Abstract
Despite the existence of an effective measles vaccine, resurgence in measles cases in the United States and across Europe has occurred, including in individuals vaccinated with two doses of the vaccine. Host genetic factors result in inter-individual variation in measles vaccine-induced antibodies, and play a role in vaccine failure. Studies have identified HLA and non-HLA genetic influences that individually or jointly contribute to the observed variability in the humoral response to vaccination among healthy individuals. In this exciting era, new high-dimensional approaches and techniques including vaccinomics, systems biology, GWAS, epitope prediction and sophisticated bioinformatics/statistical algorithms, provide powerful tools to investigate immune response mechanisms to the measles vaccine. These might predict, on an individual basis, outcomes of acquired immunity post measles vaccination.
1. Measles in Developed Countries: the Need for New Knowledge
Despite the existence of an effective measles vaccine, 266,701 measles cases were reported worldwide in 2014 with more than 146,000 measles-related deaths reported in 2013 (the majority of fatal measles cases occurring in Africa and Asia) [1]. In recent years, there has been a resurgence of measles cases in the United States and across Europe [2]. From 2010 to 2014, the European region reported 135,600 measles cases, with 26,436 and 14,059 cases in 2013 and 2014, respectively; large outbreaks were recorded (mainly among unvaccinated and individuals with unknown vaccine status) in France, Spain, Italy, Germany and Romania [2–6]. In 2014 alone, the U.S. reported 668 measles cases across 27 states—the highest number of annual cases since the U.S. measles elimination declaration in 2000 [2]. During January – April 2015, a total of 159 measles cases (of which 18% had received measles vaccine) were reported to the U.S. Centers for Disease Control and Prevention [5, 6]. The vast majority of measles cases are due to failure in administering or receiving the vaccine [6]. However, in countries with high measles vaccine coverage, outbreaks have revealed measles vaccine failure among individuals previously vaccinated with two doses of measles-containing vaccine [2, 3, 5, 7–10].
Given the ongoing public health threat of measles, it is critical to understand the development and determinants of measles vaccine immunogenicity – both those that drive initial protective responses and those that lead to vaccine failure. In this review, we examine measurements of measles-specific humoral immunity, vaccine correlates of protection, and factors associated with variability in measles-specific humoral immunity, with a focus on immunogenetics. We discuss how new “OMICS” technologies, systems biology and vaccinomics approaches to studying vaccine responses can be applied to explain the variations in immune responses to the measles vaccine. These new developments, in addition to available datasets for other vaccines at a human systems level, offer an exciting opportunity to search for evidence of common immune responses, pathways and signatures among various infectious diseases following immunization. Furthermore, these current technological advances may indeed serve to better identify specific biomarkers of vaccine immunogenicity, and/or any potential adverse reactions presented in response to one or several group(s) of vaccines.
2. Variation in Measles Vaccine Responses: General Principles
Primary vaccine failure arises when a vaccinated individual does not develop a protective immune response after immunization. Secondary failure (waning immunity) occurs when an individual develops a protective immune response after vaccination (based on the established correlates of protection), but the vaccination fails to protect the vaccinated individual from subsequent infection upon exposure. The current measles vaccines available in the U.S. contain the Edmonston-Enders-based Moraten measles strain in combination with other viruses: measles-mumps-rubella (MMR), or measles-mumps-rubella-varicella (MMRV). Other Edmonston-based strains used worldwide, with similar immunogenicity and safety profiles, include the Schwarz (produced in Brazil and Europe); the Edmonston-Zagreb (the most frequently used vaccine in the WHO immunization programs, India, Croatia, Switzerland); and the AIK-C strain (used in Japan). The non-Edmonston-based vaccines are derived independently and include the CAM-70 (produced and used in Japan and Indonesia); the Leningrad-16 (produced and used in Russia); the Changchun-47 and the Shanghai-191 strains (produced and used in China) [11]. It was anticipated that a two-dose MMR vaccination program would lead to substantial reductions in measles morbidity and measles elimination (Box 1); however, various studies have approximated that 2–10% of individuals vaccinated with two MMR doses may not develop or sustain protective measles humoral immunity, allowing a gradual accumulation of individuals susceptible to infection and subsequently, the occurrence of viral outbreaks [2–4, 6–10, 12, 13].
Box 1.
