Abstract
Macrophage (MΦ) dysregulation is increasingly becoming recognized as a risk factor for a number of inflammatory complications including atherosclerosis, cancer, and the host response elicited by biomedical devices. It is still unclear what roles the pro-inflammatory (M1) MΦ and pro-healing (M2) MΦ phenotypes play during the healing process. However, it has been shown that a local overabundance of M1 MΦs can potentially lead to a chronically inflamed state of the tissue; while a local over-exuberant M2 MΦ response can lead to tissue fibrosis and even promote tumorigenesis. These notions strengthen the argument that the tight temporal regulation of this phenotype balance is necessary to promote inflammatory resolution that leads to tissue homeostasis. In this study, we have engineered pro-inflammatory MΦ, MΦ-cTLR4 cells, which can be activated to a M1-like MΦ phenotype with a small molecule, the chemical inducer of dimerization (CID) drug. The MΦ-cTLR4 cells when activated with the CID drug, express increased levels of TNFα, IL-6, and iNOS. Activated MΦ-cTLR4 cells stay stimulated for at least 48 hours; once the CID drug is withdrawn, the MΦ-cTLR4 cells return to baseline state within 18 hours. Further, in vitro CID-activated MΦ-cTLR4 cells induce upregulation of VCAM-1 and ICAM-1 on endothelial cells (EC) in a TNFα-dependent manner. With the ability to specifically modulate the MΦ-cTLR4 cells with the presence or absence of a small molecule, we now have the tool necessary to observe a primarily M1 MΦ response during inflammation. By isolating this phase of the wound healing response, it may be possible to determine conditions for ideal healing.
Keywords: Macrophages, Toll-like receptor 4, Inflammation, Endothelial Cells, Angiogenesis
Introduction
The physiological innate inflammatory response requires a highly orchestrated series of events characterized by four basic phases: reaction, regrowth, remodeling, and resolution.[1] During the course of this healing process, MΦ play an active role in secreting chemokines and cytokines that direct the recruitment and egress of various immune cell types at the injured site. The functional MΦ phenotype depends on the microenvironment of the injured site and alters accordingly during the normal process of healing.[2] However, dysregulation of the MΦ phenotype can lead to a non-ideal healing outcome.
Monocytes are the precursor cells to MΦ, which are a main inflammatory cell type and are known to be key players in the inflammatory response. When activated, MΦ are classified into two major phenotypes that can be broadly defined as: pro-inflammatory MΦ and pro-healing MΦ. In literature, pro-inflammatory MΦ are often denoted as “classically activated” or “M1” and pro-healing MΦ are denoted as “alternatively activated” or “M2.” However, these two major MΦ phenotypes are the two extremes on the phenotype scale, as intermediate macrophage types also exist.[3] During the inflammatory reaction, M1 MΦ are the first to arrive at the inflammation site and this MΦ population subsequently shifts to a less inflammatory pro-healing M2 MΦ population during the repair phase. Pro-inflammatory MΦ release inflammatory cytokines, such as TNFα and IL-6 as well as produce reactive oxygen species (ROS).[4, 5] In comparison, the pro-healing MΦ phenotype has been shown to produce cytokines, such as IL-10 and TGFβ1, which are markers that can decrease the pro-inflammatory response and promote healing and fibrosis.[3]
Angiogenesis, or the formation of new blood vessels, is a critical step in the wound healing process. In the course of the inflammation response, pro-inflammatory MΦ secrete TNFα and IFN-γ, which regulate expression of adhesion molecules on EC. These cytokines promote leukocyte adhesion to EC and extravasation into tissues by increasing expression of both cell surface and soluble forms of vascular cell adhesion protein 1 (VCAM-1) and intercellular adhesion molecule 1 (ICAM-1).[6–9] These two adhesion molecules belong to the immunoglobulin superfamily group and have been implicated, along with integrins, in pro-angiogenic processes.[10, 11]
This study utilizes engineered pro-inflammatory M1-like cells to investigate EC activation. The engineered cells were designed by using the chemical inducer of dimerization (CID) technology to induce activation of the TLR4 receptor independent of the lipopolysaccharides (LPS) exogenous ligand, which is a well-established inducer of the M1 MΦ phenotype.[12] The CID technology has been used in several other groups to control a variety of cell signaling pathways.[13–17] For this system to function, the intracellular domain of a desired receptor is fused to a F36V protein, which is a mutated version of the FKBP12 protein. This F36V version has a binding site for a cell permeable CID drug. When CID drug is present, two F36V proteins will dimerize, bringing the desired intracellular domains in close enough proximity to activate receptor-specific pathways. These cells can be activated by the exposure to CID drug and be deactivated by the withdrawal of CID drug. Currently in literature there have been no cellular engineering approaches to control or modulate MΦ polarization to determine ideal healing conditions. Having the ability to control MΦ polarization during and after an inflammatory response could potentially allow for the manipulation of the host response and the optimal healing of multiple inflammatory conditions.
Methods and Materials
Reagents and antibodies
The monoclonal anti-human/mouse/rat FKBP12 antibody was purchased from Thermo Scientific. The following antibodies were purchased from Cell Signaling: p44/42 MAPK, Phospho -p44/42 MAPK, IRF3 and Phospho-IRF3. The anti-iNOS/NOS type II antibody was purchased from BD Biosciences. The anti-mouse CD106 (VCAM-1) PE, the anti-mouse CD54 (ICAM-1) PE, the rat IgG2b isotype, and the anti-mouse TNFα antibodies were purchased from eBioscience. The HRP-conjugated goat-anti-rabbit antibody was obtained from Jackson ImmunoResearch Laboratories, Inc. and the HRP-conjugated goat-anti-mouse antibody was obtained from Life Technologies. LPS and recombinant mouse TNFα was purchased from Sigma and recombinant mouse IL-4 was purchased from eBioscience. AP20187 (CID drug) was purchased from Clontech. Lipofectamine 2000 was purchased from Invitrogen. The Dual-Luciferase ® reporter assay system was obtained from Promega Corporation.
Plasmid construction of cTLR4
The mouse Sport6-TLR4 vector was purchased from Open Biosystems. The cytoplasmic portion of TLR4 (cTLR4) was amplified (mRNA base pairs 2207–2748) and inserted into a pBluescript II KS+ vector with an existing myristolation domain and engineered F36V domain (pBluescript-Myr-F36V) (55) following BamHI and EcoRV restriction enzyme (RE) cuts. PCR products were gel purified using a QIAEX II gel extraction kit (Qiagen) before ligations were performed. This resulted in a pBluescript-Myr-F36V-cTLR4 construct. The pCDH-EF1α-MCS-T2A-copGFP lentiviral cDNA and expression vector was purchased from System Biosciences. This vector was cut in the MCS with both NheI and EcoRI, and a PCR amplified portion of the Myr-F36V-cTLR4 sequence was ligated into this site within the pCDH-EF1α-MCS-T2A-copGFP vector (7.26 kb). This resulted in the final cTLR4 lentiviral plasmid: pCDH-EF1α-Myr-F36V-cTLR4-T2A-copGFP (8.18 kb).
Cell transduction of cTLR4 lentiviral constructs
We utilized a 3rd generation lentiviral vector, pCDH (System Biosciences), carrying the cTLR4 gene under the control of the EF-1α promoter. For stable lentiviral transductions, 5×106 HEK293T packaging cells were seeded in 10-cm cell culture dishes that were previously coated with 50 ug/mL poly-D-lysine hydrobromide (Sigma). Culture medium was changed just prior to transduction. In total, 12 µg plasmid DNA was used for each 10-cm dish (2.8 µg transfer vector (cTLR4), 0.9 µg pSL3 (vesicular stomatitis virus G envelope), 5.4 µg pSL4 (HIV-1 gag/pol packing genes), and 2.8 µg pSL5 (rev gene required for HIV-1 envelope protein expression). DNA and Lipofectamine 2000™ (Life Technologies) were diluted in Opti-MEM® medium (Gibco) separately. After a 5 minute incubation, DNA and lipofectamine were combined and incubated for 20 minutes at room temperature. The complexes were then added, drop-wise, to cell dishes with 8 mL growth media and medium was replaced after 14–16 hours. Virus supernatant was collected following an additional 48 hours by filtering through a 0.45 µm filter. Filtered virus supernatant was then added either directly or in concentrated form to previously plated RAW264.7 cells (5×105 cells per well) in 6-well plates. Cells were then sorted for GFP expression to acquire >90% transduction efficiency.
