Abstract
The human intestine contains 1014 bacteria, which outnumber the mammalian cells 10-fold. Certain other commensal or infectious agents, like helminthic parasites become members of this microbial ecosystem, especially in populations living under less hygienic conditions. Intestinal microbes, also called the microbiome or microbiota, shape the host immune reactivity to self and nonself throughout life. Changes in microbiome composition may impair the maturation of immune regulatory pathways and predispose the host to develop various forms of inflammatory disorders, like Crohn's disease or ulcerative colitis. The microbiome is also critical to successful transplantation of organs or grafts. After allogeneic hematopoietic stem cell transplantation (HSCT), when the new donor cells, such as T lymphocytes learn to discriminate “the new-self from nonself” in the transplant recipient, they need healthy microbiota-derived signals to preserve the immune homeostasis. Restoring microbiota via intestinal delivery of bacterial strains, helminths, fecal microbiota transplantation or stool substitutes have the potential to improve and correct aberrant immune reactivity in various disorders.
Soon after birth, the sterile human gut becomes populated by approximately 1014 bacteria (1, 2). Besides the huge number, intestinal bacteria have enormous diversity with different strains from various taxa being represented(3). Furthermore, certain viruses, fungi and helminthic parasites can become members of the microbial ecosystem, which is also referred to as the gut microbiome. The symbiotic relationship between the intestinal immune system and the microbiome promotes colonization by numerous, beneficial bacterial strains. The gut microbiome assists in digestion, generates essential vitamins and promotes further development of the immune system. Altered microbiome composition may predispose to diseases. Recent technologies based on large-scale nucleic acid or metabolic product identification have enabled detailed analysis of intestinal bacteria. These studies lead to discovery of critical bacterial strains in health and disease. For example, adherent-invasive Escherichia coli colonizing ileal mucosa in Crohn's disease patients has been shown to trigger severe intestinal inflammation(4). Clostridium scindens has been identified as a probiotic and beneficial strain, whose restoration in intestinal lumen appears to protect antibiotic-treated and microbiota-depleted subjects from Clostridium difficile infection(5). Additional studies, in which preclinical models or human subjects were depleted or restored of microbiota, have also classified some strains or strain combinations as protective and others as harmful, further characterizing the microbial composition of the healthy versus disease-prone intestine.
Microbial strains are believed to colonize the mammalian digestive tract in equilibrium with each other and the host. Unbalanced microbial composition or loss of microbial diversity in the gut is called dysbiosis and predisposes the host to several pathologies, such as obesity, diabetes, cardiovascular disease, cancer or even to cognitive behavioral disorders(6-12). Bacteria belonging to phyla, Firmicutes and Bacteroidetes dominate the microbiome in healthy intestine. The synergy and communication between metabolically compatible strains ensures healthy microbial commensalism(13). In equilibrium, the mammalian immune system generates a tightly controlled inflammatory response to gut commensals that is associated with intact immune regulatory and tissue renewal pathways. Dysbiosis may trigger an uncontrolled inflammatory response to microbiota and lead to allergic, autoimmune or immunological disorders.
The sterile intestine at birth inherits first the mother's microbiota and undergoes continuous changes throughout life, under the influence of genetic and environmental factors, preserving the tightly controlled intestinal inflammatory response against microbiota. Antibiotic use, poor dietary habits or illnesses modulate the microbiota strain diversity, cause dysbiosis and lead to uncontrolled inflammation. Some studies have found an association between antibiotic use during infancy or early childhood and the development of inflammatory bowel disease (IBD), suggesting that environmental factors, altering microbiota – especially in early childhood - may trigger inflammation(14, 15). Shifting the microbiota composition may induce autoimmune reactivity(16, 17).
Besides bacteria, helminthic parasites become members of the intestinal microbial ecosystem. Helminths exert immune regulation through their direct effects on host cells(18) and/or via altering the composition of intestinal commensals(19). Helminths appear to increase the number of beneficial or probiotic strains in the intestine, such as enriching the microbiota for members of the Lactobacillaceae family. Therefore, self-limited gut colonization with helminthic parasites constitutes an attractive research tool to understand intestinal immune balance and an appealing therapeutic implement to suppress aberrant immunity in various disorders.
Infection with helminths has a particular geographic distribution, with rates highest among populations living under conditions of poor hygiene, i.e. in developing countries, and lowest in landscapes where clean living conditions predominate (20). The striking inverse relationship between helminth carriage and autoimmunity has been investigated in the context of hygiene hypothesis. According to hygiene theory, modern and hygienic life style in developed countries has removed some of our natural surroundings that are critical for the maturation and maintenance of a healthy immune system. For example, pasteurization of milk that is routinely applied in developed countries prevents zoonotic infections, such as Brucellosis but also prevents exposure to beneficial commensals, like Streptococcus thermophilus(21). The immune regulatory potential of probiotic strains, including S. thermophilus has been shown in preclinical colitis models and by clinical studies in IBD patients(21, 22). Additional epidemiological studies discovered higher prevalence of inflammatory bowel disease among people with decreased frequency of infections, lending support to the hygiene hypothesis(23). Similar to S. thermophilus and other probiotic strains, helminth addback experiments have demonstrated remarkable regulation of allergic, immunological and autoimmune disorders in preclinical models (24). Moreover, clinical studies have shown clinical benefit of helminths in immunological diseases(25).
Animal models have been instrumental in understanding the pathogenesis of allergic, immunological and alloreactive pathologies, which are heterogenous disorders, characterized by multiple genetic abnormalities. Moreover, uncontrolled inflammation in the intestine or other organs is set in motion by means of various environmental factors other than dysbiosis. Preclinical model systems have been developed to address the role of an isolated variable or a group of variables in the complex pro- or dysbiotic mammalian intestine that undergoes continuous alterations under the influence of host genes and copious environmental factors. Preclinical animal models include (i) germ-free mice with no aerobic or anaerobic microbiota in the gut, (ii) mice with a controlled luminal microenvironment carrying predefined and detectable bacterial strains in their intestinal tract, (iii) animals kept at special pathogen-free (SPF) facilities, (iv) intestinal infections with certain microorganisms and (v) genetically engineered mice. All have been vital tools to address the role of a single variable or a group inconstants. These models have enabled scientists to investigate how a particular gene mutation affect the host, the immune stimulatory potential of a unique microbial strain or product.