In 1989, after recording substantial vaccine failure rates in children previously vaccinated with one dose of MMR, the American Academy of Pediatrics (AAP) and the CDC Advisory Committee on Immunization Practices (ACIP) recommended a two-dose MMR vaccine schedule.
Consistent with this, in the U.S. measles outbreaks from 1989–1991, up to 40% of children who contracted measles had previously received one dose of MMR vaccine and yet were not protected from infection.
The correlate of protection for measles is based on measles-specific humoral immunity; namely, an antibody response. The current “gold standard” is based on quantification of neutralizing antibodies against the viral hemagglutinin (H) and fusion (F) surface glycoproteins by the plaque reduction neutralization test (PRN), or its high-throughput version, the PRMN assay [12, 14, 15, 16]. Experimental evidence (serum depletion studies of H- and F-specific antibodies) suggests that H- (to a greater extent) and F-specific antibodies contribute to virus neutralization and protection [17, 18]. Thus, neutralizing antibodies are the correlate of protection which assesses disease susceptibility, with measles antibody titers ≥120 mIU/ml (or PRN/PRMN titer of 120) rendering protection against the disease, and titers ≥1,000 mIU/ml rendering protection against both infection and disease [16]. The importance of cellular immunity to vaccine-induced protection is not completely understood, though data suggest that seronegative vaccinated children may still have protection against measles, thus supporting an involvement of cellular immunity [16, 19].
In a study of 763 healthy children living in a community with no circulating natural infection (and no immune response boosting from wild type virus exposures), only 91% demonstrated protective humoral immunity, 7.4 years (median) after vaccination with two medical record-documented doses of MMR vaccine [12]. This study also demonstrated inter-individual variations in neutralizing antibody titers to measles (GMT of 832 mIU/mL [95% CIs: 776; 891]) and no correlations of antibody titer with cellular immunity measures [12, 20]. Additional information was gathered from studies demonstrating the variability and persistence of measles-specific antibodies after two measles vaccine doses, as summarized in Table 1.
Table 1.
Measles-specific antibodies, variability and persistence after two doses of MMR vaccine administration
| Study | Subjects Number | Years post second vaccination (median., range) | Antibody assay | Antibody titer (GMT, range if available, 95% CI) | Potentially susceptible (%) | Reference |
|---|---|---|---|---|---|---|
| 1. Gans HA et al.; 2001 | 73 (First dose: 6 months) 61 (First dose: 9 months) |
NA NA |
PRN | 128 mIU/mL (74–215) 426 mIU/mL (241–657) |
40% 11% % (≤120 mIU/mL) |
[94] |
| 2. Gans HA et al.; 2004 | 32 (First dose: 6 months) 23 (First dose: 9 months) |
0.5 0.5 |
PRN | 702 mIU/mL (344–1457) 1546 mIU/mL (686–3484) |
14% 10% (≤120 mIU/mL) |
[95] |
| 3. LeBaron CW et al.; 2007 | 312 Kindergarten 309 Middle school |
5.1 (4.2–6.1) 11.2 (10.1–12.5) |
PRN | 641 mIU/mL (NA) 737 mIU/mL (NA) |
4.7% (≤120 mIU/mL) | [96] |
| 4. Davidkin I et al.; 2008 | 85 younger group 6 Older group |
20 | EIA | 760 mIU/mL (75; 8400) 1113 mIU/mL (220; 5900) |
12% (EIA titer ≤200 mIU/mL) | [97] |
| 5. Haralambieva IH et al.; 2011 | 763 school children | 7.4 (5.6; 9.2). | PRMN | 832 mIU/mL (776; 891) | 8.9% (<PRMN titer of 120) | [12] |
| 6. Kakoulidou M et al.; 2013 | 33 | >7 | EIA | 2098 mIU/mL (180; 8783) | NA | [98] |
| 1097 Rochester cohort | 6.6 (5.0, 8.6) | 896 mIU/mL (423; 1744)* | 2.6% | |||
| 7. Poland GA et al.; 2015 | 970 San Diego cohort | 1.2 (0.7, 2.8) | PRMN | 848 mIU/mL (392; 1635)* | 3.7% | Unpublished data |
| 1038 US cohort | 1.8 (1.3, 2.9) | 762 mIU/mL (345; 1505)* | 4.2%(<PRMN titer of 120) |
Median (25% and 75% IQR)
3. HLA Influence on Antibody Variations to Measles Vaccination
There is a growing interest in applying novel vaccinomics approaches to vaccine studies. Vaccinomics is generally defined as “the integration of a systems biology approach with immunogenetics, immunogenomics, immune profiling and functional studies in order to understand and predict vaccine-induced immune responses” [21–23].