Cell culture
RAW264.7 and bEnd.3 cells were obtained from ATCC. RAW264.7 and bEnd.3 cells were cultured in DMEM medium from Invitrogen containing 10% (v/v) heat-inactivated FBS and 100 U/ml pen/strep (Invitrogen) and incubated at 37°C with 5% CO2.
Western Blotting
Protein from RAW264.7, MΦ-T2A (vector control cells), and MΦ-cTLR4 cell monolayers were extracted by lysis in Laemmli buffer containing 1× Halt Protease Inhibitor cocktail (Thermo Scientific). Following lysis, samples were boiled and protein concentration was determined by performing a BCA assay from Thermo Scientific. Samples (10–30 µg of lysates) were run on 4–20% Mini-PROTEAN® TGX precast polyacrylamide gels (Bio-Rad). Protein from gels were transferred onto PVDF membranes and probed with the appropriate primary antibody overnight. Membranes were washed between each antibody incubation and subsequently probed with the appropriate HRP-conjugated secondary antibody (Life Technologies). The Clarity Western ECL Substrate (Bio-Rad) was used to detect bands.
Cytokine Profile
We tested IL-6 and TNFα concentrations in supernatants of transduced RAW264.7 cells in vitro. Briefly, MΦ-cTLR4 cells (1 × 106) were plated in each well of a 6-well plate and treated with vehicle (100% EtOH), LPS (100 ng/mL), CID drug (50 nM), or left untreated in DMEM without serum. Supernatants were collected and tested using the mouse IL-6 ELISA Ready-SET-Go! and the mouse TNFα ELISA Ready-SET-Go! Kits (eBioscience) according to the manufacturer’s instructions. Plates were read at 450 nM with a 570 nM wavelength subtraction, normalized to standard solutions, and concentrations (pg/mL) were calculated.
Luciferase Assay
Two hundred thousand cells/well were seeded in 24-well plates. The following day each well was transfected with a total of 0.8 µg plasmid DNA, which consisted of a 20:1 ratio of pBIIX-LUC (NFκB reporter construct):pRL (Renilla luciferase construct). The promoterless pGL4.10 vector was also transfected in a 20:1 ratio of pGL4.10:pRL, as a control. Transfections of each well were performed with 2 µL Lipofectamine 2000 (Invitrogen). The following morning transfection reagents were replaced with fresh serum-free medium and treated with either vehicle (100% EtOH) or CID drug (50 nM) for 4 hours. Cell lysate was harvested and luciferase activity was measured using a Dual-Luciferase ® reporter assay kit (Promega) according to manufacturer’s instructions. All groups were normalized to Renilla luciferase.
Endothelial Cell Activation
MΦ-cTRL4 conditioned media, following a 6 hour treatment in 6-well plates (1×106 cells/well), was transferred to plated bEnd.3 cells in a 12-well plate (0.2×106 cells/well). Before media transfer, TNFα neutralizing and IgG isotype antibody (1 µg/mL) were incubated in media for 15 minutes. Media was then added to bEnd.3 cells for 12 hours. Following incubation, bEnd.3 cells were trypsinized and stained for ICAM-1 and VCAM-1. Cell cytometry was performed on a FACSCanto II Cell Analyzer (BD Biosciences) equipped with 488 nm and 647 nm lasers. Typically, 10,000 cells were analyzed per sample. Experiments were repeated at least three times. Non-specific staining was evaluated using a monoclonal antibody for IgG2b and IgG2a (eBioscience).
Statistical Analysis
Results are expressed as mean ± SE unless otherwise specified. Significance between groups was determined by ANOVA and p-values less than 0.05 were considered significant.
Results
Engineering MΦ-cTLR4 pro-inflammatory macrophages
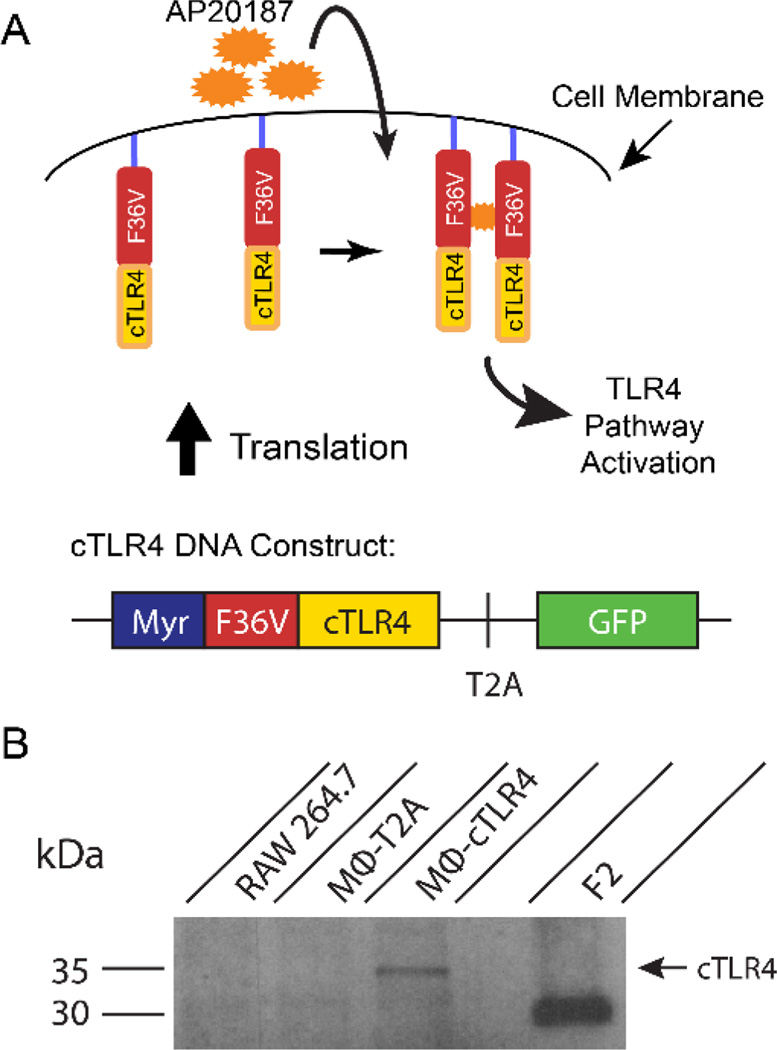
With the goal of developing inducible M1 MΦ cells, we have engineered the murine monocytic cell line RAW264.7 to express a fusion protein comprising the intracellular TLR4 signaling domain and F36V-dimerization domains that bind to a cell permeable CID drug (Figure 1A). The cTLR4 construct is in a pCDH expression system. The 5’ end of the construct starts with a myristoylation domain (Myr), which allows targeting to the membrane to mirror the spatial localization of the endogenous full length TLR4 domain. The Myr domain is followed by the engineered F36V dimerization domain, which has a binding site for the CID drug. This domain is linked to the cTLR4 domain, which is only the cytoplasmic portion of the receptor that is necessary for proper signal transduction. This design allows dimerization via a F36V-F36V interaction with the homodimerization CID drug (AP20187). Lastly, there is a T2A ribosome skipping sequence that allows the separate expression of GFP at the 3’ end of the construct.
Figure 1. Diagram of cTLR4 Construct and Engineered Receptor and Confirmation of cTLR4 construct expression.
(A) The cTLR4 construct contains: a myristolation domain (Myr), an engineered dimerization domain (F36V), the cytoplasmic portion of the TLR4 domain (cTLR4), a T2A ribosome skipping sequence, and a GFP tag. (B) Western blot probing for the FKBP12/F36V domain (35.5 kDa) containing lysates from RAW264.7 cells, MΦ-T2A negative control cells, MΦ-cTLR4 cells, and F2 positive control cells. MΦ-T2A cells have been transduced with a construct that contains only the T2A ribosome skipping sequence with the GFP tag. F2 cells have been transduced with just two adjacent F36V domains.
Delivery of the cTLR4 engineered constructs to RAW264.7 cells was achieved via lentiviral methods. Control MΦ cells were also generated. These cells were transfected with constructs lacking the cytoplasmic and engineered domain (MΦ-T2A). Confirmation that the whole engineered construct was being transcribed was validated by the expression of the GFP reporter marker in the MΦ-cTLR4 cell line (data not shown). Protein expression of the cTLR4 construct in the MΦ-cTLR4 engineered cells was verified by western blot analysis for the FKBP12/F36V domain. A corresponding 35.5 kDa band can be seen in Figure 1B.
