Abstract
The cholesterol-metabolizing enzyme sterol O-acetyltransferase (SOAT1) is implicated in an increasing number of biological and pathological processes in a number of organ systems, including the differentiation of the hair shaft. While the functional and regulatory mechanisms underlying these diverse functional roles remain poorly understood, the compartment of the hair shaft known as medulla, affected by mutations in Soat1, may serve as a suitable model for defining some of these mechanisms. A comparative analysis of mRNA and protein expression patterns of Soat1/SOAT1 and the transcriptional regulator Hoxc13/HOXC13 in postnatal skin of FVB/NTac mice indicated co-expression in the most proximal cells of the differentiating medulla. This finding combined with the significant downregulation of Soat1 expression in postnatal skin of both Hoxc13 gene-targeted and transgenic mice based on previously reported DNA microarray results suggests a potential regulatory relationship between the two genes. Non-detectable SOAT1 expression in the defective hair follicle medulla of Hoxc13tm1Mrc mice and evidence for binding of HOXC13 to the Soat1 upstream control region obtained by ChIP assay suggests that Soat1 is a downstream regulatory target for HOXC13 during medulla differentiation.
Keywords: Hoxc13, Soat1, hair follicle, lipid metabolism, Hox regulatory target
1. Introduction
Epidermal lipids are essential for barrier formation and immune function. Lipids in the pilosebaceous unit are critical for facilitating movement of the differentiating hair shaft towards the epidermal surface, and mutations affecting sebaceous gland function were linked to various cicatricial (scarring) alopecias (Stenn et al., 1999). While lipid functions in the structural formation of the hair shaft are less well understood, expression of genes involved in lipid metabolism during this process appears to be important (Evers et al., 2010). This is evident in the hair interior defect mouse mutant (HID; locus symbol hid) that exhibits defective medulla septation (Giehl et al., 2009; Trigg, 1972; Wu et al., 2010) and is linked to mutant alleles of sterol O-acyltransferase 1 (Soat1) (Wu et al., 2010). Soat1 is required for the regulation of intracellular cholesterol homeostasis (Lu et al., 2011), and SOAT1 protein is expressed in the sebaceous gland and the differentiating medulla in mice (Wu et al., 2010). Here we show that this pattern in the lower portion of the hair follicle is established at the transcriptional level where it distinctly overlaps with Hoxc13 expression, and we provide evidence for HOXC13-dependent regulation of Soat1 by utilizing Hoxc13 null mice and chromatin immunoprecipitation (ChIP) analysis.
2. Materials and methods
2.1. Mice
To facilitate histological analysis of hair unimpeded by natural pigment, Hoxc13tm1Mrc mice (Godwin and Capecchi, 1998) were crossed with albino C57BL/6J-Tyrc-2J mice to generate (B6.Cg-Tyrc-2J, Hoxc13tm1Mrc) mice. AKR/J-Soat1ald/Soat1ald (JR#648), and C57BL/6J-Tyrc-2J (JR#58) mice were obtained from The Jackson Laboratory (Bar Harbor, ME), and FVB/NTac mice were procured from Taconic, Inc. (Germantown, NY). All mice were kept in facilities of the Division of Lab Animal Resources at the Medical University of South Carolina in a humidity, temperature, and light/dark cycle (12 hrs / 12 hrs) controlled vivarium under pathogen-free conditions. All studies were done in compliance with protocols approved by the Institutional Animal Care and Use Committee.
2.2. In situ hybridization (ISH) analysis
For ISH analysis, skin samples from FVB/NTac mice at 5 days post natum (d p.n.) were fixed in 4% paraformaldehyde/PBS at 4 °C overnight followed by sequential equilibration in sucrose/PBS at 5%, 15%, and 30% sucrose concentrations before cryo-embedding into Tissue-Tek OCT compound (Fisher Scientific, St. Louis, MO) and storage at -80 °C. Probe template specific for Soat1 was generated by using cDNA derived from skin of 5 d p.n. FVB/NTac mice in PCR reactions with the following primer pair: 5’-TGAGAGACTCTGTGCCCCAC (fwd), and 5’-CGAAGAGCACCGGGTAGAAG (rev). PCR products were cloned into pCRII-TOPO vector (Invitrogen, Grand Island, NY). Hoxc13-specific probe template was generated as described (Tkatchenko et al., 2001). Synthesis of digoxigenin (DIG) (Roche Life Science, Indianapolis, IN)-labeled antisense and sense (control) RNA probes, hybridization to 10 μm cryo-sections, washing and colorimetric signal detection using alkaline phosphatase-conjugated anti-digoxigenin antibodies (Roche Life Science) was performed as previously described (Pruett et al., 2004).