It is believed that host genetic factors, including both major histocompatibility human leukocyte antigen genes (HLA) and non-HLA genes, as well as other variables (Key Figure, Fig. 1) modulate immunity following measles vaccination. A study conducted on twins revealed a strong genetic influence on the variance in measles vaccine-specific humoral immune responses (heritability 88.5%, p<0.0001) [24, 25], and we have estimated (previously described methodology [26]) that single nucleotide polymorphisms (SNPs), together with HLA alleles, explain ~30% of the inter-individual variability in measles-specific antibody titers [3]. A large body of evidence has now demonstrated that several markers in HLA genes (highly polymorphic gene region on human chromosome 6p21.31) are associated with the heterogeneity of antibody responses to measles vaccination [27–31]. A replication study using two independent cohorts (346 and 388 healthy individuals in cohorts 1 and 2, respectively) demonstrated that specific class I HLA alleles (B*35:03 and B*57:01) and class II alleles (DQA1*02:01, DQB1*03:03, DQB1*06:02, and DRB1*07:01) were associated with significant differences in antibody titers following two doses of the measles-containing vaccine [31]. Furthermore, the HLA-B7 supertype that shares common peptide binding motifs between various class IB alleles was associated with approximately 18% higher measles vaccine-induced antibodies in both cohorts [31]. There is indication that low antibody responsiveness after a first (single) dose of measles-containing vaccine is at least partially influenced by class II HLA alleles, such as DRB1*03 (p=0.004) and DQA1*02:01 (p=0.05) [27]. Conversely, very high levels of measles-specific antibodies following a first vaccine dose were found to be associated with HLA allelic variants, such as B*7 (p=0.05), DQA1*01:04 (p=0.02), and DPA1*02:02 (p=0.04) [28]. However, the second dose of measles vaccination had an extinguishing effect on these HLA allele associations, suggesting that an additional dose may overcome HLA restriction [32].
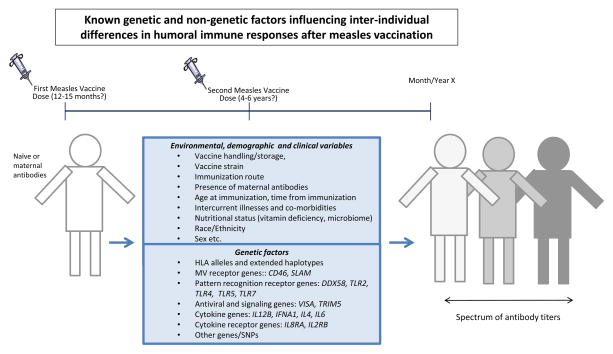
Fig. 1.
Naïve individuals (white color, no previous exposure to measles vaccine) are vaccinated with one or two doses of MMR vaccine. An array of non-genetic (upper box panel) and genetic factors (lower box panel) perturbate the development and/or persistence of measles-specific humoral immunity, leading to a spectrum (different shades of gray) of antibody titers in vaccinated individuals.
HLA haplotypes associated with measles-specific low antibody response included class I A*24-Cw*03-B*15 (p=0.04), and class II DRB1*07-DQB1*02-DPB1*02 (p=0.05) and DRB1*07-DQB1*03-DPB1*04 (p=0.001) haplotypes [31, 33]. In contrast, the class II haplotype DRB1*15/16-DQB1*06-DPB1*04 (p=0.02) was associated with high measles virus antibody titers after two doses of the measles vaccine [31, 33]. These population-based studies provided evidence that HLA genes and haplotypes, as well as immunologically relevant non-HLA class III genes (i.e., TNF, LTA, and others) [34], are important determinants in predicting an individual’s humoral response to measles vaccination.