Pro-inflammatory activation of MΦ-cTLR4 cells
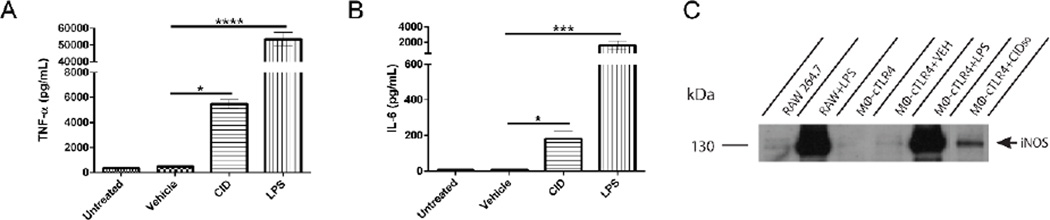
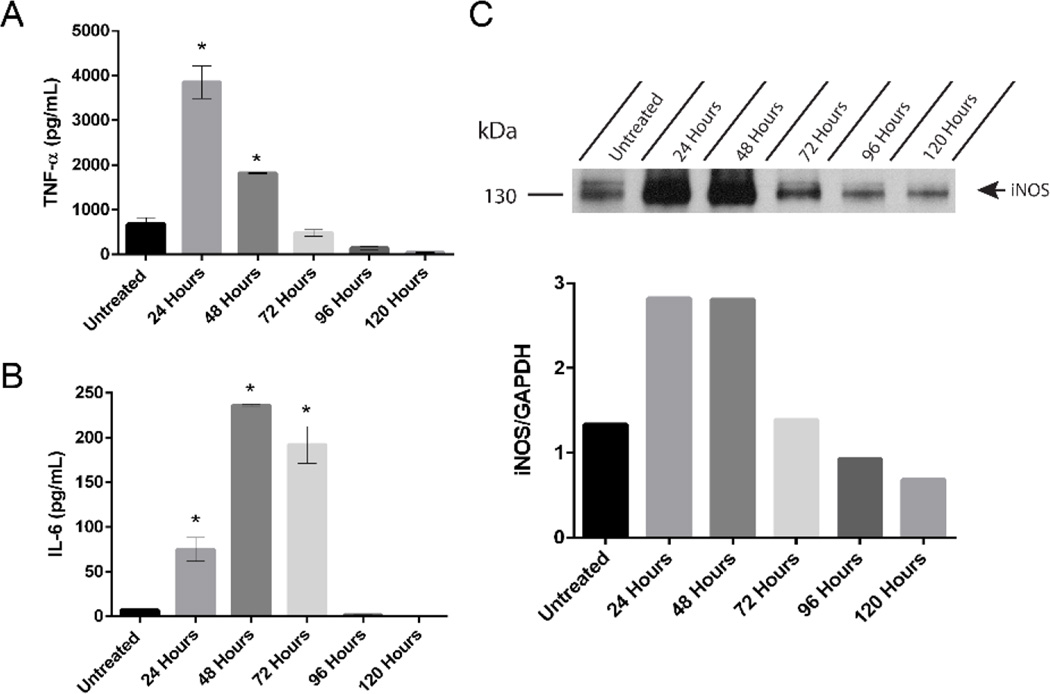
Polarized classical inflammatory MΦ are known to have increased levels of TNFα, IL-6, and iNOS.[8] Therefore the MΦ-cTLR4 cells were tested for the presence and levels of these markers. Engineered MΦ-cTLR4 cells were seeded in 6-well culture plates overnight and then treated for 24 hours with CID drug, vehicle, or LPS as a positive control. Polarization was confirmed by ELISA and western blot analyses. CID-treated MΦ-cTLR4 cells expressed increased TNFα and IL-6 levels when compared to uninduced controls (Figure 2A & 2B). However, the CID-treated MΦ-cTLR4 cell levels were not as high as the LPS-treated cells. We also tested for iNOS via a western blot and observed similar results with this marker. The positive control LPS-treated cells had a substantial increase in iNOS expression and there is a band evident at 130 kDa in the CID-treated lane, however, it has much lower intensity than the LPS-treated cells (Figure 2C). The difference in LPS-treated groups and CID-treated groups can be attributed to the fact that LPS also acts on other receptors, such as CD14 and the Macrophage Scavenger Receptor, in which both of these receptor pathways can lead to NF-κB responses. Thus, there is potentially more input signal from LPS than our CID drug, which only activates through TLR4.
Figure 2. CID-treated MΦ-cTLR4 Cells Exhibit Increased Expression of TNFα, IL-6, and iNOS.
Expression of classical inflammatory MΦ phenotype markers, determined by sandwich ELISA assay. Bar graphs show the levels of (A) TNFα and (B) IL-6 of CID-treated (50 nM) MΦ-cTLR4 cells compared to untreated, vehicle (100% EtOH), and LPS-treated cells (100 ng/mL). Cell media was collected following 24 hour treatment. (C) Western blot shows intensity of iNOS expression (130 kDa) for CID and LPS-treated MΦ-cTLR4 cells when compared to controls. Cells were lysed following 24 hour treatment.
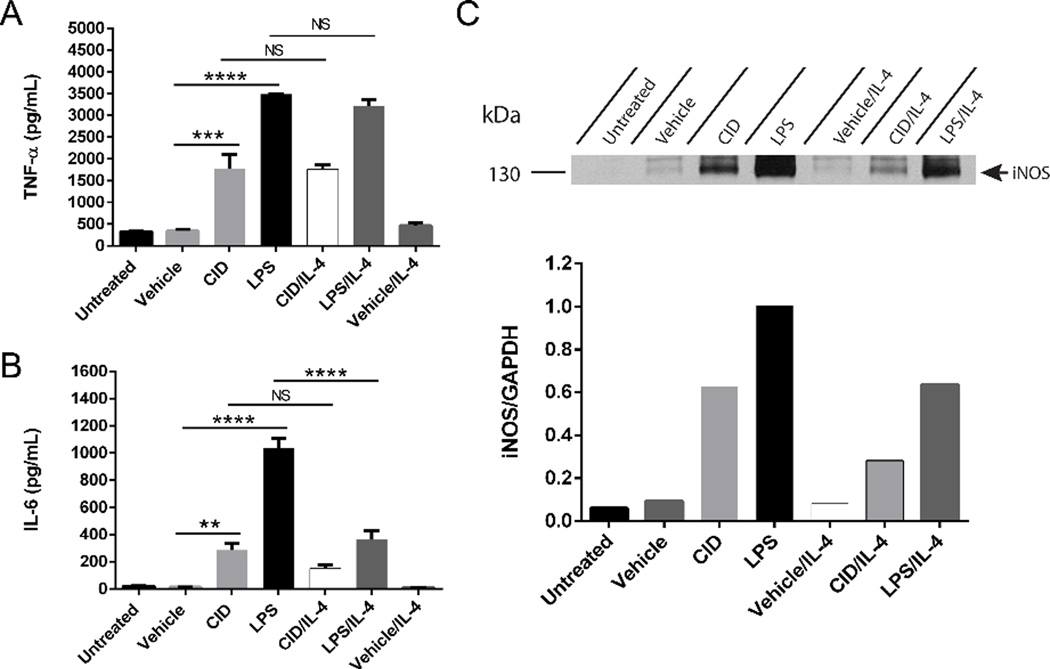
Diversity and plasticity are hallmarks of cells from the MΦ lineage and they can change phenotype depending on the surrounding microenvironment.[18] Thus, we tested how the expression of classical MΦ markers in the MΦ-cTLR4 cells were influenced by a M2 MΦ activator. Engineered cells were treated with a cocktail of either vehicle/IL-4, CID/IL-4, or LPS/IL-4, as well as the appropriate controls. The degree of polarization was assessed by ELISA and western blot analyses for IL-6, TNFα, and iNOS (Figure 3). Both CID- and LPS-treated groups responded similarly to the IL-4 cocktail treatment and exhibited about half the expression of IL-6 and iNOS when compared to treatment groups without IL-4. However, TNFα levels for both CID drug and LPS with and without IL-4 were not significantly different.
Figure 3. MΦ-cTLR4 cells are influenced by IL-4 treatment.
