Abstract
Automatic and accurate segmentation of the prostate and rectum in planning CT images is a challenging task due to low image contrast, unpredictable organ (relative) position, and uncertain existence of bowel gas across different patients. Recently, regression forest was adopted for organ deformable segmentation on 2D medical images by training one landmark detector for each point on the shape model. However, it seems impractical for regression forest to guide 3D deformable segmentation as a landmark detector, due to large number of vertices in the 3D shape model as well as the difficulty in building accurate 3D vertex correspondence for each landmark detector. In this paper, we propose a novel boundary detection method by exploiting the power of regression forest for prostate and rectum segmentation. The contributions of this paper are as follows: 1) we introduce regression forest as a local boundary regressor to vote the entire boundary of a target organ, which avoids training a large number of landmark detectors and building an accurate 3D vertex correspondence for each landmark detector; 2) an auto-context model is integrated with regression forest to improve the accuracy of the boundary regression; 3) we further combine a deformable segmentation method with the proposed local boundary regressor for the final organ segmentation by integrating organ shape priors. Our method is evaluated on a planning CT image dataset with 70 images from 70 different patients. The experimental results show that our proposed boundary regression method outperforms the conventional boundary classification method in guiding the deformable model for prostate and rectum segmentations. Compared with other state-of-the-art methods, our method also shows a competitive performance.
Keywords: CT image, prostate segmentation, rectum segmentation, regression forest, local boundary regression, deformable segmentation
1 Introduction
Prostate cancer is the second leading cause of cancer death among American males [1]. According to the American Cancer Society, approximately 233,000 new prostate cancer cases and 29,480 prostate-cancer-related deaths will occur in the United States in 2014. Image-guided radiation treatment (IGRT) is currently one of the major treatments to prostate cancer after a tumor is determined by the transrectal ultrasound guided needle biopsy [2–4]. In IGRT, the prostate and surrounding organs (e.g., rectum and bladder) are manually segmented from the planning image by physician(s). Then a treatment plan is designed, based on the manual segmentation, to determine how the high-energy x-ray beams are accurately delivered to the prostate, while effectively avoiding the surrounding healthy organs. However, manual segmentation of these pelvic organs in the planning image is a time-consuming procedure, and also suffers from large intra- and inter-observer variations [5]. As a result, inaccurate delineations of these organs could potentially lead to an improper treatment plan, thus, a wrong dose delivery. To ensure high efficiency and efficacy of the cancer treatment, automatic and accurate pelvic organ segmentation from the planning image is highly desired in IGRT. In this paper, we focus on the segmentation of the prostate and rectum.
Despite its importance in IGRT, automatic and accurate segmentation of the prostate and rectum in the CT image is still a challenging task due to the following reasons. For the prostate, although their shapes in different patients are relatively stable, the image contrast of the prostate is low (see both (a) and the blue area of (b) in Fig. 1). Besides, due to bowel gas and filling, the relative position of the prostate to its surrounding structures often changes across different patients (see the blue areas of both (b) and (c) in Fig. 1). For the rectum, its appearance and shape in different patients are highly variable due to uncertain existence of bowel gas (see the green areas of both (b) and (c) in Fig. 1).
Fig. 1.
Typical CT images. Images (a) and (b) are from the same slice of the same patient, while image (c) comes from the corresponding slice of a different patient. The blue and green areas indicate the manual segmentations (ground truth) of the prostate and rectum, respectively. The red arrows indicate the bowel gas, while the cyan arrows indicate the indistinct rectum boundary.
To address these challenges, many pelvic organ segmentation methods have been proposed for the CT image. The methods in [6–9] use patient-specific information to localize pelvic organs. In these methods, images from the same patient are exploited to facilitate organ segmentation. Feng et al. [6] leveraged both population and patient-specific image information for prostate deformable segmentation. Liao et al. [7] and Li et al. [8] enriched the training set by gradually adding segmented daily treatment images from the same patient for accurate prostate segmentation. Gao et al. [9] employed previous prostate segmentations of the same patient as patient-specific atlases to segment the prostate in CT images. Since no images previously from the same patient are ready available in the planning stage, the methods proposed in [6–9] cannot be directly applied to the pelvic organ segmentation in planning CT images. Therefore, it is critical to develop a population-based segmentation method. In this way, Costa et al. [10] presented a non-overlapping constraint (from the bladder) on coupled deformable models for prostate localization. Chen et al. [5] adopted a Bayesian framework with anatomical constraints of surrounding bones to segment the prostate. Martínez et al. [11] employed a geometrical shape model to segment the prostate, rectum and bladder under a Bayesian framework. These methods used simple image intensity or gradient as appearance information for organ segmentation. Due to low boundary contrast, it is not enough to accurately segment pelvic organs, i.e., prostate and rectum. Alternatively, Lu et al. [12] adopted a classification-based boundary detection for pelvic organ segmentation and achieved promising performance.
On the other hand, random forest [13], as an ensemble learning method, is becoming popular in image processing [14–18] due to its efficiency and robustness. Gall et al. [14] combined random forest and Hough transformation to predict a bounding box for object detection. Criminisi et al. [15] employed regression forest to vote organ locations in CT images. Chu et al. [16] adopted regression forest to detect landmarks in 2D cephalometric X-ray images. In all these methods, random forest is mainly used for rough object localization or landmark detection. Due to the robustness of random forest, these methods are often used as a robust initialization of later segmentation methods. Recently, Chen et al. [17] and Lindner et al. [18] extended the idea of landmark detection for deformable segmentation. Specifically, Chen et al. [17] applied regression forest to predict each landmark of a shape model with geometric constraint, and further exploited landmark response maps to drive the deformable segmentation of the femur and pelvis. Lindner et al. [18] trained a regression forest for each landmark of a 2D shape model to iteratively predict the optimal position of each landmark for proximal femur segmentation. Although these methods show a promising prospect in 2D segmentation, in general, they cannot be directly applied to 3D segmentation due to the following reasons: 1) the number of vertices in 3D shape is often too great to simply train one landmark detector for each vertex, as each landmark detector often requires hours to train; 2) the training of each landmark detector also requires relatively accurate landmark correspondence, which is easy to be manually annotated in 2D images, but can be very difficult in 3D images. Although Chen et al. [17] performed a preliminary study to apply the method on a 3D dataset, due to the aforementioned problem, only a small number of key landmarks are employed to infer the entire organ shape for segmentation. Since the 3D shape cannot be well represented by a small number of landmarks, especially for the complex structure such as the rectum, its accuracy for 3D segmentation can be limited. When aggregating voting results from different context locations, these methods often disregard the distances from these context locations to the target object. Since voxels closer to the target object should be more informative than those far-away voxels, their votes should be more important and their respective weights should be larger than those of the far-away voxels.
To address the limitations of these previous approaches, we propose a novel boundary detection method and further incorporate it into an active shape model for both prostate and rectum segmentation. Specifically, we first employ regression forest to train a boundary regression model, which can predict the 3D displacements from different image voxels to their nearest points on the target boundary, based on local image features of each image voxel. To determine the target boundary, regression voting is further adopted to vote the nearest boundary point from different context locations using the estimated 3D displacements. Since the voxels far away from the target boundary are not informative for boundary detection, we particularly develop a local voting strategy to restrict votes from far-away locations and only allow votes from near-boundary voxels. Then, we further integrate the regression forest with an auto-context model to increase the performance of our boundary regression. Finally, by taking the voting map of the target boundary as an external force, a deformable model is utilized for the precise organ segmentation with shape priors.