Studies on microbiota have led investigators to fruitful and provocative results not only in the direction from bench to bedside but also in reverse way, achieving clues on the cellular or molecular regulatory mechanisms involved in certain clinical applications. A good example to the latter is the use of short chain fatty acids (SCFA) in diversion colitis: gut microbiota generate SCFA that are applied to distal intestinal segments devoid of fecal stream to restore a nutrient, essential for renewal and homeostasis of the mucosa. While clinical observations have clearly demonstrated benefit and reduced rectal bleeding after short chain fatty acid enema use, recent studies in mice have shown that SCFA can induce intestinal immune regulation by promoting the generation of regulatory (suppressor) T cells (Treg) (26, 27). Whether the clinical benefit of SCFA enemas are associated with or due to Treg induction in patients after diversion surgery remains to be established. Tregs are dominant regulators of intestinal immunity and are critically important in achieving peripheral immune tolerance. Further studies in preclinical models have demonstrated the crucial contribution of gut microbiota to Tregs induction(28, 29). These studies have become classical examples of microbiome-induced immune regulation.
Regulatory T cells (Tregs) that express the transcription factor FoxP3 have recently emerged as a separate peripheral T lymphocyte lineage over the last ~15 years (30). Most Tregs are generated in the thymus and they are called thymic regulatory T cells (tTregs). Besides the group originating in the thymus, FoxP3 negative naïve T cells can be converted to FoxP3 positive Tregs in the periphery (peripheral; pTregs). Tregs exert dominant immune regulation in the gut as well as other organ systems. FoxP3 deficiency results in intestinal and systemic inflammation in patients as well as in preclinical models. Adoptive transfer of Tregs regulates intestinal inflammation in different animal models.
Intestinal microbiota appear critical in peripheral induction and maintenance of Tregs. Germ-free mice without microorganisms in their gut but with an intact thymus have reduced numbers of Tregs in their colon(28). Colonic Treg numbers in germ-free mice can be restored by colonizing their gut with bacterial strains of SPF facilities(28). T cell receptor chain analyses have shown that colonic Tregs populating the gut in these mice are peripheral Tregs(31), suggesting that gut microbiota drive conversion of FoxP3 negative T cells into FoxP3 positive Tregs besides possibly promoting the expansion and maintenance of tTregs. Generation of pTreg in the gut appears to contribute to peripheral immune tolerance because mice with normal number of tTregs but inability to generate pTregs due to a genetic defect develop intestinal inflammation(32). These findings attest to a crucial role of intestinal microorganisms in inducing peripheral Treg generation and regulation of inflammation in various conditions. In exchange and to the benefit of the microbial ecosystem, Tregs promote the maintenance of bacterial diversity(33), an essential element of intestinal symbiosis.
Specific microorganisms or bacterial products derived from the gut microbiome have been shown to increase the Treg number and enhance Treg function. Colonizing the gut of germ-free mice with spore forming Clostridia or Bacteroides was sufficient for Treg generation(29, 34). Further research has shown that a single product of Bacteroides, polysaccharide A (35, 36) was sufficient to promote Treg generation and stimulate Treg function. Similarly, helminths have been shown to promote Treg survival and stimulate their regulatory function(24, 37). Helminthic parasites may also exert immune regulation through engaging innate immune pathways independent of regulatory T and B cell subsets. Studies on gut microbiota-induced immune regulation have led to a provocative idea in clinical medicine whether one can stimulate Tregs or other immune regulatory pathways to control inflammation in various allergic or autoimmune disorders. In the era of stool transplantation, delivery of fecal microbiota - as a whole or as selected microbial strains - into the intestine is an appealing tool to restore a healthy gut microbiome, stimulate regulatory T cell responses and suppress aberrant immune reactivity. This is especially true for disorders with impaired or insufficient Treg function, such as IBD, as well as immune pathologies with decreased Treg number, such as graft-versus-host disease (GVHD). We will address the similarities between host-microbiome interactions in IBD and GVHD in the next sections.
Inflammatory bowel disease
Before the analysis of microbiota became available, several preclinical and clinical observations attested to the importance of gut commensals in the pathogenesis of IBD. A good clinical example is diversion surgery where the surgeons have diverted fecal stream to treat therapy resistant terminal ileal Crohn's disease(38). As Rutgeerts et al. reported repeat colonoscopy showed no disease recurrence in ileal segments, devoid of the fecal stream. By contrast, disease recurrence occurred in patients whose terminal ilea were exposed to fecal material after takedown surgery. Complementing this clinical observation, intestinal inflammation did not occur in mice raised in germ-free facilities(39). Thus fecal elements, that stimulate intestinal tissue renewal, repair and regulatory responses, can trigger inflammation in IBD.
Inflammatory responses to microbiota occur in IBD due to genetic and environmental factors. Genome-wide association studies (GWAS) indicated more than 100 gene variants that predispose to or protect from IBD. One of the best-characterized genes in IBD is CARD15/NOD2. It was first described as a result of gene association studies. Subsequent research has shown that CARD15/NOD2 is the intracellular receptor of bacterial muramyl dipeptide(40). CARD15/NOD2 is expressed by several tissues in the intestine but distinctively in Paneth cells and contributes to antibacterial gut immunity. Antimicrobial responses in the intestine prevent bacterial translocation and influence the gut microbiota composition, preserving diversity. Indeed, CARD15/NOD2 deficient mice display a different pattern of intestinal flora suggesting that host immune factors determine the quality and quantity of intestinal microbiota(41).
