Abstract
The alphavirus membrane protein E1 mediates low pH-triggered fusion of the viral and endosome membranes during virus entry. During virus biogenesis E1 associates as a heterodimer with the transmembrane protein p62. Late in the secretory pathway, cellular furin cleaves p62 to the mature E2 protein and a peripheral protein E3. E3 remains bound to E2 at low pH, stabilizing the heterodimer and thus protecting E1 from the acidic pH of the secretory pathway. Release of E3 at neutral pH then primes the virus for fusion during entry. Here we used site-directed mutagenesis and revertant analysis to define residues important for the interactions at the E3-E2 interface. Our data identified a key residue, E2 W235, which was required for E1 pH protection and alphavirus production. Our data also suggest additional residues on E3 and E2 that affect their interacting surfaces and thus influence the pH protection of E1 during alphavirus exit.
Keywords: Alphavirus, membrane fusion, virus entry, fusion protein biogenesis, class II fusion protein
INTRODUCTION
The alphavirus genus includes a number of important human and animal pathogens such as Chikungunya virus (CHIKV) and the encephalitic alphaviruses (reviewed in Kuhn, 2013). Alphaviruses are small, spherical enveloped viruses with a positive-sense single-stranded RNA genome. The viral genome is surrounded by a capsid protein shell, which is enveloped by a membrane bilayer containing trimeric spikes of E1 and E2 glycoprotein heterodimers (Kuhn, 2013). The E2 membrane protein is responsible for receptor binding at the host cell surface, which then mediates uptake of the virus by clathrin-mediated endocytosis (Helenius et al., 1980; Kielian et al., 2010). E1 is the virus membrane fusion protein, and is triggered by the low pH environment of the endosome. Upon exposure to low pH, the E2-E1 heterodimer dissociates (Wahlberg et al., 1989; Wahlberg and Garoff, 1992). Dissociation of the heterodimer is a key early step in the fusion reaction, releasing the hydrophobic fusion loop of E1, which inserts into the endosome membrane. E1 then forms an extended homotrimer that refolds to a hairpin-like conformation, driving the merger of the viral and endosome membranes and releasing the virus genome into the cytoplasm (reviewed in Harrison, 2015; Kielian, 2014).
During alphavirus biogenesis the envelope proteins are translocated into the endoplasmic reticulum (ER), with E2 initially produced as a precursor termed p62 (or PE2) (Kuhn, 2013). p62 and E1 form a heterodimer within the ER, and dimerization is necessary for the proper folding and transport of E1 to the plasma membrane, where budding takes place. During transport p62 is cleaved by the cellular enzyme furin to the mature E2 protein and a smaller peripheral protein E3 (deCurtis and Simons, 1988; Zhang et al., 2003). The p62-E1 dimer has a more acidic fusion threshold (~pH 5.0) than that of the mature E2-E1 dimer (~pH 6.2) (Salminen et al., 1992; Smit et al., 2001; Zhang et al., 2003). The acid-stable p62-E1 dimer protects E1 from premature fusion as it is transported through the acidic pH (~5.5–6.0) of the late secretory pathway (Demaurex et al., 1998). Even after furin cleavage, E3 remains bound to the E2-E1 dimer and pH protection is maintained (Sjoberg et al., 2011). The release of E3 in the neutral pH extracellular environment completes the maturation process and primes the virus for fusion during its entry into a new host cell.
The structure of the CHIKV p62-E1 heterodimer before and after cleavage shows that E3 interacts only with E2, and thereby stabilizes the E2-E1 dimer (Voss et al., 2010). We previously used site-directed mutagenesis of the alphavirus Semliki Forest virus (SFV) to destabilize the E3-E2 interface and demonstrate the critical role of E3 in pH protection (Uchime et al., 2013). Our studies identified a conserved tyrosine on E3, Y47, that is required for maintenance of dimer stability during exposure to exocytic low pH, transport of E1 to the plasma membrane, and virus particle production. Here we address key residues in the interacting surfaces of E3-E2. Using site-directed mutagenesis and revertant analysis, we define residues on E3 and E2 that promote pH protection of the alphavirus fusion protein.
MATERIALS AND METHODS
Cell and viruses
BHK-21 cells were maintained at 37°C in complete BHK medium (Dulbecco’s modified Eagle medium containing 5% fetal bovine serum, 10% tryptose phosphate broth, 100 U penicillin/ml, and 100 μg streptomycin/ml). The furin-deficient FD-11 CHO cell line and its parental cell line, CHO-K1, were kindly provided by Dr. Stephen H. Leppla at the National Institutes of Health (Gordon et al., 1995) and have been previously used to demonstrate the role of furin in p62 processing (Zhang et al., 2003). The cells were cultured in α-minimal essential medium containing 10% fetal bovine serum, 100 U penicillin/ml, and 100 μg streptomycin/ml at 37°C.
