Abstract
Age-related disease risk has been linked to short sleep duration and sleep disturbances; however, the specific molecular pathways linking sleep loss with diseases of aging are poorly defined. Key cellular events seen with aging, which are thought to contribute to disease, may be particularly sensitive to sleep loss. We tested whether one night of partial sleep deprivation (PSD) would increase leukocyte gene expression indicative of DNA damage responses (DDR), the senescence-associated secretory phenotype (SASP), and senescence indicator p16INK4a in older adult humans, who are at increased risk for cellular senescence. Community-dwelling older adults aged 61-86 years (n=29; 48% male) underwent an experimental partial sleep deprivation (PSD) protocol over 4 nights, including adaptation, an uninterrupted night of sleep, partial sleep deprivation (sleep restricted 3 AM to 7 AM), and a subsequent full night of sleep. Blood samples were obtained each morning to assess peripheral blood mononuclear cell (PBMC) gene expression using Illumina HT-12 arrays. Analyses of microarray results revealed that SASP (p < .05) and DDR (p = .08) gene expression were elevated from baseline to PSD nights. Gene expression changes were also observed from baseline to PSD in NFKB2, NBS1 and CHK2 (all p's < .05). The senescence marker p16INK4a (CDKN2A) was increased one day after PSD compared to baseline (p < .01), however confirmatory RT-PCR did not replicate this finding. One night of partial sleep deprivation activates PBMC gene expression patterns consistent with biological aging in this older adult sample. PSD enhanced the SASP and increased the accumulation of damage that initiates cell cycle arrest and promotes cellular senescence. These findings causally link sleep deprivation to the molecular processes associated with biological aging.
Keywords: older adults, sleep deprivation, cellular aging, senescence, DNA damage, inflammation, gene expression
Introduction
Aged adults experience more sleep complaints than younger individuals, with estimates that as much as 50% of adults aged 65 years or more experience insomnia symptoms1 and other sleep difficulties.2,3 Moreover, sleep loss is thought to interact with numerous regulatory systems to influence health and chronic disease risk,4–10 findings that are particularly salient for older adults with sleep difficulties. Disease morbidity and mortality risk are elevated among those with short sleep duration and poor sleep quality.1,11–13 However, the specific molecular pathways altered by sleep loss, which impact human disease, are poorly defined. Potential pathways include an increase in unrepaired cellular stress and accumulation of damage,14–18 which is thought to contribute to biological aging.19–24 Initial evidence has linked poor sleep quality and short sleep duration with greater cellular aging, as indicated by shorter leukocyte telomere length.19–24
The accumulation of damage is part of the pathology of common chronic diseases of aging,25 and is suspected to be a mechanism by which aging itself promotes disease.26 Accumulated damage from chronic low grade inflammation,27,28 for example, leads to imbalance in the redox system, mitochondrial dysfunction, and reduced ability to repair.25 Driven in part by these processes, aging is also associated with an accrual of senescent cells.29–31 Cellular senescence is a state of cell cycle arrest, commonly reached by replicative (e.g., critically short telomeres) or cell stress (e.g., DNA damage) pathways, and preceded by signaling of DNA damage response (DDR) elements derived from telomeric or non-telomeric damage.29,31–34
Accrual of senescent cells is thought to effect the aging process, as removal of senescent cells in mice slows aging.35 These senescent cells have a unique secretory pattern, termed the senescence associated secretory phenotype (SASP), which is characterized by increased release of inflammatory cytokines and chemokines (e.g., IL-6, IL-8, monocyte chemotactic protein (MCP)-2, MCP-4, chemokine C-X-C motif ligand (CXCL)-1, CXCL-2, CXCL-3, granulocyte macrophage colony-stimulating factor (GM-CSF), intracellular adhesion molecule(ICAM)-1) that promote disease.31,34,36–38 Removing the senescent cells may reduce the development of the aging phenotype in mice by eliminating the inflammatory secretory patterns that contribute to the pathology. Indeed, genetically modified mice (NFKB1−/−) that have an enhanced inflammatory signal show accelerated aging through accumulation of senescent cells and enhancement of the SASP.28 Furthermore, accumulation of senescence predicts shortened lifespan.28
Given these findings that aging is associated with an increase in DNA damage, accumulation of senescent cells, and enhancement of the inflammatory secretome, it is possible that these pathways serve as molecular links between short sleep duration and increases in age-related chronic disease burden in late life. In this experimental study, we test the hypothesis that sleep loss alters these molecular pathways by examining whether partial night sleep deprivation (PSD) in aged humans increases peripheral blood mononuclear cell (PBMC) expression of genes indicative of rises in DNA damage responses (DDR), increases in proinflammatory senescent associated secretory phenotype (SASP), and increases in cellular senescent signal expression (p16INK4a). PSD replicates the kind of sleep loss that is ubiquitous in the general population and also resembles the reduction of sleep duration that is often found in persons with age-related medical disorders, however, its effects on physiological process has been examined almost exclusively in adult,39–44 as opposed to older adult populations.45
Materials and Methods
Participants
All subjects gave informed consent and the University of California, Los Angeles (UCLA) institutional review board approved the protocols. Initial medical interview, physical examination, and screening laboratory tests determined eligibility for the PSD. Subjects were invited to participate in the experimental session if they were deemed physically healthy including no past history of cancer or inflammatory disorders, were non-smokers, and had a body mass index (BMI) <40 (calculated as weight (kg) divided by the square of height (m)). Participants completed a 2 week sleep diary, and were excluded if their normal sleep pattern was less than 7 hours nightly and or they showed signs of circadian phase shifting (early or delayed sleep onset by more than 2 hours); had atypical sleeping hours not occurring between 11 PM and 7 AM. Additional exclusion criteria included: current diagnosis of mental illness based on the Diagnostic and Statistical Manual of Mental Disorders (Editions IV (revised) or V); sleep apnea, restless leg syndrome, or other sleep disorders as identified during the night of adaptation to the sleep laboratory; chronic or acute (< 2 weeks) infection; and comorbid uncontrolled chronic disease. The present analysis includes 29 participants who underwent the PSD protocol and had samples collected for RNA analyses.
