Abstract
Sarcomas arising in the sinonasal region are uncommon and encompass a wide variety of tumor types, including the newly described biphenotypic sinonasal sarcoma (BSNS), which is characterized by a monomorphic spindle cell proliferation with dual neural and myogenic phenotypes. Most BSNSs harbor a pathognomonic PAX3-MAML3 fusion driven by t(2;4)(q35;q31.1), while the alternative fusion partner gene remains unidentified in a subset of PAX3-rearranged cases. As NCOA1 on 2p23 is a known partner in PAX3-related fusions in other tumor types (i.e. alveolar rhabdomyosarcoma), we investigated its status by FISH and RT-PCR assays in 2 BSNS cases showing only PAX3 gene rearrangements. Novel PAX3-NCOA1 fusions were identified in these two index cases showing an inv(2)(q35p23) by FISH and confirmed by RT-PCR. Five additional BSNS cases with typical morphology were studied by FISH, revealing a PAX3-MAML3 fusion in 4 cases and only PAX3 rearrangement in the remaining case without abnormalities in MAML3 or NCOA1 gene. Except for one case with surface ulceration, all other tumors lacked increased mitotic activity or necrosis, and all cases immunohistochemically co-expressed S100 protein and actin, but lacked SOX10 reactivity. Interestingly, the two PAX3-NCOA1-positive cases showed desmin reactivity and displayed a small component of rhabdomyoblastic cells, which were not seen in the more common PAX3-MAML3 fusion cases. In conclusion, we report a novel PAX3-NCOA1 fusion in BSNS, which appears to be associated with focal rhabdomyoblastic differentiation and should be distinguished from PAX3-NCOA1-positive alveolar rhabdomyosarcoma or malignant Triton tumor. SOX10 immunohistochemistry is a useful marker in distinguishing BSNS from peripheral nerve sheath tumors.
Keywords: biphenotypic sinonasal sarcoma, low-grade sinonasal sarcoma with neural and myogenic features, PAX3, MAML3, NCOA1
INTRODUCTION
Head and neck sarcomas are relatively uncommon, accounting for 5–15% of all soft tissue sarcoma.1,2 Sinonasal tract sarcomas comprise less than 10% of head and neck sarcomas and tend to be more aggressive due to their proximity to the eyes and brain.3,4 The histologic classification is an important prognostic factor, with rhabdomyosarcoma (RMS) conferring the most dismal outcome, with 30% survival rate at 5 years.3,4 Given the rarity and wide morphologic spectrum of sinonasal sarcomas, the histologic diagnosis is often challenging, especially on limited and fragmented biopsies.
Aside from the well-defined sarcomas, such as RMS, malignant peripheral nerve sheath tumor (MPNST), leiomyosarcoma, synovial sarcoma, angiosarcoma, etc, a significant proportion of sinonasal sarcomas remain unclassified.1,4 Recently, Lewis and colleagues described a new pathologic entity using the terminology: ‘low grade sinonasal sarcoma with neural and myogenic features (LGSSNMF)’, since it histologically resembled adult fibrosarcoma or monophasic synovial sarcoma, but expressed a dual neural and myogenic immunophenotype.5 LGSSNMF was subsequently renamed by the same group as biphenotypic sinonasal sarcoma (BSNS), being characterized by a recurrent t(2;4)(q35;q31.1) translocation, resulting in a PAX3-MAML3 fusion.6 This genetic abnormality, identified by transcriptome sequencing, was found in 76% of BSNS cases, but not in other tumor types. However, a subset (20%) of BSNS showed only PAX3 rearrangement without a known fusion partner.
Based on two index cases exhibiting PAX3 gene rearrangements without the canonical MAML3 fusion, we screened alternative PAX3-candidate gene partners by FISH and RT-PCR. Additional 5 cases were collected and investigated for clinicopathologic features and genetic alterations of BSNS.
MATERIALS AND METHODS
Case Collection and Immunohistochemistry
We identified 7 cases of BSNS from the Surgical Pathology files of MSKCC, the Johns Hopkins Hospital, and the Kaohsiung and Linkou Chang Gung Memorial Hospital. The diagnostic criteria included a highly cellular spindle cell neoplasm with monomorphic cytomorphology and at least focal S100 protein and actin immunoreactivity.5 The relevant clinical information was collected from the medical records or communications with the referring pathologists. The H&E slides and the corresponding immunohistochemical stains were reviewed to better analyze the histologic features and immunoprofile of BSNS. In addition, all cases were tested for SOX10 immunostaining (Cell Marque, 1:50, Rocklin, CA). Study was approved by the individual IRB at each participating institution.