Expression of classical inflammatory MΦ phenotype markers following cocktail treatment of IL-4 (60 ng/mL) and CID drug (50 nM), LPS (100 ng/mL) or vehicle. Bar graphs show the levels of (A) TNFα and (B) IL-6 of cocktail-treated cells compared to controls. Cell media was collected following 24 hour treatment. (C) Western blot shows intensity of iNOS expression (130 kDa) for cocktail-treated MΦ-cTLR4 cells when compared to controls. Cells were lysed following 24 hour treatment.
MΦ-cTLR4 cells response to CID drug dose, withdrawal, and length of treatment
For MΦ-cTLR4 cells, IL-6 levels are elevated in CID-treated cells when compared to controls. In order to find the optimal in vitro dosage, an IL-6 ELISA was performed to test for the maximum signal of this cytokine in a CID drug titration experiment. The optimal dose of CID drug corresponds to the lowest dose that induces the highest level of IL-6 expression. The IL-6 ELISA results are seen in Supplemental Figure 1. These results suggest that a dose of at least 50 nM, produces the maximum activation of MΦ-cTLR4 cells in the range from 50 nM – 250 nM.
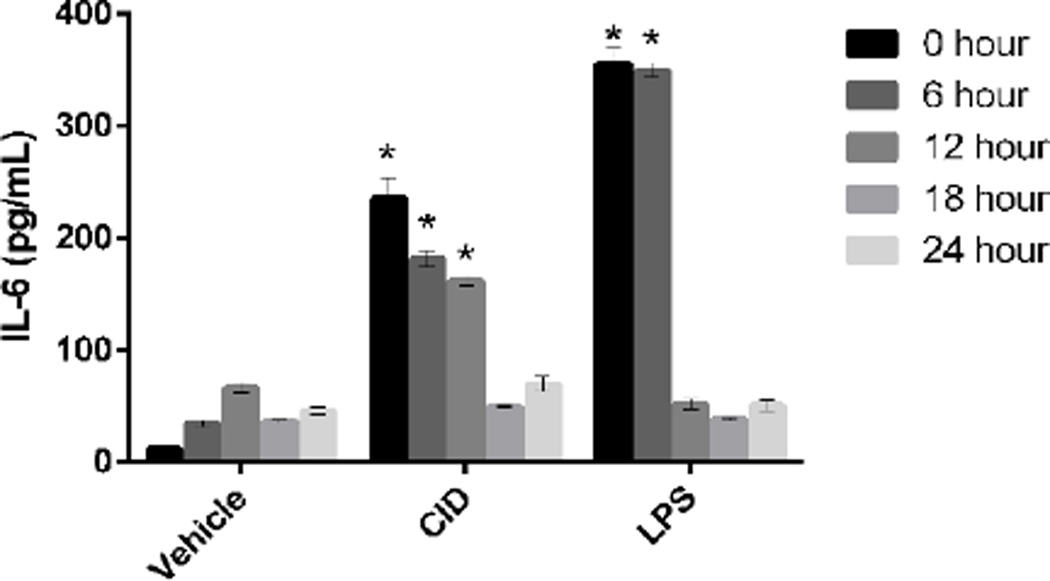
A withdrawal experiment was also performed to determine the time in which the cells would revert to a baseline state following CID drug withdrawal. MΦ-cTLR4 cells were seeded in a 6-well culture plate (1×106 cells/well). Cells were treated with vehicle, CID drug, or LPS for 24 hours. Timepoints were collected after complete CID drug withdrawal and IL-6 levels were measured at each timepoint to determine activation intensity. Results showed that cells converged to their baseline state at approximately 18 hours (Figure 4).
Figure 4. MΦ-cTLR4 Cells Return to Baseline Levels 18 Hours Following CID Drug Withdrawal.
CID drug withdrawal experiment, determined by IL-6 expression. Cells were treated with CID drug (50 nM), LPS (100 ng/mL), or vehicle for 24 hours and then left untreated for up to 24 hours. Media was collected at the indicated timepoints after complete CID drug withdrawal and IL-6 levels were measured at each timepoint to determine activation intensity.
In order to determine how long the engineered MΦ-cTLR4 cells would stay “on” or activated, we performed a longevity study for TNFα, IL-6, and iNOS. With constant CID drug presence in the media, we found that the MΦ-cTLR4 cells maintain considerable elevated levels of all three pro-inflammatory markers for at least 48 hours (Figure 5A–5C). The IL-6 levels stayed activated the longest for 72 hours.
Figure 5. CID-treated MΦ-cTLR4 Cells Remain Activated for At Least 48 Hours.
Longevity study determined by expression of (A) TNFα, (B) IL-6, and (C) iNOS in MΦ-cTLR4 cells. Media containing CID drug (50 nM) was changed every 24 hours. Cell media was collected at timepoints indicated.
Lastly, the MΦ-cTLR4 cells were optimized for maximal signal to baseline activation by sorting four different GFP intensity populations: dim, midlow, midhigh, and high. An IL-6 ELISA was performed to determine activation of these populations compared to unsorted MΦ and MΦ-T2A populations (Supplemental Figure 2). As signal intensity increased, the baseline activation of MΦ-cTLR4 cells also increased. A potential explanation for the high baseline activation as GFP intensity increases might be that some cells have more cTLR4 constructs integrated into their genome, thus resulting in higher GFP intensity. This higher integration will yield a greater concentration of the engineered cTLR4 construct on the cell surface and might result in self-dimerization, if the constructs are in close enough proximity. Ultimately, we determined that the midlow MΦ-cTLR4 population had similar CID and LPS activation, as well as the highest signal to noise ratio, so we used this sorted population for the remaining experiments.
MyD88-dependent and MyD88-independent signaling pathway activation in MΦ-cTLR4 cells
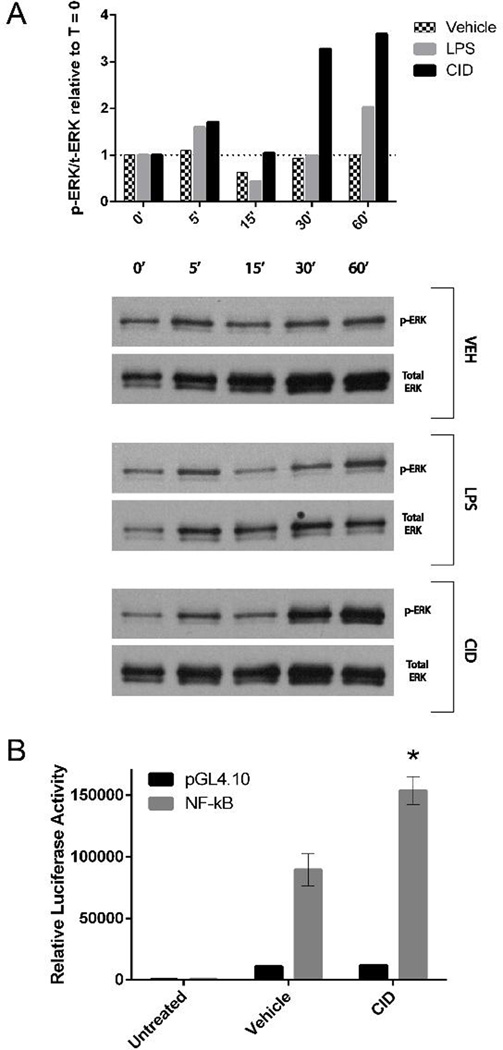
Following LPS stimulation and subsequent TNFα production, the TLR4 pathway leads to activation of NF-κB and the three MAPK pathways through the MyD88-dependent pathway. Both NF-κB and MAPK pathways directly control the transcription of the IL-6 and iNOS inflammatory genes, as well as control the mRNA stability of those transcripts. For the activated MΦ-cTLR4 cells, ERK1/2 phosphorylation is expected if the MyD88 dependent pathway and subsequent downstream TRAF6 activation has occurred. Therefore, we performed a western blot to probe for phosphorylated-ERK (p-ERK) and total ERK and compare the p-ERK/total ERK ratio relative to the zero timepoint (Figure 6A). As time increases from 0 minutes to 60 minutes, the CID-treated MΦ-cTLR4 cells exhibit an upregulation of ERK1/2 phosphorylation at the 5 minute timepoint, a subsequent decrease for the 15 minute timepoint, and then a significant increase for the last two timepoints. The LPS-treated MΦ-cTLR4 cells exhibited a similar ERK1/2 phosphorylation pattern with lower maximum phosphorylation. The NF-κB transcription factor has also been shown to be activated following TLR4 dimerization. Thus, MΦ-cTLR4 cells were tested for NF-κB promoter activation via a Dual-Luciferase reporter assay. Cells were transduced with a NF-kB responsive promoter element driving the luciferase gene. Measurement of the luciferase activity following CID treatments (Figure 6B) shows that CID-treated MΦ-cTLR4 cells have increased NF-κB promoter activation when compared to the vehicle. These results suggest that the CID-treated cells signal through the MyD88 dependent pathway.