Procedures
After eligibility and medical evaluation, subjects were invited for a four night stay at the UCLA Clinical Translational Research Center (CTRC) where they underwent experimental procedures. Following the first night, which served as an adaptation night, subjects had an uninterrupted night of sleep from 11 PM to 7 AM (baseline). On the third night, the partial sleep deprivation night was assigned. On this night, subjects were not allowed to sleep from 11 PM to 3 AM, sleep occurred between 3 AM to 7 AM (PSD), and awakening occurred regardless of sleep stage. The fourth night (1 day after the PSD) subjects were allowed to sleep uninterrupted from 11 PM to 7 AM. Blood samples were obtained each morning prior to eating and following baseline, PSD, and recovery nights for the assessment of cellular gene expression. Sleep patterns were monitored using polysomnography (PSG) recordings of sleep each night, and are reported separately.45
Measures
PBMC gene expression patterns
Samples of quality-verified RNA extracted (RNeasy; Qiagen) from approximately 2 million PBMC were assayed by Illumina HT-12 arrays (Illumina Inc., San Diego, CA) in the UCLA Neuroscience Genomics Core Laboratory as previously described.46 Briefly, RNA was tested for integrity and converted to fluorescent cRNA for hybridization to Illumina Human HT-12 v4 BeadArrays, following the manufacturer's standard protocol. Quintile-normalized gene expression values were generated for more than 35,000 probes derived from the National Center for Biotechnology Information Reference Sequence (NCBI) and other sources, and provides genome-wide transcriptional coverage of well-characterized genes, gene candidates, and splice variants.
Senescence Associated Secretory Phenotype (SASP): Genes representative of key components of the SASP were selected a priori based on prior work showing differential gene expression patterns, with confirmatory protein quantification, in replicative and damage induced senescent cells.47,48 These included the following genes: IL6, CSF2, CCL8, IL8, CCL13, ICAM1, CXCL1, CXCL2, CXCL3, which were z-transformed and then summed to create a composite measure of SASP. Signals known to be upregulated during the DNA damage response (DDR)49 were selected to create a composite index by z-transforming and summing the following genes: GADD45GIP1, TP53BP1, CHEK1, TP53, TERF2, SIRT1, TERT, GADD45A, CDK7, DDB1, HUS1, NTHL1, MRE11A, ERCC2, PCNA, ERCC1, RAD51, OGG1, BRCA1, H2AFX, NBN, DDB2, RAD50, XPC, RPA1, MLH1, LIG1, MDC1, GADD45B, XPA. Further selection was made of additional individual genes important in signaling senescence and initiating the inflammatory cytokine response after DDR: ATM, NBS1 (NBN), and CHK2.47,50 We also selected key genes that play a vital role in our pathways of interest: p16INK4a (CDKN2A) was selected as an established senescent cell signal,35,51 and NFKB1 and NFKB2 gene expression were selected as the key transcriptional elements of both inflammation and SASP.52 A complete list of gene names, along with correlation tables of each gene within the DDR and SASP composites can be found in the supplementary material, Tables S1-S3.
Analytic Strategy
SPSS statistical software v.22 was used to perform all statistical analyses. To test for the effect of PSD on gene expression, general linear model analyses were performed, controlling for baseline differences in gene expression and batch effects, along with BMI and sex. Adjusting for multiple comparisons, the Least Significant Difference (LSD) test was used for planned pairwise comparisons that examined estimated marginal mean differences between baseline and PSD, baseline and recovery nights, and PSD and recovery nights.
Results
A total of 29 older adults (48.3% male) aged 61-86, M(SD)=71(7.5) years, and average body mass index (BMI) of M(SD)=27.3(4.4) kg/m2) completed the 4 night study and had blood collected for the examination of gene expression profiles upon waking.
