Segmentation results of different methods on a randomly picked patch. From left to right: original image, MS, ISO, and GCC, and Level Set, and ours.
Acknowledgments
This work is supported in part by NIH R01 AR06547901A1 and Oak Ridge Associated Universities.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akakin CH, Kong H, Elkins C, Hemminger J, Miller B, Ming J, Plocharczyk E, Roth R, M., W., Ziegler R, Lozanski G, Gurcan M. Automated detection of cells from immunohistochemically-stained tissues: application to ki-67 nuclei staining. SPIE. 2012 Feb;8315 [Google Scholar]
- Al-Kofahi Y, Lassoued W, Lee W, Roysam B. Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Trans. on Biomedical Engineering. 2010 Apr;57(4):841–852. doi: 10.1109/TBME.2009.2035102. [DOI] [PubMed] [Google Scholar]
- Al-Lahham HZ, Alomari RS, Hiary H, Chaudhary V. Automating proliferation rate estimation from ki-67 histology images. SPIE Medical Imaging. 2012:83152A–83152A. [Google Scholar]
- Ali S, Madabhushi A. An integrated region-, boundary-, shape-based active contour for multiple object overlap resolution in histological imagery. Medical Imaging, IEEE Transactions on. 2012 Jul;31(7):1448–1460. doi: 10.1109/TMI.2012.2190089. [DOI] [PubMed] [Google Scholar]
- Ali S, Veltri R, Epstein JI, Christudass C, Madabhushi A. Adaptive energy selective active contour with shape priors for nuclear segmentation and gleason grading of prostate cancer. MICCAI. 2011:661–669. doi: 10.1007/978-3-642-23623-5_83. [DOI] [PubMed] [Google Scholar]
- Andoni A, Indyk P. IEEE Symposium on Foundations of Computer Science (FOCS) Berkeley, CA.: Oct. 2006. Near-optimal hashing algorithms for approximate nearest neighbor in high dimensions. [Google Scholar]
- Bernardis E, Yu S. Finding dots: Segmentation as popping out regions from boundaries. CVPR. 2010:199–206. [Google Scholar]
- Boykov Y, Funka-Lea G. Graph cuts and efficient n-d image segmentation. Inter. J. Comput. Vision (IJCV) 2006 Nov;70(2):109–131. [Google Scholar]
- Cai H, Xu X, Lu J, Lichtman JW, Yung SP, Wong STC. Repulsive force based snake model to segment and track neuronal axons in 3d microscopy image stacks. NeuroImage. 2006;32(4):1608–1620. doi: 10.1016/j.neuroimage.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Chang H, Han J, Spellman PT, Parvin B. Multireference level set for the characterization of nuclear morphology in glioblastoma multiforme. Biomedical Engineering, IEEE Transactions on. 2012;59(12):3460–3467. doi: 10.1109/TBME.2012.2218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Li H, Zhou X, Wong S. Constraint factor graph cut–based active contour method for automated cellular image segmentation in rnai screening. Journal of microscopy. 2008;230(2):177–191. doi: 10.1111/j.1365-2818.2008.01974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Rajapakse J. Segmentation of clustered nuclei with shape markers and marking function. IEEE Trans. on Biomedical Engineering. 2009 Mar;56(3) doi: 10.1109/TBME.2008.2008635. [DOI] [PubMed] [Google Scholar]
- Cohen LD. On active contour models and balloons. CVGIP: Image understanding. 1991;53(2):211–218. [Google Scholar]
- Comaniciu D, Meer P, Foran DJ. Image-guided decision support system for pathology. Machine Vision and Applications. 1999;11(4):213–224. [Google Scholar]
- Dalal N, Triggs B. Histograms of oriented gradients for human detection. CVPR. 2005;1:886–893. [Google Scholar]
- Datar M, Immorlica N, Indyk P, Mirrokni VS. SoCG. ACM; 2004. Locality-sensitive hashing scheme based on p-stable distributions. pp. 253–262. [Google Scholar]
- Doyle S, Agner S, Madabhushi A, Feldman M, Tomaszewski J. Automated grading of breast cancer histopathology using spectral clustering with textural and architectural image features. ISBI. 2008:496–499. [Google Scholar]