Figure 6. CID-treated MΦ-cTLR4 Cells Activate the MyD88-dependent Pathway.
(A) Western blot probing for p-ERK and total ERK. Cells were lysed at the time points indicated following CID drug, LPS, or vehicle treatment. Top panel shows phosphorylated over total ERK ratio relative to the zero timepoint for each subsequent timepont. Bottom panel shows corresponding blots (B) A Dual-luciferase® assay was used to determine NF-κB activity in MΦ-cTLR4 cells. Luciferase activity was measured 4 hours following treatment of MΦ-TLR4 cells with CID drug, LPS, or vehicle treatment. Luciferase activity was normalized to baseline Renilla luciferase activity.
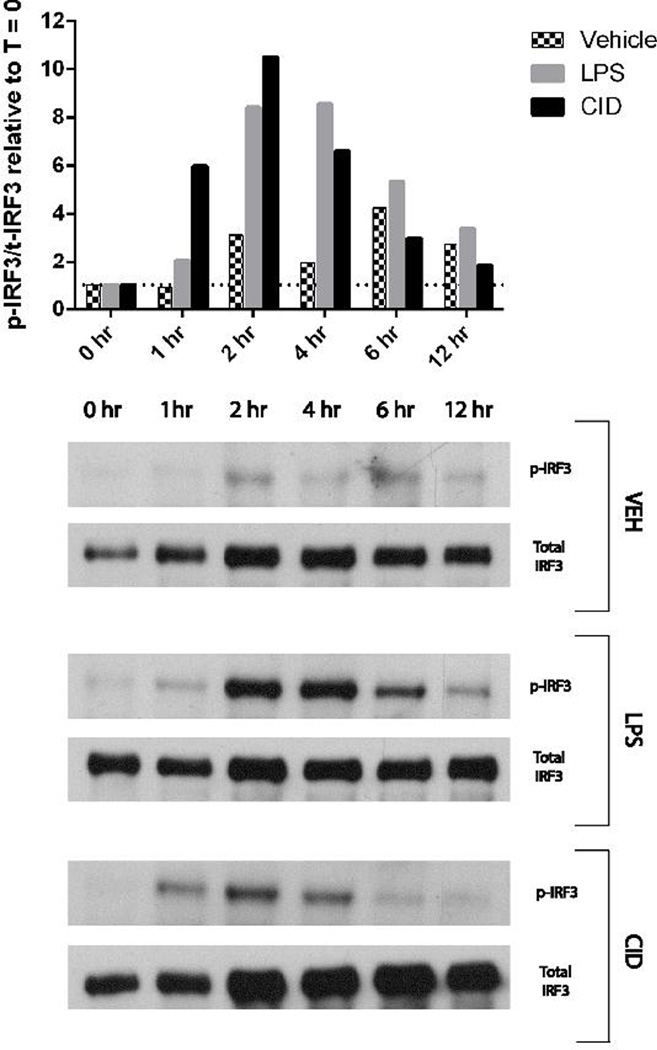
To determine if the MΦ-cTLR4 cells were signaling through the MyD88-independent pathway, we tested for phosphorylated IRF3. This protein is downstream of the MyD88-independent pathway and has been shown to translocate into the nucleus and regulate type I interferon responses.[19] Western blot analysis of the p-IRF3/total IRF3 ratio relative to the zero timepoint (Figure 7) shows a pronounced activation peak at 2 hours for both CID- and LPS-treated MΦ-cTLR4 cells when compared to vehicle. The IRF3 phosphorylation of the CID- and LPS-treated MΦ-cTLR4 cells starts to decrease following the 2 hour timepoint and subsequently reaches similar levels as vehicle at the 6 and 12 hour timepoints.
Figure 7. CID-treated MΦ-cTLR4 Cells Activate the MyD88-independent Pathway.
Western Blot for p-IRF3 and total IRF3. Cells were lysed at timepoints indicated following CID drug (50 nM), LPS (100 ng/mL), or vehicle treatment. Top panel shows phosphorylated over total IRF3 ratio relative to the zero timepoint for each subsequent timepoint. Bottom panel shows corresponding blots.
MΦ-cTLR4 endothelial cell activation
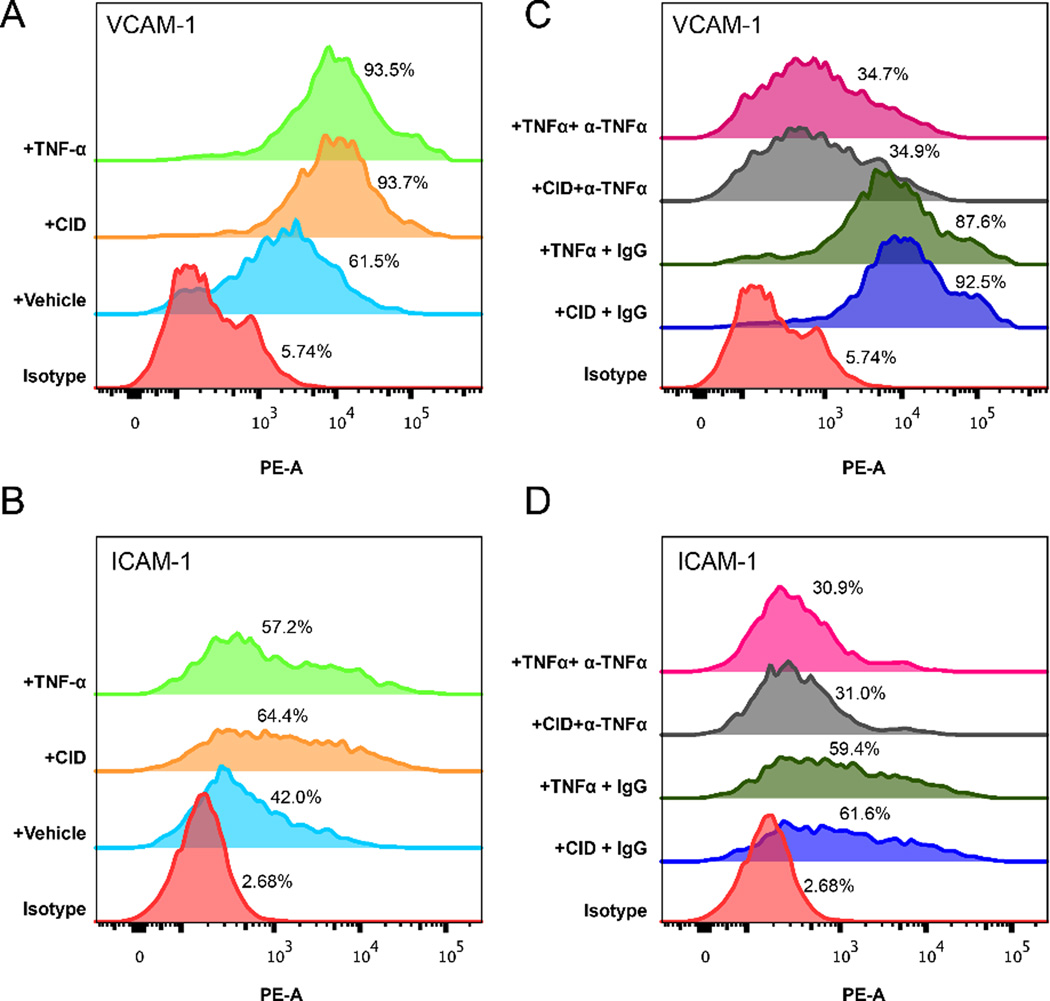
Better wound healing outcomes have been correlated with increased angiogenesis.[20] Furthermore, areas containing almost entirely pro-inflammatory MΦ have been shown to correlate with angiogenesis, in specific cases.[20, 21] Thus, we tested whether engineered MΦ-cTLR4 cells-derived factors were able to induce EC activation by measuring the expression of the VCAM-1 and ICAM-1 adhesion molecules. EC incubated with media from MΦ-cTLR4 treated with TNFα and CID drug both had increased expression of VCAM1 and ICAM-1 (Figure 8A & 8B) when compared to vehicle alone. For the TNFα-treatment group, 93.5% of EC were positive for VCAM-1 and 57.2% of EC were positive for ICAM-1. The CID drug-treatment group was very similar, with 93.7% of the EC being positive for VCAM-1 and 64.4% of the EC being positive for ICAM-1. The vehicle group had a baseline VCAM-1 and ICAM-1 expression when compared to the isotype control.