2.4. Transcriptome array analysis (Hoxc13 null mouse)
Inter-scapular dorsal skin from three, 5 d p.n. male Hoxc13 null (Hoxc13tm1Mrc) mice and three age- and sex-matched wild type littermates was dissected for RNA isolation and subsequent differential gene expression analysis using the MOE430v2.0 GeneChip arrays (Affymetrix, Santa Clara, CA) including statistical analysis of as previously described in detail (Potter et al., 2011) (complete DNA microarray data set is accessible at the GEO database under GSE23759). Differentially expressed genes were detected by using Fs, a modified F-statistic incorporating shrinkage estimates of variance components from within the R/MAANOVA package (Cui et al., 2005). Statistical significance levels of the pair wise comparison were calculated by permutation analysis (1000 permutations) and adjusted for multiple testing using the false discovery rate (FDR), q-value method (Storey, 2002). Differentially expressed genes were declared at an FDR q-value threshold of 0.05.
2.5. Immunofluorescence analysis (IFA)
Skin from 5 d p.n. mice was embedded in OCT medium (Andwin Scientific, Schaumburg, IL) and allowed to freeze at -20 °C. Cryo-sections (8 μm) were transferred to Superfrost Plus (Fisher Scientific, St. Louis, MO) slides and dried for 2 hrs at room temperature prior to rehydration in PBS and fixation for 10 min in 4% paraformaldehyde. After washing in PBS (3 × 5 min) slides were permeabilized in 0.1% Triton-x100 (Roche Applied Science, Indianapolis, IN) washed in PBS and air-dried for 30 min. Antigen retrieval was achieved by heating in 0.01 M sodium citrate for 10 min. Sections were then blocked with 3% donkey serum (Jackson ImmunoResearch, West Grove, PA) in PBS for 2 hrs at room temperature before incubation with rabbit polyclonal anti-SOAT1 antibody (Abcam, Cambridge, MA) at a 1:500 dilution, and/or guinea pig polyclonal anti-HOXC13 antibody (Acris Antibodies, Herford, Germany) at a 1:100 dilution in 0.3% BSA/PBS overnight at 4 °C. Cy3 donkey anti-rabbit or Cy5 donkey anti-guinea pig were used as secondary antibodies (Jackson ImmunoResearch, West Grove, PA) as appropriate. Hoechst (Sigma Aldrich, St. Louis, MO) was used as counter-stain before mounting in Gel Mount Aqueous Mounting Medium, (Sigma Aldrich, St. Louis, MO). The slides were imaged with a confocal fluorescent microscope (TCS SP5 confocal microscope, Leica Microsystems Inc., Bannockburn, IL).
2.6. Chromatin immunoprecipitation (ChIP)
ChIP assays were performed using methods from Ren and Dynlacht (Ren and Dynlacht, 2004) with the following modifications. C2C12 mouse myoblast cells were plated at a density of 3 × 105 in 100 mm plates. After 48 hrs, cells were transfected with a Hoxc13 expression vector where the Hoxc13 coding sequence was fused to a FLAG epitope at the amino terminus. After an additional 24 hours, cells were re-suspended using trypsin/EDTA (Sigma, St. Louis, MO) and cross-linked using formaldehyde solution. After 10 min, glycine (2.5 M) was added to stop the cross-linking reaction. Cells were then removed with a rubber cell scraper. Cells were lysed to extract chromatin and sonicated to generate DNA fragments of approximately 500 base pairs. Immuno-precipitation of FLAG-tagged protein/DNA complexes, was achieved by incubating the lysate overnight at 4 °C with EZview Red Anti-FLAG M2 Affinity gel (Sigma, St. Louis, MO). After 3 washes with PBS the FLAG-tagged complexes were eluted with 2.5 M glycine, pH 3.5. Cross-links were reversed with 1% SDS and protein digested with proteinase K. DNA was purified by phenol/chloroform extraction. Precipitated DNA was detected by PCR using the following forward and reverse primers, respectively: Soat1BS1: 5’-CGGGAAGAGAGCTAACCAAG, 5’-CATTCGGTCTCCTCTCTAGCC; Foxq1BS1: 5’-TCCTCCATCCTCCTCTCCTC, 5’-ACTTCATCCGTCACCACCTC; Prrx1BS1: 5’-CCTGAGTTACCTGCACTCTG, 5’-AGGACTGAGGAGGATTCTTG. Prrx1 genomic region not containing HOXC13 consensus binding sequences was used as control. Foxq1 was used because mutations in this gene cause different abnormalities in the hair medulla of Foxq1sa mutant mice (Hong et al., 2001). PCR was performed using 60 °C as annealing temperature with 36 cycles under standard conditions.