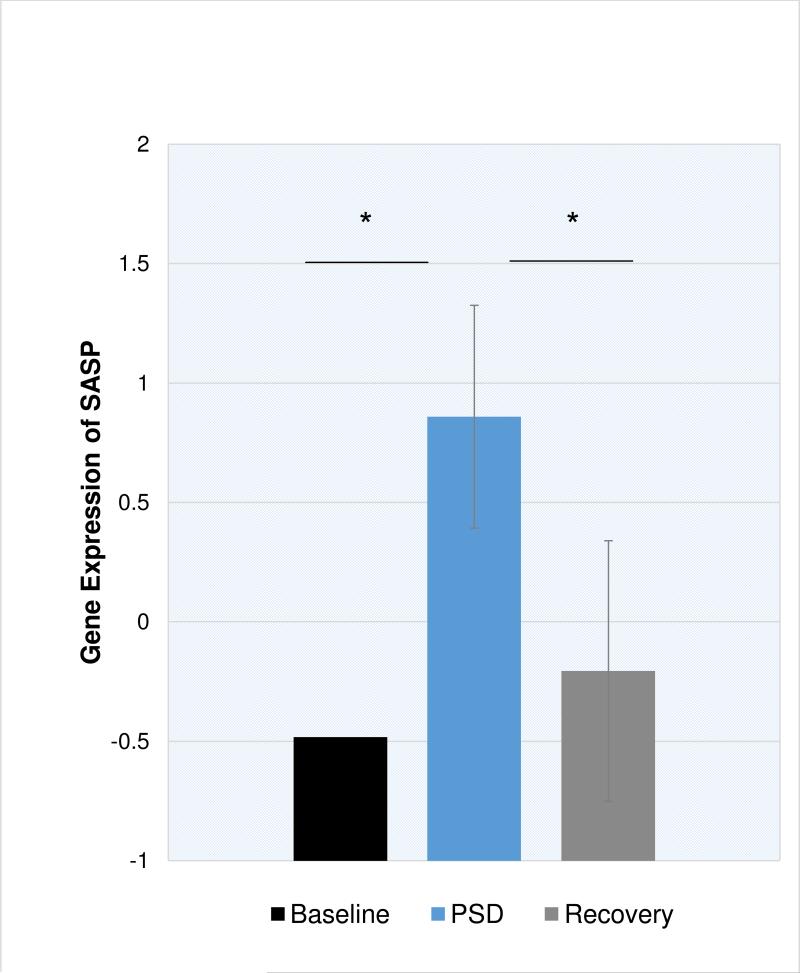
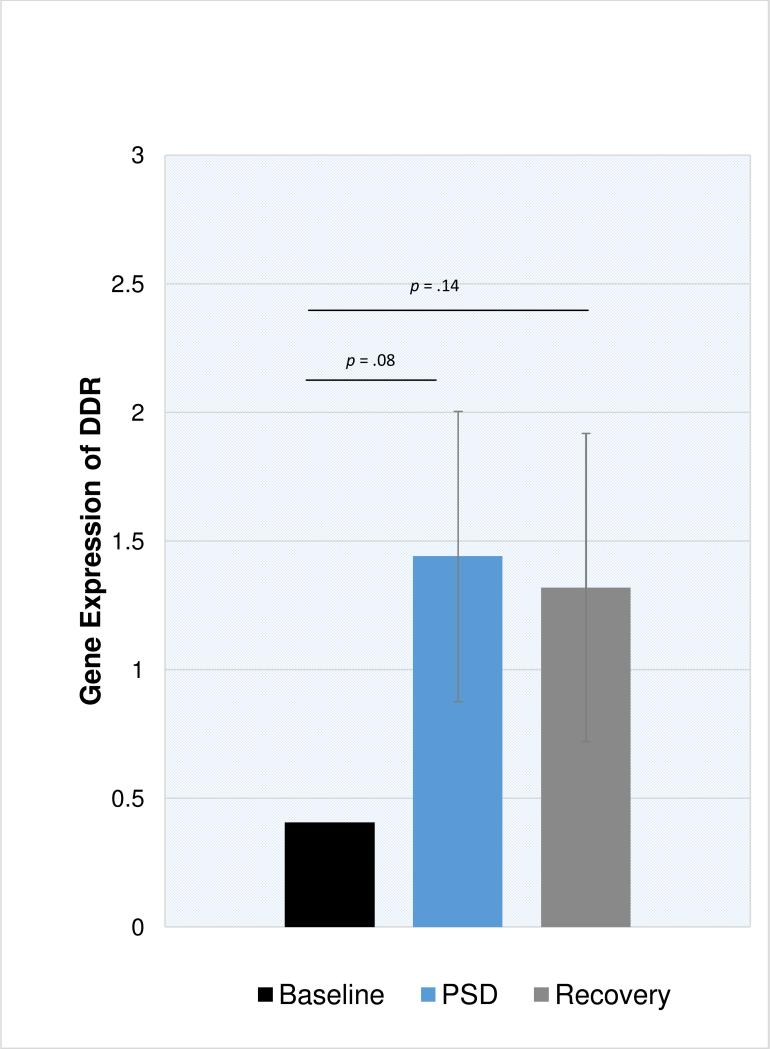
As compared to baseline, PSD induced increases in SASP expression (Figure 1; p = .01), NFKB2 (p = .008), and NFKB1 (p = .07). As compared to baseline, expression of SASP and NFKB1 were similar after a night of recovery sleep, however NFKB2 remained slightly elevated at recovery (p = .06). The composite DNA damage response (DDR) showed a similar pattern, with increases in DDR gene expression after PSD compared to baseline (Figure 2; p = .08), which remained elevated after a night of recovery sleep, however this was not significantly different from baseline (p = .14). Finally, PSD altered genes involved in signaling senescence. As compared to baseline, PSD induced increases in NBS1 (p = .004), CHK2 (p < .001) and ATM (p = .09). In contrast, increases in the senescent signal marker p16INK4a (CDKN2A) on microarray were delayed and found after a night of recovery sleep (p < .01). PSD did not significantly alter MRE11 or RAD50 gene expression. Confirmatory RT-PCR verified significant PSD-induced differences in gene expression for CXCL1, CXCL2, and IL8 (all p < .001) and a trend toward differential expression for NFKB2 (p = .07). RT-PCR failed to verify any significant PSD-induced difference in ICAM1, IL6, or CDKN2A (all p > .18).