Fluorescence In Situ Hybridization
FISH on interphase nuclei from paraffin-embedded 4-micron sections was performed applying custom probes using bacterial artificial chromosomes (BACs), flanking PAX3, MAML3 and NCOA1 genes. BAC clones were chosen according to UCSC genome browser (http://genome.ucsc.edu), see Supplementary Table 1. The BAC clones were obtained from BACPAC sources of Children’s Hospital of Oakland Research Institute (CHORI) (Oakland, CA) (http://bacpac.chori.org). DNA from individual BACs was isolated according to the manufacturer’s instructions, labeled with different fluorochromes in a nick translation reaction, denatured, and hybridized to pretreated slides. Slides were then incubated, washed, and mounted with DAPI (4′,6-diamidino-2-phenylindole) in an antifade solution, as previously described.7 The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes. Two hundred successive nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems, Newton, MA). A positive score was interpreted when at least 20% of the nuclei showed a break-apart signal. Nuclei with incomplete set of signals were omitted from scoring.
Reverse Transcription-Polymerase Chain Reaction
Three 10-μm-thick tissue scrolls were cut from a representative paraffin block of cases #2 and #4 for RNA extraction using RecoverAll Total Nucleic Acid isolation kit (Ambion, Austin, TX). Briefly, the tissue scrolls were deparaffinized, digested with proteinases, and incubated at 50°C until completely dissolved. ImPromII RT System (Promega, Madison, WI) was used to synthesize the first-strand cDNA and the cDNA product was subjected to PCR amplification by using Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA) and touchdown thermal conditions. The mRNA of phosphoglycerate kinase (PGK) gene was in parallel reverse transcribed and amplified as an internal control.
Through multiple combinations of different PAX3 forward primers and NCOA1 reverse primers, a single amplicon was identified using the PAX3 exon 7 forward (Ex7F) primer (5′-GATTCCTTCCAACCCAGACA-3′) and NCOA1 exon 14 reverse (Ex14R) primer (5′-ATAAGCCTGGCAACTGTGCT-3′). In addition, the PAX3 Ex7F primer and the MAML3 exon 3 reverse (Ex3R) primer (5′-CCATCACAAGCACCATTCTG-3′) were used to amplify the canonical PAX3-MAML3 chimeric fusion. The PCR products were examined on agarose gels and sent to direct sequencing on an automated sequencer (Applied Biosystems 3730 DNA Analyzer) with BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems).
RESULTS
The clinical, pathologic and genetic features of the 7 cases are summarized in Tables 1 and 2. There were 4 males and 3 females, with a mean age of 50 years (median 47, range 37–70). Three cases occurred in the frontal and ethmoid sinus, 2 cases were restricted to the nasal cavity, and 2 cases involved both the nasal cavity and the ethmoid sinus. The average size was 4.1 cm (range 2.8–6.5 cm). Among the 4 cases with available follow-up data no cancer-related mortality was noted. One patient (case #4) developed local recurrence 3 years after diagnosis and had no evidence of disease (NED) after 1 year follow-up. One patient (case #7) received chemotherapy and radiation therapy postoperatively and was NED after 11 years follow-up.
Table 1.
Clinical features and genetic alterations of biphenotypic sinonasal sarcoma
| Case | Age/Sex | Location | Size (cm) | Genetic alteration | Recurrence | Status |
|---|---|---|---|---|---|---|
| 1 | 70/M | Frontal sinus | 3.5 | PAX3-NCOA1 | NA | |
| 2 | 37/F | Upper nasal meatus | 2.8 | PAX3-NCOA1 | None | NED for 3 months |
| 3 | 48/F | Nasal cavity/ethmoid sinus | NA | PAX3-MAML3 | NA | |
| 4 | 46/F | Nasal septum | 4.3 | PAX3-MAML3 | Yes (3 years) | NED for 1 year after recurrence |
| 5 | 44/M | Ethmoid sinus | NA | PAX3-MAML3 | NA | |
| 6 | 64/M | Nasal cavity/ethmoid sinus | 6.5 | PAX3-MAML3 | None | NED for 10 years |
| 7 | 44/M | Frontal sinus | 3.3 | PAX3-rearranged | None | NED for 11 years |
NED, no evidence of disease; NA, not available
Table 2.
Immunohistochemical results of biphenotypic sinonasal sarcoma
| Case | S-100 | SMA | MSA | SOX10 | Desmin | MyoD1 | Myogenin | AE1/AE3 | CD34 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | +++ | NA | ++ | 0 | ++ | + | 0 | NA | NA |
| 2 | +++ | ++ | NA | 0 | + | + | + | + | 0 |
| 3 | + | NA | ++ | 0 | 0 | 0 | 0 | 0 | + |
| 4 | ++ | ++ | NA | 0 | 0 | 0 | 0 | + | 0 |
| 5 | + | + | NA | 0 | + | 0 | 0 | 0 | NA |
| 6 | + | +++ | +++ | 0 | + | + | 0 | NA | 0 |
| 7* | + | ++ | + | 0 | 0 | 0 | 0 | 0 | 0 |
+++: diffuse and strong positivity (>70%); ++: patchy positivity (30–70%); +: focal positivity (<30%); 0: negative; NA: not available
This case was patchy positive for calponin.