- Dy JG, Brodley CE, Kak A, Broderick LS, Aisen AM. Unsupervised feature selection applied to content-based retrieval of lung images. TPAMI. 2003;25(3):373–378. [Google Scholar]
- Edwards S, Roberts C, McKean M, Cockburn J, Jeffrey R, Kerr K. Preoperative histological classification of primary lung cancer: accuracy of diagnosis and use of the non-small cell category. Am J Clin Path. 2000;53(7):537–540. doi: 10.1136/jcp.53.7.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Naqa I, Yang Y, Galatsanos NP, Nishikawa RM, Wernick MN. A similarity learning approach to content-based image retrieval: application to digital mammography. IEEE Transactions on Medical Imaging. 2004;23(10):1233–1244. doi: 10.1109/TMI.2004.834601. [DOI] [PubMed] [Google Scholar]
- Faustino GM, Gattass M, Rehen S, de Lucena C. Automatic embryonic stem cells detection and counting method in fluorescence microscopy images. ISBI. 2009:799–802. [Google Scholar]
- Felzenszwalb PF, Huttenlocher DP. Efficient graph-based image segmentation. IJCV. 2004;59(2):167–181. [Google Scholar]
- Foran DJ, Yang L, et al. Imageminer: a software system for comparative analysis of tissue microarrays using content-based image retrieval, high-performance computing, and grid technology. JAMIA. 2011;18(4):403–415. doi: 10.1136/amiajnl-2011-000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman DL. Harrison's principles of internal medicine. JAMA: The Journal of the American Medical Association. 2001;286(8):506. [Google Scholar]
- Grady L, Schwartz EL. Isoperimetric graph partitioning for image segmentation. T-PAMI. 2006;28(3):469–475. doi: 10.1109/TPAMI.2006.57. [DOI] [PubMed] [Google Scholar]
- Grala B, Markiewicz T, Kozłowski W, Osowski S, Słodkowska J, Papierz W. New automated image analysis method for the assessment of ki-67 labeling index in meningiomas. Folia Histochemica et Cytobiologica. 2009;47(4) doi: 10.2478/v10042-008-0098-0. [DOI] [PubMed] [Google Scholar]
- Grau V, Mewes AUJ, Alcaniz M, Kikinis R, Warfield S. Improved watershed transform for medical image segmentation using prior information. IEEE Trans. on Medical Imaging. 2004;23(4):447–458. doi: 10.1109/TMI.2004.824224. [DOI] [PubMed] [Google Scholar]
- Greenspan H, Pinhas AT. Medical image categorization and retrieval for pacs using the gmm-kl framework. IEEE Transactions on Information Technology in BioMedicine. 2007;11(2):190–202. doi: 10.1109/titb.2006.874191. [DOI] [PubMed] [Google Scholar]
- Hanbury A, Müller H, Langs G, Menze BH. FIA book 2013. Springer; LNCS: 2013. Cloud-based evaluation framework for big data. [Google Scholar]
- Janowczyk A, Chandran S, Singh R, Sasaroli D, Coukos G, Feldman MD, Madabhushi A. High-throughput biomarker segmentation on ovarian cancer tissue microarrays via hierarchical normalized cuts. IEEE Trans. on Biomedical Engineering. 2012;59(5):1240–1252. doi: 10.1109/TBME.2011.2179546. [DOI] [PubMed] [Google Scholar]
- Jung C, Kim C. Segmenting clustered nuclei using h-minima transform-based marker extraction and contour parameterization. IEEE Trans. on Biomedical Engineering. 2010 Oct;57(10):2600–2604. doi: 10.1109/TBME.2010.2060336. [DOI] [PubMed] [Google Scholar]
- Kårsnäs A, Dahl AL, Larsen R. Learning histopathological patterns. Journal of pathology informatics. 2011;2 doi: 10.4103/2153-3539.92033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser G, Riede U, Werner M, Hufnagl P, Kayser K. Towards an automated morphological classification of histological images of common lung carcinomas. Elec J Pathol Histol. 2002;8:022–03. [Google Scholar]
- Kolmogorov V, Zabih R. What energy functions can be minimized via graph cuts? T-PAMI. 2004;26(2):147–159. doi: 10.1109/TPAMI.2004.1262177. [DOI] [PubMed] [Google Scholar]