Residue numbering for site-directed mutagenesis is based on the CHIKV envelope protein sequence and structure throughout. Site-directed mutagenesis of the DG-1 subgenomic plasmid encoding the SFV envelope proteins was performed as previously described (Liu et al., 2010). Briefly, the E2 K254A and E2 W235A mutations (CHIKV numbering) were introduced into the DG-1 plasmid by circular mutagenesis using PrimeSTAR HS DNA polymerase (Takara Bio Inc., Madison, WI). The mutated DG-1 NsiI/SpeI fragments were then subcloned into the pSP6-SFV4 infectious clone (Liljeström et al., 1991). Mutant infectious clones were sequenced to confirm the sequence of the mutated region and the absence of other mutations in the structural proteins (Genewiz Inc., North Brunswick, NJ). Infectious RNAs were generated by in vitro transcription and electroporated into BHK-21 cells to produce virus infection (Liljeström et al., 1991). All plaque and infectious center assays were performed on BHK cells. As indicated, viruses were quantitated by infectious center assays in cases where they produced very small plaques.
To determine the effects of neutralization of the exocytic pathway on virus production, 100 nM bafilomycin A-1 was added 2 hours post-electroporation and virus-containing media samples were collected after continued incubation at 37°C for 8 hours. Expression of E1 and E2 glycoproteins was examined by fixation in 3% formaldehyde (nonpermeabilized) or methanol (permeabilized) 12 hours post-electroporation. Indirect immunofluorescence staining was performed using monoclonal antibody (mAb) E1-1 against E1, or mAb E2-1 against E2 (Kielian et al., 1990).
Fusion infection assay
Serial dilutions of mature or immature virus stocks were prebound to BHK cells on ice to prevent endocytosis. Virus fusion with the plasma membrane was then triggered by treatment for 3 minutes at 37°C with medium buffered at the indicated pH value. Cells were then cultured overnight at 28°C in medium containing 20 mM NH4Cl to prevent secondary infection, and virus-infected cells were quantitated by immunofluorescence, all as described previously (Liao and Kielian, 2005). Dilutions of virus that produced ~200 infected cells were used for quantitation. The pH treatment that produced the maximal number of infected cells was used to calculate “100% fusion efficiency.”
Isolation of E3 Y47A and E3 Y47/48A revertants
Mutant viral RNAs were electroporated into BHK cells. Electroporated cells were mixed with non-electroporated cells at varying ratios and plated in 6 well plates. The cells were cultured until cytopathic effects were observed, typically after 4 days at 37°C or 6 days at 28°C. Medium containing revertants was harvested, and the virus amplified by growth on BHK cells for 24 hours at 37°C and pelleted by ultracentrifugation. Viral RNA was extracted from the pelleted virus and reverse-transcribed, and the cDNA was sequenced, all as previously reported (Chanel-Vos and Kielian, 2006). Note that independent revertant isolates were generated by selecting for growth on independent wells/plates. Such serial selections for efficient growth of a lethal or highly deleterious mutation have been confirmed to identify “rescue” mutations (Zheng et al., 2014).
RESULTS
Generation and characterization of E2 K254A mutant
Our previous studies indicated that E3 Y47 is a key residue that promotes the pH protection of the alphavirus membrane fusion protein (Uchime et al., 2013). An E3 Y47/48A mutation destabilizes the heterodimer in the low pH of the late exocytic pathway, resulting in a block in E1 transport to the plasma membrane and inhibition of virus particle production. Dimer stability, E1 transport and virus production are all rescued by neutralization of exocytic pH. E3 Y47 is critical for the mutant phenotype, while E3 Y48 is not required. We also found that E3 Y47 could be functionally replaced by phenylalanine, and we hypothesized a mechanism involving a cation-π interaction between an aromatic residue at E3 position 47 and E2 lysine 254, which lies within 3.5 Å of E3 Y47 (Voss et al., 2010). We here used site-directed mutagenesis to test the importance of E2 K254. The lysine residue was changed to alanine (K254A) in the WT SFV infectious clone, and viral RNA was transcribed in vitro and electroporated into BHK cells to characterize the mutant phenotype.
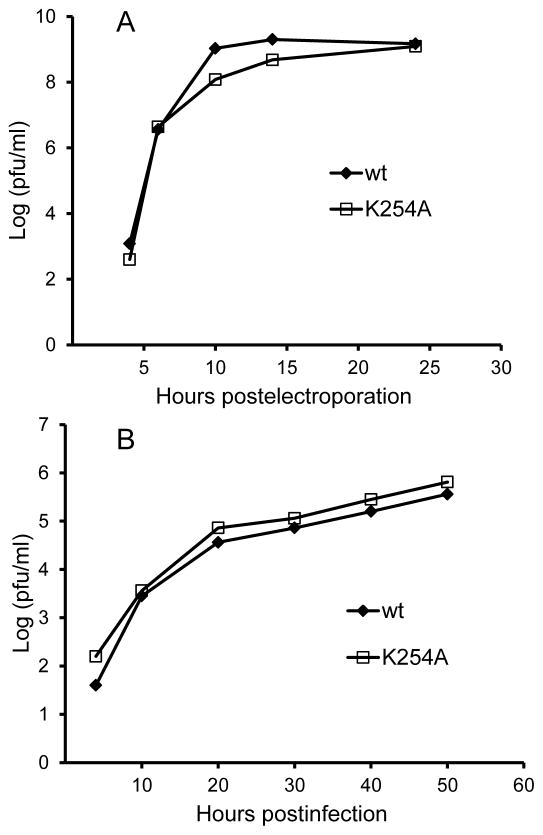
In contrast to our previous results with the lethal E3 Y47A mutation (Uchime et al., 2013), cells infected with the E2 K254A mutant efficiently expressed both E1 and E2 on the plasma membrane and produced secondary infection of co-cultured non-electroporated BHK cells (data not shown). We compared the growth kinetics of WT SFV and the K254A mutant in BHK cells, where both viruses grew efficiently and with similar kinetics (Fig. 2A). We also compared virus growth kinetics in furin-deficient FD-11 CHO cells (Fig. 2B). FD-11 cells have a complete block in p62 processing and show less efficient alphavirus growth (Zhang et al., 2003), while mutations that destabilize the p62-E1 heterodimer promote increased virus growth (Zhang and Kielian, 2004). As expected, WT virus growth was less efficient in the absence of furin cleavage (Fig. 2B), but no rescue of growth was observed by the E2 K254A mutation (Fig. 2B).