Figure 1. Gene expression of the senescence associated secretory phenotype (SASP) at baseline, partial sleep deprivation (PSD), and one day after PSD (Recovery) in older adults.
SASP is a composite score created from a sum of 9 z-transformed genes. Error bars represent standard error of estimated marginal mean, adjusting for BMI and sex. *p<.05
Figure 2. Gene expression of the DNA damage response (DDR) at baseline, partial sleep deprivation (PSD), and one day after PSD (Recovery) in older adults.
DDR is a composite score created from a sum of 30 z-transformed genes. Error bars represent standard error of estimated marginal mean, adjusting for BMI and sex.
Discussion
One night of partial sleep deprivation activated gene expression patterns in PBMCs consistent with the hypothesis that sleep loss promotes biological aging in older adults. We observed increases in expression of the NF-κB related gene (NFKB2) and an enhanced expression of representative genes that give rise to the senescent associated secretome. Moreover, we observed an increase in DNA damage response signals, along with the expression of NBS1-CHK2 genes implicated in DDR initiated cell cycle arrest that promotes senescent and pre-senescent increases in inflammatory cytokines.47,50 PSD altered genes involved in signaling senescence including NBS1 (p = .004), CHK2 (p < .001) and ATM (p = .09). And one day following sleep deprivation we observed on microarray an increase in the gene expression of the senescent marker p16INK4a, however confirmatory RT-PCR did not replicate this finding. Together, these findings indicate that sleep deprivation increases the DNA damage response, increases senescence associated secretory phenotypic expression pattern, and promotes pathways involved in the initiation of cellular senescence in aged adults. Future work should consider whether this effect is more pronounced after repeated days of sleep restriction or in aged individuals with chronic sleep disturbances such as insomnia.
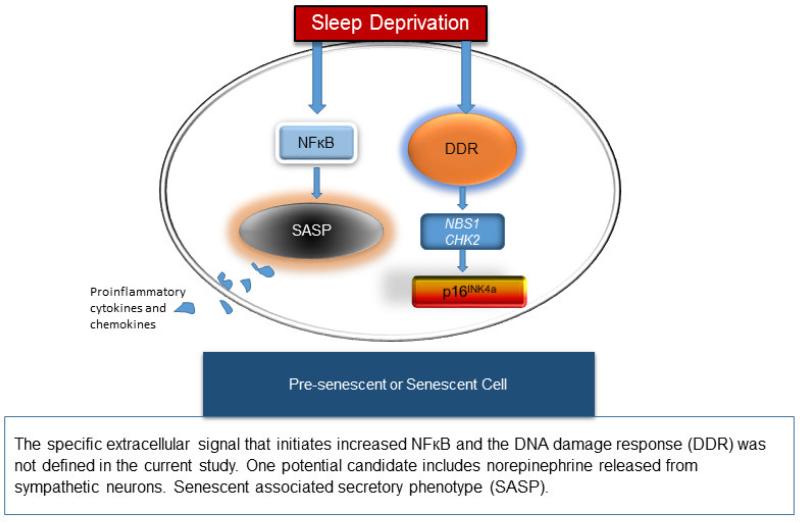
The enhanced expression of SASP may occur in pre-existing senescent cells, which are prevalent in aged adults, and which is enhanced through NF-κB signaling. However, it also is possible that pre-senescent cells exhibit gene expression patterns similar to senescent cells after a sustained DDR has been initiated,50 which in the present sample occurred after sleep deprivation. These proposed pathways are characterized in Figure 3. Further work is necessary to distinguish the cell source for these responses after sleep deprivation.
Figure 3.
Proposed molecular pathways promoting aging after sleep deprivation.
Consistent with our results, prior work has reported that sleep deprivation induces increases in NF-κB signaling within immune cells of young adults.39 Although the current study was unable to assess the extracellular signal that might initiate the intracellular changes, several proposed mechanisms may work together to drive this effect. The sympathetic nervous system is highly responsive to sustaining wakefulness during periods of high demand for sleep.40,53–55 The release of norepinephrine stimulates β-adrenergic receptors on immune cells to activate transcriptional elements, including NF-κB.54,56 Although this transcription factor is known to regulate proinflammatory immune response genes,54,57 recent evidence has also implicated NF-κB as a major regulator of the senescent associated secretory phenotype within cells expressing senescent markers.52,58,59 Our previous research has demonstrated PSD increases release of catecholamines,53 as well as activation of these same proinflammatory NF-κB transcriptional control pathways within immune cells in young adults.41 Knowing this, we speculate that sleep deprivation may enhance SASP in existing senescent cells of aged adults via norepinephrine ligation of β-adrenergic receptors on the cell surface, which signals transcriptional regulation via NF-κB. Future work is necessary to identify whether norepinephrine is a key neurotransmitter that can activate the SASP in senescent cells, with implications for both sleep loss as well as other social adversities known to activate the sympathetic nervous system response.