Another group of hereditary abnormalities associated with Crohn's disease affect genes associated with autophagy. Autophagy involves sequestration and recycling of intracellular organelles and other structures. It is a cellular regenerative process critical in intestinal tissue repair, renewal and antibacterial host defense. Intestinal microbiota trigger autophagy that is important in maintaining the immune homeostasis and preventing bacterial translocation. In IBD, autophagy defects, such as ATG16L1 mutations, result in impaired tissue renewal, bacterial translocation and inflammatory responses(42).
GWAS also identified interleukin 23 receptor (IL23R) variants to protect from Crohn's disease. Signaling through IL23R is believed to trigger inflammatory T helper 17 responses and cause inflammation(43). Additional data showed that IL23R signaling controls the abundance of certain strains and, through the induction of IL22, promotes barrier repair(44).
Research on IBD genetics may have impact on treatment. Besides showing the importance of genes in controlling the gut microbial diversity and helping us better understand the disease pathogenesis, these studies contributed to the development of a monoclonal antibody targeting IL23 p40 subunit, namely ustekinumab, for clinical use. Therapeutic induction of autophagy may also lead to innovative treatment modalities of IBD in the future. Immune modulators, biologics or medications that induce autophagy are potent drugs, though with concerning short- and long-term toxicities. By contrast, restoring microbiota with stool transfer or helminth colonization may have a safer side effect profile. Clinical data on the use of stool transplantation, fecal microbial transfer and helminths are accumulating, as we discuss below.
Besides genetic, environmental factors including intestinal microbiota play a role in IBD pathogenesis. One approach in the search for environmental factors that influence IBD has focused on changes during the second half of 20th century, as the prevalence of this condition and other autoimmune diseases has increased significantly during that period (23). For example, the use of antibiotics and the refrigeration of food became frequent practices. Whereas both have clear benefits, they also have long term unappreciated negative effects. In the case of antibiotics, the positives are that they cure infections; the negatives are that they reduce intestinal bacterial load and change the quality of microbiome. Similarly, although refrigeration suppresses bacterial growth and prevents food from spoiling, it permits the growth of certain bacteria in food before it is ingested, again shifting the balance of the microbiome. For example, refrigeration selectively permits the growth of Listeria that is frequently isolated from the gut of inflammatory bowel disease patients(45).
The microbiota as a whole may stimulate or regulate inflammation in IBD. Early antibiotic use during childhood is associated with a predisposition to ulcerative colitis and Crohn's disease, suggesting that the intestinal flora during early childhood is critical for peripheral immune system development and plays a role as a protective factor from IBD(14, 15). By contrast, antibiotics have a limited and well-defined therapeutic role in certain forms of inflammatory bowel disorders, such as fistulizing Crohn's disease(46). Thus, altering the balance between microbiota strains during early childhood may predispose to dysbiosis and IBD, whereas possible effector bacterial strains of a dysbiotic intestine may be eradicated by antibiotic use in IBD patients. Similarly, pouchitis can be treated using probiotics, such as Lactobacillus rhamnosus or VSL #3 (a combination of 8 probiotic strains) as well as antibiotics(47, 48). Preclinical models in germ-free mice or in mice raised at SPF facilities showed microbiota-driven induction of Tregs and Treg-mediated regulation of colitis. Whether bacteria, helminths or their products, such as SCFA induce immune regulation by stimulating Tregs in IBD patients remains to be established. As adoptive Treg transfer has limitations in clinical practice, expanding and stimulating Tregs in vivo by means of manipulating luminal microenvironment is a provocative idea for clinical IBD research.
Graft versus host disease
A severe and devastating form of intestinal inflammation with similarities to IBD in preclinical models and bone marrow transplant recipients is graft versus host disease (GVHD). It is caused by donor cells, especially T lymphocytes, administered with the hematopoietic stem cells to the recipient to enhance marrow engraftment and suppress cancer growth(49-51). In GVHD, donor T cells attack mainly the skin, gut and liver because of major or minor histocompatibility antigen mismatching. Controlled donor T cell alloreactivity to recipient tissues is a desired endpoint in allogeneic stem cell transplantation, as it suppresses cancer growth. However, uncontrolled alloreactive activation of the graft leads to severe and potentially lethal GVHD-related inflammation. Thus, in hematopoietic stem cell transplantation (HSCT), donor cell alloreactivity is desired to be sufficient to suppress cancer growth, in patients with malignant disorders, but also limited to prevent overwhelming GVHD. Studies have shown the intestine to be a primal organ in the induction, maintenance and regulation of alloreactive donor T cell activation that leads to GVHD (52, 53). GVHD-related gut inflammation is difficult to treat and is a bad outcome sign in clinical medicine(54). Patients do not see a great benefit from glucocorticoids or other immune suppressives. However, they incur severe toxicity. Therefore, regulation of gut inflammation using microbiota or helminths is an attractive research idea in the management of lethal and devastating GVHD. In mice, helminths expand Tregs in vivo(37), a cell population that regulates GVHD preserving the anti-tumor immunity(55).
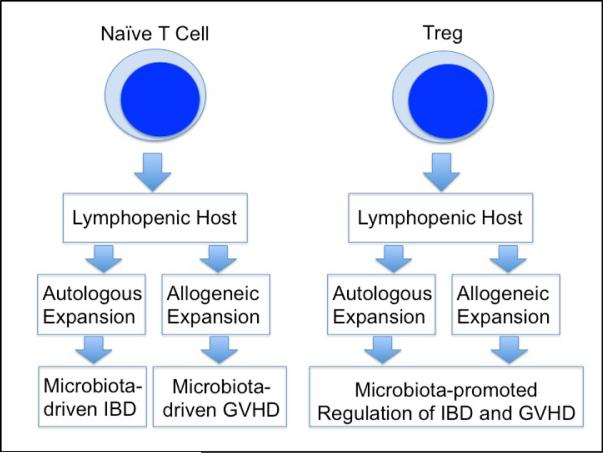
The endoscopic appearance of GVHD may mimic IBD. Moreover, colitis in certain preclinical IBD and GVHD models is similar and based on colitogenic effects of expanding naïve T cells in an immunodeficient host (Figure 1). The host is usually T and B cell deficient recombinase activation gene (RAG) knockout or immune compromised severe combined immunodeficient (scid) mouse in the case of IBD and, most often, a lethally irradiated recipient in the case of GVHD. Intestinal inflammation relies on autologous or syngeneic T cell activation in inflammatory bowel disease, while donor T cell alloreactivity and MHC mismatch drive the immune pathology in GVHD. Inflammation does not occur in mice raised in a germ-free environment attesting to the importance of gut microbiota in driving the inflammation in either group of disorders(39, 56).