Figure 2. Growth kinetics of WT and mutant SFV on control and furin-deficient cells.
BHK cells (A) were electroporated with WT SFV or K254A mutant viral RNA and allowed to recover at 37°C for two hours. FD11 furin-deficient CHO cells (B) were infected with WT SFV or the K254A mutant at a multiplicity of infection of 0.01 PFU/cell for 90 minutes at 37°C. Cells were then washed and cultured at 37°C. Viruses secreted into the media were collected at the indicated time points and the titers were determined by plaque assay on BHK cells. Data in A and B are each representative examples of two independent experiments.
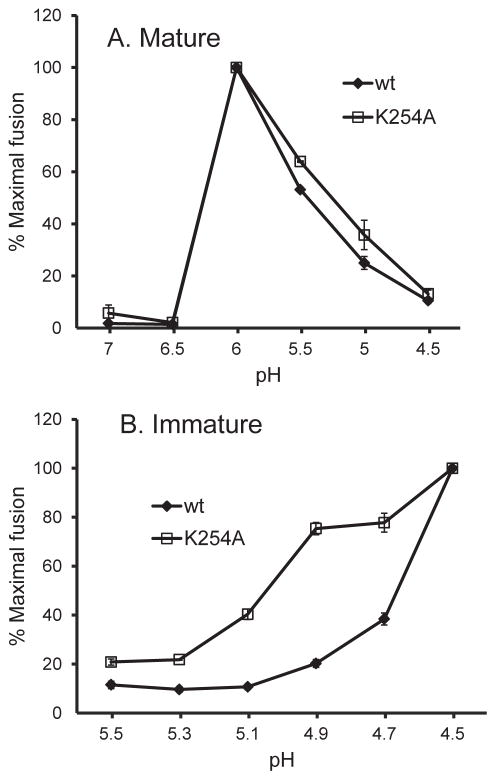
We then compared the pH dependence of membrane fusion of the WT SFV and the E2 K254A mutant. Since the immature virus requires a much lower pH to trigger fusion, it provides a sensitive test for mutations that destabilize the p62-E1 heterodimer (Zhang et al., 2003; Zhang and Kielian, 2005). We therefore produced mature virus stocks in control cells and immature virus stocks in FD-11 cells. The mature forms of WT and K254A SFV showed comparable pH dependence, with maximal fusion at ~pH 6.0 (Fig. 3A). The immature WT SFV showed maximal fusion at pH 4.5 and a fusion threshold of ~4.9 (Fig. 3B). The immature E2 K254A mutant showed a modest increase in the pH threshold of fusion compared to the WT, with 75% maximal fusion at pH 4.9 and 40% at pH 5.1 (Fig. 3B). Taken together, the very different phenotypes of the E3 Y47A and E2 K254A mutants suggest that K254 does not play a significant role in pH protection and heterodimer stability.
Figure 3. pH dependence of fusion of WT and K254A mutant SFV.
Mature, E2-containing virus produced by BHK cells (A) and immature, p62-containing virus produced by furin-deficient FD11 cells (B) were adsorbed to BHK cells on ice. Virus-plasma membrane fusion was then triggered by treatment at the indicated pH for 3 min at 37°C. Cells were cultured in the presence of 20 mM NH4Cl to prevent further infection. Virus-infected cells were quantitated by immunofluorescence. Results are graphed as the percentage of maximal fusion for each virus, and show the mean and standard deviation of three independent experiments.