The contributions of our work are threefold. 1) We present a novel local boundary regression method by training only one regression forest to detect the entire target boundary. In this way, our method avoids training for a large number of landmark detectors in guiding the deformable segmentation, as well as the construction of 3D landmark correspondences for the shape model across training images, which is often difficult to be achieved in practice as mentioned above. 2) An auto-context model is adopted to exploit contextual structure information of images in order to improve the performance of our boundary regression. 3) Based on the predicted organ boundary, a deformable model is applied to incorporate shape priors for precise segmentation of each pelvic organ. In the experimental section, we compared our boundary regression method with the boundary classification method. The results show that our boundary regression method can achieve much higher segmentation accuracy than the boundary classification method for both prostate and rectum.
The remainder of the paper is organized as follows. Section 2 gives an overview of the local boundary regression method, followed by details of each component in our method. Section 3 presents the experimental results. Finally, the paper concludes with Section 4.
2 Method
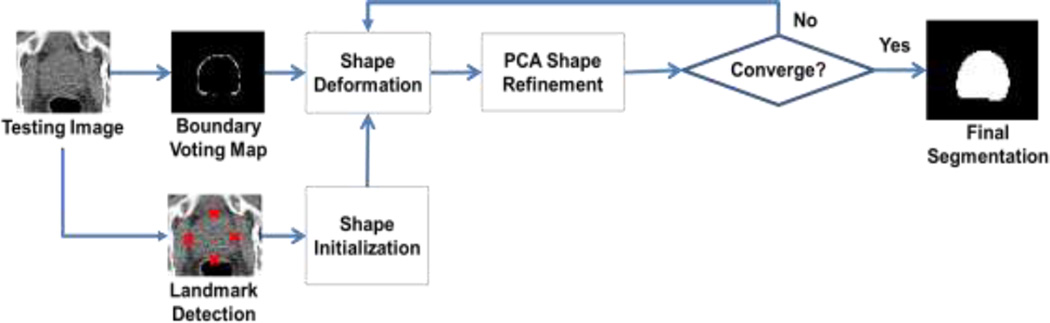
Our method aims to accurately segment pelvic organs from the planning CT images via a novel boundary detection method. Specifically, we first automatically detect several key landmarks on the organ boundary (by a regression-based detection method [19]) to initialize the organ shape (as demonstrated in Fig. 2(a)). Based on the initialization, we know the rough location of the organ boundary and define a local region near that potential organ boundary. Second, we propose a local boundary regression method (as shown in Fig. 2(b)) to vote the organ boundary from the image voxels within the estimated local region. Thus, a boundary voting map can be obtained to enhance the entire organ boundary in the CT image (as indicated in Fig. 2(c)). Third, to boost the performance of our boundary regression method, we further combine our regression forest with the auto-context model [20] to improve the quality of the boundary voting map (as shown in Fig. 2(d)). Finally, a deformable model, learned in the training stage, is applied to the obtained boundary voting map for the final organ segmentation (as demonstrated in Fig. 2(e)).
Fig. 2.
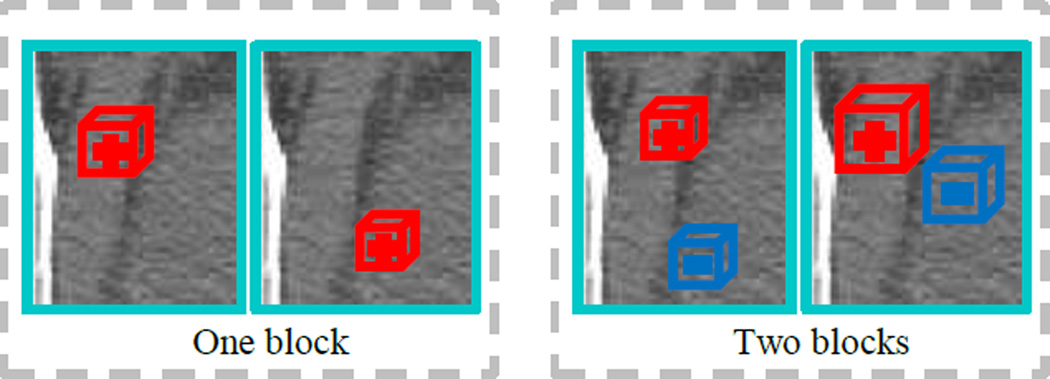
A demonstration of our segmentation method for the prostate. (a) shows the original image overlaid with an initial shape (green curve), which is warped from the mean shape according to the detected key landmarks (as detailed in Section 2.2). (b) demonstrates the local boundary regression by the near-boundary image patches (blue cubes), where three red arrows indicate the displacement vectors (regression targets) from the centers of the image patches to their corresponding nearest boundary points. (c) shows the boundary voting map by our local boundary regression method using only image appearance features. (d) shows the refined boundary voting map by the auto-context model, integrating the context features. (e) shows the final prostate segmentation (green region) by the deformable model.
2.1 Regression Forest
Recently, regression forest achieved promising results in image processing [14, 15, 17, 18, 21–23] due to its efficiency and robustness. Essentially, regression forest is a non-linear regression model that can learn the mapping embedded between input variables and continuous targets. In this paper, the inputs to regression forest are local appearance features (e.g., average intensities of blocks, or their differences, within a CT image patch), and the outputs (i.e., regression targets) are the 3D displacements from image voxels to an organ boundary or a specific boundary point. In general, regression forest consists of a set of decision trees. Based on the bootstrapping of training samples, each decision tree grows by recursively splitting the selected training samples into left and right child nodes. Here, the splitting of each node is determined by an optimal feature among local appearance features and a corresponding threshold, which are achieved by exhaustively searching over a large number of randomly-selected features and randomly-generated thresholds. The best splitting amounts to the maximum variance reduction of regression targets after node splitting, as defined below.
| (1) |
| (2) |
where Gi is the variation reduction of the i-th node after splitting, Oi is the i-th node, which contains displacements (regression targets) of training samples falling into this node, Oi,j (jε{L, R}) is the left and right child nodes of Oi, H(·) indicates the displacement variance of training samples in a node, d is one of the displacements in Oi, and d̅ is the mean of displacements in Oi. The node splitting stops when the maximum depth of a decision tree or the minimum number of training samples in a node is reached. In this paper, each leaf node of a decision tree stores the mean displacement of training samples in that leaf.
Given a testing sample, each decision tree of a trained regression forest individually predicts a 3D displacement to the potential target boundary. The predictions of all decision trees are then averaged to make a final prediction.
| (3) |
where D is the number of decision trees, and d̂i is the prediction from the i-th decision tree.
2.2 Shape Initialization by Landmark Detection
To effectively detect organ boundaries and segment pelvic organs by deformable models, we need to perform shape initialization for each organ on a new image. The shape initialization consists of two parts: landmark detection and shape initialization, which are detailed in the following sub-sections.
2.2.1 Landmark Detection
To locate the prostate and rectum, we first automatically detect six prostate landmarks (positioned at six utmost points, namely superior, inferior, left, right, anterior, and posterior points of the prostate) and five rectum landmarks (positioned evenly along the central line of the rectum) by a regression-based landmark detection method [19]. Specifically, in the training stage, a regression forest Rl (l = 1,2, …, 11) is trained for each landmark to predict the landmark location from image voxels. The regression target of each regression forest is the 3D displacement vector from an image voxel to the interested landmark. Each image voxel is represented by the extended Haar features, which are obtained by convoluting the image with randomized Haar-like filters in a local patch (Fig. 3).
Fig. 3.
Illustration of typical Haar-like filters. The cyan rectangle indicates an image patch. The red/blue cube indicates a block in the image patch with positive/negative polarities.