Novel PAX3-NCOA1 fusions in a subset of BSNS with focal rhabdomyoblastic differentiation
The study was initiated by 2 BSNSs (cases #1 and 2) lacking the typical PAX3-MAML3 fusion. As both cases showed PAX3 gene abnormalities by FISH, further FISH and RT-PCR assays were performed to identify potential novel partners. The FISH analysis showed a distinct pattern of rearrangement in both cases, suggestive of an intra-chromosomal inversion, being characterized by fixed small gaps, rather than wide-apart split signals at random distances, typical for inter-chromosomal translocations. Thus, further screening was focused mainly on potential gene partners located on chromosome 2. In parallel, published literature was reviewed for all PAX3-fusion partners reported, revealing PAX3-NCOA1 and PAX3-NCOA2 fusions have been previously described in a small subset of alveolar rhabdomyosarcoma (ARMS).8 As NCOA1 is located on 2p23, custom BAC probes were designed flanking this gene, as well as NCOA1 reverse primers were designed for RT-PCR.
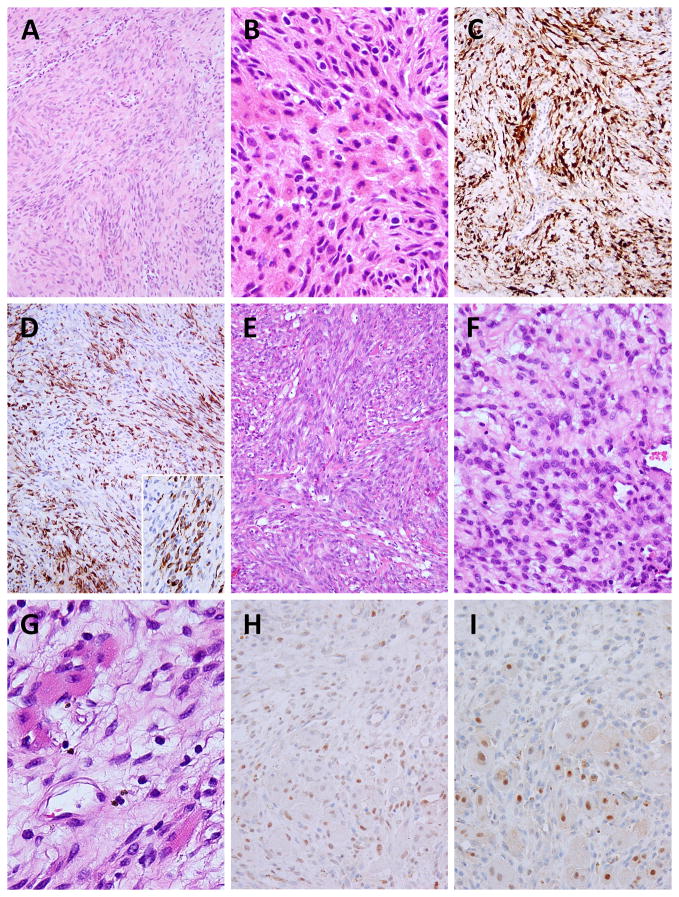
Case #1 occurred in the frontal sinus of a 70 year-old man, while case #2 arose in the nasal cavity of a 37 year-old woman. Both tumors demonstrated infiltrative borders and were composed of a hypercellular proliferation of uniform spindle cells, with indistinct cell borders and scant cytoplasm. The tumor cells had mostly blunt-ended nuclei, with finely granular chromatin and inconspicuous nucleoli. Some tumor cells in case #2 exhibited round cell cytomorphology (Fig. 1F). A focal (~10%) component of plump to epithelioid cells with moderate amount of eosinophilic cytoplasm, reminiscent of rhabdomyoblastic differentiation, was seen interspersed in both cases, distributed either centrally or at the periphery of the tumor (Fig. 1B, 1G). In addition, scattered well-differentiated rhabdomyoblasts displaying cross-striations were identified in case #2 (Fig. 1G). The extracellular collagenous stroma was relatively scarce, without prominent vascularity (Fig. 1A, 1E). Mitotic figures were very difficult to find (<1/50 HPF) and no necrosis was observed. Both cases displayed diffuse and strong S100 protein staining as well as actin reactivity, but no SOX10 expression was present. The epithelioid/polygonal cell component was highlighted by the desmin immunostain (Fig. 1D). In case #2 the rhabdomyoblast cells showed focal nuclear MyoD1 and myogenin expression (Fig. 1H–I).
Figure 1. Pathologic features of PAX3-NCOA1-related biphenotypic sinonasal sarcoma.
Case #1 displays interlacing fascicles of monomorphic spindle cells (A), with a focal component of epithelioid cells, displaying rhabdomyoblastic-like features (B). Tumor cells are diffusely positive for S100 protein (C) and show patchy desmin expression (D). Desmin immunostain highlights the epithelioid cell component (D, inset). Case #2 exhibits a cellular spindle cell proliferation with interspersed thick collagen fibers (E), focal round cell features (F) and scattered well-differentiated rhabdomyoblasts displaying cytoplasmic cross-striations (G) in a loose-textured stroma. MyoD1 (H) and myogenin (I) positivity was observed in the rhabdomyoblastic elements.