Generation and characterization of E2 W235A mutant
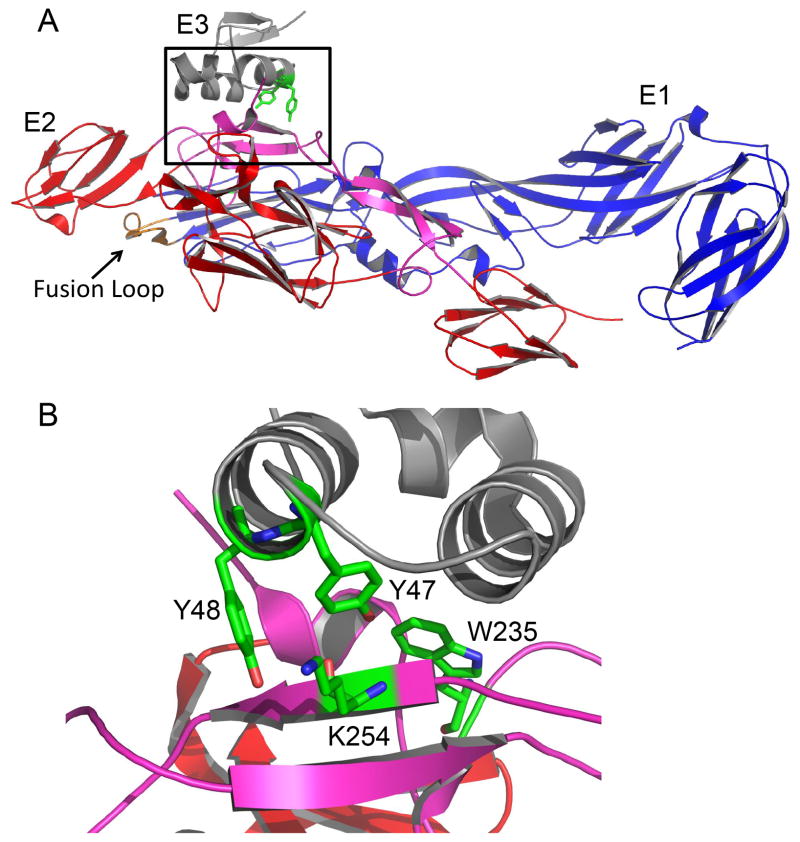
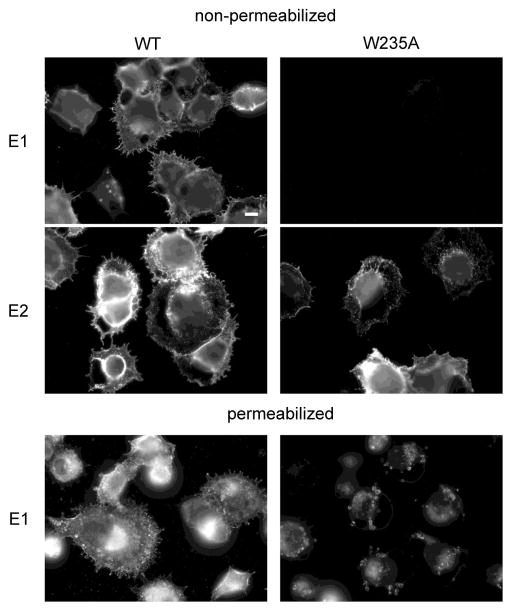
Further examination of the E3-E2 interface identified a tryptophan residue at E2 position 235 that is within 3.3Å of E3 Y47 (Fig. 1B) and is conserved in all arthropod-borne alphaviruses. We generated an E2 W235A mutation in the SFV infectious clone and analyzed its effects on virus biogenesis. The E2 W235A mutation inhibited the transport of E1 to the cell surface (Fig. 4). Instead, the E1 protein was localized primarily in intracellular compartments, with an immunofluorescence staining level lower than that of WT SFV E1. This block in transport and decreased level of the E1 protein is similar to the phenotype of the E3 Y47/48A mutant (Uchime et al., 2013). Although transport of some alphavirus envelope protein mutants can be rescued by incubation at 28°C (Chatterjee et al., 2002; Duffus et al., 1995), these conditions did not rescue the cell surface expression of E1 in W235A-infected cells (data not shown). As expected from the E1 transport defect, no infectious virus was produced from cells electroporated with W235A viral RNA (data not shown, and Fig. 5A).
Figure 1. Alphavirus envelope protein structure and interactions at the E3-E2 interface.
A. The structure of the CHIKV E3-E2-E1 complex. E3 is shown in grey, E2 is in red with the β-ribbon connector region in purple, and E1 is in blue with the fusion loop in orange (Protein Data Bank accession number 3N42) (Voss et al., 2010). The E3-E2 interface is highlighted by the black box. B. A close up view of the E3-E2 interface (rotated from panel A) showing E3 Y47 and Y48 and E2 K254 and W235 in green stick view. Colors are as in panel A. Figure was prepared using the program PYMOL (DeLano, 2002).
Figure 4. Cell surface expression of E1 and E2 in WT- and W235A-infected cells.
WT or W235A mutant SFV RNA was electroporated into BHK cells. Cells were cultured at 37°C for 12 h, fixed with either paraformaldehyde (nonpermeabilized) or methanol (permeabilized), and stained with mAbs to the E1 or E2 protein. Fluorescence microscopy images were all acquired with the same exposure times. Images are representative examples of three independent experiments. Scale bar, 10 μm.