Our findings showing rises in DDR after sleep loss are consistent with animal research showing extended periods of sleep deprivation induce cellular stress and oxidative damage to DNA.14,15,18 We demonstrated that in humans a partial night of sleep loss increases the cellular stress - DNA damage response (DDR) pathway, as well as specific signaling molecules responsive to the DDR: ATM which phosphorylates CHK2 (checkpoint kinase-2) to promote growth arrest, and NBS1, which can participate in DNA repair, especially in the telomeric region, when part of the MRE11-RAD50-NBS1 (MRN) complex.34 In light of the increase in NBS1, we examined individually MRE11 and RAD50 gene expression. We did not observe a significant increase in MRE11 or RAD50, suggesting the cells were likely not in growth arrest in order to repair damage, and then exit growth arrest. Rather we hypothesize that, in response to PSD, these cells were on a trajectory towards a senescent state. In addition to growth arrest, NBS1, ATM, and CHK2 are thought to combine to stimulate proinflammatory activity associated with the SASP in damaged and/or senescent cells.34,50 These results support our conclusion that DDR signaling was induced by sleep deprivation and sufficient to signal growth arrest, which can drive senescence.
Strengths and Limitations
Limitations to the current study design should be noted. First, the analysis of molecular signaling patterns using gene expression analysis within human peripheral blood mononuclear cells is an upstream measure of protein levels and cellular function. Future research should identify senescent cells to confirm the SASP is originating from this cell source after PSD, and that the molecular products (i.e., cytokines, chemokines) of gene expression are also noticeably increased. The present analysis does not account for any potential redistribution of leukocyte subsets that might be induced by PSD. Although we have previously reported that PSD does not induce significant change in the percentage of monocytes, it does appear to influence lymphocyte populations.42 Observed patterns suggest a need for future work using models where cells are isolated, specific genes experimentally manipulated (e.g., silencing using siRNA, enhancing with viral vectors), and signaling pathways pharmacologically manipulated to definitively identify the causal mediators involved. Secondly, although we speculate that β-adrenergic signaling and related cascade effects on NF-κB pathways were involved in the observed changes after a night of partial sleep deprivation, and previous work has reported this53, we did not directly test β-adrenergic signaling activity in the present study. Future research should consider whether β-adrenergic blockade modifies biological aging dynamics induced by sleep deprivation. An alternative explanation for the current findings is that one night of PSD alters gene expression by delaying onset of the sleep-wake pattern, and the observed differences are a result of capturing gene expression at a different point in the sleep cycle rather than a PSD-induced alteration in daytime DDR and SASP. However, prior work has not found that one night of PSD alters the onset or amplitude of circadian hormones, cortisol and melatonin.60 Several nights of chronic sleep restriction can induce gene expression consistent with increases in cellular stress and inflammation,61 suggesting the findings may not be due to a shift in amplitude timing. These results are concordant with findings in an animal model of chronic sleep loss.14 Moreover, it is important to note that the findings in the current study are in an older adult sample, which may have a different response to sleep loss than younger subjects, particularly given the reduced capacity of an aging system to respond to perturbations (e.g., drug clearance, tissue healing, recovery speed, vaccine antibody responses),62,63 and existing evidence that sleep loss impacts older adults more strongly than younger individuals (e.g., increased circulating inflammation).45,64 Future work should determine whether the present results generalize to a younger population.
Several strengths of the current research exist. First, the methods employed include a well-controlled experimental design lasting 4 nights at a clinical translational research center where patients were regularly monitored by trained nursing staff. Participants were carefully screened for exclusion criteria to eliminate influences of excessive chronic disease burden, mental health problems, substance abuse, and sleep problems (i.e., short sleep duration, sleep apnea, or phase shifting). The present research applied gene expression analyses to identify key molecular changes induced by sleep deprivation that are implicated in aging biology, representing a novel approach to assessing biological aging processes in humans.
Conclusions
One night of partial sleep loss in older adults induced gene expression changes consistent with an increase in the DNA damage response and the promotion of the senescent associated secretory phenotype characterized by a proinflammatory profile, initiated via NF-κB transcriptional regulation. One night of sleep deprivation was also followed by a subsequent increase in gene expression suggesting the promotion of cellular senescence and an indication of cellular aging.51 Our data support the hypothesis that sleep loss in aged adults activates these biological pathways. Hence, loss of sleep in late life may promote biological aging due to increased accumulation of damage that initiates cell cycle arrest and enhanced expression of the senescence associated secretory phenotype, which together increase susceptibility to cellular senescence. These findings causally link sleep deprivation to the biological processes intrinsic to aging, and further support the hypothesis that insufficient sleep may contribute to chronic disease risk through the activation of molecular pathways that drive biological aging.
Supplementary Material
Experimental partial sleep deprivation (PSD) in older adults increased expression of genes associated with the senescence associated secretory phenotype.
PSD modestly increased expression of 30 genes associated with the DNA damage response.
Results link one night of partial sleep deprivation to molecular processes involved in biological aging.
Acknowledgements
This study was supported by the National Institute on Aging (NIA) R01 AG034588, UCLA Older Americans Independence Center (P30 AG028748), UCLA Clinical and Translational Science Institute (National Center for Advancing Translational Sciences, UL1TR000124), the National Institute of Mental Health (T32 MH019925) to JEC, the NIA K01 AG044462 to JEC, the American Sleep Medicine Foundation Bridge to K Award to JEC, the Cousins Center for Psychoneuroimmunology, the USC/UCLA Center on Biodemography & Population Health (NIA, P30 AG017265), and the UCLA Neuroscience Genomics Core Laboratory. Additional support to MRI from R01 AG026364; R01 CA160245; R01 HL095799; R01 DA032922.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare no competing financial interests.