Figure 8. Media From CID-treated MΦ-cTLR4 Cells Upregulate VCAM-1 and ICAM-1 on Endothelial Cells.
Endothelial activation, determined by flow cytometry. Conditioned media from MΦ-cTLR4 cells treated for 6 hours with TNFα, CID, and vehicle was transferred to plated bEnd.3 endothelial cells and left to incubate for 12 hours. Following the 12 hour incubation, bEnd.3 cells were then trypsinized and stained for both VCAM-1 and ICAM-1. (A & B) Histograms showing intensity of VCAM-1 and ICAM-1 expression on bEnd.3 cells incubated with MΦ-TLR4 conditioned media treated with TNFα (20 ng/mL), CID (50 nM), or vehicle (C & D) Histograms showing intensity of VCAM-1 and ICAM-1 expression on bEnd.3 cells incubated with MΦ-TLR4 conditioned media treated with TNFα (20 ng/mL), CID (50 nM), or vehicle, as well as with or without TNFα neutralizing antibody (α-TNFα).
To determine if the TNFα was the main driver of the EC activation, we repeated the experiment with a neutralizing antibody for TNFα (Figure 8C & 8D). Before adding the MΦ-cTLR4 conditioned media to the EC, a TNFα neutralizing antibody was incubated in the media for 15 minutes. The CID and the TNFα groups with the neutralizing antibody had severely decreased levels of both VCAM-1 and ICAM-1 when compared to the IgG control. The VCAM-1 expression was decreased by a factor of three and the ICAM-1 expression was decreased by a factor of two and fell below vehicle/baseline treatment, thus indicating that some of the activity at baseline is TNFα-dependent.
Discussion
In this study we engineered RAW264.7 cells to have the ability to polarize into pro-inflammatory MΦ, by using the CID system for the TLR4 receptor. We confirmed that the engineered cTLR4 receptor was being expressed in the stably transduced RAW264.7 cell line. Additionally we determined that both MyD88-dependent and MyD88-independent pathways were activated by CID drug treatment in MΦ-cTLR4 cells. The CID-treated MΦ-cTLR4 cells displayed M1-like MΦ characteristics, such as increased IL-6, TNFα, and iNOS expression. MΦ-cTLR4 cells were influenced by IL-4 cocktail treatment, however, the engineered cells still displayed M1 MΦ characteristics, albeit at lower expression levels. The MΦ-cTLR4 cells remained polarized in response to CID drug for at least 48 hours and CID drug withdrawal experiments suggest that the engineered cells became deactivated 18 hours after drug withdrawal. Lastly, we showed that these engineered cells have functional properties by performing a MΦ-cTLR4 conditioned-media experiment with EC. CID-polarized MΦ-cTLR4 conditioned-media had the ability to activate EC by upregulating both VCAM-1 and ICAM-1 expression on the cell surface, which are two cell adhesion molecules associated with angiogenic processes.[22, 23] Further, the activation of EC by CID-treated MΦ-cTLR4 was determined to be dependent on TNFα.
The CID system has been successfully used in literature to trigger a variety of signal transduction cascades. In vitro immunology studies using this system have been mostly focused on downstream effects of a specific engineered receptor’s signaling pathway.[24–26] For instance, Kuenzel et al. transfected HeLaS3 cells with a nucleotide-binding oligomerization domain-like receptor 5 (NLRC5)-FKBP fusion protein and determined that induced oligomerization of this receptor activated certain IFN signaling pathways that contributed to an antiviral defense mechanism.[25] In another study, Fooksman et al. transiently transfected T2 cell lines with a dimerizable mouse class I H2-Kb H chain-FKBP fusion protein and determined that induced dimerization, and thus clustering of this class I MHC construct, enhanced lymphoblast recognition by T cells.[26] In contrast to using the CID system to examine cause and effect relationships within a specific pathway, our study is the first to use this system to regulate the phenotype of a cell by polarizing RAW264.7 cells into a specific pro-inflammatory MΦ. Further, our lab has previously demonstrated that this system can be used to engineer inducible bone resorbing osteoclasts from the monocyte-macrophage RAW264.7 cell line, in which it is important to note that monocytes are a common precursor to both macrophages and osteoclasts.[27]
Other groups have attempted to engineer macrophages to control the inflammatory response. For example, Wu et al. transduced MΦs in vitro with the IFN-γ gene and delivered them intratracheally to immunodeficient mice.[28] These MΦs restored immune function in the lungs of the immunodeficient mice. However, these constitutively active IFNγ-expressing pro-inflammatory MΦs probably have limited applications, since the cells were not engineered to be tunable. Additionally, Oxford BioMedica has engineered human MΦs to express cytochrome P450, which can convert a cancer prodrug into its active form during hypoxic tumor conditions. When delivered into an avascular spheroid model, the human engineered P450 MΦs were able to induce tumor cell death following the addition of the prodrug.[29] The success of this study was dependent on the hypoxia-driven expression of cytochrome P450 in MΦs. The engineered MΦ-cTLR4 in our study, on the other hand, can be controlled temporally and specifically with the addition or withdrawal of the CID drug and activation is independent of the local environment. Indeed, we observe an upregulation of key pro-inflammatory markers from these engineered MΦs as soon as 6 hours after CID drug addition and then we observe return to baseline conditions in 18 hours following drug withdrawal. The ability to tune the engineered MΦs with respect to selective activation provides a large added benefit, since the engineered MΦ-cTLR4 cells could be turned on or off when and if necessary.
Several studies have elucidated that temporal expression of key angiogenic cytokines, such as TNFα, is necessary for tip formation in EC.[30] Sainson et al. showed that 2- to 3-day pulses of TNFα in vitro and in vivo stimulates angiogenesis, as opposed to the inhibition of angiogenesis with continuous administration. We observe robust TNFα expression in our engineered pro-inflammatory MΦ-cTLR4 cells, which may possibly be utilized to promote angiogenesis, if controlled in a time-based manner. Indeed, the MΦ-cTLR4 engineered cells may be tailored to exhibit pulse behavior with the simple addition and withdrawal of CID drug at certain timepoints. In addition, we do observe that the MΦ-cTLR4-conditioned media stimulates EC activation by increasing VCAM-1 and ICAM-1 adhesion molecule expression in an in vitro setting and in a TNFα-dependent manner, which suggests that our engineered MΦ may be able to promote angiogenesis. Further, iNOS levels directly correlate with VCAM-1 expression.[31] We do see similar activation patterns with both TNFα and iNOS in our MΦ-cTLR4 cells, so both of these factors could be working in concert to upregulate adhesion molecule expression. Activation of VCAM-1 and ICAM-1 has been shown to destabilize endothelial junctions resulting in leaky vessels, a first step in the angiogenesis process. It has been suggested that a subsequent M2 MΦ phase may be necessary for the process of angiogenesis to continue and come to completion, as the M2 MΦ phenotype has been hypothesized to bridge and stabilize newly formed vessels.[32] Thus, CID drug activated MΦ-cTLR4 cells may provide the required priming step for angiogenesis to initiate.
In addition to iNOS and TNFα, our CID-treated MΦ-cTLR4 cells also produce increased levels of IL-6. The IL-6 cytokine has been closely associated with promotion of angiogenesis. Increased IL-6 mRNA levels correlated with the development of ovarian follicles and the uterine lining, which are two independent physiological angiogenic processes.[33] Moreover, IL-6 treatment has been shown to promote tubule formation in brain microvessel EC in an in vitro setting. This correlated with increased IL-6 and VEGF mRNA expression in the healing adult murine brain tissue following injury.[34] These studies suggest that IL-6 may play a role in normal physiological angiogenesis as well as angiogenesis related to inflammatory remodeling of tissue. Studies in IL-6 KO mice showed that the IL-6 deletion resulted in delayed wound healing, accompanied with both delayed angiogenesis and collagen deposition.[35] The direct mechanism of IL-6 and its influence on pro-angiogenic behavior is still not completely understood, however, IL-6 seems to be a key player in this process. Our engineered MΦ-cTLR4 cells produce IL-6, along with two other factors implicated with pro-angiogenic behavior. This strongly suggests that our MΦ-cTLR4 cells may have the ability to aid in the priming of the endothelium for early stage angiogenesis.