Each randomized Haar-like filter consists of one or more blocks with different polarities defined as follows:
| (4) |
| (5) |
where U is the number of blocks, and B(·) represents a block, the size of which is 𝕫. ai and ci are the polarity and center of the i-th block, respectively. To fully utilize all information within the patch, the number of blocks (U), the center of each block (ci), and the size of each block (𝕫) are all randomly determined. In our implementation, the extended Haar features extract two types of appearance information from a patch. The first type is one-block Haar features (U = 1), which extract the average intensity around a particular location within the patch. The second type is two-block Haar features (U = 2), which extract the average intensity difference between two particular locations within the patch. Since the convolution of a Haar-like filter with the image will sum the voxel intensities covered by each block, the large size of each block cannot accurately localize the organ boundary, while the small size of each block could be disturbed by strong noises in CT images. Hence, the size 𝕫 of each block is randomly selected as either 3 or 5 with the organ size considered.
The reason we adopt the extended Haar features is that, the tissue in CT images appears to be more homogenous and less textural than that in other modalities, such as MRI. The extended Haar features can characterize both intensity and gradient information robustly by taking average intensities and average intensity differences of image blocks. Moreover, the extended Haar features can be efficiently computed by using the integral image. Even though the extended Haar features may include invalid features, these uninformative features will be filtered out during the supervised training of random forest; therefore, they will not affect the boundary displacement regression later.
During the testing stage, the learned regression forest Rl is used to predict the displacement vectors from image voxels to the potential location of landmark l. Then, these displacement vectors are used to vote for the l-th landmark location based on the respective image voxels. The position with the maximum vote in the landmark voting map is considered as the target landmark location. To ensure the robustness and efficiency of shape initialization, we adopt a multi-resolution strategy to detect the landmarks. Specifically, in the coarse resolution, we predict 3D displacement for every voxel in a down-sampled image. In the fine resolution, we only predict 3D displacements of the voxels near the initialization determined by the coarse resolution. (The details of the multi-resolution strategy are descripted in [24].) The landmark detection results are demonstrated in Fig. 4.
Fig. 4.
Illustration of landmark detection for prostate (image (a), in transverse view) and rectum (image (b), in sagittal view). The green + crosses indicate the ground-truth landmarks manually annotated by an expert, while the red × crosses are the detected landmarks by the regression based landmark detection method.
2.2.2 Landmark-Based Shape Initialization
To initialize the organ shape with detected landmarks, we first compute a mean organ shape based on all training shapes. Specifically, a typical training shape is selected as a reference shape. Then, coherent point drift (CPD) [25] is used to warp this reference shape onto other training shapes for building the vertex correspondence across training shape meshes. For the rectum, there are several cases where CPD surface registration fails. For those cases, we use an in-house tool to deform the reference rectum mesh onto the rectum mask image, with human interaction, for building the vertex correspondence. Once the vertex correspondence is built for all training shapes, we affine align all these training shapes to a common space and then average the aligned training shapes to obtain a mean organ shape 𝒮̅. (See Section 2.5 for more details) In the testing stage, the mean organ shape 𝒮̅ is affine transformed onto the testing image based on the detected landmarks. The affine transformation is computed between the detected landmarks, in the testing image, and their counterparts in the mean organ shape as below:
| (6) |
where LM denotes landmark, ALM ∈ ℝ4×11 denotes the coordinate matrix of the detected landmarks in the testing image, T ∈ ℝ4×4 is an affine transformation matrix to be estimated, ELM ∈ ℝ4×11 is the coordinate matrix of corresponding landmarks from the mean organ shape. Therefore, the initial organ shape for the testing image can be obtained by applying the estimated affine transformation TLM on the mean organ shape.
| (7) |
where Sinit is the estimated initial organ shape for the testing image, and 𝒮̅ is the mean organ shape.
2.3 Local Boundary Regression
Motivated by point regression in deformable segmentation [21], we propose a local boundary regression to vote pelvic organ boundaries. Specifically, a regression forest is used to learn the non-linear mapping from local patch of each image voxel to its nearest target boundary point. Note that, compared with the landmark detection, each image voxel is still being represented by the extended Haar features extracted from its local patch (see Section 2.2); however, the regression target is being defined as a 3D displacement vector from an image voxel to its nearest boundary point on a specific target organ.
In the training stage, from each training image, we first randomly sample a large number of image voxels pi (i = 1,2, …, M) around the manually-delineated organ boundary. Each sampled voxel pi is characterized by the randomly-generated extended Haar features F, extracted from its W × W × W local image patch. The corresponding regression target di is also defined as the 3D displacement from the sampled point pi to its nearest boundary point. Since the image voxels near the boundary are more informative than those far away from the boundary, we sample the image voxel pi using a Gaussian distribution around the target boundary as follows:
| (8) |
| (9) |
where q is a boundary point randomly selected on the manually-delineated organ boundary, N(q) is the normal of q at the manually-delineated organ boundary, and r is a random offset along the normal direction, which is determined by a Gaussian distribution 𝒩(0, τ). In this way, most sampled voxels are located around the target boundary, which makes our regression model specific to the target boundary. In this paper, we take 10000 training samples/voxels for each training image with τ = 16mm. Fig. 5(a) shows one example of our sampling pattern.
Fig. 5.
Illustration of both our sampling scheme for a target organ (a) and the region of interest for local boundary regression (b). In (a), the green curve indicates the manually-delineated organ boundary, and the white points show the randomly sampled voxels. In (b), the green curve indicates the near-boundary initial shape of an organ, and the white region represents a ring-shaped local region.
Up to this stage, we can learn a regression forest ℜ0 by recursive node splitting (as described in Section 2.1), based on all pairs of Haar feature representation and 3D displacement, i.e., < F(pi), di > from all training images. For the learned regression forest, the features F̃(pi) ⊂ F(pi), which are used in the best splitting of decision trees, are considered as optimal features and then recorded tree-by-tree for the later testing stage.
In the testing stage, given a testing image, we use all image voxels in a region of interest (ROI) for local boundary regression. The ROI is defined as a ring-shaped local region on the testing image, centered at the initial shape of an organ, as shown in Fig. 5(b). Here, the initial shape of the organ is achieved by warping the mean organ shape onto the testing image, according to the previously detected landmarks (see Section 2.2). Since regression-based landmark detection could achieve robust shape initialization with reasonable overlap ratio (i.e., the overlap ratios [26] between the initial shapes and the manual segmentations of the prostate and rectum are 0.78 and 0.71, respectively, according to our experiment), the ring-shaped local region can be considered near the organ boundary. Then, a boundary voting map of the testing image can be obtained by voting from all image voxels within this local region. Specifically, for each image voxel p̂ in the local region, its optimal Haar features F̃ (p̂) can be extracted, as recorded in the training stage. Then, the respective displacement d̂ = ℜ0 (F̃(p̂)) can be predicted by the trained regression forest ℜ0. Based on all estimated 3D displacements, we can transform the testing image into a displacement map. Finally, a voting strategy is used to vote the target boundary from different context locations. For each image voxel p̂ and its corresponding estimated displacement d̂, a weighted vote will be accumulated at the position p̂ + d̂ in a 3D boundary voting map V.
| (10) |
| (11) |
where ω(p̂) is a weighted vote from the image voxel positioned at p̂, DIST (p̂) is the Euclidean distance from p̂ to the potential target boundary determined by the initial shape (mentioned in Eq.(7)), δ is the radius of the ring-shape region, and σ is a control coefficient. In this way, we can get a boundary voting map for a target pelvic organ. A typical example of the prostate is shown in Fig. 2(c).