Figure 1. Similar effector and regulatory immune pathways are involved in mouse GVHD besides IBD.
Expansion of naïve T cells in syngeneic host leads to IBD, whereas allogeneic activation and expansion of naïve T cells in an MHC-mismatched host results in GVHD. Treg co-administration regulates IBD- and GVHD-related inflammation. The lymphopenic host, that receives naïve T cells or Tregs, is a RAG-/- or scid mouse in IBD models and is usually an irradiated recipient in GVHD experiments.
Additional surprising similarities between GVHD and IBD came via further genetic association studies that showed that autophagy or CARD15/NOD2 gene defects predispose to and IL23R mutations protect from GVHD (Table 1). Studies have demonstrated more frequent and more severe GVHD in HSCT recipients with CARD15/NOD2 mutations in donor/recipient pairs(57). CARD15/NOD2 regulates GVHD through its inhibitory effect on antigen presenting cell function. This knowledge opens a new area of therapeutic approaches. Whether CARD15/NOD2 mutations predispose to GVHD by influencing the gut flora composition or by modulating various immune pathways remains to be established.
Table 1.
Common gene mutations in IBD and GVHD
| Mutation | Effect on IBD | Effect on GVHD |
|---|---|---|
| CARD15/Nod2 | Increases risk | Increases risk |
| ATG16L | Increases risk | Increases risk |
| IL23R | Reduces risk | Reduces risk |
Association studies have shown that IL23R mutations protect from GVHD similar to IBD(58). In mice, autophagy gene defects, whose homologues predispose to IBD in humans, predispose the bone marrow transplant recipients to GVHD(59). According to some studies, mutations of toll-like receptors (TLRs), a predominant group of microbiota sensors in the intestine, also predispose to graft versus host or inflammatory bowel disease.
Surprising similarities between inflammatory bowel and graft versus host diseases have motivated researchers to investigate the role of gut microbiota in GVHD. Loss of bacterial strain diversity – not as a result of antibiotic use but as a consequence of alloreactive inflammation – has been shown in GVHD(53). Loss of strain diversity is accompanied by loss of Clostridia and enrichment for Lactobacillae and Enterobacteriaceae in intestinal lumen. Bacterial translocation from the gut is a major trigger for systemic inflammation after chemo- or radiotherapy prior hematopoietic stem cell transplantation and worsens alloreactive donor cell activation. Broad-spectrum antibiotic use is routine in clinical bone marrow transplantation (BMT). This prevents bacterial infections in severely immune compromised patients and interferes with bacterial translocation. Besides antibiotics, probiotics may also reduce bacterial translocation, as shown in mice(60). Whether restoration of bacterial diversity by probiotics or fecal microbiota transplantation suppresses GVHD-related inflammation in patients remains to be established. Colonizing the gut with helminths can also regulate GVHD(37), where helminths suppress inflammation through their direct immune regulatory effects on host cells or through modulating microbiota. Helminths have been shown to enrich for Lactobacillae in murine gut. Although Lactobacillae enrichment appears to accompany loss of microbiota strain diversity and can be associated with worsening GVHD(53), addback of certain Lactobacillus strains into the intestine alleviates graft-versus-host reactivity in mice. Better definition of microbiota and their interactions with helminths or host cells will help us design better treatment strategies and safer environments for bone marrow transplant patients.
It will be also important to characterize in molecular and cellular detail the impact of antibiotics on intestinal dysbiosis. A study from Sweden has demonstrated a decrease in infectious complications and a lower incidence of GVHD in patients undergoing hematopoietic stem cell transplantation at home(61). The home environment with different antibiotic practices and microbial communities may prevent bacterial dysbiosis in HSCT patients.
Administration of enriched donor or ex vivo propagated Tregs can prevent GVHD(62). However, this is a technically challenging and costly practice in clinical medicine. Strategies also are being developed to in vivo expand Tregs in an attempt to prevent GVHD(63). With well-established stimulatory effects on Tregs in mice, gut bacteria or helminths may provide novel treatments to patients with GVHD in future.
HSCT is mostly performed as an immunotherapy against certain malignant diseases, such as leukemia, lymphoma and myeloma. The direct effects of the microbiome against tumor cells is heavily investigated not only in these hematological malignancies where HSCT is frequently applied but also in other forms of cancer.
Cancer
The gut immune system and the intestinal microbiota communicate to maintain a balance between tolerance and activation. Since there is such a close interaction between these two players, it is not surprising that cancers of the immune system, e.g., lymphomas such as mucosal-associated lymphoid tissue (MALT) lymphomas have been shown to be caused by bacteria. These lymphomas originate in the marginal zone and 90% of MALT lymphomas are associated with the presence of Helicobacter; elimination of Helicobacter leads to complete remission in approximately 80% of all cases(64). The causative effect of Helicobacter in MALT lymphomas has been demonstrated in animal models. Other bacteria such as Campylobacter jejuni, Borrelia burgdorferi and Chlamydia psittaci may also play a role in lymphoma development and associations with these bacteria have been demonstrated in humans. Bacteria can cause changes in T-cell activity eliciting increases in inflammatory cytokines. Besides increasing cellular turnover in the intestine through inflammation, oxidative stress caused by intestinal bacteria can also affect carcinogenesis. Helicobacter pylori has also been implicated in esophageal cancer, but its role remains unclear(65).