3. Results and discussion
Hoxc13 gene-targeted mice (Hoxc13tm1Mrc) lack external hair and vibrissae (Godwin and Capecchi, 1998) due to structural defects within the developing hair shaft that result in its disintegration before emerging from the skin surface (Potter et al., 2011). Likewise, Hoxc13-overexpressing transgenic mice (Tg(Hoxc13)61B1Awg) suffer from similar, albeit less dramatic, hair growth defects (Tkatchenko et al., 2001) including abnormal septation patterns within the medulla. Transcriptome analysis of both Hoxc13 transgenic and gene-targeted mice identified a large number of differentially expressed genes known to be involved in hair follicle differentiation, and several of these were shown to be transcriptionally regulated by HOXC13 (Peterson et al., 2005; Potter et al., 2006; Potter et al., 2011; Pruett et al., 2004), thus supporting the idea that Hoxc13 is a key regulator of hair follicle morphogenesis and cycling. Soat1 was among the genes differentially expressed in both mutants, being 3.14 -fold down-regulated in Hoxc13 overexpressing skin (Q=0.0015) and 2.02 -fold downregulated in Hoxc13 null skin (Q=0.001). Comparative analysis of the Soat1 and Hoxc13 expression patterns in fully differentiated hair follicles by in situ hybridization showed Soat1 expression in two separate locations including the reserve cells of the sebaceous gland, where Hoxc13 is not expressed, and in the proximal region of the medulla, where it overlaps with the Hoxc13 pattern (Figure 1a-c). Within this region of the differentiating medulla SOAT1 and HOXC13 apparently are co-expressed in the same cells as revealed by double-immunolabeling with SOAT1 and HOXC13-specific antibodies (Figure 2a). In Hoxc13 null mice, SOAT1 expression is barely detectable in the lower portion of the hair follicle (Figure 2b), which is consistent with the downregulation of Soat1 expression in skin of Hoxc13tm1Mrc mice as determined by DNA microarray analysis (Potter et al., 2011). Notably, SOAT1 was still expressed in the Hoxc13tm1Mrc sebaceous glands that should not directly be affected by the loss of Hoxc13 function. Combined, these results infer regulation of Soat1 expression in the hair follicle by HOXC13.
Fig. 1.

Comparative ISH analysis of Hoxc13 and Soat1 expression in anagen hair follicles. Comparison of the Hoxc13 (a) and Soat1 (b) expression patterns in fully differentiated hair follicles (scapular skin) of FVB mice at 5 days post natum reveals co-expression in the medulla-forming compartment (blue arrowheads). In addition, Soat1 is expressed in cells restricted to the proximal region of the sebaceous gland (red arrow in panel b), whereas Hoxc13 is not expressed in these cells (black arrow in panel a). (c) Schematic summary of the Soat1 expression pattern in two spatially separate subpopulations of cells located in the pre-medulla, where it overlaps with Hoxc13 expression (blue) and the sebaceous gland (red). DP, dermal papilla; IRS, internal root sheath; ORS, external root sheath. Space bars: 50 μm.
Fig. 2.