- Kong H, Gurcan M, Belkacem-Boussaid K. Partitioning histopatho-logical images: An integrated framework for supervised color-texture segmentation and cell splitting. IEEE Trans. on Medical Imaging. 2011;30(9):1661–1677. doi: 10.1109/TMI.2011.2141674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulis B, Grauman K. Kernelized locality-sensitive hashing for scalable image search. CVPR. 2009 doi: 10.1109/TPAMI.2011.219. [DOI] [PubMed] [Google Scholar]
- Langs G, Müller H, Menze BH, Hanbury A. MCBR-CDS MICCAI workshop. Vol. 7723. Springer; LNCS: 2013. Visceral: Towards large data in medical imaging - challenges and directions. [Google Scholar]
- Li G, Liu T, Tarokh A, Nie J, Guo L, Mara A, Holley S, Wong ST. 3d cell nuclei segmentation based on gradient flow tracking. BMC cell biology. 2007;8(1):40. doi: 10.1186/1471-2121-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Adiga U, Olson K, Guzowski JF, Barnes CA, Roysam B. A hybrid 3d watershed algorithm incorporating gradient cues and object models for automatic segmentation of nuclei in confocal image stacks. Cytometry Part A. 2003;56A(1):23–36. doi: 10.1002/cyto.a.10079. [DOI] [PubMed] [Google Scholar]
- Liu W, Wang J, Ji R, Jiang Y-G, Chang S-F. Supervised hashing with kernels. CVPR. 2012:2074–2081. [Google Scholar]
- Liu W, Wang J, Kumar S, Chang S-F. Hashing with graphs. Proceedings of the 28th International Conference on Machine Learning (ICML-11) 2011:1–8. [Google Scholar]
- Lou X, Koethe U, Wittbrodt J, Hamprecht F. Learning to segment dense cell nuclei with shape prior. CVPR. 2012:1012–1018. [Google Scholar]
- Loukas CG, Wilson GD, Vojnovic B, Linney A. An image analysis-based approach for automated counting of cancer cell nuclei in tissue sections. Cytometry part A. 2003;55(1):30–42. doi: 10.1002/cyto.a.10060. [DOI] [PubMed] [Google Scholar]
- Lucchi A, Smith K, Achanta R, Lepetit V, Fua P. A fully automated approach to segmentation of irregularly shaped cellular structures in em images. MICCAI. 2010;6362:463–471. doi: 10.1007/978-3-642-15745-5_57. [DOI] [PubMed] [Google Scholar]
- Mao K, Zhao P, Tan P. Supervised learning-based cell image segmentation for p53 immunohistochemistry. IEEE Trans. on Biomedical Engineering. 2006 Jun;53(6):1153–1163. doi: 10.1109/TBME.2006.873538. [DOI] [PubMed] [Google Scholar]
- Markiewicz T, Jochymski C, Koktysz R, Kozlowski W. Automatic cell recognition in immunohistochemical gastritis stains using sequential thresholding and svm network. ISBI. 2008:971–974. [Google Scholar]
- Markiewicz T, Wisniewski P, Osowski S, Patera J, Kozlowski W, Koktysz R. Comparative analysis of methods for accurate recognition of cells through nuclei staining of ki-67 in neuroblastoma and estrogen/progesterone status staining in breast cancer. Anal. and Quant. Cytol. Histol. 2009;31(1):49. [PubMed] [Google Scholar]
- Mijović Ž, Mihailović D, Kostov M. Discriminant analysis of nuclear image variables in lung carcinoma. Facta universitatis-series: Medicine and Biology. 2008;15(1):28–32. [Google Scholar]
- Monaco J, Hipp J, Lucas D, Smith S, Balis U, Madabhushi A. Image segmentation with implicit color standardization using spatially constrained expectation maximization: Detection of nuclei. MICCAI. 2012:365–372. doi: 10.1007/978-3-642-33415-3_45. [DOI] [PubMed] [Google Scholar]
- Müller H, Geissbühler A, Ruch P. Multilingual Information Access for Text, Speech and Images. Springer; 2005. Imageclef 2004: Combining image and multi-lingual search for medical image retrieval. pp. 718–727. [Google Scholar]
- Nath SK, Palaniappan K, Bunyak F. Cell segmentation using coupled level sets and graph-vertex coloring. MICCAI. 2006:101–108. doi: 10.1007/11866565_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Cancer Institute The cancer genome atlas. 2013 retrieved from https://tcga-data.nci.nih.gov.