The immune response to major measles virus structural proteins is controlled by T lymphocytes that recognize measles virus-derived endogenous and exogenous antigens in connection with specific HLA class I and class II molecules, respectively. Mass spectrometry studies have allowed the isolation and identification of naturally processed as well as HLA-presented measles virus-derived peptides from HLA class I alleles (A*02:01, B*27:05), and the class II (DRB1*03:01) allele [35, 36], associated with measles vaccine non-responses [27]. By isolating HLA-restricted peptides, novel promiscuous multi-peptide HLA supertype vaccines administered with specific directed adjuvants can be developed and used with greater efficacy [21]. These immunogenic measles virus peptides are being utilized in the design of novel measles vaccine candidates to overcome HLA polymorphic restrictions [37].
4. Non-HLA and Multigenic Influences on Antibody Variations to Measles Vaccination
Recent candidate gene association studies have shown that genetic variation in non-HLA genes is linked to variations in humoral immune responses after measles vaccination. Polymorphisms in genes involved in viral recognition, viral binding/entry (SLAM, CD46) and pathogen-associated molecular patterns (PAMPs) sensing (TLRs, CD209/DC-SIGN); as well as cytokine/cytokine receptor genes (IL2, IL10, IL12B, IL4RA, IL12RB1, IL7R, IL6, TNFA); antiviral genes (TRIM5, DDX58/RIG-I, OAS1, ADAR, MX2, OAS3, VISA); vitamin A and vitamin D receptor genes (RARA, RARB, RARG RXRA), and a growing set of other innate immunity-related or signaling genes, have been associated with a variation in measles-specific neutralizing antibodies or IgG titers after vaccination [3, 21, 22, 31, 37–52].
Host antiviral defenses and immune response activation after live viral vaccination and/or infection requires efficient recognition (related to viral cell entry and sensing) to trigger innate and adaptive immunity. Evidence from recent reports points to an association of genetic polymorphisms in the measles virus cell entry receptors CD46 and SLAM (and in particular CD46) with variations in humoral immunity after vaccination, as well as with adverse events following administration of the measles virus-containing vaccine [38, 40, 42, 49, 51, 53, 54] (Table 2). For example, CD46 is an ubiquitously expressed type I membrane cofactor protein and a complement regulatory factor acting as a cellular-entry receptor for measles virus vaccine strains. A candidate gene association study of 339 schoolchildren (12–18 years of age, 94% Caucasians) receiving two doses of the measles-mumps-rubella (MMR, Merck) vaccine demonstrated a significant association (p=0.01) of an intronic SNP in the CD46 gene (rs2724384) with measles vaccine-specific IgG antibody titers [40]. This result was also replicated in two subsequent independent studies [42, 49] (Fig. 1, Table 2). Increased representation of the minor G allele for this CD46 SNP was significantly associated (p = 0.0007) with a two-fold decrease in neutralizing antibody titers in a cohort of 745 individuals (80.3 % Caucasians; median age at enrollment = 15 years), following two doses of MMR vaccine [42]. Concomitantly, CD46 SNP rs2724384 was also associated with variations in IFN-α (p = 0.006), IL-6 (p = 0.027) and TNF-α (p = 0.0006) secretion [42]. This SNP was also significantly associated with neutralizing antibody responses in a multivariate/multigenic assessment model (p=0.0006) [38]. In a cohort of 137 Australian children (12 to 14 months old) following a primary dose of measles vaccination, the CD46 polymorphism was significantly associated with measles virus-specific IgG levels (p=0.018) [49]. Notably, a recently published state-of-the-art genome-wide association-replication study in a Danish population revealed a high correlation between the CD46 (1q32.2) genetic region and the intronic SNP rs2724384 with MMR-related febrile seizures (p=1.7E-10), where the risk allele [A] for febrile seizures is related to higher antibody titers [54]. Furthermore, data from the 1000 Genomes Project, based on genome and transcriptome sequencing and analysis of lymphoblastoid cell lines of 462 subjects, revealed an association of the CD46 SNP rs2724384 [A] with increased CD46 gene expression [55], and provided evidence of the functional consequences of this and/or other tagged causal CD46 SNPs. A coding synonymous genetic variant (rs164288, Thr210Thr) in the SLAM gene has also been associated with measles vaccine-induced antibody levels [40] (Table 2). Although this genetic association was not successfully replicated in two independent studies [42, 51], the same SLAM polymorphism was found to be associated with a measles-specific IFNγ ELISPOT response after measles vaccination [42]. The genetic influence of the newly discovered measles virus receptor PVRL4 (nectin-4) [56, 57] on antibody responses has yet to be elucidated. In summary, several genetic association and replication-validation studies implicate the CD46 genetic region in the regulation of humoral immunity after measles vaccination, and in susceptibility to adverse events (febrile seizures) after vaccination. Further fine mapping and functional studies are warranted to identify the causal genetic risk factors (variants), and to elucidate the downstream immunologic consequences of measles vaccination.