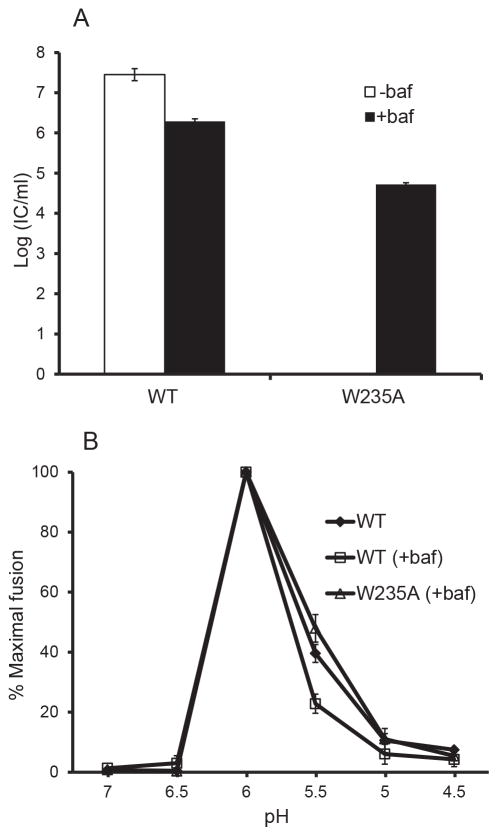
Figure 5. Neutralization of exocytic pH rescues growth of W235A SFV mutant.
BHK cells were electroporated with WT or W235A mutant viral RNA and incubated for 2 hours at 37°C. Culture medium with (+) or without (−) bafilomycin (baf) was added, and the incubation continued for an additional 8 h. The culture medium was then collected and titered by infectious center assay on BHK cells (A). These virus stocks were then used to determine the pH dependence of fusion with the plasma membrane of BHK cells (B) as in Fig. 3. The graphs in each panel show the mean and standard deviation of three independent experiments.
If E2 W235 is important in the interaction with E3, the observed inhibition of E1 transport and lower level of E1 protein in the mutant could be caused by the loss of the E3 clamp that stabilizes the E2-E1 dimer and protects E1 from exocytic low pH. If so, then the phenotype of the E2 W235A mutant should be rescued by neutralization of exocytic pH with bafilomycin, similar to the rescue we observed for the E3 Y47/48A mutant (Uchime et al., 2013). Bafilomycin neutralizes the pH in the exocytic pathway without affecting p62 processing (Uchime et al., 2013; Zhang et al., 2003). To test for rescue, cells were electroporated with WT or W235A mutant viral RNA and cultured in the presence or absence of bafilomycin for eight hours. The culture media were then collected and progeny virus titers were determined by infectious center assays on BHK cells (Fig. 5A). While no infectious virus was produced from untreated mutant-infected cells, virus production was significantly rescued by bafilomycin treatment. Mutant rescue occurred even though neutralization of the secretory pathway decreases virus production overall (see WT virus in Fig. 5A and reference Uchime et al., 2013). Fusion-infection assays showed that WT and W235A viruses produced in the presence of bafilomycin have similar pH profiles for virus fusion (Fig. 5B). Thus, once E1 was rescued from the low pH of the secretory pathway and packaged into virus particles, the E2 W235A mutation had no detectable effects on the pH dependence of fusion.
Isolation of Y47A and Y47/48A revertants
We also used a less targeted approach to obtain further information on important residues in the E3-E2 interface. We selected for revertants that rescued the lethal phenotype of E3 Y47A and Y47/48A mutants. These include true revertants of the original mutations and second-site revertants; we will refer to them collectively as revertants here.
Seven distinct types of revertants were isolated from E3 Y47A-infected cells after 4 days growth at 37 °C (Table 1). All were second-site revertants containing the same amino acid substitution in E2, a change from aspartic acid 168 to glutamic acid. This E2 D168E revertant was isolated independently 5 times, and was also isolated 6 times in conjunction with additional second site mutations in E1 and/or E2. Although D168 is located near the E3-E2 interface (Fig. 6B), it was surprising that such a conservative amino acid change could rescue the lethal phenotype of E3 Y47. Although strong, rescue was not complete, with the E2 D168E revertant producing viral titers ~2 logs lower than WT SFV (Fig. 6A).
Table 1.