2.4 Refinement of Boundary Voting Map by Auto-Context Model
Recently, the auto-context model [20] is becoming popular, due to its capability to iteratively refine the classification/labeling result. Under the auto-context model, for a typical binary classification, a classifier can be trained to label each image point based on its local appearance. Then, the learned classifier can be employed on a testing image to obtain a probability map, where each image point has its probabilities belonging to the foreground and background. This probability map can be treated as another image modality for extracting new types of local features. Based on both the new features (namely the context features) from the probability map and the conventional appearance features from the intensity image, a new classifier can be trained to label the image again. This procedure can be repeated based on the iteratively-updated probability map.
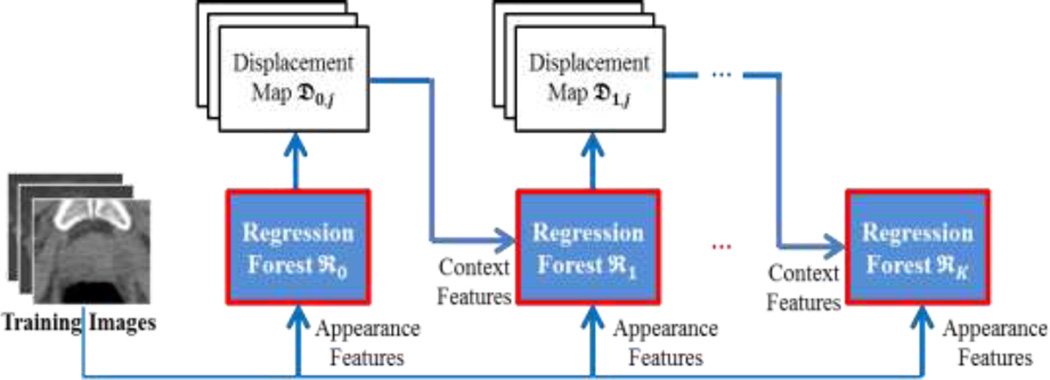
In our work, we extend this idea to regression for refining our estimated displacement map. Specifically, we adopt the auto-context model to iteratively train a sequence of regression forests, by integrating both the appearance features extracted from the intensity image and the context features extracted from the intermediate displacement map from the previous regression forest. Here, the type of context features is also the extended Haar features, as shown in Fig. 3. The iterative procedure of the auto-context model is shown in Fig. 6. Since the testing stage of an auto-context model follows the same pipeline as the training stage, we take the training stage of the auto-context model as example (see Fig. 6).
Fig. 6.
Flowchart of the auto-context model in our proposed method.
In the training stage, we learn a sequence of regression forests ℜt (t = 0,1, …, K) by using the same technique as Section 2.3. Specifically, the regression forest ℜ0 is trained by using only appearance features (i.e., extended Haar features) extracted from intensity training images Ij (j = 1,2, …, N) (i.e., the original training images). Then, to learn the successive regression forests ℜt (t = 1,2, …, K), we extract appearance features from the intensity training images Ij (j = 1,2, …, N), and context features from the displacement maps 𝔇t−1,j (j = 1,2, …, N) of those training images in the previous iteration. These two types of features are further combined to learn a new regression forest. In an iterative way, a set of regression forests can be trained to refine the boundary regression.
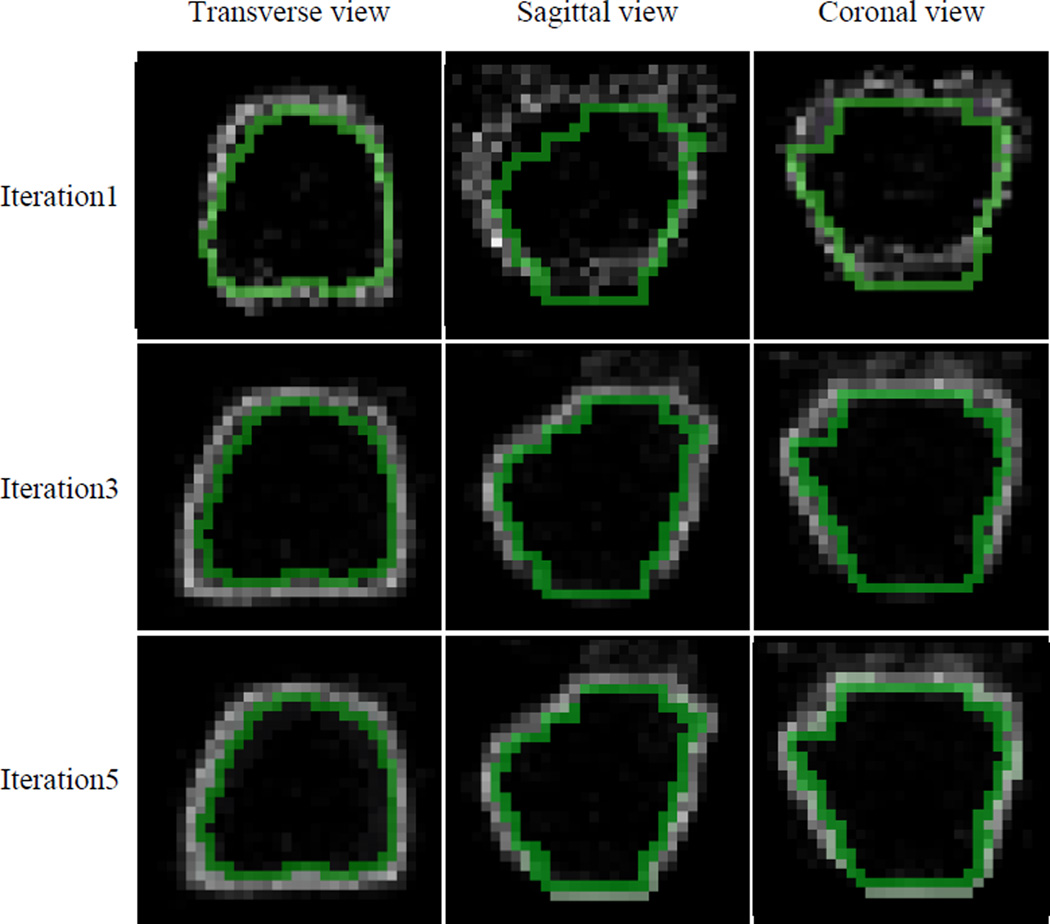
In the testing stage, given a testing image, the learned regression forests are sequentially employed to estimate refined displacement maps to achieve an accurate boundary voting map. Specifically, the regression forest ℜ0 is first adopted to predict the first displacement map, using only local appearance features from the testing image. Then, appearance features are extracted from the intensity testing image, while context features are extracted from the displacement map 𝔇t−1 (t = 1,2, …, K) of the testing image in the previous iteration. By combining these features, the learned regression forest ℜt (t = 1,2, …, K) can gradually refine the boundary displacement map, which leads to a better boundary voting map in the end. The main reason why auto-context works is that, by using context features, the correlation between the displacement of the current voxel and the displacements of neighboring voxels can be captured in the learned regression model. This structure information helps improve the 3D displacement estimation, which then refine the boundary voting map. The refined boundary voting maps are shown in Fig. 7 (where we take the prostate as an example). To determine the iteration number K of the auto-context model during this procedure, initially we set K to a relatively large number, i.e., 10. Then, we observed that the training performance of our method converges by 5 iterations. Thus, we finally adjusted K to 5 iterations.
Fig. 7.
Boundary voting maps of a patient generated by the auto-context model. The green curves indicate the manually-delineated organ boundaries. With iterative refinement, we can see a more and more accurate boundary voting map.