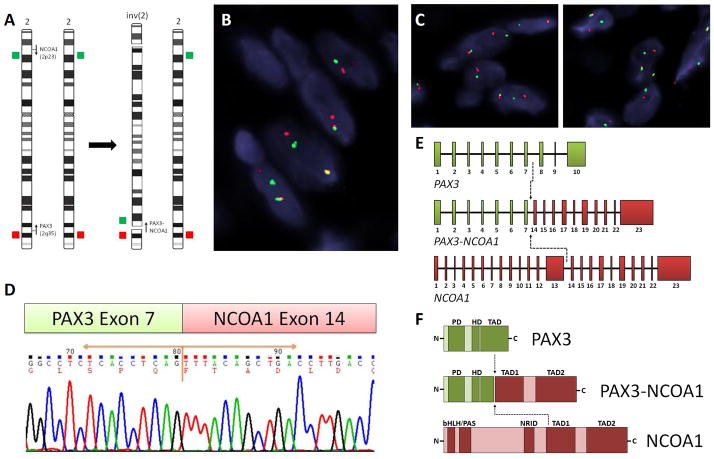
FISH fusion assays confirmed a novel PAX3-NCOA1 fusion in both cases (Fig. 2A–C). Furthermore, RT-PCR confirmed the PAX3 exon 7 fusion with NCOA1 exon 14 in case #2 with available material (Fig. 2D–E). The predicted fusion protein contained the DNA binding domain (DBD) of PAX3 protein, including paired domain (PD) and homeodomain (HD), and the transactivation domains (TADs) of NCOA1 protein (Fig. 2F), similar to the type II of PAX3-NCOA1 fusion described in ARMS.8
Figure 2. Novel PAX3-NCOA1 fusion in BSNS.
Schematic diagram of inv(2)(p23q35) and the corresponding FISH probes applied, flanking the PAX3 and NCOA1 genes (arrows indicate the direction of transcription)(A). FISH fusion assay showed the PAX3 (red) and NCOA1 (green) signals joined together (B), as well as the individual break-apart assays showing PAX3 (C, left) and NCOA1 (C, right) rearrangements in case #1 (red, centromeric; green, telomeric). RT-PCR chromatogram of case #2 demonstrates exon 7 of PAX3 gene joined to exon 14 of NCOA1 gene (D), resulting in a PAX3 ex1-7 - NCOA1 ex14-23 chimeric transcript (E). The predicted fusion protein retained the DNA-binding domain (DBD), including PD and HD of PAX3, and the transcriptional activation domain (TAD) of NCOA1 (F). PD, paired domain; HD, homeodomain; bHLH/PAS, basic helix-loop-helix-Per/Ah receptor nuclear translocation/Sim motif; NRID, nuclear receptor interacting domain.
BSNS with the canonical PAX3-MAML3 fusion
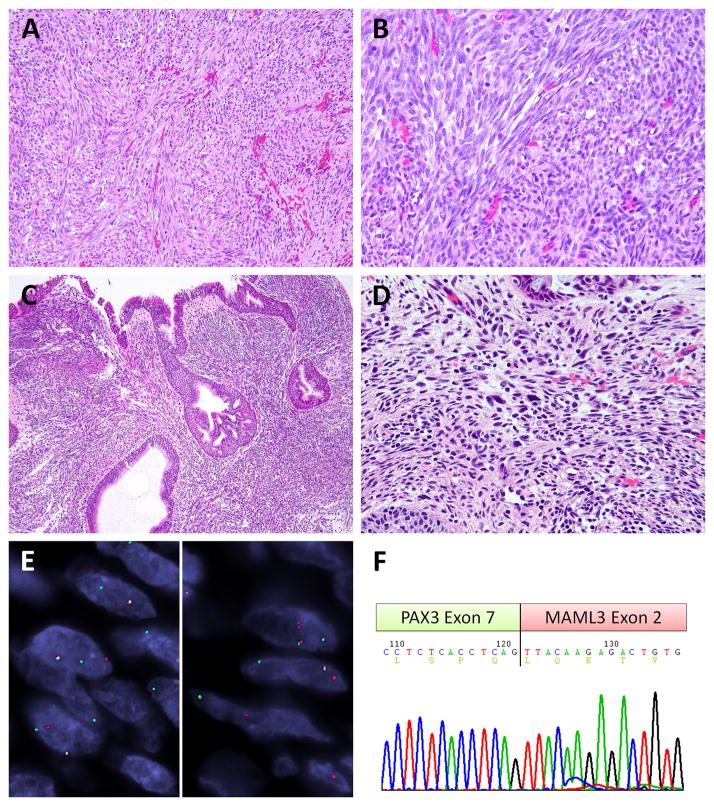
Case #3–6 were BSNSs showing the canonical PAX3-MAML3 fusions. Case #3 was a nasal and ethmoid sinus tumor in a 48-year-old woman presenting with nasal obstruction, case #4 was a nasal septal tumor in a 46-year-old woman suffering from recurrent epistaxis, and case #5 was an ethmoid polypoid lesion in a 44 year-old male. The case #6 was a hypervascular and expansile nasoethmoid tumor in a 65 year-old male with symptoms of purulent rhinorrhea and epistaxis. The typical histologic features of infiltrative borders, monotonous spindle cells arranged in a vague fascicular or whorling pattern were noted in all cases and inert mitotic activity was observed in case #3–5 (Fig. 3A–B). Surface ulceration with necrosis and increased mitotic figures (4/10HPFs) was found in case #6. The respiratory epithelium showed immature squamous metaplasia in these cases and case #5 and #6 exhibited respiratory-type glandular proliferation and invaginations (Fig. 3C). Case #5 exhibited obvious hemosiderin deposits, mast cell infiltrates and scattered pleomorphic hyperchromatic cells, suggestive of degenerative atypia (Fig. 3D). Case #4 and #6 had congested and dilated blood vessels. The immunohistochemical studies showed focal, patchy S100 expression as well as actin staining. Case #5 and #6 demonstrated focal desmin immunoreactivity and the latter also showed rare MyoD1 expression. No SOX10 or myogenin immunoreactivity was noted in these 4 cases.