References
- 1.Ohayon MM. Epidemiology of insomnia: what we know and what we still need to learn. Sleep Med Rev. 2002;6(2):97–111. doi: 10.1053/smrv.2002.0186. doi:10.1053/smrv.2002.0186. [DOI] [PubMed] [Google Scholar]
- 2.Petit L, Azad N, Byszewski A, Sarazan FF- A, Power B. Non-pharmacological management of primary and secondary insomnia among older people: review of assessment tools and treatments. Age Ageing. 2003;32(1):19–25. doi: 10.1093/ageing/32.1.19. [DOI] [PubMed] [Google Scholar]
- 3.Foley DJ, Monjan AA, Brown SL, Simonsick EM, Wallace RB, Blazer DG. Sleep complaints among elderly persons: an epidemiologic study of three communities. Sleep. 1995;18(6):425–32. doi: 10.1093/sleep/18.6.425. [DOI] [PubMed] [Google Scholar]
- 4.McEwen BS. Sleep deprivation as a neurobiologic and physiologic stressor: Allostasis and allostatic load. Metabolism. 2006;55(10 Suppl 2):S20–3. doi: 10.1016/j.metabol.2006.07.008. doi:10.1016/j.metabol.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Robles TF, Carroll JE. Restorative biological processes and health. Soc Personal Psychol Compass. 2011;5(8):518–837. doi: 10.1111/j.1751-9004.2011.00368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, Redline S, Shields AE, Williams DR, Williams MA. Associations of allostatic load with sleep apnea, insomnia, short sleep duration, and other sleep disturbances: findings from the National Health and Nutrition Examination Survey 2005 to 2008. Ann Epidemiol. 2014;24(8):612–9. doi: 10.1016/j.annepidem.2014.05.014. doi:10.1016/j.annepidem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullington JM, Haack M, Toth M, Serrador JM, Meier-Ewert HK. Cardiovascular, Inflammatory, and Metabolic Consequences of Sleep Deprivation. Prog Cardiovasc Dis. 2009;51(4):294–302. doi: 10.1016/j.pcad.2008.10.003. doi:10.1016/j.pcad.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Cauter E, Holmback U, Knutson K, et al. Impact of sleep and sleep loss on neuroendocrine and metabolic function. Horm Res. 2007;67(Suppl 1):2–9. doi: 10.1159/000097543. doi:10.1159/000097543. [DOI] [PubMed] [Google Scholar]
- 9.Ju S- Y, Choi W- S. Sleep duration and metabolic syndrome in adult populations: a meta-analysis of observational studies. Nutr Diabetes. 2013;3:e65. doi: 10.1038/nutd.2013.8. doi:10.1038/nutd.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palesh O, Aldridge-Gerry A, Zeitzer JM, et al. Actigraphy-measured sleep disruption as a predictor of survival among women with advanced breast cancer. Sleep. 2014;37(5):837–42. doi: 10.5665/sleep.3642. doi:10.5665/sleep.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Center for Disease Control [August 4, 2011];CDC Data & Statistics | Feature: Insufficient Sleep Is a Public Health Epidemic. 2011 Available at: http://www.cdc.gov/Features/dsSleep/.
- 12.Colten HR, Altevogt BM. Sleep disorders and sleep deprivation: an unmet public health problem. National Academies Press; 2006. Research I of MUS. C on SM and. [PubMed] [Google Scholar]
- 13.Motivala SJ. Sleep and Inflammation: Psychoneuroimmunology in the Context of Cardiovascular Disease. Ann Behav Med. 2011;42(2):141–152. doi: 10.1007/s12160-011-9280-2. doi:10.1007/s12160-011-9280-2. [DOI] [PubMed] [Google Scholar]
- 14.Everson CA, Henchen CJ, Szabo A, Hogg N. Cell injury and repair resulting from sleep loss and sleep recovery in laboratory rats. Sleep. 2014;37(12):1929–40. doi: 10.5665/sleep.4244. doi:10.5665/sleep.4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naidoo N. Cellular stress/the unfolded protein response: relevance to sleep and sleep disorders. Sleep Med Rev. 2009;13(3):195–204. doi: 10.1016/j.smrv.2009.01.001. doi:10.1016/j.smrv.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naidoo N. Roles of Endoplasmic Reticulum and Energetic Stress in Disturbed Sleep. Neuromolecular Med. 2012 doi: 10.1007/s12017-012-8179-9. doi:10.1007/s12017-012-8179-9. [DOI] [PubMed] [Google Scholar]
- 17.Naidoo N, Ferber M, Master M, Zhu Y, Pack AI. Aging impairs the unfolded protein response to sleep deprivation and leads to proapoptotic signaling. J Neurosci. 2008;28(26):6539–48. doi: 10.1523/JNEUROSCI.5685-07.2008. doi:10.1523/JNEUROSCI.5685-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown MK, Naidoo N. The UPR and the anti-oxidant response: relevance to sleep and sleep loss. Mol Neurobiol. 2010;42(2):103–13. doi: 10.1007/s12035-010-8114-8. doi:10.1007/s12035-010-8114-8. [DOI] [PubMed] [Google Scholar]