Joint Boundary Regression of the Prostate and Rectum
Since the prostate and rectum are nearby organs, in the auto-context model, we extract context features, not only from the previous displacement map of itself, but also from the previous displacement map of its nearby organ. Specifically, to estimate the prostate displacement map in the i -th iteration, we extract context features from the prostate displacement map in the (i − 1) -th iteration, and context features from the rectum displacement map in the (i − 1)-th iteration. The same situation applies to the rectum. In this way, the displacement maps of both organs are jointly predicted. The joint boundary regression can enforce non-overlapping constraint on the displacement maps, hence, alleviating the overlapping issues on the segmentation of nearby organs. The details are shown in Algorithm 1.
Algorithm 1.
Joint boundary regression on Prostate and Rectum by Auto-Context Model
| Input: | Itest - a testing image |
| ℜt - trained regression model of the prostate at the t-th iteration | |
| ℜ′t - trained regression model of the rectum at the t-th iteration | |
| Sinit - initial shape of the prostate for the testing image (Section 2.2) | |
| S′init - initial shape of the rectum for the testing image (Section 2.2) | |
| K - number of iterations in the auto-context model | |
| Output: Refined Boundary Voting Maps | |
| Determine a ring-shaped local region for the prostate on the testing image Itest, based on the initial shape Sinit. | |
| Determine a ring-shaped local region for the rectum on the testing image Itest, based on the initial shape S′init. | |
| for each t in {0,1, …, K} do | |
for each p̂ in voxel set within the ring-shaped local region of the prostate do
| |
| end for | |
| Achieve the current prostate displacement map 𝔇t. | |
for each p̃ in voxel set within the ring-shaped local region of the rectum do
| |
| end for | |
| Achieve the current rectum displacement map 𝔇′t. | |
| end for | |
| Return: Final Boundary Voting Maps VK and V′K | |
| Here, 𝔇−1 and 𝔇′−1 are both null, which means that, if t = 0, the displacement vector prediction depends only on Itest. | |
Note that, instead of the traditional radiation-like features [20] (as shown in Fig. 8), the context features in our paper are randomly-generated extended Haar features (as shown in Fig. 3), extracted from the local patch of the estimated intermediate displacement map. This is because the traditional radiation-like features only consider the appearance characteristics of some fixed positions along radiation directions, while the extended Haar features can consider the appearance characteristics of any position within a local image patch. Besides, the randomly-generated extended Haar features provide much richer feature representations for learning the regression forest, since the extended Haar features take into account a large number of feature patterns by randomly selecting the parameters in Eqs. (4)–(5). Thus, by combining the appearance features (from the original training images) and the context features (from the displacement map of previous regression forest), the auto-context model can iteratively refine the prediction of 3D displacement vectors for the testing image, and finally obtain a more accurate boundary voting map.
Fig. 8.
Traditional radiation-like context features. For a specific point (the cyan rectangle), multiple rays in equal intervals are extended from the point in different directions, and the context locations are sparsely sampled on these rays (as the colorful points show). The average (probability) of a small block centered at each context location is usually considered as a feature to represent this context location.
2.5 Deformable Segmentation based on the Boundary Voting Map
Up to this stage, the boundary voting map for the pelvic organ is achieved by using the local boundary regression with the auto-context model. Since the estimated boundary voting map highlights the potential organ boundary by votes, it can be used as an external force to guide the deformable model [27, 28] for pelvic organ segmentation.
In the training stage, we first extract the dense surface meshes for the prostate and rectum from manually-delineated binary images by the marching cube algorithm [29]. Then, one typical surface mesh is selected as a reference mesh, which is further smoothed and decimated to obtain good quality. In this paper, the number of surface vertices for the prostate and rectum are 931 and 1073, respectively. Afterwards, the surface registration method [25] is used to warp this surface onto other dense surface meshes to build vertex correspondence. Once the vertex correspondence is built, all surfaces have the same number of vertices, and vertex correspondences. All these surfaces of training images can be affine aligned into a common space. By taking the vertex-wise average of all aligned surfaces, we can obtain a mean shape 𝒮̅. Furthermore, by using PCA on the aligned shapes with 98% variation, we can also achieve a PCA shape space.
where 𝒮 is a shape in the PCA shape space, 𝒮̅ is the mean shape of all aligned training surfaces, 𝒫 includes a set of major modes of shape variation, and b is the parameter of the shape space to control the shape 𝒮.
In the testing stage, based on the initial shape inferred by the detected landmarks in Section 2.2, each vertex in the shape model can be independently deformed on the boundary voting map. Since the appearance of each image is simplified by the boundary regression voting, the shape model deforms along each model point’s surface normal, to a position with the maximum boundary votes. In the meantime, the deformed shape Sdfm is refined by the learned PCA shape model. Specifically, we project the deformed shape Sdfm into the parameter space, i.e., b = 𝒫T (T−1 Sdfm − 𝒮̅), and then limit each eigen-coefficient bi of b into a range of , to obtain a new parameter b̂. Here, λi is the eigenvalue regarding the i-th mode of the shape variation. T−1 is a transformation to project the deformed shape into the PCA shape space. With an inverse transformation T, a refined shape Sref can be achieved by Sref = T(𝒮̅ + 𝒫 b̂). The weighted sum of the refined shape Sref and the deformed shape Sdfm is considered as the finally deformed shape of this iteration. During the segmentation, the weight of the PCA refined shape is gradually decreased from 1 to 0 with the increase of iteration. Initially, we solely rely on the shape constraint. When linearly combining the tentatively deformed shape and the PCA refined shape, we use only the PCA refined shape. In the later iterations, the weight for the PCA refined shape is linearly decreased to make the model more adaptive to the boundary voting map. In the last iterations, when the model is close to the target boundary, the weight for the shape constraint becomes zero, and, thus, the model is purely guided by the boundary voting map. This weight-adaptive strategy is especially important for those not-well-initialized cases, since the initial focus of shape constraint can largely relieve the dependency on good initialization of the deformable model. By alternating the model deformation and shape refinement, the shape model can be gradually driven onto the organ boundary under guidance from both the boundary voting map and the PCA shape space. If the intermediate deformed shape no longer changes, the deformable model will converge.
3 Experiments
To evaluate the performance of our proposed method, we conduct experiments on a pelvic organs dataset acquired from the North Carolina Cancer Hospital. The dataset consists of 70 planning CT images from 70 different patients, all with prostate cancer. Each image has voxel size 0.938×0.938×3.0 mm3, and was isotropically resampled to 2.0 × 2.0 × 2.0 mm3 for the experiment. A clinical expert manually delineated the prostate and rectum in all 70 images, which we use as the ground truth in our experiment.
To quantitatively evaluate our proposed method, we employ five commonly used metrics:
- Dice similarity coefficient (DSC) [26] is an overlap measure between the segmented organ and the manual ground-truth.
where TP is the number of correctly labeled organ voxels, FP is the number of falsely labeled organ voxels, and FN is the number of falsely labeled background voxels.(12) - False Positive Ratio (FPR) is the proportion of wrongly labeled voxels in the automatic segmentation.
(13) - Positive Predictive Value (PPV) is the proportion of correctly labeled voxels in the automatic segmentation.
(14) - Sensitivity (SEN) is the proportion of correctly labeled voxels in the organ.