Figure 3. PAX3-MAML3 fusion positive BSNS.
All three cases displayed similar histology with sweeping or intersecting bundles (A) of monotonous and mitotically inert spindle cells (B) as exemplified by case #3. Case #5 showed respiratory type glandular hyperplasia and invaginations, mimicking inverted papilloma (C), and had scattered mast cells and rare pleomorphic hyperchromatic cells (D). The canonical PAX3-MAML3 fusion is illustrated by the break-apart signals of PAX3 (E, left) and MAML3 (E, right) by FISH in case #3 (red, centromeric; green, telomeric). The RT-PCR chromatogram of case #4 confirmed the classic fusion variant of PAX3 exon 7 to MAML3 exon 2 (F).
FISH in these 4 cases showed break-apart signals in both PAX3 and MAML3 genes (Fig. 3E). The RT-PCR of case #4 with available tissue confirmed the classic PAX3 exon 7 fused to MAML3 exon 2 (Fig. 3F), indicating a projected chimeric protein of PAX3 DBD and MAML3 TAD, identical to the previous report.6
BSNS with only PAX3 gene rearrangement
Case #7 was a BSNS harboring PAX3 rearrangement without MAML3 or NCOA1 abnormalities in a 44-year-old man manifesting proptosis. Histologically, this case was similar to the other cases, showing an infiltrating and hypercellular tumor composed of irregular fascicles of spindle cells with monotonous nuclear features, and lacking mitotic figures or necrosis. S100 protein, actin, and calponin stains showed focal, patchy expression. In spite of the negative pancytokeratin (AE1:AE3), this tumor was focally positive for EMA, Cam 5.2, and CK7 immunostains. The PAX3 abnormality by FISH showed a similar pattern of intra-chromosomal inversion, however, the fusion assay of PAX3 and NCOA1 genes suggested the involvement of chromosome 2p outside the NCOA1 gene in this case.
DISCUSSION
BSNS involves with predilection the upper nasal cavity and ethmoid sinus of middle-aged female patients and shows a characteristic histologic appearance of infiltrative and hypercellular proliferation of monomorphic spindle cells arranged in fascicles and whorling pattern.5,9 Overt features of a high-grade lesion, such as brisk mitotic activity, nuclear pleomorphism, or necrosis are typically lacking. Other histologic features, including hyperplastic respiratory-type glands (71%, 20/28), staghorn vessels, and focal rhabdomyoblastic differentiation (11%, 3/28) have been described.5 The characteristic immunoprofile is the co-expression of S100 protein and actin in most cases. Focal cytokeratin immunoreactivity has been reported and raises the possibility of a monophasic synovial sarcoma, however, FISH for SS18 abnormalities are consistently negative.5 Nearly half of the patients may experience local recurrence, but none so far have developed metastasis or died of disease.5
The current case series confirms the middle-aged presentation, but shows instead an equal gender distribution rather than a female predominance. All present cases demonstrate the classic histologic features previously described in BSNS, but also some minor variations. First, the glandular hyperplasia was not a prevailing feature and was seen only in two cases, however, one drawback was that in some of our cases the tissue was limited and fragmented. Second, one case showed scattered hyperchromatic and pleomorphic cells, reminiscent of degenerative nuclear atypia seen in neurogenic tumors. Moreover, both PAX3-NCOA1 positive tumors displayed a minor but distinctive rhabdomyoblastic component. The rhabdomyoblastic cells were plump to epithelioid, with eccentric nuclei and moderate amount of fibrillary eosinophilic cytoplasm, focally displaying cross striations. These cells were highlighted by desmin and MyoD1, and in case #2 were also focally positive for myogenin. Although the presence of rhabomyoblastic differentiation and focal desmin and MyoD1 expression was described, no myogenin reactivity was previously reported.5,6,9 None of our 4 PAX3-MAML3-positive tumors showed this component, and only two of them displayed focal desmin and in one some rare MyoD1 reactivity, compared to both cases with PAX3-NCOA1 fusion. Desmin and MyoD1 expression has been described in about 20% of BSNSs with PAX3-MAML3 fusions, however, none of the 4 reported cases with only PAX3 rearrangement were positive for desmin, MyoD1, or myogenin.6 Nonetheless, this study did not examine the potential correlation between the presence of a rhabdomyoblastic component and the fusion type.6 Due to the small number of cases it is difficult to draw conclusions to a potential relationship between the presence of rhabdomyoblastic differentiation / skeletal muscle marker expression and the type of chimeric protein in BSNS.