Table 2.
Single nucleotide polymorphisms (SNP) associated with measles vaccine-induced antibody responses
| SNP ID | Gene Symbol | Location | References |
|---|---|---|---|
| rs2724384 | CD46 | intron | [38, 40, 42, 49, 54] |
| rs164288 | SLAM | coding Thr210Thr | [40, 42] |
| rs3212227 | IL12B | 3′UTR | [3, 39, 44] |
| rs669260 | DDX58 | intron | [38, 60] |
| rs28383797 | IFNA1 | 3′UTR | [38, 44] |
| rs3218266 | IL2RB | intron | [38] |
| rs2243248 | IL4 | 5′UTR | [38, 44] |
| rs2069824 | IL6 | 5′UTR | [38, 44] |
| rs2069835 | IL6 | intron | [38] |
| rs2854386 | IL8RA | 3′UTR | [38, 44] |
| rs1816702 | TLR2 | intron | [38] |
| rs11536897 | TLR4 | 3′UTR | [38] |
| rs851178 | TLR5 | intron | [38] |
| rs864058 | TLR7 | coding | [38] |
| rs6037678 | VISA | intron | [38, 43] |
| rs7122620 | TRIM5 | 3′UTR | [38, 99] |
Given the profound importance of cytokines and their receptors in innate and adaptive immunity, these genes have been the focus of several measles vaccine immunogenetic studies, as reviewed in Haralambieva, et al. [3]. For example, a significant association of the functional genetic variant rs3212227 (TaqI polymorphism; located in the 3′UTR region of the IL12B gene) with variations in measles vaccine-induced IgG antibody titers (p =0.01) has been reported [39] (Fig. 1; Table 2). This genetic association has been subsequently validated in a larger genetic association study of 994 common cytokine and cytokine receptor SNPs in 764 subjects (p=0.037) [44]. Interleukin-12 (IL-12) is a proinflammatory cytokine with a key role in Th1 differentiation, IFNγ secretion, and induction of Th1 immune responses. It can thus modulate the Th1/Th2 cytokine balance and indirectly have an effect on antibody responses. The replicated 3′UTR TaqI polymorphism (rs3212227) can potentially influence transcriptional activity, mRNA stability and protein abundance, and was previously reported to correlate with IL-12 protein secretion, gene expression and antibody titers following HBsAg immunization [58, 59]; therefore, it is a promising target for future functional studies.
The discovery of gene polymorphisms having a potential impact on measles vaccine-induced humoral immunity offers a unique opportunity to assess high-priority candidates in vaccination strategies. Focused fine-mapping and functional studies could subsequently aim to validate the significance and biological relevance of given polymorphisms in measles vaccination. This research may also increase our understanding of the mechanisms through which such polymorphisms affect gene function and, ultimately, the responses and ensuing immunological memory to a measles vaccine. As such, functional characterization studies of measles-specific gene associations are being performed, and are reviewed elsewhere [3].
Genetic data from candidate gene panels has provided the identification of SNP combinations that conjointly contribute to variability in antibody responses using a multigenic analysis approach [3, 38]. This approach identified genetic variants in the CD46 (rs2724384), innate pattern recognition receptor genes (DDX58, TLR2, TLR4, TLR5, TLR7), antiviral effector and signaling genes (e.g., TRIM5, VISA), and cytokine/cytokine receptor genes (IFNA1, IL4, IL6, IL8RA, IL2RB) that cooperatively contribute to the observed variability in measles-specific antibody responses in a multigenic fashion [3, 38] (Fig. 1; Table 2). It is important to note that while most of these findings are specific to the measles component of the MMR vaccine (the immune outcomes in these analyses were measles-specific IgG or neutralizing antibodies), some of the genetic associations were described together with the antibody responses against the rubella component of the vaccine (e.g., DDX58) [60].