Revertants of E3 Y47A and Y47/48A mutants
| Revertant groupa | revertant Y47/48 sequence | Second site mutation(s)b | No. of independent isolates |
|---|---|---|---|
| Y47A revertants | A/Y | E2 D168E | 5 |
| A/Y | E2 D168E, H99Y | 1 | |
| A/Y | E2 D168E, D261G, E1 T253S | 1 | |
| A/Y | E2 D168E, A121V, D261G | 1 | |
| A/Y | E2 D168E, T219S | 1 | |
| A/Y | E2 D168E, E1 V113A | 1 | |
| A/Y | E2 D168E, E1 S395R | 1 | |
| Y47/48A revertants | Y/Y | none | 3 |
| Y/Y | E2 R336K | 1 | |
| Y/Y | E1 E183D | 1 | |
| A/A | E2 H256P | 3 | |
| A/A | E2 H256P, N238V | 1 | |
| A/A | E2 H256P, D231N | 1 |
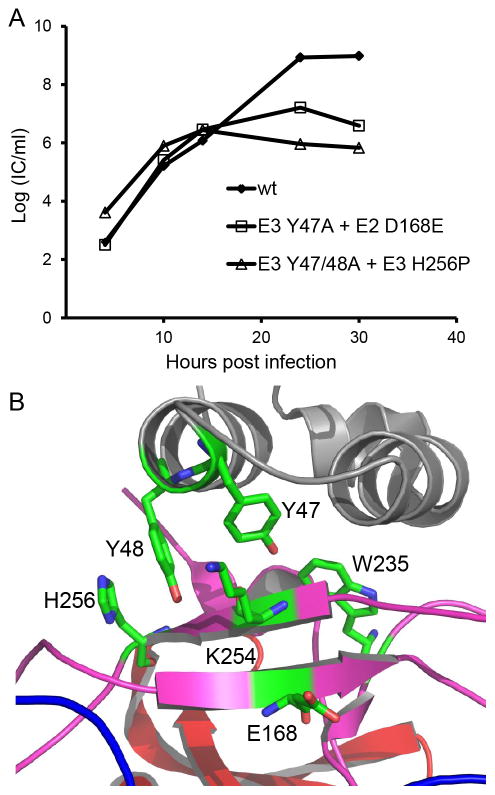
Figure 6. Properties of E3 Y47A and Y47/48A second site revertants.
A. Growth kinetics of the second site revertants E3 Y47A+E2 D168E and E3 Y47/48A+E2 H256P. BHK cells were infected with WT or mutant virus at a multiplicity of infection of 0.01 PFU/cell for 90 minutes at 37°C, cultured for the indicated times, and progeny virus quantitated by infectious center assay on BHK cells. Graph shows the mean of two independent experiments. B. Zoom view of the CHIKV E3-E2 interface region colored as in Figure 1. The location of important residues identified by mutagenesis and revertant analysis is shown (Protein Data Bank accession number 3N42) (Voss et al., 2010). Note that SFV D168 is E in CHIKV. Figure was prepared using the program PYMOL (DeLano, 2002).
We then selected for revertants of the double mutant E3 Y47/48A, with the expectation that the absence of both tyrosines could produce a different and informative set of revertants (Table 1). It was more difficult to isolate revertants of the double mutant, but successful isolation was achieved by growth of the electroporated cells at 28°C for ~ 6 days. True revertants in which E3 Y47/48A were both changed back to the original tyrosine were isolated multiple times, either alone or in conjunction with additional second site mutations in E1 or E2. The remaining revertants all contained the Y47/48A sequence, but also contained the same second site mutation, a change from a highly conserved histidine at E2 residue 256 to a proline. In two revertants, the E2 H256P mutation was accompanied by an additional mutation in E2, N238V or D231N. The E2 H256P revertant produced strong but incomplete rescue of infectious virus production, with titers ~3 logs lower than WT (Fig. 6A). Thus, the revertants identified two additional SFV E2 residues, D168 and H256, that can help to stabilize the E2-E1 dimer and thus affect pH protection.
DISCUSSION
Biogenesis of the alphavirus fusion protein takes place by translocation of the polyprotein precursor into the ER and signal peptidase processing to produce p62 and E1 (Kuhn, 2013). p62 and E1 rapidly dimerize within the ER and the dimer is transported to the plasma membrane where budding occurs. Typically most of the p62 protein is matured by furin cleavage in the late secretory pathway. While virus particles are produced with comparable efficiency with or without p62 processing (Salminen et al., 1992), in the absence of cleavage the immature virus particles have reduced infectivity due to their significantly more acidic fusion threshold (Salminen et al., 1992; Zhang et al., 2003). E3 remains bound to the mature dimer in the low pH of the secretory pathway and is released at the extracellular neutral pH. Bound E3 thus plays a critical role in silencing the E1 fusion protein during exit while its release converts the heterodimer to the low pH-responsive form.
Our previous data demonstrated a key role for E3 Y47 in maintaining the interaction of E3 with E2 during exit, thus stabilizing the E2-E1 heterodimer and conferring pH protection to E1 (Uchime et al., 2013). An aromatic substitution Y47F supported robust virus growth, while no role for the adjacent tyrosine E3 Y48 was observed. Based on these data and the details of the E3-E2 interface in the crystal structure (Voss et al., 2010), we hypothesized a critical interaction between E3 Y47 and E2 K254. Here we show that K254 plays at most a minor role in pH protection and dimer stability. In contrast, we found that E2 W235 was required for pH protection, as evinced by the effect of the W235A mutant on E1 transport and its rescue by bafilomycin treatment. As was the case for E3 Y47 (Uchime et al., 2013), no role of E2 W235 was observed in the fusion phenotype of the mature virus particle. The E3-E2-E1 structure suggests that E2 W235 forms an important component of E2’s interaction with the E3 surface, interacting with a pocket formed by E3 residues Y47, L35, L38, L51, and E39 (Voss et al., 2010).