Commensal bacteria play a major role in the development of colorectal cancer (CRC). It was recently proposed that selected commensals polarize colon macrophages to produce endogenous mutagens that initiate chromosomal instability (CIN), which leads to expression of cancer stem cell markers in non-cancer stem cell malignant cells and drive CRC through a bystander effect(66). This hypothesis was validated in immunodeficient mice. The same group also showed that Enterococcus faecalis induced aneuploidy and tetraploidy in colonic epithelial cells in mice(67). An association between the salivary microbiota and pancreatic cancer has been observed. Two bacteria (Neisseria elongata and Streptococcus mitis) showed significant association with pancreatic cancer with a 96.4% sensitivity and 82.1% specificity(68).
Recent studies have also suggested that intestinal bacteria can modulate cancer development in distant non-intestinal tissues, such as prostate cancer and breast cancer due to Helicobacter hepaticus(12, 69).
The beneficial effects of the microbiota can be enhanced by probiotics and microbiota transplants to increase butyrate-producing bacteria and by decreasing inflammation-causing and Th17-inducing bacteria. Butyrate acts as a histone deacetylase inhibitor and anti-inflammatory agent; it also improves the oxidative status(70). Curcumin is a natural phenol that decreases inflammatory response in human intestinal epithelial cells(71).
Chemotherapy-or radiation-induced mucositis
Patients receiving cytotoxic and radiation therapy exhibit marked changes in the intestinal microbiota with most frequently a decrease in Bifidobacterium, Clostridium cluster XIVa, Faecalibacterium pra-usnitzii, and increase in Enterobacteriaceae and Bacteroides. These modifications contribute to diarrhea and bacteremias. There have been some promising results with probiotics to prevent cancer therapy-induced mucositis(72). Results showing benefit in utilizing microbiota in cancer or IBD patients and Clostridium difficile-infected individuals initiated studies that may lead to wider use of microbiota or its products in clinical medicine.
Fecal Microbiota Transplantation: Current and Future Applications
Over the past few years there has been a heightened interest in fecal microbiota transplantation (FMT), primarily due to the increasing incidence of Clostridium difficile infection. FMT is increasingly viewed as an established effective therapy for recurrent C. difficile infection. These patients have been shown to have loss of intestinal bacterial diversity, which is reversed following engraftment of donor stool(73). Three randomized, open label trials have demonstrated cure rates of 90-94%(74-76). Two of the three trials compared FMT to oral vancomycin(74, 76), while the third compared FMT delivered by the nasogastric route to delivery via colonoscopy(75). Another emerging therapeutic application of FMT is for multidrug resistant pathogen decolonization of the GI tract. Two case reports demonstrate successful decolonization of highly resistant gram-negative pathogens in humans(77), and decolonization of vancomycin resistant enterococci has been demonstrated in a mouse model(78).
FMT is also under active evaluation for the treatment of IBD. A meta-analysis of nine cohort studies determined that the pooled estimate of clinical remission for ulcerative colitis was 22%, with results of individual studies varying from 0% to 75%(79). More recently, two randomized, placebo controlled trials of FMT for ulcerative colitis have been published. In the first study (70 patients randomized to FMT or placebo administered weekly via enema for 6 weeks), 24% of patients in the FMT arm were in remission at 7 weeks versus 5% in the placebo arm (P=0.03)(80). In the second study, 50 patients were randomized to FMT from healthy donors versus autologous donation (placebo) administered via nasoduodenal tube at two time points (0 and 3 weeks). Remission was achieved in 30% in the active treatment arm versus 20% in the placebo arm (P=.51)(81). Moreover, as of May 19, 2015, there are an additional 18 clinical trials for patients with IBD actively recruiting patients (https://clinicaltrials.gov).
FMT has also been studied in human subjects for several other diseases. In a small, randomized, double blind, placebo controlled trial, 18 men received either FMT from a lean male donor or autologous FMT (placebo). Those receiving FMT from a lean donor showed a significant improvement in peripheral insulin sensitivity(82). Another potential role for FMT may be GVHD following allogeneic HSCT, given the loss of bacterial diversity (83) and elevation of fecal inflammatory biomarkers (calprotectin and α1-antitrypsin) (84) seen in these patients. Indeed, loss of intestinal bacterial diversity has been shown to be an independent predictor for mortality in patients following allogeneic HSCT (85). Lastly, case reports on patients with multiple sclerosis (MS) or Parkinson's disease that underwent FMT, were reported. Of the 3 patients with MS, who underwent FMT for chronic constipation, all experienced a significant durable improvement in neurologic findings (86).
After helminth infection models in mice showed very promising data on the regulation of aberrant immune reactivity, clinical trials have been initiated, that addressed the role of helminths in patients(24, 25). Early observations showed clinical benefit in various diseases, such as IBD or MS(25). Currently, six clinical trials on the effect of helminths on disease outcome in autoimmunity and other pathological conditions are actively recruiting patients (https://clinicaltrials.gov).
Given the concerns regarding transmission of pathogens during FMT, there has been interest in the development of synthetic stool substitutes. Two studies evaluated the use of the same mixture of 10 intestinal bacteria, each of which was grown in pure culture, then reconstituted in normal saline and delivered via enema for the treatment of recurrent C. difficile infection. The combined results demonstrated cure in 12 of 13 patients (92%)(87, 88). In another study, 33 bacterial strains were chosen from the stool of a healthy donor, grown in pure culture, reconstituted in normal saline, and delivered via colonoscopy to two patients with recurrent C. difficile infection. Both patients were cured(89). It is anticipated that synthetic stool substitutes may ultimately replace FMT.