At this point, due to the limited number of consistent findings, it is difficult to integrate the current knowledge into a more comprehensive view of the mechanisms leading to hyper- and hypo-responsiveness to the measles vaccine. However, it is obvious that initial infection stages (infection of susceptible cells and activation of innate immunity) are crucial for the genesis of an adaptive immune response to measles vaccination. Protective immunity against measles begins with (and is greatly influenced by) the infection of susceptible cells through measles-specific cellular surface receptors (e.g., CD46) as well as innate receptor mediated-sensing of pathogen-associated molecular patterns, such as through Toll-like receptors and RIG-I (DDX58). Together, innate receptors trigger various intracellular signaling cascades and IFN responses. Secretion of type I interferons (e.g., IFNα), pro-inflammatory cytokines (e.g., IL-6) and chemokines, initiate innate antiviral responses (e.g., VISA, TRIM5), induce HLA expression, differentiation and activation of antigen-presenting cells, and augment cellular chemotaxis and inflammation. It is well known that these events trigger and amplify measles-specific adaptive immunity.
A next step in vaccinology is to uncover the yet unknown genetic (or other) factors influencing measles vaccine-induced immunity. This could be accomplished in part, by performing larger genome-wide association/replication studies (such as those performed for other viral vaccines, [61–63]) to interrogate genome associations amongst millions of SNPs and specific measles vaccine-induced phenotypes of interest (many studies are currently in progress). While such studies pose analytic challenges, they also present a unique opportunity for comprehensive gene set- and pathway-based assessments of genetic associations [3].
Sets of identified genetic markers and data serve as a starting point for future genetic and functional studies. Functional information will further enhance our understanding of the correlation between expression and function of the genetic variants likely to regulate inter-individual immune response variations in antibody responses following measles vaccination. Ultimately, such studies may delineate biomarkers of protective immunity or risk (adverse events or low/non-response to vaccines). This may assist the development of new or improved measles vaccines.
5. New Technologies and Approaches for Studying Immune Responses to Vaccines
As we move further away from empirical vaccine development and toward more rational and directed approaches, several critical elements are necessary for success. First, we must have a detailed understanding of the pathogenesis of the measles virus. Second, we need to define better correlates of protection—correlates that go beyond measuring antibody titers. Third, it is essential to understand what drives a vaccine response, a vaccine non-response, and even adverse events following vaccination. Fourth, it is important to develop a complete picture of vaccine-induced immunity. This picture must accurately cover both innate and adaptive immunity, as well as capture as many complex interactions between immune function-related components as possible [64].
Over the last decade, there have been multiple advances in immunology, virology, molecular biology, bioinformatics, and related research fields that have provided new tools and technologies aimed at enhancing our understanding of measles vaccine-induced immunity and directly address the aforementioned points. Although this review focuses on specific areas of measles vaccine immunity, these approaches can be, and in many cases have been, applied to other viral vaccines as well.
We have seen vast improvements in our ability to simultaneously capture high dimensional data in biology. The “OMIC” technologies are capable of measuring gene expression, protein levels, epigenetic events, and metabolic processes at a comprehensive level [65]. Thus, it is now possible to simultaneously examine changes in essentially every gene, protein, or biological product in response to a given stimulus. Indeed, sequencing experiments can now provide large gene sequence datasets, identify different isoforms, quantify and evaluate differential mRNA transcript usage, measure miRNA species, and assess DNA methylation status across the entire genome.
These advances in data acquisition have, of necessity, spurred the development of sophisticated bioinformatics algorithms capable of handling and evaluating immense datasets [66]. Many of these tools and databases are online, providing the research community with publicly available large dataset information and easily accessible tools for data mining capabilities [67]. Epitope-prediction algorithms have moved beyond simply defining potential immunogenic T and B cell epitopes, to complex sequence analysis and structural vaccinology-modeling tools that identify putative epitopes, model 3D structures of antibody-antigen interfaces, evaluate energy and binding requirements, or optimize antigen backbones, side chains and antibody interaction surfaces [68, 69]. Such approaches have not been used for measles, but have been successfully applied to the development of broadly neutralizing antibodies to other viruses such as influenza and RSV, among others [70, 71]. These online tools cover a range of applications including viral sequencing data, comparative analyses, RNA folding, miRNA and siRNA functional studies, protein-protein interactions, structural analyses, and host-pathogen interactions.