(15) - Average surface distance (ASD) is the average distance between the surface of automatic segmented organ (SEG) and that of the manual ground-truth (GT).
where d (z, GT) is the minimum distance of voxel z on the automatic segmented organ surface SEG to the voxels on the ground-truth surface GT, d (u, SEG) is the minimum distance of voxel u on the ground-truth surface GT to the voxels on the automatic segmented organ surface SEG, and |․ | is the cardinality of a set.(16)
In the experiments, we use four-fold cross-validation to evaluate our method. In each fold, the landmark detectors, boundary regression forests, and PCA shape models are trained with images from different folds. The accuracies of four folds are finally averaged to achieve the reported results. The parameter setting of our method is as follows: the number of trees D in each regression forest is 10; the maximum depth of each tree is 15; the number of candidate features for node splitting is 1000; the minimum number of training samples in each leaf is 5; the patch size W for extracting Haar features is 60mm; the number of samples drawn near the pelvic organ boundary in each training image is M = 10000; the number of iterations in the auto-context model is 5 (i.e., K = 4); the number Y of iterations used for deformable model is 20. The sigma in Gaussian weighted local voting strategy, (i.e., σ in Eq. (11)) is 8. The local region for the boundary regression voting is determined by a ring of 16mm width near the initial shape of the deformable model.
3.1 Parameter Setting
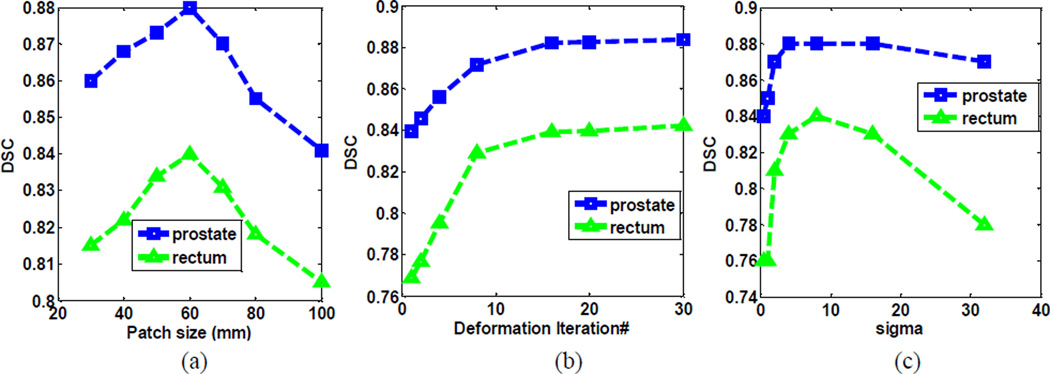
In this paper, most parameters in random forest are learned from [15, 18, 21, 30]. In [25], the authors provided a comprehensive sensitivity analysis of random-forest parameters, which we found also applicable to our method. Besides, we also perform sensitivity analyses of other key parameters, such as patch size, deformation iterations, and sigma in the Gaussian weighting. Fig. 10 shows the segmentation accuracy of our method with respect to 1) the patch size of Haar features, 2) the number of deformation iterations, and 3) sigma values in Gaussian-weighted local voting, respectively. As we can see, the optimal patch size is 60 × 60 × 60mm, and both smaller and larger patch sizes lead to inferior performances. The number of deformation iterations should be set to be greater than 15, to ensure convergence. The optimal sigma is between 4 and 10. Smaller sigma causes insufficient votes, which leads to unreliable boundary voting maps, while larger sigma introduces noisy votes from far-away regions, which lowers the specificity of boundary voting maps.
Fig. 10.
Segmentation accuracy with respect to (a) the path size of Haar features, (b) the number of iterations in deformable model, and (c) sigma values in Gaussian-weighted local voting, respectively.
3.2 Intensity Image vs Boundary Voting Map
To validate the effectiveness of our boundary voting map for deformation segmentation, we apply the deformable model on the intensity image and the boundary voting map to compare their segmentation results. On the intensity image, the deformable model searches the maximal gradient magnitude along the surface normal of each model point; while on the boundary voting map, the deformable model finds the maximum boundary votes along the surface normal of each model point. Note that, the deformable model on the intensity image uses the same initial shape, same shape constraint, and same number of iterations as those on the boundary voting map.
From Table 1, we can see that, the segmentation results on the intensity image are significantly worse than those on the boundary voting map. The reason is that the image contrast of pelvic organs is low in CT images, which tends to result in boundary leakage of the segmentation results on the intensity images. In contrast, our boundary regression method provides a much clearer boundary voting map for guiding deformable segmentation, which produces much higher segmentation accuracy.
Table 1.
Quantitative comparison of deformable segmentations on the intensity image and boundary voting map, respectively.
| Method | DSC | ASD (mm) | |
|---|---|---|---|
| Intensity image | Prostate | 0.80±0.06 | 2.58±0.67 |
| Rectum | 0.74±0.10 | 3.58±1.09 | |
| Boundary voting map | Prostate | 0.88±0.02 | 1.86±0.21 |
| Rectum | 0.84±0.05 | 2.21±0.50 | |
3.3 Boundary Regression vs Boundary Classification
Classification and regression are the two common ways to detect anatomy [31]. To evaluate the effectiveness of our boundary regression method, we conduct a comparative experiment with the boundary classification method [12, 32].
In the boundary classification method, we employ classification forest as a classifier, by using the same setting as boundary regression, to estimate the probability of each voxel belonging to the target boundary. We dilate the manually-delineated boundary by one voxel to take into account the near-boundary region. In this region, we randomly draw samples to get positive samples. To separate the positive and negative samples, we set a neutral region between the positive and negative samples where no samples are drawn. Then, in another region, such as 3 to 8 voxels from the manually-delineated boundary, we randomly select the same number of negative samples as that of positive samples. Finally, we randomly draw a few negative samples in the rest region. Consequently, the total proportion between the positive and negative samples is 1:1.5. Since the majority of samples are close to the target boundary, the learned classification model is specific to the target boundary detection. After classification, the generated boundary classification map can be used to guide a deformable model onto the target boundary by finding the maximum response along the normal direction of each model point. It is worth noting that, in our boundary classification method, the image patch sampled at each voxel is not aligned with the shape normal. While, there are other boundary classification methods [33, 34] that can leverage the available shape normal information for sampling image patches. This facilitates the training of a better boundary detector using steerable features, since boundary voxel patterns are much better aligned with the shape normal.
In our boundary regression, we just use the sampling scheme, as shown in Eqs. (8)–(9). Here, the proportion between positive and negative samples is 1:1. After regression voting, the obtained boundary voting map is employed to drive the deformable segmentation by searching the maximum boundary votes along the normal direction of each model point.
Note that, both methods use the same features, same shape models, and same auto-context strategy.
To visually compare our boundary classification and regression methods, Fig. 11 shows the boundary classification maps and the boundary voting maps for the prostate and rectum, respectively. Due to the ambiguous boundaries of both organs (especially at the apex and base), the boundary classification maps of both organs do not agree well with the manually-delineated boundaries (as shown in column (d) of Fig. 11), while the boundary voting maps of both organs agree better with the manually-delineated boundaries (as shown in column (e) of Fig. 11). This is because the boundary classification methods aim to voxel-wisely label the target boundary according to its local appearance, which is difficult due to the ambiguous boundaries of both the prostate and rectum, and possibly results in the incorrect classification of boundary voxels. In contrast, our boundary regression employs voxels near the target boundary to vote the boundary location, which provides rich context information to overcome the limitation of indistinct boundary appearances.
Fig. 11.
Results of boundary classification and boundary regression for the prostate (top row) and rectum (bottom row) in sagittal view. The red and green contours denote the automatically-segmented and manually-delineated boundaries, respectively.
Table 2 quantitatively compares the segmentation accuracies of these two methods. As we can see, our boundary regression method gives better segmentations, in terms of higher DSC and lower ASD, than the boundary classification method. It is worth noting that, the much inferior accuracies of rectum segmentation by the classification method is because the boundary appearance is complex at the apex and base of the rectum, which results in a great challenge for the classification method. In summary, both qualitative and quantitative results show that our regression method is more effective than the classification method for boundary segmentations of the prostate and rectum in CT images.