Conclusions
The intestine has emerged as a crucial immune organ influencing inflammatory reactivity in allergic, autoimmune and alloreactive disorders. Intestinal immune regulatory pathways, like Tregs are stimulated by intestinal microbiota or helminths. Live organisms or their products trigger intestinal and systemic immune regulation. Dysbiosis of microbiota and immune dysregulation accompany each other in several diseases, with pathogenesis mechanisms overlapping significantly in IBD and GVHD. Preclinical models and early clinical trials suggest that various pathological conditions with aberrant immune reactivity can be managed by restoring healthy and nondysbiotic microbiota. Future research on microbiota or helminths colonizing the gut may lead to better treatment modalities of IBD as well as similar devastating disorders, such as GVHD.
Acknowledgments
Supported by R56 AI116715, R01 HL56067, R01 AI112613 and R01 AI 34495
References
- 1.Kau AL, Ahern PP, Griffin NW, et al. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annual review of immunology. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahern PP, Faith JJ, Gordon JI. Mining the human gut microbiota for effector strains that shape the immune system. Immunity. 2014;40:815–823. doi: 10.1016/j.immuni.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darfeuille-Michaud A, Boudeau J, Bulois P, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 5.Buffie CG, Bucci V, Stein RR, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 7.Tilg H, Kaser A. Gut microbiome, obesity, and metabolic dysfunction. The Journal of clinical investigation. 2011;121:2126–2132. doi: 10.1172/JCI58109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rak K, Rader DJ. Cardiovascular disease: the diet-microbe morbid union. Nature. 2011;472:40–41. doi: 10.1038/472040a. [DOI] [PubMed] [Google Scholar]
- 9.Musso G, Gambino R, Cassader M. Obesity, diabetes, and gut microbiota: the hygiene hypothesis expanded? Diabetes care. 2010;33:2277–2284. doi: 10.2337/dc10-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwabe RF, Jobin C. The microbiome and cancer. Nature reviews Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsiao EY, McBride SW, Hsien S, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang YJ, Li S, Gan RY, et al. Impacts of gut bacteria on human health and diseases. International journal of molecular sciences. 2015;16:7493–7519. doi: 10.3390/ijms16047493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamont RJ, Hajishengallis G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends in molecular medicine. 2015;21:172–183. doi: 10.1016/j.molmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilat T, Hacohen D, Lilos P, et al. Childhood factors in ulcerative colitis and Crohn's disease. An international cooperative study. Scandinavian journal of gastroenterology. 1987;22:1009–1024. doi: 10.3109/00365528708991950. [DOI] [PubMed] [Google Scholar]
- 15.Wurzelmann JI, Lyles CM, Sandler RS. Childhood infections and the risk of inflammatory bowel disease. Digestive diseases and sciences. 1994;39:555–560. doi: 10.1007/BF02088342. [DOI] [PubMed] [Google Scholar]
- 16.Alkanani AK, Hara N, Gottlieb PA, et al. Alterations in Intestinal Microbiota Correlate with Susceptibility to Type 1 Diabetes. Diabetes. 2015 doi: 10.2337/db14-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yurkovetskiy LA, Pickard JM, Chervonsky AV. Microbiota and Autoimmunity: Exploring New Avenues. Cell host & microbe. 2015;17:548–552. doi: 10.1016/j.chom.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grainger JR, Smith KA, Hewitson JP, et al. Helminth secretions induce de novo T cell Foxp3 expression and regulatory function through the TGF-beta pathway. JExpMed. 2010;207:2331–2341. doi: 10.1084/jem.20101074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walk ST, Blum AM, Ewing SA, et al. Alteration of the murine gut microbiota during infection with the parasitic helminth Heligmosomoides polygyrus. InflammBowelDis. 2010;16:1841–1849. doi: 10.1002/ibd.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yazdanbakhsh M, Kremsner PG, van RR. Allergy, parasites, and the hygiene hypothesis. Science. 2002;296:490–494. doi: 10.1126/science.296.5567.490. [DOI] [PubMed] [Google Scholar]
- 21.Del Carmen S, Miyoshi A, Azevedo V, et al. Evaluation of a Streptococcus thermophilus strain with innate anti-inflammatory properties as a vehicle for IL-10 cDNA delivery in an acute colitis model. Cytokine. 2015;73:177–183. doi: 10.1016/j.cyto.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 22.Tursi A, Brandimarte G, Papa A, et al. Treatment of relapsing mild-to-moderate ulcerative colitis with the probiotic VSL#3 as adjunctive to a standard pharmaceutical treatment: a double-blind, randomized, placebo-controlled study. The American journal of gastroenterology. 2010;105:2218–2227. doi: 10.1038/ajg.2010.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. NEnglJMed. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 24.Weinstock JV, Elliott DE. Helminth Infections Decrease Host Susceptibility to Immune-Mediated Diseases. Journal of immunology. 2014;193:3239–3247. doi: 10.4049/jimmunol.1400927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleming JO, Weinstock JV. Clinical trials of helminth therapy in autoimmune diseases: rationale and findings. Parasite immunology. 2015;37:277–292. doi: 10.1111/pim.12175. [DOI] [PubMed] [Google Scholar]