Many advanced “OMICS” technologies, combined with cutting-edge bioinformatics tools, have made it possible to study vaccine responses from a systems-level perspective; however, systems-level findings are still lacking for measles-specific vaccination and immunity [64] (Box 2). Other techniques such as flow cytometry, are readily amenable to single-cell analysis, and high-dimensional “OMICS” technologies are also being coupled to single cell analysis. Mass cytometry, or CyTOF (cytometry time of flight), utilizes mass spectrometry to dramatically enhance the number of distinguishable markers compared to conventional flow cytometry. CyTOF technology has allowed complex phenotyping of lymphocyte subsets, [72] and detailed T cell epitope mapping [73]. Others have combined flow cytometry with single-cell PCR, or next-generation sequencing technologies in order to study single cell transcriptomes [74]. In terms of immune response profiling and vaccine development, single-cell transcriptomics has provided insights into the control of dendritic cell heterogeneity and function, and can be used to assess transcriptomic changes in specific cell populations relevant to the development and persistence of protective immune responses to measles vaccines [75, 76].
Box 2.
New approaches (i.e., vaccinomics, systems vaccinology, vaccine response profiling) seek to provide an integrated picture of the immune system as it responds to vaccination. The systems-level data (transcriptomic, epigenomic, metabolomic, proteomic, lipidomic, glycomic, and others) is examined using computational approaches and predictive modeling that identify signatures or profiles correlated with immune outcomes. In turn, these signatures or biomarkers are used to experimentally manipulate vaccine formulations and vaccination protocols (e.g., adjuvants, dose, route and schedule of administration, etc.) in order to elicit optimal protective immunity. Studies such as these have provided critical insights into vaccines for yellow fever [79], influenza [80], polysaccharide and conjugate vaccines [81], as well as diseases such as tuberculosis[89]. New technologies and approaches applied to vaccinology may have a positive effect on vaccine evaluation, discovery and development [77, 78, 90].
Understanding the role of genetic variation in measles vaccine immunity has led to the identification of genetic polymorphisms associated with a measles vaccine response or non-response. Identifying the specific activation or suppression “genotype” of viral receptors (e.g., CD46), would potentially allow the development of new vaccine candidates – with the use of adjuvants (and/or different moieties), it might be possible to induce “correction” of an underlying genetic defect responsible for an immunosuppression phenotype. One can also consider (with regard to current knowledge) taking advantage of the RIG-I (DDX58) pathway and/or TLR agonists to boost innate immunity and elicit optimal protection (neutralizing antibody levels and cellular immunity) after measles immunization. The aforementioned immunogenetics, vaccinomics and systems vaccinology published reports [23, 65, 77–81], provide examples of how such studies might lead to new vaccine development (Box 2).
There are reports that the route of vaccine administration has a significant impact on the resulting immune response, with many studies focused on the mobilization of skin-resident APCs in order to enhance antigen presentation and subsequent adaptive immunity [82]. Therefore, there has been considerable interest in evaluating different routes of administration. A recent clinical trial found that seroconversion rates were 10% lower for the aerosolized measles vaccine than for the conventional injected vaccine [83, 84]. However, the measles vaccine administered via microneedle patch has shown promise in non-human primate studies [85]. The evaluation of immune responses has also been improved considerably by the introduction of new assays and more sophisticated animal models, to name some of the various novel vaccine approaches (Box 3). Collectively, these new techniques provide advantageous tools, allowing investigators to study the mechanisms of measles vaccine immune responses with far greater power and precision than previously possible.
Box 3.
A recent measles microarray-based study (assessing all proteins targeted by humoral responses) indicated that antibody reactivity toward a set of four viral proteins (P, N, F, L) was highly associated with, and predictive of, neutralizing antibody responses following measles vaccination [91]. Nucleic Acid Programmable Protein Arrays bind cDNA encoding pathogen proteins of interest, directly on the array, thus allowing in situ protein translation and reliable quantification of antibody responses to antigens from multiple viruses (e.g. 761 antigens from 25 different viruses); in turn, the identification of unique and common antibody response patterns in humans can be assessed [92]. This technology can be applied to simultaneously study all MMR vaccine components, and to provide rapid and easily tested biomarkers of MMR vaccine responses, and/or to obtain more complete correlates of protection.