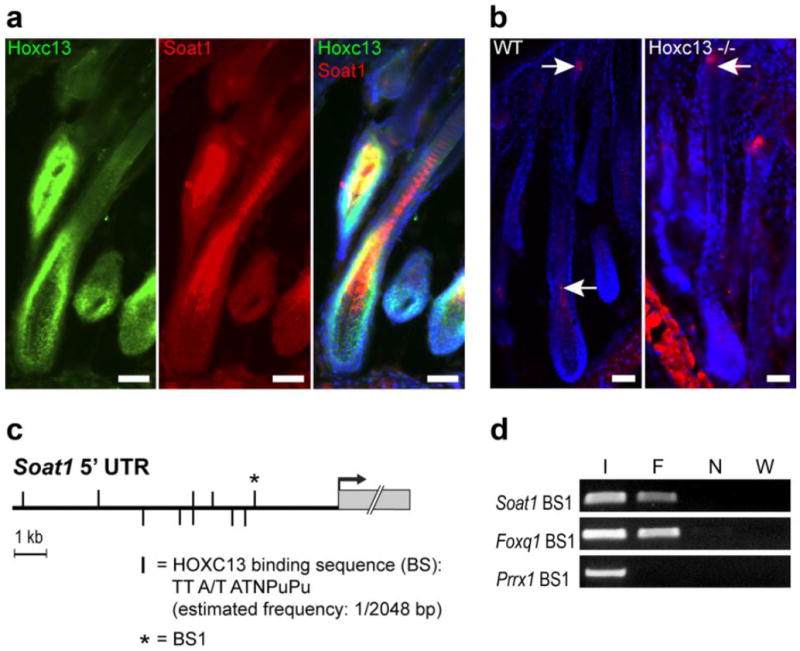
Evidence for Soat1 regulation by HOXC13 in hair. (a) Co-localization (yellow) of HOXC13 (Green) and SOAT1 protein expression (Red) by IFA in the medulla-forming compartment. (b) SOAT1 (red) expression in hair follicle bulb and sebaceous gland of wild type (WT) B6(Cg)-Tyrc-2J/J mice as indicated by arrows; SOAT1 expression is lost in the pre-medulla of Hoxc13 null (Hoxc13-/-) mice but remains in the sebaceous gland (arrow); scale bars: 50 μm. (c) Map of putative HOXC13 binding sites (vertical bars) within the 10 kb of the Soat1 5’ non-transcribed region (5’ UTR) upstream of the transcriptional start (arrow); the most proximal binding sequence (BS1) that has been subjected to ChIP analysis is marked by a star. (d) PCR analysis of HOXC13-immunoprecipitated chromatin containing Soat1 BS1 (F) and unprecipitated input DNA as positive control (I) yielded the predicted amplification products whereas ChIP DNA from untransfected cells (N) and water (W) used in negative control reactions did not. Using the same batch of immunoprecipitated DNA and a primer set specific for sequences containing a previously identified HOXC13 binding site located in the Foxq1 promoter region (Foxq1 BS1) served as a further positive control, whereas control reactions using primer sets specific for genomic regions of Prrx1 that contain no putative HOXC13binding sites failed to yield PCR products.
In silico analysis of the Soat1 genomic region revealed the presence of 10 copies of the HOXC13 consensus binding sequence (TT A/T ATNPuPu) (Jave-Suarez et al., 2002) located within 10 kb upstream of the Soat1 coding sequence (Figure 2c). ChIP analysis suggests in vivo interaction of HOXC13 with at least one of these putative binding sites located 2.6 kb upstream of the Soat1 transcriptional start codon, (Figure 2d). Combined with the other data this suggests that Soat1 is a direct regulatory target of HOXC13. These results are consistent with the finding of similar structural defects seen in the hair shaft medullae of both Hoxc13 and Soat1 mutant mice (Peterson et al., 2005; Wu et al., 2010) and support the concept that both genes are involved in a regulatory network that controls lipid metabolism within the hair shaft.
4. Conclusion
The transcriptional activities of Soat1 in the hair follicle are restricted to separate small subsets of cells of different lineages that match its translational expression in the reserve cells of the sebaceous gland (Sundberg et al., 2014) and in cells of the most proximal region of the hair follicle medulla (Wu et al., 2010). This suggests complex transcriptional mechanisms of great precision that due to the data presented in this study apparently involve Hoxc13 as an upstream regulator exclusively in the medulla. The HOXC13 transcription factor has been shown to play a pivotal role in hair shaft differentiation by affecting different pathways through interactions with target genes of diverse functional categories including hair keratins and keratin-associated proteins (Jave-Suarez et al., 2002; Pruett et al., 2004; Tkatchenko et al., 2001), transcription factors of the Fox family (Potter et al., 2006; Potter et al., 2011), desmosomal cadherins (Bazzi et al., 2009), and the gene encoding the epididymal cysteine-rich secretory protein 1 (Crisp1) (Peterson et al., 2005). Accordingly, our data suggesting regulation of the gene encoding the lipid-metabolizing enzyme SOAT1 further expand the diversity and complexity of the HOXC13-dependent regulatory network in the hair follicle.
Acknowledgments
This work was supported by National Institutes of Health grants R21AR053639, 2R01AR47204, and R01 AR056635.The Jackson Laboratory Shared Scientific Services were supported in part by a Basic Cancer Center Core Grant from the National Cancer Institute (CA34196).