The canonical PAX3-MAML3 fusions have been identified in the majority (76%) of BSNSs, with PAX3 rearrangements being the consistent genetic alteration in nearly all (96%) BSNSs.6 The PAX3-MAML3 chimeric protein combining the DBD of PAX3 and the TAD of MAML3 has been shown to act as a transactivator of PAX3 response elements with comparable potency of PAX3-FOXO1 fusion, the main genetic alteration of ARMS. No other fusion was identified in the previous study in the PAX3-rearranged BSNS cases, using RT-PCR for all potential fusion partners, such as FOXO1, MAML1, MAML2, NCOA1, and NCOA2.6 In the current study, we took a combined approach using FISH and RT-PCR with different primer pairs to identify the PAX3-NCOA1 fusions. Nevertheless, there still remain unknown PAX3 fusion partners in BSNS as illustrated by our case #7. PAX3 is a member of the paired box transcription factor family featured by the N-terminal DBD including PD, octapeptide, and HD.10,11 PAX family plays an important role in tissue and organ development during embryogenesis. PAX3 determines the cell fate of melanocytic, neuronal, and skeletal muscle differentiation and regulates normal myogenesis and postnatal muscular regeneration.6,10 All BSNSs tested were positive for PAX3 immunohistochemistry using a rabbit polyclonal antibody (Invitrogen), including one case lacking a PAX3 gene rearrangement.6
NCOA1 (nuclear coactivator factor, also known as SRC1) is the first member described of the p160/steroid receptor coactivator (SRC) family, which also includes NCOA2 (SRC2) and NCOA3 (SRC3).12,13 This protein, encoded by the NCOA1 gene at chromosome 2p23.3, mediates the transcriptional function of steroid nuclear hormone receptors by recruiting other coactivators to form a steroid-receptor-directed transcriptional activation complex upon binding with hormone nuclear factors.12 The protein complex remodels the chromatin structure by its histone acetyltransferase and methyltransferase and facilitates the assembly of general transcription factors to initiate transcriptional activities. The PAX3-NCOA1 fusion, previously described in a small subset of ARMS, behaves as a transcriptional activator with similar capability to PAX3-FOXO1 fusion8,14 In ARMS, two reported fusion transcripts have been identified: PAX3ex6-NCOA1ex12 (type I) and PAX3ex7-NCOA1ex11 (type II).8 The current PAX3ex7-NCOA1ex14 chimeric transcript found in one of the BSNS is identical to the type II fusion (using a different exon annotation), retaining the last 10 exons of NCOA1 gene.8 Similar with ARMS, the predicted PAX3-NCOA1 fusion protein in BSNS retains the DBD of PAX3 and the TAD1/2 of NCOA1, the latter being critical for transforming activity in ARMS.8 Furthermore, the gene expression profiles of BSNS and ARMS share a significant number of myogenic genes, but differ in neurogenic and cytokine-related genes, metalloproteinase genes, and MYOCD.6 Although MYOD1 transcriptional up-regulation was noted in BSNS, only focal and weak MyoD1 immunoexpression was demonstrated in a subset of BSNS. 6
Moreover, our current study demonstrates consistent lack of SOX10 immunoexpression in BSNS. SOX10, known as SRY (sex-determinant region Y) box 10, is a transcription factor essential for development of neural crest-derived cells.15,16 Nuclear SOX10 expression is observed in Schwann cells, melanocytes, oligodendrocytes, mast cells, myoepithelial cells, and acinar cells of the salivary gland.17,18 Non-neural crest-derived soft tissue neoplasms with infrequent S100 immunostaining, such as Ewing sarcoma, RMS, extraskeletal myxoid chondrosarcoma, and synovial sarcoma, are typically negative for SOX10 immunohistochemistry.17–21 It remains unclear the reason for discrepancy between S100 protein/SOX10 expression, but it might be related to the multiple specificities against both S100A and S100B or the cross-reactivity of the current polyclonal S100 antibody with equivalent epitopes in similar proteins. Based on the gene expression profiling, BSNS shows enriched gene expression in neural development pathways, including NTRK3, ALX1, ALX3, ALX4, DBX1, GREM1, and NEUROG2.6
Although BSNS needs to be differentiated from a large spectrum of spindle cell neoplasms,5 in addition BSNS exhibiting rhabdomyoblastic differentiation can be mistaken for an MPNST with rhabdomyoblastic differentiation (malignant Triton tumor) or a spindle cell RMS (SC-RMS). Malignant Triton tumors commonly occur in patients with neurofibromatosis type 1 (NF1) and follow an aggressive clinical course, with examples being documented in the sinonasal region.22–24 However, malignant Triton tumors exhibit features of a high grade MPNST with hyperchromasia, brisk mitotic activity and geographic areas of necrosis. In contrast, BSNS has bland cytomorphology, with no increased mitotic activity or necrosis. In this regard, SOX10 immunoreactivity may be helpful since 50–70% of MPNSTs express this marker.17–21 It is likely that the previously reported ‘low-grade MPNSTs’ from the sinonasal tract represent in fact BSNS with or without the rhabdomyoblastic differentiation.5,22,25 In an analysis of 53 cases of malignant Triton tumor in the head and neck,24 sinonasal malignant Triton tumor accounted for 13 cases (24%), only second to neck (34%), and none of them being associated to NF1, while half of neck cases were NF1-related. Intriguingly, the sinonasal cases pursued an excellent outcome. These findings argue that most of the so-called ‘sinonasal malignant Triton tumors’ were likely misdiagnosed BSNS with rhabdomyoblastic differentiation. Demonstration of PAX3 gene abnormalities by FISH can serve as a useful diagnostic tool in the differential diagnosis of a sinonasal low-grade spindle cell tumor with neural and myogenic features, especially in patients without NF1.