Despite the possible use of the MΦ-cTLR4 cells as angiogenesis priming agents, in which a following M2 MΦ response might need to be necessary, these cells could also be used in certain diseases to skew the balance of a M2 MΦ-abundant process. For example, diseases characterized by excessive fibrosis could benefit from this technology, as there is often a local abundance of M2 MΦs present during fibrotic events. Fibrosis occurs due to the abundance of these M2 MΦs over-producing TGFβ, which in turn recruits fibroblasts. The recruitment of fibroblasts then leads to the overproduction of collagen, thus leading to a fibrotic state. This dysregulated process is often associated with the dense collagen fibrous capsule that surrounds an implanted material, as well as with cardiac fibrosis that plagues congestive heart failure patients.[36] A few studies have suggested that a proper balance of M1 and M2 MΦ is necessary to achieve a reduction in the extent of fibrosis.[37, 38] Thus, CID-activated MΦ-cTLR4 cells may provide a tool to reestablish the proper M1 vs M2 MΦ equilibrium and decrease the excessive collagen deposition. Another possible application of the MΦ-cTLR4 cells could be tumor inhibition. Tumor-induced angiogenesis is essential for cancer cell survival, tumor growth and metastasis propagation. An abundance of pro-angiogenic, anti-inflammatory M2 MΦs, known as tumor-associated MΦ (TAMs), is normally present in the tumor environment thus aiding tumor progression. In contrast, very few M1 MΦs able to activate NK cells and TH1 responses are present in and around the growing tumor mass[39]. Thus, the delivery of tunable MΦ-cTLR4 cells to the tumor may halt progression by activating a more pro-inflammatory immune response.
We have shown that the MΦ-cTLR4 cells become activated with the addition of CID drug and conversely that these engineered cells return to baseline conditions once CID drug has been withdrawn in an in vitro environment. However, it is still not known what will occur in vivo with the addition or withdrawal of CID drug. The IL-4 cocktail results showed that the MΦ-cTLR4 cells had decreased IL-6 and iNOS levels when compared to CID drug or LPS treatment alone, however, IL-4 did not seem to affect TNF-α levels. This suggests that the engineered cells are influenced by competing M2-like MΦ signals. These competing signals may be changing the phenotype of the engineered MΦ-cTLR4 cells into an intermediate phenotype or even skewing the cells toward a M2-like MΦ phenotype. These results are not completely surprising, as MΦ are known to be very plastic cells and can change phenotypes depending on the surrounding microenvironment. Future in vivo studies will be necessary to determine if elevated TNF-α levels, or other increased pro-inflammatory MΦ markers, are adequate enough to maintain a pro-inflammatory surrounding environment with M2 MΦ competing signals present. In a physiological setting, the CID-treated and subsequently CID-withdrawn engineered MΦs could potentially: 1) be primed to polarize into M2 pro-healing MΦs, 2) remain in a pro-inflammatory MΦ phenotype state due to the surrounding environment, 3) develop into an intermediate phenotype state due to M2 MΦ competing signals, 4) undergo apoptosis or, 5) migrate out of the inflammation site. Future studies will determine the degree of plasticity of the engineered cells, as well as how precise we can control these cells in vivo.
Conclusion
It has been shown previously that MΦ drive the wound healing response.[11, 40, 41] In this study, we have engineered tunable pro-inflammatory MΦ that could possibly be used to better regulate inflammation. By utilizing these MΦ-cTLR4 cells to control the host response, it might be possible to increase angiogenesis for better healing outcomes. Additionally, these engineered cells could be used as a tool to better understand and better regulate M1 MΦ-like dynamics. While RAW264.7 cells are suitable to use during in vitro inflammation studies, future investigations will focus on using a more physiological engineered primary cell type, such as bone marrow derived MΦ. [42] While ongoing studies continue to unravel MΦ-cTLR4 cell possibilities in both in vitro and in vivo settings, currently these engineered cells serve as a platform technology that could be applied to various inflammatory diseases including the FBR, fibrosis, atherosclerosis, and cancer.
Supplementary Material
Acknowledgments
KVE supported by-Bioengineering Cardiovascular NIH Training Grant (1T32 EB001650-06A2), University of Washington Royalty Research Fund (A83365), John H. Tietze Stem Cell Scientist Research Award and University of Washington College of Engineering Fellowship.
The authors would like to thank Melissa Jackson and Matt Coons for assistance with this work.
List of Abbreviations
- MΦ
Macrophage/s
- M1 MΦ
pro-inflammatory MΦ
- M2 MΦ
pro-healing MΦ
- CID
chemical inducer of dimerization
- cTLR4
cytoplasmic portion of TLR4
- MΦ-cTLR4
engineered pro-inflammatory MΦ
- MΦ-T2A
engineered control MΦ
- VCAM-1
vascular cell adhesion protein 1
- ICAM-1
intercellular adhesion molecule 1
- ROS
reactive oxygen species
- TNFα
tumor necrosis factor alpha
- IL-6
interleukin 6
- iNOS
inducible nitric oxide synthase
- EC
endothelial cell/s
- LPS
lipopolysaccharides
- F36V
engineered dimerization domain
- RE
restriction enzyme
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
KVE and MS designed the study, KVE designed the constructs, HYLY created the final constructs, KVE performed the experiments, KVE wrote the paper.
Conflict of Interest:
The authors declare that there are no conflicts of interest.
Contributor Information
KV Eaton, Email: eatonium@uw.edu.
HYL Yang, Email: hly@u.washington.edu.
CM Giachelli, Email: ceci@u.washington.edu.
M Scatena, Email: mscatena@u.washington.edu.
References
- 1.Larsen GL, Henson PM. Mediators of inflammation. Annual review of immunology. 1983;1:335–359. doi: 10.1146/annurev.iy.01.040183.002003. [DOI] [PubMed] [Google Scholar]
- 2.Delavary BM, van der Veer WM, van Egmond M, Niessen FB, Beelen RH. Macrophages in skin injury and repair. Immunobiology. 2011;216:753–762. doi: 10.1016/j.imbio.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nature Reviews Immunology. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez A, Meyerson H, Anderson JM. Quantitative in vivo cytokine analysis at synthetic biomaterial implant sites. Journal of Biomedical Materials Research Part A. 2009;89:152–159. doi: 10.1002/jbm.a.31939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schutte RJ, Xie L, Klitzman B, Reichert WM. In vivo cytokine-associated responses to biomaterials. Biomaterials. 2009;30:160–168. doi: 10.1016/j.biomaterials.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakao S, Kuwano T, Ishibashi T, Kuwano M, Ono M. Synergistic effect of TNF-α in soluble VCAM-1-induced angiogenesis through α4 integrins. The Journal of Immunology. 2003;170:5704–5711. doi: 10.4049/jimmunol.170.11.5704. [DOI] [PubMed] [Google Scholar]
- 7.Stewart RJ, Kashour TS, Marsden PA. Vascular endothelial platelet endothelial adhesion molecule-1 (PECAM-1) expression is decreased by TNF-alpha and IFNgamma. Evidence for cytokine-induced destabilization of messenger ribonucleic acid transcripts in bovine endothelial cells. The Journal of Immunology. 1996;156:1221–1228. [PubMed] [Google Scholar]
- 8.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends in immunology. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Mackay F, Loetscher H, Stueber D, Gehr G, Lesslauer W. Tumor necrosis factor alpha (TNF-alpha)-induced cell adhesion to human endothelial cells is under dominant control of one TNF receptor type, TNF-R55. The Journal of experimental medicine. 1993;177:1277–1286. doi: 10.1084/jem.177.5.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garmy-Susini B, Jin H, Zhu Y, Sung R-J, Hwang R, Varner J. Integrin α4β1–VCAM-1–mediated adhesion between endothelial and mural cells is required for blood vessel maturation. The Journal of clinical investigation. 2005;115:1542–1551. doi: 10.1172/JCI23445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behm B, Babilas P, Landthaler M, Schreml S. Cytokines, chemokines and growth factors in wound healing. Journal of the European Academy of Dermatology and Venereology. 2012;26:812–820. doi: 10.1111/j.1468-3083.2011.04415.x. [DOI] [PubMed] [Google Scholar]