- 19.Cribbet MR, Carlisle M, Cawthon RM, et al. Cellular aging and restorative processes: subjective sleep quality and duration moderate the association between age and telomere length in a sample of middle-aged and older adults. Sleep. 2014;37(1):65–70. doi: 10.5665/sleep.3308. doi:10.5665/sleep.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackowska M, Hamer M, Carvalho LA, Erusalimsky JD, Butcher L, Steptoe A. Kiechl S, editor. Short Sleep Duration Is Associated with Shorter Telomere Length in Healthy Men: Findings from the Whitehall II Cohort Study. PLoS One. 2012;7(10):e47292. doi: 10.1371/journal.pone.0047292. doi:10.1371/journal.pone.0047292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prather AA, Puterman E, Lin J, et al. Shorter leukocyte telomere length in midlife women with poor sleep quality. J Aging Res. 2011;2011:721390. doi: 10.4061/2011/721390. doi:10.4061/2011/721390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang G, Schernhammer E, Qi L, Gao X, De Vivo I, Han J. Associations between Rotating Night Shifts, Sleep Duration, and Telomere Length in Women. PLoS One. 2011;6(8):e23462. doi: 10.1371/journal.pone.0023462. doi:10.1371/journal.pone.0023462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen S, Lin J, Matsuguchi T, et al. Short leukocyte telomere length predicts incidence and progression of carotid atherosclerosis in American Indians: The Strong Heart Family Study. Aging (Albany NY) 2014;6(5):414–27. doi: 10.18632/aging.100671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prather AA, Gurfein B, Moran P, et al. Tired telomeres: Poor global sleep quality, perceived stress, and telomere length in immune cell subsets in obese men and women. Brain Behav Immun. 2015;47:155–162. doi: 10.1016/j.bbi.2014.12.011. doi:10.1016/j.bbi.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–217. doi: 10.1016/j.cell.2013.05.039. doi:10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennedy BK, Berger SL, Brunet A, et al. Geroscience: Linking Aging to Chronic Disease. Cell. 2014;159(4):709–713. doi: 10.1016/j.cell.2014.10.039. doi:10.1016/j.cell.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci. 2014;69(Suppl 1):S4–9. doi: 10.1093/gerona/glu057. doi:10.1093/gerona/glu057. [DOI] [PubMed] [Google Scholar]
- 28.Jurk D, Wilson C, Passos JF, et al. Chronic inflammation induces telomere dysfunction and accelerates ageing in mice. Nat Commun. 2014;2:4172. doi: 10.1038/ncomms5172. doi:10.1038/ncomms5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120(4):513–22. doi: 10.1016/j.cell.2005.02.003. doi:10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Sanoff HK, Cho H, et al. Expression of p16(INK4a) in peripheral blood T-cells is a biomarker of human aging. Aging Cell. 2009;8(4):439–48. doi: 10.1111/j.1474-9726.2009.00489.x. doi:10.1111/j.1474-9726.2009.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Effros RB. The role of CD8 T cell replicative senescence in human aging. Discov Med. 2005;5(27):293–7. [PubMed] [Google Scholar]
- 32.Blackburn EH. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 2005;579(4):859–62. doi: 10.1016/j.febslet.2004.11.036. doi:10.1016/j.febslet.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 33.Blackburn EH. Telomere states and cell fates. Nature. 2000;408(6808):53–6. doi: 10.1038/35040500. doi:10.1038/35040500. [DOI] [PubMed] [Google Scholar]
- 34.Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8(9):729–40. doi: 10.1038/nrm2233. doi:10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 35.Baker DJ, Wijshake T, Tchkonia T, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479(7372):232–6. doi: 10.1038/nature10600. doi:10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erusalimsky JD. Vascular endothelial senescence: From mechanisms to pathophysiology. J Appl Physiol. 2009;106(1):326–32. doi: 10.1152/japplphysiol.91353.2008. doi:10.1152/japplphysiol.91353.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coppé J-P, Desprez P-Y, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. doi:10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freund A, Orjalo A V, Desprez P- Y, Campisi J. Inflammatory networks during cellular senescence: causes and consequences. Trends Mol Med. 2010;16(5):238–46. doi: 10.1016/j.molmed.2010.03.003. doi:10.1016/j.molmed.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irwin MR, Wang M, Ribeiro D, et al. Sleep loss activates cellular inflammatory signaling. Biol Psychiatry. 2008;64(6):538–540. doi: 10.1016/j.biopsych.2008.05.004. doi:10.1016/j.biopsych.2008.05.004.Sleep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irwin MR, Ziegler M. Sleep deprivation potentiates activation of cardiovascular and catecholamine responses in abstinent alcoholics. Hypertension. 2005;45(2):252–257. doi: 10.1161/01.HYP.0000153517.44295.07. doi:10.1161/01.HYP.0000153517.44295.07. [DOI] [PubMed] [Google Scholar]
- 41.Irwin MR, Wang M, Campomayor CO, Collado-Hidalgo A, Cole S. Sleep deprivation and activation of morning levels of cellular and genomic markers of inflammation. Arch Intern Med. 2006;166(16):1756–62. doi: 10.1001/archinte.166.16.1756. doi:10.1001/archinte.166.16.1756. [DOI] [PubMed] [Google Scholar]
- 42.Irwin M, McClintick J, Costlow C, Fortner M, White J, Gillin JC. Partial night sleep deprivation reduces natural killer and cellular immune responses in humans. FASEB J. 1996;10(5):643–53. doi: 10.1096/fasebj.10.5.8621064. [DOI] [PubMed] [Google Scholar]
- 43.Irwin M, Gillin JC, Dang J, Weissman J, Phillips E, Ehlers CL. Sleep deprivation as a probe of homeostatic sleep regulation in primary alcoholics. Biol Psychiatry. 2002;51(8):632–41. doi: 10.1016/s0006-3223(01)01304-x. [DOI] [PubMed] [Google Scholar]