SC-RMS has a bimodal incidence in young children and adults, with head and neck being the most common affected site in adults.26–28 This RMS variant is characterized by a cellular spindle cell proliferation arranged in fascicles, occasionally demonstrating a herringbone pattern and very focal rhabdomyoblastic differentiation. However, SC-RMS exhibits increased mitotic activity and necrosis, and displays more diffuse expression of myogenic markers. In contrast, the MyoD1/myogenin reactivity in BSNS is restricted to the focal rhabdomyoblastic region. Furthermore, S100 protein expression is an unusual feature for SC-RMS and no PAX3 or NCOA1 rearrangements have been identified.29 Interestingly, a subset of infantile/congenital SC-RMS harbors NCOA2 rearrangements, being fused with either SRF or TEAD1 gene.29
In conclusion, this is the second molecular characterization of BSNS, reporting on additional 7 cases, two of them harboring a novel PAX3-NCOA1 fusion and exhibiting focal rhabdomyoblastic features. In spite of S100 expression, all cases lacked SOX10 immunoreactivity. BSNS with rhabdomyoblastic differentiation should be distinguished from malignant Triton tumor and SC-RMS. A comprehensive immunohistochemical panel and FISH analysis for PAX3 gene rearrangements should be considered in difficult cases, especially in limited tissue biopsies. Our findings suggest that PAX3-NCOA1 fusions can be seen in a small subset of both BSNS and ARMS, and might be implicated in the rhabdomyoblastic phenotype shared by these two distinct tumor types. These findings further emphasize the functional diversity of gene fusions in sarcomagenesis.
Supplementary Material
Acknowledgments
Supported in part by: P01CA47179 (CRA), P50CA140146-01 (CRA), Cycle for Survival (CRA)
Footnotes
Conflict of interest: none
References
- 1.Sturgis EM, Potter BO. Sarcomas of the head and neck region. Curr Opin Oncol. 2003;15:239–252. doi: 10.1097/00001622-200305000-00011. [DOI] [PubMed] [Google Scholar]
- 2.de Bree R, van der Waal I, de Bree E, et al. Management of adult soft tissue sarcomas of the head and neck. Oral Oncol. 2010;46:786–790. doi: 10.1016/j.oraloncology.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Wu AW, Suh JD, Metson R, et al. Prognostic factors in sinonasal sarcomas: analysis of the surveillance, epidemiology and end result database. Laryngoscope. 2012;122:2137–2142. doi: 10.1002/lary.23442. [DOI] [PubMed] [Google Scholar]
- 4.Szablewski V, Neuville A, Terrier P, et al. Adult sinonasal soft tissue sarcoma: analysis of 48 cases from the French Sarcoma Group database. Laryngoscope. 2015;125:615–623. doi: 10.1002/lary.24910. [DOI] [PubMed] [Google Scholar]
- 5.Lewis JT, Oliveira AM, Nascimento AG, et al. Low-grade sinonasal sarcoma with neural and myogenic features: a clinicopathologic analysis of 28 cases. Am J Surg Pathol. 2012;36:517–525. doi: 10.1097/PAS.0b013e3182426886. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Bledsoe KL, Graham RP, et al. Recurrent PAX3-MAML3 fusion in biphenotypic sinonasal sarcoma. Nat Genet. 2014;46:666–668. doi: 10.1038/ng.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antonescu CR, Zhang L, Chang NE, et al. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49:1114–1124. doi: 10.1002/gcc.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sumegi J, Streblow R, Frayer RW, et al. Recurrent t(2;2) and t(2;8) translocations in rhabdomyosarcoma without the canonical PAX-FOXO1 fuse PAX3 to members of the nuclear receptor transcriptional coactivator family. Genes Chromosomes Cancer. 2010;49:224–236. doi: 10.1002/gcc.20731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powers KA, Han LM, Chiu AG, et al. Low-grade sinonasal sarcoma with neural and myogenic features-diagnostic challenge and pathogenic insight. Oral Surg Oral Med Oral Pathol Oral Radiol. 2015;119:e265–269. doi: 10.1016/j.oooo.