Table 2.
Quantitative comparison of boundary classification and boundary regression.
| Method | DSC | ASD | |
|---|---|---|---|
| Boundary classification | Prostate | 0.82±0.07 | 2.37±0.95 |
| Rectum | 0.76±0.06 | 3.36±0.09 | |
| Boundary regression | Prostate | 0.88±0.02 | 1.86±0.21 |
| Rectum | 0.84±0.05 | 2.21±0.50 | |
The unit of ASD is mm.
3.4 Effectiveness of the Auto-context Model
To evaluate the importance of the auto-context model, we refine the boundary voting map using both the appearance features of intensity training images and the context features of intermediate displacement maps iteratively. In Fig. 7 above, we took the prostate as an example to show the boundary voting maps estimated with the auto-context model at the 1st, 3rd and 5th iteration, respectively. As we can see, with the increase of iteration, the estimated prostate boundary becomes much clearer and closer to the manually-delineated prostate boundary.
To quantitatively evaluate the auto-context model, at each iteration, we perform deformable segmentation on the boundary voting map, and then measure the performance using the average segmentation accuracy across four-fold cross-validation. It is worth noting that, according to our observation, the training performance of our method converges with 5 iterations. As shown in Fig. 12, the DSC values of both the prostate and rectum increase with iterations, while the ASD values of both the prostate and rectum decrease with iterations. These results also show the iterative improvement with the auto-context model, which demonstrates the effectiveness of the auto-context model in enhancing the boundary voting map.
Fig. 12.
Iterative improvement of organ segmentation accuracy (using the DSC and ASD metrics) with the use of the auto-context model. The unit of ASD is mm.
3.5 Comparison with Other State-of-the-art Methods
To justify the effectiveness of our proposed method in prostate and rectum segmentations, we also compare our method with other pelvic organ segmentation methods. However, since none of the state-of-the-art methods [5, 10–12] published their source codes or binary executables, it is difficult to conduct fair comparisons using the same dataset. To obtain a rough accuracy assessment of our method in CT pelvic organ segmentation, we list the performances achieved by the state-of-the-art methods [5, 10–12] in Table 3 and Table 4, respectively. Since different methods use different metrics to evaluate their segmentation results, in this section, we have computed seven metrics to evaluate our method and perform comparison with other methods.
Table 3.
Quantitative comparison between our method and other methods on the prostate segmentation.
| Method |
Mean SEN |
Median SEN |
Mean ASD |
Median ASD |
Mean PPV |
Median FPR |
Mean DSC |
|---|---|---|---|---|---|---|---|
| Chen et al. [5] | NA | 0.84 | NA | 1.10 | NA | 0.13 | NA |
| Costa et al. [10] | 0.75 | NA | NA | NA | 0.80 | NA | NA |
| Martínez et al. [11] | NA | NA | NA | NA | NA | NA | 0.87 |
| Lu et al. [12] | NA | NA | 2.37 | 2.15 | NA | NA | NA |
| Multi-atlas with majority voting | 0.79 | 0.82 | 2.24 | 1.91 | 0.82 | 0.07 | 0.84 |
| Our method | 0.84 | 0.87 | 1.86 | 1.85 | 0.86 | 0.06 | 0.88 |
NA indicates that the respective metric was not reported in the publication. The unit of ASD is mm.
Table 4.
Quantitative comparison between our method and other methods on the rectum segmentation.
| Method |
Mean SEN |
Median SEN |
Mean ASD |
Median ASD |
Mean PPV |
Median FPR |
Mean DSC |
|---|---|---|---|---|---|---|---|
| Chen et al. [5] | NA | 0.71 | NA | 2.20 | NA | 0.24 | NA |
| Martínez et al. [11] | NA | NA | NA | NA | NA | NA | 0.82 |
| Lu et al. [12] | NA | NA | 4.23 | 4.09 | NA | NA | NA |
| Multi-atlas with majority voting | 0.67 | 0.68 | 3.40 | 3.38 | 0.72 | 0.26 | 0.75 |
| Our method | 0.72 | 0.75 | 2.21 | 2.26 | 0.76 | 0.18 | 0.84 |
NA indicates that the respective metric was not reported in the publication. The unit of ASD is mm.
As can be seen from Table 3 and Table 4, our method achieves competitive performance on both the prostate and rectum to other segmentation methods under comparison. Since Costa et al. [10] did not included results on the rectum segmentation, this method is not listed in Table 4.
To further justify the effectiveness of our method, we also implement a multi-atlas based segmentation method on the same dataset for comparison. Specifically, we first uniformly select 26 atlases from training images. Given a testing image, we detect landmarks for each organ (i.e., 6 landmarks for the prostate and 5 landmarks for the rectum) by the method described in Section 2.2, and then warp those 26 atlases onto the testing image with their affine transformations, estimated between the detected landmarks and the corresponding vertices of prostate and rectum surfaces in the atlas images. After all atlases are affine aligned onto the testing image, a majority voting strategy is then adopted to fuse the labels from the warped atlases. From Table 3 and Table 4, we can see that, our method outperforms the multi-atlas based segmentation method for both the prostate and rectum on the same dataset.
4 Conclusions
We have presented a novel boundary detection method for both prostate and rectum segmentations in planning CT images. Compared with previous regression methods, which train one regression forest for each point on the deformable model, we learn a local boundary regression to predict the entire boundary of each target organ. To boost the boundary regression performance, we further combine the regression forest with the auto-context model to iteratively refine the boundary voting map for both the prostate and rectum. Finally, the deformable model is adopted to segment the prostate and rectum under guidance from both the boundary voting map and the learned shape space. Validated on 70 planning CT images from 70 different patients, our boundary regression method achieves 1) better segmentation performance than the boundary classification method, and 2) competitive segmentation accuracy to the state-of-the-art methods under comparison.
In the future, we will further perform the following investigations:
In our current implementation, the prostate and rectum are not completely jointly segmented. We manually checked all of our segmentation results, and found that only 15 out of 70 images have slight overlapping issue for the segmented prostate and rectum, with at most 1–2 voxels overlapping in the touching boundary. It may be caused by the use of joint boundary regression, which takes context features of both organs into account for boundary localization and has less overlapping issue. In future works, we plan to explicitly address this overlapping issue as what Kohlberger et al. [35] did in their paper.
In this paper, we voted the nearest boundary point from image voxels by utilizing the local appearance. If there is a deep concave on the organ boundary, e.g., rectum boundary, it is likely to cast no votes at the valley of the concave, which results in the disconnection of the voted organ boundary. This is the main reason that we employed the shape-constrained deformable model to achieve the final segmentation. A potential solution is to vote multiple nearest boundary points (instead of only 1 boundary point) for each image voxel to alleviate the issue.
So far, we mainly focused on the segmentation of the prostate and rectum in CT images. We also tested our method on segmenting the bladder in CT images, and obtained 0.86±0.08 for DSC and 2.22±1.01 mm for ASD. However, due to the variable shape of the bladder, it is difficult to define anatomical landmarks with reliable appearance on the bladder, which prevents us from achieving robust initialization for the deformable model. This also accounts for the large standard deviation in our segmentation results on the bladder. In future works, we will investigate the joint segmentation of all three pelvic organs to address this limitation.
Fig. 9.
Flowchart of boundary-voting-guided deformable segmentation. Here, we take the prostate as an example. The red crosses indicate the detected landmarks.
Highlights.
We present a novel boundary detection method to segment prostate and rectum in CT images with deformable model.
This is achieved by using local boundary regression to vote the target boundary from a near-boundary local region.
Auto-context model is integrated to iteratively improve the voted target boundary.