- 12.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Frontiers in bioscience: a journal and virtual library. 2008;13:453. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 13.Blau CA, Peterson KR, Drachman JG, Spencer DM. A proliferation switch for genetically modified cells. Proceedings of the National Academy of Sciences. 1997;94:3076. doi: 10.1073/pnas.94.7.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin L, Siritanaratkul N, Emery DW, Richard RE, Kaushansky K, Papayannopoulou T, Blau CA. Targeted expansion of genetically modified bone marrow cells. Proceedings of the National Academy of Sciences. 1998;95:8093. doi: 10.1073/pnas.95.14.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin L, Asano H, Blau CA. Stimulating cell proliferation through the pharmacologic activation of c-kit. Blood. 1998;91:890–897. [PubMed] [Google Scholar]
- 16.Zeng H, Masuko M, Jin L, Neff T, Otto KG, Blau CA. Receptor specificity in the self-renewal and differentiation of primary multipotential hemopoietic cells. Blood. 2001;98:328–334. doi: 10.1182/blood.v98.2.328. [DOI] [PubMed] [Google Scholar]
- 17.Zhao S, Zoller K, Masuko M, Rojnuckarin P, Yang XO, Parganas E, Kaushansky K, Ihle JN, Papayannopoulou T, Willerford DM. JAK2, complemented by a second signal from c-kit or flt-3, triggers extensive self-renewal of primary multipotential hemopoietic cells. The EMBO journal. 2002;21:2159–2167. doi: 10.1093/emboj/21.9.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. The Journal of clinical investigation. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: a lesson in detente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 20.Guo S, DiPietro LA. Factors affecting wound healing. Journal of dental research. 2010;89:219–229. doi: 10.1177/0022034509359125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sussman EM, Halpin MC, Muster J, Moon RT, Ratner BD. Porous implants modulate healing and induce shifts in local macrophage polarization in the foreign body reaction. Annals of biomedical engineering. 2013:1–9. doi: 10.1007/s10439-013-0933-0. [DOI] [PubMed] [Google Scholar]
- 22.Wu Y, Ip JE, Huang J, Zhang L, Matsushita K, Liew C-C, Pratt RE, Dzau VJ. Essential role of ICAM-1/CD18 in mediating EPC recruitment, angiogenesis, and repair to the infarcted myocardium. Circulation Research. 2006;99:315–322. doi: 10.1161/01.RES.0000235986.35957.a3. [DOI] [PubMed] [Google Scholar]
- 23.Koch AE, Halloran MM, Haskell CJ, Shah MR, Polverini PJ. Angiogenesis mediated by soluble forms of E-selectin and vascular cell-adhesion molecule-1. 1995 doi: 10.1038/376517a0. [DOI] [PubMed] [Google Scholar]
- 24.Sun M, Ames KT, Suzuki I, Fink PJ. The cytoplasmic domain of Fas ligand costimulates TCR signals. The Journal of Immunology. 2006;177:1481–1491. doi: 10.4049/jimmunol.177.3.1481. [DOI] [PubMed] [Google Scholar]
- 25.Kuenzel S, Till A, Winkler M, Häsler R, Lipinski S, Jung S, Grötzinger J, Fickenscher H, Schreiber S, Rosenstiel P. The nucleotide-binding oligomerization domain-like receptor NLRC5 is involved in IFN-dependent antiviral immune responses. The journal of immunology. 2010;184:1990–2000. doi: 10.4049/jimmunol.0900557. [DOI] [PubMed] [Google Scholar]
- 26.Fooksman DR, Grönvall GK, Tang Q, Edidin M. Clustering class I MHC modulates sensitivity of T cell recognition. The Journal of Immunology. 2006;176:6673–6680. doi: 10.4049/jimmunol.176.11.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rementer CW, Wu M, Buranaphatthana W, Yang H-YL, Scatena M, Giachelli CM. An Inducible, Ligand-Independent Receptor Activator of NF-κB Gene to Control Osteoclast Differentiation from Monocytic Precursors. PloS one. 2013;8:e84465. doi: 10.1371/journal.pone.0084465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu M, Hussain S, He Y-H, Pasula R, Smith PA, Martin WJ. Genetically engineered macrophages expressing IFN-γ restore alveolar immune function in scid mice. Proceedings of the National Academy of Sciences. 2001;98:14589–14594. doi: 10.1073/pnas.251451498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffiths L, Binley K, Iqball S, Kan O, Maxwell P, Ratcliffe P, Lewis C, Harris A, Kingsman S, Naylor S. The macrophage-a novel system to deliver gene therapy to pathological hypoxia. Gene therapy. 2000;7:255–262. doi: 10.1038/sj.gt.3301058. [DOI] [PubMed] [Google Scholar]
- 30.Sainson RC, Johnston DA, Chu HC, Holderfield MT, Nakatsu MN, Crampton SP, Davis J, Conn E, Hughes CC. TNF primes endothelial cells for angiogenic sprouting by inducing a tip cell phenotype. Blood. 2008;111:4997–5007. doi: 10.1182/blood-2007-08-108597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klug F, Prakash H, Huber PE, Seibel T, Bender N, Halama N, Pfirschke C, Voss RH, Timke C, Umansky L. Low-dose irradiation programs macrophage differentiation to an iNOS+/M1 phenotype that orchestrates effective T cell immunotherapy. Cancer cell. 2013;24:589–602. doi: 10.1016/j.ccr.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt T, Carmeliet P. Blood-vessel formation: Bridges that guide and unite. Nature. 2010;465:697–699. doi: 10.1038/465697a. [DOI] [PubMed] [Google Scholar]
- 33.Motro B, Itin A, Sachs L, Keshet E. Pattern of interleukin 6 gene expression in vivo suggests a role for this cytokine in angiogenesis. Proceedings of the National Academy of Sciences. 1990;87:3092–3096. doi: 10.1073/pnas.87.8.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fee D, Grzybicki D, Dobbs M, Ihyer S, Clotfelter J, Macvilay S, Hart MN, Sandor M, Fabry Z. Interleukin 6 promotes vasculogenesis of murine brain microvessel endothelial cells. Cytokine. 2000;12:655–665. doi: 10.1006/cyto.1999.0599. [DOI] [PubMed] [Google Scholar]
- 35.Lin Z-Q, Kondo T, Ishida Y, Takayasu T, Mukaida N. Essential involvement of IL-6 in the skin wound-healing process as evidenced by delayed wound healing in IL-6-deficient mice. Journal of leukocyte biology. 2003;73:713–721. doi: 10.1189/jlb.0802397. [DOI] [PubMed] [Google Scholar]
- 36.Stempien-Otero A, Plawman A, Meznarich J, Dyamenahalli T, Otsuka G, Dichek DA. Mechanisms of cardiac fibrosis induced by urokinase plasminogen activator. J Biol Chem. 2006;281:15345–15351. doi: 10.1074/jbc.M512818200. [DOI] [PubMed] [Google Scholar]
- 37.Meneghin A, Hogaboam CM. Infectious disease, the innate immune response, and fibrosis. Journal of Clinical Investigation. 2007;117:530. doi: 10.1172/JCI30595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anders H-J, Ryu M. Renal microenvironments and macrophage phenotypes determine progression or resolution of renal inflammation and fibrosis. Kidney international. 2011;80:915–925. doi: 10.1038/ki.2011.217. [DOI] [PubMed] [Google Scholar]
- 39.Allavena P, Sica A, Solinas G, Porta C, Mantovani A. The inflammatory micro-environment in tumor progression: the role of tumor-associated macrophages. Critical reviews in oncology/hematology. 2008;66:1–9. doi: 10.1016/j.critrevonc.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Hunt T, Knighton D, Thakral K, Goodson W, 3rd, Andrews W. Studies on inflammation and wound healing: angiogenesis and collagen synthesis stimulated in vivo by resident and activated wound macrophages. Surgery. 1984;96:48–54. [PubMed] [Google Scholar]
- 41.Koh TJ, DiPietro LA. Inflammation and wound healing: the role of the macrophage. Expert reviews in molecular medicine. 2011;13:e23. doi: 10.1017/S1462399411001943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chamberlain LM, Godek ML, Gonzalez-Juarrero M, Grainger DW. Phenotypic non-equivalence of murine (monocyte-) macrophage cells in biomaterial and inflammatory models. Journal of Biomedical Materials Research Part A. 2009;88A:858–871. doi: 10.1002/jbm.a.31930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.