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Buckingham M, Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol. 2007;23:645–673. doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q, Fang WH, Krupinski J, et al. Pax genes in embryogenesis and oncogenesis. J Cell Mol Med. 2008;12:2281–2294. doi: 10.1111/j.1582-4934.2008.00427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, Wu RC, O’Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9:615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walsh CA, Qin L, Tien JC, et al. The function of steroid receptor coactivator-1 in normal tissues and cancer. Int J Biol Sci. 2012;8:470–485. doi: 10.7150/ijbs.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wachtel M, Dettling M, Koscielniak E, et al. Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;p23) translocation fusing PAX3 to NCOA1. Cancer Res. 2004;64:5539–5545. doi: 10.1158/0008-5472.CAN-04-0844. [DOI] [PubMed] [Google Scholar]
- 15.Paratore C, Goerich DE, Suter U, et al. Survival and glial fate acquisition of neural crest cells are regulated by an interplay between the transcription factor Sox10 and extrinsic combinatorial signaling. Development. 2001;128:3949–3961. doi: 10.1242/dev.128.20.3949. [DOI] [PubMed] [Google Scholar]
- 16.Kelsh RN. Sorting out Sox10 functions in neural crest development. Bioessays. 2006;28:788–798. doi: 10.1002/bies.20445. [DOI] [PubMed] [Google Scholar]
- 17.Nonaka D, Chiriboga L, Rubin BP. Sox10: a pan-schwannian and melanocytic marker. Am J Surg Pathol. 2008;32:1291–1298. doi: 10.1097/PAS.0b013e3181658c14. [DOI] [PubMed] [Google Scholar]
- 18.Karamchandani JR, Nielsen TO, van de Rijn M, et al. Sox10 and S100 in the diagnosis of soft-tissue neoplasms. Appl Immunohistochem Mol Morphol. 2012;20:445–450. doi: 10.1097/PAI.0b013e318244ff4b. [DOI] [PubMed] [Google Scholar]
- 19.Ordonez NG. Value of SOX10 immunostaining in tumor diagnosis. Adv Anat Pathol. 2013;20:275–283. doi: 10.1097/PAP.0b013e318297a9d0. [DOI] [PubMed] [Google Scholar]
- 20.Miettinen M, McCue PA, Sarlomo-Rikala M, et al. Sox10-A Marker for Not Only Schwannian and Melanocytic Neoplasms But Also Myoepithelial Cell Tumors of Soft Tissue: A Systematic Analysis of 5134 Tumors. Am J Surg Pathol. 2015 Feb 25; doi: 10.1097/PAS.0000000000000398. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang Y, Pekmezci M, Folpe AL, et al. Diagnostic utility of SOX10 to distinguish malignant peripheral nerve sheath tumor from synovial sarcoma, including intraneural synovial sarcoma. Mod Pathol. 2014;27:55–61. doi: 10.1038/modpathol.2013.115. [DOI] [PubMed] [Google Scholar]
- 22.Heffner DK, Gnepp DR. Sinonasal fibrosarcomas, malignant schwannomas, and “Triton” tumors. A clinicopathologic study of 67 cases. Cancer. 1992;70:1089–1101. doi: 10.1002/1097-0142(19920901)70:5<1089::aid-cncr2820700513>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 23.Stasik CJ, Tawfik O. Malignant peripheral nerve sheath tumor with rhabdomyosarcomatous differentiation (malignant triton tumor) Arch Pathol Lab Med. 2006;130:1878–1881. doi: 10.5858/2006-130-1878-MPNSTW. [DOI] [PubMed] [Google Scholar]
- 24.Terzic A, Bode B, Gratz KW, et al. Prognostic factors for the malignant triton tumor of the head and neck. Head Neck. 2009;31:679–688. doi: 10.1002/hed.21051. [DOI] [PubMed] [Google Scholar]
- 25.Shajrawi I, Podoshin L, Fradis M, et al. Malignant triton tumor of the nose and paranasal sinuses: a case study. Hum Pathol. 1989;20:811–814. doi: 10.1016/0046-8177(89)90079-8. [DOI] [PubMed] [Google Scholar]
- 26.Nascimento AF, Fletcher CD. Spindle cell rhabdomyosarcoma in adults. Am J Surg Pathol. 2005;29:1106–1113. [PubMed] [Google Scholar]
- 27.Mentzel T, Kuhnen C. Spindle cell rhabdomyosarcoma in adults: clinicopathological and immunohistochemical analysis of seven new cases. Virchows Arch. 2006;449:554–560. doi: 10.1007/s00428-006-0284-4. [DOI] [PubMed] [Google Scholar]
- 28.Carroll SJ, Nodit L. Spindle cell rhabdomyosarcoma: a brief diagnostic review and differential diagnosis. Arch Pathol Lab Med. 2013;137:1155–1158. doi: 10.5858/arpa.2012-0465-RS. [DOI] [PubMed] [Google Scholar]
- 29.Mosquera JM, Sboner A, Zhang L, et al. Recurrent NCOA2 gene rearrangements in congenital/infantile spindle cell rhabdomyosarcoma. Genes Chromosomes Cancer. 2013;52:538–550. doi: 10.1002/gcc.22050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.