This is the first work to vote the whole target boundary by one regression forest.
Our method is competitive to the state-of-the-art methods under comparison.
Acknowledgement
This work was supported in part by the National Institutes of Health (NIH) under Grant CA140413, in part by the National Basic Research Program of China under Grant 2010CB732506, and in part by the National Natural Science Foundation of China (NSFC) under Grants (61473190, 61401271, and 81471733).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cancer Facts and Figures 2014. American Cancer Society. 2014 [Google Scholar]
- 2.RODRIGUEZ LV, TERRIS MK. Risks and complications of transrectal ultrasound guided prostate needle biopsy: a prospective study and review of the literature. The Journal of urology. 1998;160(6):2115–2120. doi: 10.1097/00005392-199812010-00045. [DOI] [PubMed] [Google Scholar]
- 3.Shen D, et al. Optimized prostate biopsy via a statistical atlas of cancer spatial distribution. Medical Image Analysis. 2004;8(2):139–150. doi: 10.1016/j.media.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Zhan Y, et al. Targeted prostate biopsy using statistical image analysis. Medical Imaging, IEEE Transactions on. 2007;26(6):779–788. doi: 10.1109/TMI.2006.891497. [DOI] [PubMed] [Google Scholar]
- 5.Chen S, Lovelock DM, Radke RJ. Segmenting the prostate and rectum in CT imagery using anatomical constraints. Medical Image Analysis. 2011;15(1):1–11. doi: 10.1016/j.media.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Feng Q, et al. Segmenting CT prostate images using population and patient-specific statistics for radiotherapy. Medical Physics. 2010;37(8):4121–4132. doi: 10.1118/1.3464799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao S, et al. Sparse patch-based label propagation for accurate prostate localization in CT images. Medical Imaging, IEEE Transactions on. 2013;32(2):419–434. doi: 10.1109/TMI.2012.2230018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W, et al. Learning image context for segmentation of the prostate in CT-guided radiotherapy. Physics in Medicine and Biology. 2012;57(5):1283. doi: 10.1088/0031-9155/57/5/1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Y, Liao S, Shen D. Prostate segmentation by sparse representation based classification. Medical physics. 2012;39(10):6372–6387. doi: 10.1118/1.4754304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa MJ, et al. Automatic segmentation of bladder and prostate using coupled 3D deformable models, in Medical Image Computing and Computer-Assisted Intervention–MICCAI 2007. Springer; 2007. pp. 252–260. [DOI] [PubMed] [Google Scholar]
- 11.Martínez F, et al. Segmentation of pelvic structures for planning CT using a geometrical shape model tuned by a multi-scale edge detector. Physics in Medicine and Biology. 2014;59(6):1471. doi: 10.1088/0031-9155/59/6/1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu C, et al. Precise segmentation of multiple organs in CT volumes using learning-based approach and information theory, in Medical Image Computing and Computer-Assisted Intervention–MICCAI 2012. Springer; 2012. pp. 462–469. [DOI] [PubMed] [Google Scholar]
- 13.Breiman L. Random forests. Machine learning. 2001;45(1):5–32. [Google Scholar]
- 14.Gall J, Lempitsky V. Class-specific hough forests for object detection, in Decision Forests for Computer Vision and Medical Image Analysis. Springer; 2013. pp. 143–157. [Google Scholar]
- 15.Criminisi A, et al. Regression forests for efficient anatomy detection and localization in CT studies, in Medical Computer Vision. Recognition Techniques and Applications in Medical Imaging. Springer; 2011. pp. 106–117. [Google Scholar]
- 16.Chu C, et al. Fully Automatic Cephalometric X-Ray Landmark Detection Using Random Forest Regression and Sparse shape composition. submitted to Automatic Cephalometric X-ray Landmark Detection Challenge. 2014 [Google Scholar]
- 17.Chen C, et al. Automatic X-ray landmark detection and shape segmentation via data-driven joint estimation of image displacements. Medical Image Analysis. 2014;18(3):487–499. doi: 10.1016/j.media.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Lindner C, et al. Fully automatic segmentation of the proximal femur using random forest regression voting. Medical Imaging, IEEE Transactions on. 2013;32(8):1462–1472. doi: 10.1109/TMI.2013.2258030. [DOI] [PubMed] [Google Scholar]
- 19.Gao Y, Shen D. Context-Aware Anatomical Landmark Detection: Application to Deformable Model Initialization in Prostate CT Images, in Machine Learning in Medical Imaging. Springer; 2014. pp. 165–173. [Google Scholar]
- 20.Tu Z, Bai X. Auto-context and its application to high-level vision tasks and 3d brain image segmentation. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 2010;32(10):1744–1757. doi: 10.1109/TPAMI.2009.186. [DOI] [PubMed] [Google Scholar]
- 21.Cootes TF, et al. Robust and accurate shape model fitting using random forest regression voting, in Computer Vision–ECCV 2012. Springer; 2012. pp. 278–291. [Google Scholar]
- 22.Fanelli G, et al. Random forests for real time 3d face analysis. International journal of computer vision. 2013;101(3):437–458. [Google Scholar]
- 23.Zhou SK. Shape regression machine and efficient segmentation of left ventricle endocardium from 2D B-mode echocardiogram. Medical Image Analysis. 2010;14(4):563–581. doi: 10.1016/j.media.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Gao Y, Zhan Y, Shen D. Incremental learning with selective memory (ILSM): Towards fast prostate localization for image guided radiotherapy. Medical Imaging, IEEE Transactions on. 2014;33(2):518–534. doi: 10.1109/TMI.2013.2291495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myronenko A, Song X. Point set registration: Coherent point drift. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 2010;32(12):2262–2275. doi: 10.1109/TPAMI.2010.46. [DOI] [PubMed] [Google Scholar]
- 26.Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. [Google Scholar]
- 27.Shen D, Herskovits EH, Davatzikos C. An adaptive-focus statistical shape model for segmentation and shape modeling of 3-D brain structures. Medical Imaging, IEEE Transactions on. 2001;20(4):257–270. doi: 10.1109/42.921475. [DOI] [PubMed] [Google Scholar]
- 28.Shen D, Ip HH. A Hopfield neural network for adaptive image segmentation: An active surface paradigm. Pattern Recognition Letters. 1997;18(1):37–48. [Google Scholar]
- 29.Lorensen WE, Cline HE. Marching cubes: A high resolution 3D surface construction algorithm. SIGGRAPH Comput. Graph. 1987;21(4):163–169. [Google Scholar]
- 30.Wang L, et al. LINKS: learning-based multi-source integration framework for segmentation of infant brain images. Neuroimage. 2015;108:160–172. doi: 10.1016/j.neuroimage.2014.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou SK. Discriminative anatomy detection: Classification vs regression. Pattern Recognition Letters. 2014;43:25–38. [Google Scholar]
- 32.Lang A, et al. Retinal layer segmentation of macular OCT images using boundary classification. Biomedical optics express. 2013;4(7):1133–1152. doi: 10.1364/BOE.4.001133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ling H, et al. IEEE Conference on. IEEE; 2008. Hierarchical, learning-based automatic liver segmentation. in Computer Vision and Pattern Recognition, 2008. CVPR 2008. [Google Scholar]
- 34.Lay N, et al. Rapid multi-organ segmentation using context integration and discriminative models. in Information Processing in Medical Imaging. Springer; 2013. [DOI] [PubMed] [Google Scholar]
- 35.Kohlberger T, et al. Automatic multi-organ segmentation using learning-based segmentation and level set optimization, in Medical Image Computing and Computer-Assisted Intervention–MICCAI 2011. Springer; 2011. pp. 338–345. [DOI] [PubMed] [Google Scholar]