- Ozolek JA, Tosun AB, Wang W, Chen C, Kolouri S, B., S., Huang H, Rohde GK. Accurate diagnosis of thyroid follicular lesions from nuclear morphology using supervised learning. MedIA. 2014;18(5):772–780. doi: 10.1016/j.media.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Huang JZ, Ji JX, Ding Y. Segmentation, inference and classification of partially overlapping nanoparticles. T-PAMI. 2013;35(3):669–681. doi: 10.1109/TPAMI.2012.163. [DOI] [PubMed] [Google Scholar]
- Qi X, Xing F, Foran D, Yang L. Robust segmentation of overlapping cells in histopathology specimens using parallel seed detection and repulsive level set. IEEE Trans. on Biomedical Engineering. 2012 Mar.59(3):754–765. doi: 10.1109/TBME.2011.2179298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt O, Hasse M. Radial symmetries based decomposition of cell clusters in binary and gray level images. Journal of Pattern Recognition. 2008 Jun;41(6):1905–1923. [Google Scholar]
- Sertel O, Kong J, Catalyurek UV, Lozanski G, Saltz JH, Gurcan MN. Histopathological image analysis using model-based intermediate representations and color texture: Follicular lymphoma grading. Journal of Signal Processing Systems. 2009;55(1-3):169–183. [Google Scholar]
- Shen F, Shen C, Liu W, Shen H. Supervised discrete hashing. In: Computer Vision and Pattern Recognition. IEEE. 2015 [Google Scholar]
- Shen F, Shen C, Shi Q, Van Den Hengel A, Tang Z. Computer Vision and Pattern Recognition (CVPR), 2013 IEEE Conference on. IEEE; 2013. Inductive hashing on manifolds. pp. 1562–1569. [Google Scholar]
- Shi J, Malik J. Normalized cuts and image segmentation. T-PAMI. 2000;22(8):888–905. [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CAJC. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Song Y, Cai W, Feng D. International ACM Workshop on Medical Multimedia Analysis and Retrieval. ACM; 2011. Hierarchical spatial matching for medical image retrieval. pp. 1–6. [Google Scholar]
- Tabesh A, Teverovskiy M, Pang H-Y, Kumar VP, Verbel D, Kotsianti A, Saidi O. Multifeature prostate cancer diagnosis and gleason grading of histological images. TMI. 2007;26(10):1366–1378. doi: 10.1109/TMI.2007.898536. [DOI] [PubMed] [Google Scholar]
- Thunnissen F, Diegenbach P, Van Hattum A, Tolboom J, van der Sluis D, Schaafsma W, Houthoff H, Baak JR. Further evaluation of quantitative nuclear image features for classification of lung carcinomas. Pathology-Research and Practice. 1992;188(4):531–535. doi: 10.1016/s0344-0338(11)80050-6. [DOI] [PubMed] [Google Scholar]
- Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger K, Yatabe Y, Powell CA, Beer D, Riely G, Garg K, et al. International association for the study of lung cancer/american thoracic society/european respiratory society: international multidisciplinary classification of lung adenocarcinoma: executive summary. Proceedings of the American Thoracic Society. 2011;8(5):381–385. doi: 10.1513/pats.201107-042ST. [DOI] [PubMed] [Google Scholar]