Abbreviations
- ChIP
chromatin immunoprecipitation
- GEO
gene expression omnibus
- HID
hair interior defect
- Hoxc13/HOXC13
homeobox C13, gene/PROTEIN
- IFA
indirect immunofluorescence assay
- ISH
in situ hybridization
- Soat1
sterol O-acyltransferase 1 gene
Footnotes
Conflicts of Interests: The authors state that they have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bazzi H, et al. Desmoglein 4 is regulated by transcription factors implicated in hair shaft differentiation. Differentiation. 2009;78:292–300. doi: 10.1016/j.diff.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, et al. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics. 2005;6:59–75. doi: 10.1093/biostatistics/kxh018. [DOI] [PubMed] [Google Scholar]
- Evers BM, et al. Hair growth defects in Insig-deficient mice caused by cholesterol precursor accumulation and reversed by simvastatin. J Invest Dermatol. 2010;130:1237–48. doi: 10.1038/jid.2009.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl KA, et al. Hair interior defect in AKR/J mice. Clin Exp Dermatol. 2009;34:509–17. doi: 10.1111/j.1365-2230.2008.03135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin AR, Capecchi MR. Hoxc13 mutant mice lack external hair. Genes Dev. 1998;12:11–20. doi: 10.1101/gad.12.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong HK, et al. The winged helix/forkhead transcription factor Foxq1 regulates differentiation of hair in satin mice. Genesis. 2001;29:163–71. doi: 10.1002/gene.1020. [DOI] [PubMed] [Google Scholar]
- Jave-Suarez LF, et al. HOXC13 is involved in the regulation of human hair keratin gene expression. J Biol Chem. 2002;277:3718–26. doi: 10.1074/jbc.M101616200. [DOI] [PubMed] [Google Scholar]
- Lu Z, et al. Identification of Soat1 as a quantitative trait locus gene on mouse chromosome 1 contributing to hyperlipidemia. PLoS One. 2011;6:e25344. doi: 10.1371/journal.pone.0025344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RL, et al. Epididymal cysteine-rich secretory protein 1 encoding gene is expressed in murine hair follicles and downregulated in mice overexpressing Hoxc13. J Investig Dermatol Symp Proc. 2005;10:238–42. doi: 10.1111/j.1087-0024.2005.10114.x. [DOI] [PubMed] [Google Scholar]
- Potter CS, et al. Evidence that the satin hair mutant gene Foxq1 is among multiple and functionally diverse regulatory targets for Hoxc13 during hair follicle differentiation. J Biol Chem. 2006;281:29245–55. doi: 10.1074/jbc.M603646200. [DOI] [PubMed] [Google Scholar]
- Potter CS, et al. The nude mutant gene Foxn1 is a HOXC13 regulatory target during hair follicle and nail differentiation. J Invest Dermatol. 2011;131:828–37. doi: 10.1038/jid.2010.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruett ND, et al. Krtap16, characterization of a new hair keratin-associated protein (KAP) gene complex on mouse chromosome 16 and evidence for regulation by Hoxc13. J Biol Chem. 2004;279:51524–33. doi: 10.1074/jbc.M404331200. [DOI] [PubMed] [Google Scholar]
- Ren B, Dynlacht BD. Use of chromatin immunoprecipitation assays in genome-wide location analysis of mammalian transcription factors. Methods Enzymol. 2004;376:304–15. doi: 10.1016/S0076-6879(03)76020-0. [DOI] [PubMed] [Google Scholar]
- Stenn KS, et al. Hair follicle biology, the sebaceous gland, and scarring alopecias. Arch Dermatol. 1999;135:973–4. doi: 10.1001/archderm.135.8.973. [DOI] [PubMed] [Google Scholar]
- Storey J. A direct approach to false discovery rates. Journal of the Royal Statistical Society. Series B. 2002:479–498. [Google Scholar]
- Sundberg JP, et al. Immune status, strain background, and anatomic site of inoculation affect mouse papillomavirus (MmuPV1) induction of exophytic papillomas or endophytic trichoblastomas. PLoS One. 2014;9:e113582. doi: 10.1371/journal.pone.0113582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkatchenko AV, et al. Overexpression of Hoxc13 in differentiating keratinocytes results in downregulation of a novel hair keratin gene cluster and alopecia. Development. 2001;128:1547–58. doi: 10.1242/dev.128.9.1547. [DOI] [PubMed] [Google Scholar]
- Trigg MJ. Hair growth in mouse mutants affecting coat texture. J Zool, Lond. 1972;168:165–98. [Google Scholar]
- Wu B, et al. Mutations in sterol O-acyltransferase 1 (Soat1) result in hair interior defects in AKR/J mice. J Invest Dermatol. 2010;130:2666–8. doi: 10.1038/jid.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]