Revertant analysis identified several other residues that presumably promote dimer stability and thus rescue the lethal phenotype resulting from the loss of pH protection (Table 1). The E2 D168E mutation was present in all 11 independent revertants of the E3 Y47A mutant. This relatively conservative mutation is located in the “acid-sensitive region”, a part of the β-ribbon that connects the three domains of E2 and is in contact with E3 (Voss et al., 2010)(Fig. 6B). The acid-sensitive region of the connector was not visualized in the structure of the Sindbis virus E2-E1 dimer crystallized at low pH, suggesting that at low pH it can move to allow uncapping of the E1 fusion loop (Li et al., 2010). Mutations that rescue p62 cleavage mutants map to the acid-sensitive region (Voss et al., 2010), supporting the role of this region in dimer stability.
Selection for growth of the E3 Y47/48A double mutant resulted in a number of true revertants that had acquired tyrosine at both positions. Our previous data showed that an E3 Y48A mutant was viable and had growth properties similar to WT (Uchime et al., 2013). The revertant analysis thus suggests that either the path towards viability of the double-mutant required first the acquisition of Y48, or that Y48A remained less fit than WT and was lost in the selection. We also isolated 5 second-site revertants of E3 Y47/48A, all of which contained a mutation of E2 histidine 256 to proline. E2 H256 is highly conserved among the alphaviruses although it is a proline in several fish alphaviruses. In keeping with effects on dimer stability, E2 H256 is also located in the acid-sensitive region of the β-ribbon connector and has van der Waals contacts with E3 Y48 (Voss et al., 2010). In contrast to the E3 Y47A revertants, mutations at E2 D168 were not isolated as revertants of the E3 Y47/48A double mutant.
In some cases the E2 D168E and E2 H256P rescue mutations were isolated along with other second site mutations (Table 1). Since these additional mutations were not required for rescue, it is not known if they promote rescue or simply reflect the relatively high mutation rate of RNA viruses. The additional second site mutations in the Y47/48A revertants, E2 N238V and D231N, are located along the E1-E2 interface and lie within the acid sensitive region. The asparagine at E2 position 238 is quite conserved and is part of a network of interactions with E1 S57 and E2 H170 (Voss et al., 2010). The S57-H170 interaction was shown to be important in regulating E2-E1 dimer stability (Fields and Kielian, 2013), and thus the N238V mutation could help to rescue the E3 Y47/48A mutant by stabilizing the dimer. Independent analysis of N238 would be required to determine this.
Together our data indicate that E2 W235 is involved in an important stabilizing interaction with the E3 protein. The overall E3-E2 interaction is stabilized at low pH, thus clamping E2 and E1 and preventing their dissociation during transport through the secretory pathway. Identification of pH-dependent interactions can be difficult due to redundant contributions, with the loss of one interaction not necessarily producing a significant change in pH dependence or viral fitness (Harrison, 2008; Sanchez-San Martin et al., 2009). Our data suggest that the pH-dependent stability of the E3-E2-E1 complex can also be influenced by residues including E2 D168 and E2 H256. These complex interactions thus help to mediate the pH protection of E1 during viral exit.
Acknowledgments
We thank Niloy Iqbal for his contributions to the generation of the K254A mutant and the isolation of Y47A revertants. We thank all the members of our lab for helpful discussions, Youqing Xiang for her excellent technical assistance and Katie Stiles and Guadalupe Martinez for critical reading of the manuscript. The data in this paper are from a thesis submitted by W.F. in partial fulfillment of the requirements for the Degree of Doctor of Philosophy in the Graduate Division of Medical Sciences of the Albert Einstein College of Medicine.
This work was supported by a grant to MK from the National Institute of Allergy and Infectious Diseases (R01-AI075647) and by Cancer Center Core Support Grant NIH/NCI P30-CA13330. WF was supported in part by the Training Program in Cellular and Molecular Biology and Genetics, T32 GM007491.