- 26.Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 28.Geuking MB, Cahenzli J, Lawson MA, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 29.Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 30.Hoeppli RE, Wu D, Cook L, et al. The environment of regulatory T cell biology: cytokines, metabolites, and the microbiome. Frontiers in immunology. 2015;6:61. doi: 10.3389/fimmu.2015.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lathrop SK, Bloom SM, Rao SM, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Josefowicz SZ, Niec RE, Kim HY, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482:395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawamoto S, Maruya M, Kato LM, et al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity. 2014;41:152–165. doi: 10.1016/j.immuni.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 34.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazmanian SK, Liu CH, Tzianabos AO, et al. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Chen HL, Bannick N, et al. Intestinal helminths regulate lethal acute graft-versus-host disease and preserve the graft-versus-tumor effect in mice. Journal of immunology. 2015;194:1011–1020. doi: 10.4049/jimmunol.1303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rutgeerts P, Goboes K, Peeters M, et al. Effect of faecal stream diversion on recurrence of Crohn's disease in the neoterminal ileum. Lancet. 1991;338:771–774. doi: 10.1016/0140-6736(91)90663-a. [DOI] [PubMed] [Google Scholar]
- 39.Stepankova R, Powrie F, Kofronova O, et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflammatory bowel diseases. 2007;13:1202–1211. doi: 10.1002/ibd.20221. [DOI] [PubMed] [Google Scholar]
- 40.Strober W, Watanabe T. NOD2, an intracellular innate immune sensor involved in host defense and Crohn's disease. Mucosal immunology. 2011;4:484–495. doi: 10.1038/mi.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biswas A, Petnicki-Ocwieja T, Kobayashi KS. Nod2: a key regulator linking microbiota to intestinal mucosal immunity. Journal of molecular medicine. 2012;90:15–24. doi: 10.1007/s00109-011-0802-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen HT, Lapaquette P, Bringer MA, et al. Autophagy and Crohn's disease. Journal of innate immunity. 2013;5:434–443. doi: 10.1159/000345129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duerr RH, Taylor KD, Brant SR, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shih VF, Cox J, Kljavin NM, et al. Homeostatic IL-23 receptor signaling limits Th17 response through IL-22-mediated containment of commensal microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:13942–13947. doi: 10.1073/pnas.1323852111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huijsdens XW, Linskens RK, Taspinar H, et al. Listeria monocytogenes and inflammatory bowel disease: detection of Listeria species in intestinal mucosal biopsies by real-time PCR. Scandinavian journal of gastroenterology. 2003;38:332–333. [PubMed] [Google Scholar]
- 46.Vavricka SR, Rogler G. Fistula treatment: The unresolved challenge. Digestive diseases. 2010;28:556–564. doi: 10.1159/000320416. [DOI] [PubMed] [Google Scholar]
- 47.Ghouri YA, Richards DM, Rahimi EF, et al. Systematic review of randomized controlled trials of probiotics, prebiotics, and synbiotics in inflammatory bowel disease. Clinical and experimental gastroenterology. 2014;7:473–487. doi: 10.2147/CEG.S27530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chowdhry S, Katz JA. Update on the pathogenesis and management of pouchitis. Current infectious disease reports. 2014;16:442. doi: 10.1007/s11908-014-0442-9. [DOI] [PubMed] [Google Scholar]
- 49.Macdonald KP, Shlomchik WD, Reddy P. Biology of graft-versus-host responses: recent insights. BiolBlood Marrow Transplant. 2013;19:S10–S14. doi: 10.1016/j.bbmt.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shlomchik WD. Graft-versus-host disease. NatRevImmunol. 2007;7:340–352. doi: 10.1038/nri2000. [DOI] [PubMed] [Google Scholar]
- 51.Socie G, Blazar BR. Acute graft-versus-host disease: from the bench to the bedside. Blood. 2009;114:4327–4336. doi: 10.1182/blood-2009-06-204669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murai M, Yoneyama H, Ezaki T, et al. Peyer's patch is the essential site in initiating murine acute and lethal graft-versus-host reaction. NatImmunol. 2003;4:154–160. doi: 10.1038/ni879. [DOI] [PubMed] [Google Scholar]
- 53.Jenq RR, Ubeda C, Taur Y, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. JExpMed. 2012;209:903–911. doi: 10.1084/jem.20112408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gooley TA, Chien JW, Pergam SA, et al. Reduced mortality after allogeneic hematopoietic-cell transplantation. NEnglJMed. 2010;363:2091–2101. doi: 10.1056/NEJMoa1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kohrt HE, Pillai AB, Lowsky R, et al. NKT cells, Treg, and their interactions in bone marrow transplantation. EurJImmunol. 2010;40:1862–1869. doi: 10.1002/eji.201040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Penack O, Holler E, van den Brink MR. Graft-versus-host disease: regulation by microbe-associated molecules and innate immune receptors. Blood. 2010;115:1865–1872. doi: 10.1182/blood-2009-09-242784. [DOI] [PubMed] [Google Scholar]
- 57.Penack O, Holler E, van den Brink MR. Graft-versus-host disease: regulation by microbe-associated molecules and innate immune receptors. Blood. 2010;115:1865–1872. doi: 10.1182/blood-2009-09-242784. [DOI] [PubMed] [Google Scholar]
- 58.Elmaagacli AH, Koldehoff M, Landt O, et al. Relation of an interleukin-23 receptor gene polymorphism to graft-versus-host disease after hematopoietic-cell transplantation. Bone Marrow Transplant. 2008;41:821–826. doi: 10.1038/sj.bmt.1705980. [DOI] [PubMed] [Google Scholar]
- 59.Hubbard-Lucey VM, Shono Y, Maurer K, et al. Autophagy gene Atg16L1 prevents lethal T cell alloreactivity mediated by dendritic cells. Immunity. 2014;41:579–591. doi: 10.1016/j.immuni.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gerbitz A, Schultz M, Wilke A, et al. Probiotic effects on experimental graft-versus-host disease: let them eat yogurt. Blood. 2004;103:4365–4367. doi: 10.1182/blood-2003-11-3769. [DOI] [PubMed] [Google Scholar]