- Vincent L, Soille P. Watersheds in digital spaces: an efficient algorithm based on immersion simulations. T-PAMI. 1991;13(6):583–598. [Google Scholar]
- Wang J, Kumar S, Chang S-F. Semi-supervised hashing for large-scale search. T-PAMI. 2012;34(12):2393–2406. doi: 10.1109/TPAMI.2012.48. [DOI] [PubMed] [Google Scholar]
- Weiss Y, Torralba A, Fergus R. Spectral hashing. NIPS. 2008 [Google Scholar]
- Wu Z, Gurari D, Wong JY, Betke M. Hierarchical partial matching and segmentation of interacting cells. MICCAI. 2012:389–396. doi: 10.1007/978-3-642-33415-3_48. [DOI] [PubMed] [Google Scholar]
- Xie Y, Ji Q. A new effcient ellipse detection method. ICPR. 2002;2:957–960. [Google Scholar]
- Xing F, Su H, Neltner J, Yang L. Automatic ki-67 counting using robust cell detection and online dictionary learning. IEEE Transactions on Biomedical Engineering. 2014 Mar;61(3):859–870. doi: 10.1109/TBME.2013.2291703. [DOI] [PubMed] [Google Scholar]
- Xu C, Prince JL. Snakes, shapes, and gradient vector flow. TIP. 1998;7(3):359–369. doi: 10.1109/83.661186. [DOI] [PubMed] [Google Scholar]
- Xu N, Ahuja N, Bansal R. Object segmentation using graph cuts based active contours. CVIU. 2007;107:210–224. [Google Scholar]
- Yan P, Zhou X, Shah M, Wong S. Automatic segmentation of high-throughput rnai fluorescent cellular images. T-ITB. 2008 Jan;12(1):109–117. doi: 10.1109/TITB.2007.898006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Lee HK, Hariharan S, Bu W, Ahmed S. Quantitative neu-rite outgrowth measurement based on image segmentation with topological dependence. Cytometry Part A. 2009;75A(4):289–297. doi: 10.1002/cyto.a.20664. [DOI] [PubMed] [Google Scholar]
- Yu W, Lee HK, Hariharan S, Bu W, Ahmed S. Evolving generalized voronoi diagrams for accurate cellular image segmentation. Cytometry Part A. 2010;77(4):379–386. doi: 10.1002/cyto.a.20876. [DOI] [PubMed] [Google Scholar]
- Zhang C, Yarkony J, Hamprecht FA. Cell detection and segmentation using correlation clustering. MICCAI. 2014a;8673:9–16. doi: 10.1007/978-3-319-10404-1_2. [DOI] [PubMed] [Google Scholar]
- Zhang X, Liu W, Dundar M, Badve S, Zhang S. Towards large-scale histopathological image analysis: Hashing-based image retrieval. IEEE Transactions on Medical Imaging. 2015a Feb;34(2):496–506. doi: 10.1109/TMI.2014.2361481. [DOI] [PubMed] [Google Scholar]
- Zhang X, Su H, Yang L, Zhang S. Fine-grained histopatho-logical image analysis via robust segmentation and large-scale retrieval. The IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2015b Jun; [Google Scholar]
- Zhang X, Yang L, Liu W, Su H, Zhang S. Medical Image Computing and Computer-Assisted Intervention. Springer; 2014b. Mining histopathological images via composite hashing and online learning. pp. 479–486. [DOI] [PubMed] [Google Scholar]