Footnotes
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chanel-Vos C, Kielian M. Second-site revertants of a Semliki Forest virus fusion-block mutation reveal the dynamics of a class II membrane fusion protein. J Virol. 2006;80:6115–6122. doi: 10.1128/JVI.00167-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee PK, Eng CH, Kielian M. Novel mutations that control the sphingolipid and cholesterol dependence of the Semliki Forest virus fusion protein. J Virol. 2002;76:12712–12722. doi: 10.1128/JVI.76.24.12712-12722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deCurtis I, Simons K. Dissection of Semliki Forest virus glycoprotein delivery from the trans-Golgi network to the cell surface in permeabilized BHK cells. Proc Natl Acad Sci USA. 1988;85:8052–8056. doi: 10.1073/pnas.85.21.8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL User’s Manual. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- Demaurex N, Furuya W, D’Souza S, Bonifacino JS, Grinstein S. Mechanism of acidification of the trans-Golgi network (TGN). In situ measurements of pH using retrieval of TGN38 and furin from the cell surface. J Biol Chem. 1998;273:2044–2051. doi: 10.1074/jbc.273.4.2044. [DOI] [PubMed] [Google Scholar]
- Duffus WA, Levy-Mintz P, Klimjack MR, Kielian M. Mutations in the putative fusion peptide of Semliki Forest virus affect spike protein oligomerization and virus assembly. J Virol. 1995;69:2471–2479. doi: 10.1128/jvi.69.4.2471-2479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields W, Kielian M. A key interaction between the alphavirus envelope proteins responsible for initial dimer dissociation during fusion. J Virol. 2013;87:3774–3781. doi: 10.1128/JVI.03310-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon VM, Klimpel KR, Arora N, Henderson MA, Leppla SH. Proteolytic activation of bacterial toxins by eukaryotic cells is performed by furin and by additional cellular proteases. Infect Immun. 1995;63:82–87. doi: 10.1128/iai.63.1.82-87.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC. The pH sensor for flavivirus membrane fusion. J Cell Biol. 2008;183:177–179. doi: 10.1083/jcb.200809175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC. Viral membrane fusion. Virol. 2015;479–480C:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Kartenbeck J, Simons K, Fries E. On the entry of Semliki Forest virus into BHK-21 cells. J Cell Biol. 1980;84:404–420. doi: 10.1083/jcb.84.2.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian M. Mechanisms of Virus Membrane Fusion Proteins. Annual Review of Virology. 2014;1:171–189. doi: 10.1146/annurev-virology-031413-085521. [DOI] [PubMed] [Google Scholar]
- Kielian M, Chanel-Vos C, Liao M. Alphavirus entry and membrane fusion. Viruses. 2010;2:796–825. doi: 10.3390/v2040796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian M, Jungerwirth S, Sayad KU, DeCandido S. Biosynthesis, maturation, and acid-activation of the Semliki Forest virus fusion protein. J Virol. 1990;64:4614–4624. doi: 10.1128/jvi.64.10.4614-4624.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RJ. Togaviridae. In: Knipe DM, Howley PM, editors. Fields Virology. 6. Lippincott, Williams and Wilkins; Philadelphia, PA: 2013. pp. 629–650. [Google Scholar]
- Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. Structural changes of envelope proteins during alphavirus fusion. Nature. 2010;468:705–708. doi: 10.1038/nature09546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Kielian M. Domain III from class II fusion proteins functions as a dominant-negative inhibitor of virus-membrane fusion. J Cell Biol. 2005;171:111–120. doi: 10.1083/jcb.200507075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljeström P, Lusa S, Huylebroeck D, Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Besanceney C, Song Y, Kielian M. Pseudorevertants of a Semliki Forest virus fusion-blocking mutation reveal a critical interchain interaction in the core trimer. J Virol. 2010;84:11624–11633. doi: 10.1128/JVI.01625-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A, Wahlberg JM, Lobigs M, Liljeström P, Garoff H. Membrane fusion process of Semliki Forest virus II: Cleavage- dependent reorganization of the spike protein complex controls virus entry. J Cell Biol. 1992;116:349–357. doi: 10.1083/jcb.116.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-San Martin C, Liu CY, Kielian M. Dealing with low pH: entry and exit of alphaviruses and flaviviruses. Trends in Microbiol. 2009;17:514–521. doi: 10.1016/j.tim.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoberg M, Lindqvist B, Garoff H. Activation of the alphavirus spike protein is suppressed by bound E3. J Virol. 2011;85:5644–5650. doi: 10.1128/JVI.00130-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit JM, Klimstra WB, Ryman KD, Bittman R, Johnston RE, Wilschut J. PE2 cleavage mutants of Sindbis virus: correlation between viral infectivity and pH-dependent membrane fusion activation of the spike heterodimer. J Virol. 2001;75:11196–11204. doi: 10.1128/JVI.75.22.11196-11204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchime O, Fields W, Kielian M. The Role of E3 in pH Protection during Alphavirus Assembly and Exit. J Virol. 2013;87:10255–10262. doi: 10.1128/JVI.01507-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 2010;468:709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- Wahlberg JM, Boere WAM, Garoff H. The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to low pH during virus maturation. J Virol. 1989;63:4991–4997. doi: 10.1128/jvi.63.12.4991-4997.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg JM, Garoff H. Membrane fusion process of Semliki Forest virus I: Low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J Cell Biol. 1992;116:339–348. doi: 10.1083/jcb.116.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Fugere M, Day R, Kielian M. Furin processing and proteolytic activation of Semliki Forest virus. J Virol. 2003;77:2981–2989. doi: 10.1128/JVI.77.5.2981-2989.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Kielian M. Mutations that promote furin-independent growth of Semliki Forest virus affect p62-E1 interactions and membrane fusion. Virol. 2004;327:287–296. doi: 10.1016/j.virol.2004.06.037. [DOI] [PubMed] [Google Scholar]
- Zhang X, Kielian M. An interaction site of the envelope proteins of Semliki Forest virus that is preserved after proteolytic activation. Virol. 2005;337:344–352. doi: 10.1016/j.virol.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Zheng A, Yuan F, Kleinfelter LM, Kielian M. A toggle switch controls the low pH-triggered rearrangement and maturation of the dengue virus envelope proteins. Nat Commun. 2014:5. doi: 10.1038/ncomms4877. [DOI] [PMC free article] [PubMed] [Google Scholar]