- 61.Ringden O, Remberger M, Holmberg K, et al. Many days at home during neutropenia after allogeneic hematopoietic stem cell transplantation correlates with low incidence of acute graft-versus-host disease. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2013;19:314–320. doi: 10.1016/j.bbmt.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 62.Brunstein CG, Miller JS, Cao Q, et al. Infusion of ex vivo expanded T regulatory cells in adults transplanted with umbilical cord blood: safety profile and detection kinetics. Blood. 2011;117:1061–1070. doi: 10.1182/blood-2010-07-293795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koreth J, Matsuoka K, Kim HT, et al. Interleukin-2 and regulatory T cells in graft-versus-host disease. NEnglJMed. 2011;365:2055–2066. doi: 10.1056/NEJMoa1108188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamamoto ML, Schiestl RH. Intestinal microbiome and lymphoma development. Cancer journal. 2014;20:190–194. doi: 10.1097/PPO.0000000000000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang L, Chaudhary N, Baghdadi J, et al. Microbiome in reflux disorders and esophageal adenocarcinoma. Cancer journal. 2014;20:207–210. doi: 10.1097/PPO.0000000000000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang X, Yang Y, Huycke MM. Commensal bacteria drive endogenous transformation and tumour stem cell marker expression through a bystander effect. Gut. 2015;64:459–468. doi: 10.1136/gutjnl-2014-307213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang X, Allen TD, May RJ, et al. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer research. 2008;68:9909–9917. doi: 10.1158/0008-5472.CAN-08-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farrell JJ, Zhang L, Zhou H, et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2012;61:582–588. doi: 10.1136/gutjnl-2011-300784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lakritz JR, Poutahidis T, Mirabal S, et al. Gut bacteria require neutrophils to promote mammary tumorigenesis. Oncotarget. 2015;6:9387–9396. doi: 10.18632/oncotarget.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kusaczuk M, Bartoszewicz M, Cechowska-Pasko M. Phenylbutyric Acid: simple structure - multiple effects. Current pharmaceutical design. 2015;21:2147–2166. doi: 10.2174/1381612821666150105160059. [DOI] [PubMed] [Google Scholar]
- 71.Cho JA, Park E. Curcumin utilizes the anti-inflammatory response pathway to protect the intestine against bacterial invasion. Nutrition research and practice. 2015;9:117–122. doi: 10.4162/nrp.2015.9.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Touchefeu Y, Montassier E, Nieman K, et al. Systematic review: the role of the gut microbiota in chemotherapy- or radiation-induced gastrointestinal mucositis - current evidence and potential clinical applications. Alimentary pharmacology & therapeutics. 2014;40:409–421. doi: 10.1111/apt.12878. [DOI] [PubMed] [Google Scholar]
- 73.Weingarden A, Gonzalez A, Vazquez-Baeza Y, et al. Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome. 2015;3:10. doi: 10.1186/s40168-015-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. The New England journal of medicine. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 75.Youngster I, Sauk J, Pindar C, et al. Fecal microbiota transplant for relapsing Clostridium difficile infection using a frozen inoculum from unrelated donors: a randomized, open-label, controlled pilot study. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2014;58:1515–1522. doi: 10.1093/cid/ciu135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cammarota G, Masucci L, Ianiro G, et al. Randomised clinical trial: faecal microbiota transplantation by colonoscopy vs. vancomycin for the treatment of recurrent Clostridium difficile infection. Alimentary pharmacology & therapeutics. 2015;41:835–843. doi: 10.1111/apt.13144. [DOI] [PubMed] [Google Scholar]
- 77.Singh R, van Nood E, Nieuwdorp M, et al. Donor feces infusion for eradication of Extended Spectrum beta-Lactamase producing Escherichia coli in a patient with end stage renal disease. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2014;20:O977–978. doi: 10.1111/1469-0691.12683. [DOI] [PubMed] [Google Scholar]
- 78.Ubeda C, Bucci V, Caballero S, et al. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infection and immunity. 2013;81:965–973. doi: 10.1128/IAI.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Colman RJ, Rubin DT. Fecal microbiota transplantation as therapy for inflammatory bowel disease: a systematic review and meta-analysis. Journal of Crohn's & colitis. 2014;8:1569–1581. doi: 10.1016/j.crohns.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moayyedi P, Surette MG, Kim PT, et al. Fecal Microbiota Transplantation Induces Remission in Patients With Active Ulcerative Colitis in a Randomized Controlled Trial. Gastroenterology. 2015;149:102–109. e106. doi: 10.1053/j.gastro.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 81.Rossen NG, Fuentes S, van der Spek MJ, et al. Findings From a Randomized Controlled Trial of Fecal Transplantation for Patients With Ulcerative Colitis. Gastroenterology. 2015;149:110–118. e114. doi: 10.1053/j.gastro.2015.03.045. [DOI] [PubMed] [Google Scholar]
- 82.Vrieze A, Van Nood E, Holleman F, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913–916. e917. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 83.Holler E, Butzhammer P, Schmid K, et al. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2014;20:640–645. doi: 10.1016/j.bbmt.2014.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.O'Meara A, Kapel N, Xhaard A, et al. Fecal calprotectin and alpha1-antitrypsin dynamics in gastrointestinal GvHD. Bone marrow transplantation. 2015 doi: 10.1038/bmt.2015.109. [DOI] [PubMed] [Google Scholar]
- 85.Taur Y, Jenq RR, Perales MA, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124:1174–1182. doi: 10.1182/blood-2014-02-554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Borody TJ, Paramsothy S, Agrawal G. Fecal microbiota transplantation: indications, methods, evidence, and future directions. Current gastroenterology reports. 2013;15:337. doi: 10.1007/s11894-013-0337-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tvede M, Rask-Madsen J. Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. Lancet. 1989;1:1156–1160. doi: 10.1016/s0140-6736(89)92749-9. [DOI] [PubMed] [Google Scholar]
- 88.Emanuelsson F, Claesson BE, Ljungstrom L, et al. Faecal microbiota transplantation and bacteriotherapy for recurrent Clostridium difficile infection: a retrospective evaluation of 31 patients. Scandinavian journal of infectious diseases. 2014;46:89–97. doi: 10.3109/00365548.2013.858181. [DOI] [PubMed] [Google Scholar]
- 89.Petrof EO, Gloor GB, Vanner SJ, et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: 'RePOOPulating' the gut. Microbiome. 2013;1:3. doi: 10.1186/2049-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]