- 44.Irwin MR, Olmstead R, Carroll JE. Sleep Disturbance, Sleep Duration, and Inflammation: A Systematic Review and Meta-Analysis of Cohort Studies and Experimental Sleep Deprivation. Biol Psychiatry. 2015 doi: 10.1016/j.biopsych.2015.05.014. doi:10.1016/j.biopsych.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carroll JE, Carrillo C, Olmstead R, et al. Sleep deprivation and divergent toll-like receptor-4 activation of cellular inflammation in aging. Sleep. 2015;38(2):205–11. doi: 10.5665/sleep.4398. doi:10.5665/sleep.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fredrickson BL, Grewen KM, Coffey KA, et al. A functional genomic perspective on human well-being. Proc Natl Acad Sci U S A. 2013;110(33):13684–9. doi: 10.1073/pnas.1305419110. doi:10.1073/pnas.1305419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coppé J-P, Patil CK, Rodier F, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6(12):2853–68. doi: 10.1371/journal.pbio.0060301. doi:10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laberge R- M, Zhou L, Sarantos MR, et al. Glucocorticoids suppress selected components of the senescence-associated secretory phenotype. Aging Cell. 2012;11(4):569–78. doi: 10.1111/j.1474-9726.2012.00818.x. doi:10.1111/j.1474-9726.2012.00818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–8. doi: 10.1038/nature08467. doi:10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodier F, Coppé J-P, Patil CK, et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11(8):973–9. doi: 10.1038/ncb1909. doi:10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krishnamurthy J, Torrice C, Ramsey MR, et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114(9):1299–307. doi: 10.1172/JCI22475. doi:10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salminen A, Kauppinen A, Kaarniranta K. Emerging role of NF-kB signaling in the induction of senescence-associated secretory phenotype (SASP). Cell Signal. 2012;24(4):835–845. doi: 10.1016/j.cellsig.2011.12.006. doi:10.1016/j.cellsig.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 53.Irwin M, Thompson J, Miller C, Gillin JC, Ziegler M. Effects of sleep and sleep deprivation on catecholamine and interleukin-2 levels in humans: clinical implications. J Clin Endocrinol Metab. 1999;84(6):1979–85. doi: 10.1210/jcem.84.6.5788. [DOI] [PubMed] [Google Scholar]
- 54.Irwin MR, Cole SW. Reciprocal regulation of the neural and innate immune systems. Nat Rev Immunol. 2011;11(9):625–632. doi: 10.1038/nri3042. doi:10.1038/nri3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Irwin M, Clark C, Kennedy B, Christian Gillin J, Ziegler M. Nocturnal catecholamines and immune function in insomniacs, depressed patients, and control subjects. Brain Behav Immun. 2003;17(5):365–72. doi: 10.1016/s0889-1591(03)00031-x. [DOI] [PubMed] [Google Scholar]
- 56.Cole SW, Arevalo JMG, Takahashi R, et al. Computational identification of gene-social environment interaction at the human IL6 locus. Proc Natl Acad Sci U S A. 2010;107(12):5681–6. doi: 10.1073/pnas.0911515107. doi:10.1073/pnas.0911515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salminen A, Huuskonen J, Ojala J, Kauppinen A, Kaarniranta K, Suuronen T. Activation of innate immunity system during aging: NF-kB signaling is the molecular culprit of inflamm-aging. Ageing Res Rev. 2008;7(2):83–105. doi: 10.1016/j.arr.2007.09.002. doi:10.1016/j.arr.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 58.Rovillain E, Mansfield L, Caetano C, et al. Activation of nuclear factor-kappa B signalling promotes cellular senescence. Oncogene. 2011;30(20):2356–66. doi: 10.1038/onc.2010.611. doi:10.1038/onc.2010.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chien Y, Scuoppo C, Wang X, et al. Control of the senescence-associated secretory phenotype by NF-B promotes senescence and enhances chemosensitivity. Genes Dev. 2011;25(20):2125–36. doi: 10.1101/gad.17276711. doi:10.1101/gad.17276711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Redwine L. Effects of Sleep and Sleep Deprivation on Interleukin-6, Growth Hormone, Cortisol, and Melatonin Levels in Humans. J Clin Endocrinol Metab. 2000;85(10):3597–3603. doi: 10.1210/jcem.85.10.6871. doi:10.1210/jc.85.10.3597. [DOI] [PubMed] [Google Scholar]
- 61.Moller-Levet CS, Archer SN, Bucca G, et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc Natl Acad Sci. 2013;110(12):E1132–E1141. doi: 10.1073/pnas.1217154110. doi:10.1073/pnas.1217154110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sahin E, Colla S, Liesa M, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470(7334):359–365. doi: 10.1038/nature09787. doi:10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taffett GE. Physiology of Aging. In: Cassel CK, editor. Geriatric Medicine: An Evidence-Based Approach. 4th ed. Springer Science & Business Media; 2003. pp. 27–35. [Google Scholar]
- 64.Vgontzas AN, Zoumakis M, Bixler EO, et al. Impaired nighttime sleep in healthy old versus young adults is associated with elevated plasma interleukin-6 and cortisol levels: physiologic and therapeutic implications. J Clin Endocrinol Metab. 2003;88(5):2087–95. doi: 10.1210/jc.2002-021176. doi:10.1210/jc.2002-021176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.