The discovery of the three cellular receptors (CD46, SLAM, PVRL4) for measles virus paved the way for the creation of more sophisticated animal models for studying measles infection susceptibility and immunity (e.g., transgenic mice that express specific receptors of interest), not covered by this review. Nevertheless, together with non-human primate models, studies such as these have provided invaluable information regarding measles pathogenicity, cellular tropism, viral spread, and immunosuppression [93].
To date, the nature of inter-individual variability in humoral immune responses to live attenuated measles vaccine, and the mechanisms behind it, remain only partially explained, with much yet to discover. Future research avenues and scientific questions are further summarized in this review (see Box 4 and Outstanding Questions).
Box 4. A Glimpse of Future Predictive Mathematical Applications and Bioinformatics in Vaccinology.
Certain genes and SNPs are associated with both high and low anti-measles neutralizing antibody levels but, at this point, it is not possible to predict on an individual basis, humoral antibody responses. A mathematical equation for predicting putatively the humoral immune response to a vaccine has been reported, but all the factors constituting this equation remain to be fully identified for measles [77]. A general form of this equation is
where y indicates the humoral immune response measure (e.g., a person’s antibody titer after vaccination), fv indicates a linear or nonlinear function of the variables for the vth vaccine, HF designates human demographic factors, VF designates vaccine variables, GF designates genomics factors, etc. [77] Once such factors (particularly genetic) are identified, it will be necessary to perform a series of functional studies designed to understand the mechanism(s) behind their causal effects. In this manner, we may learn the individual, cumulative, and interactive effects of genetic and non-genetic factors that together compose humoral adaptive responses to a viral vaccine such as the measles vaccine. We believe that predicting these responses is theoretically possible, but the major obstacles remaining include the need for additional high-throughput, high-dimensional assays allowing to study large numbers of subjects, such that phenotype:genotype databases can be constructed for data mining. In addition, more advanced bioinformatic algorithms that can handle and identify patterns within terabyte-size experiments are needed to perform large systems biology analyses. This composes the field of vaccinomics, a method useful in both personalized vaccinology and in the design of novel vaccine candidates.
Outstanding Questions.
What are the yet unknown genetic factors influencing measles vaccine-induced antibody responses?
What is the quantifiable contribution of specific genetic factors (from a genome-wide perspective) and their association to antibody variability after measles vaccination?
What is the quantifiable contribution of specific non-genetic factors (i.e., environmental, demographic and clinical factors) to antibody variability after measles vaccination?
What are the mechanisms (behind specific genetic and/or other factors) leading to hypo- or hyper- antibody responsiveness after measles vaccination?
6. Limitations of genetic association analyses in vaccine research
A major issue with genetic association studies is being able to adjust for all confounding variables and to control false positive associations by using stringent p-values. For agnostic GWAS, a common practice is to use a p-value threshold of 5×10−8[86]. The choice of critical p-values should consider the number of statistical tests performed, but also, the prior probability/evidence that the association between the SNP/HLA allele and the outcome is real [87]. Performing analyses that appropriately account for the large number of tests in candidate gene studies requires a Bonferroni correction (or other multiple testing approaches) to correct for false discovery rate (FDR) for the number of tested SNPs or HLA alleles, which was not performed in some of the reported findings in this review. With that in mind, the current review summarizes and interprets consistent and/or replicated findings, with the understanding that false positives are less likely to demonstrate consistent associations. In addition, due to the high dimensionality of many new technologies applicable to vaccines (e.g., mRNA-Seq, Mass spectrometry, etc.), FDRs need to be computed to assess statistical significance, as is standard practice in the field [88]. Furthermore, filtering strategies and analytical methods can be designed for data reduction and for incorporating a priori knowledge from outside sources. This would allow a knowledge-driven analytical approach to minimize false discoveries [64, 66].
7. Concluding Remarks
Recent genetic association and replication studies implicate HLA alleles, CD46 and other genetic variants of innate pattern recognition receptors, cytokine/cytokine receptors, as well as antiviral effector and signaling genes in the regulation of humoral immunity (Fig. 1), and in the susceptibility to febrile seizures following measles vaccination. Future genome-wide, systems biology and functional studies are warranted to further expand our knowledge of the genetic influences on measles vaccine-induced antibody responses. Technological and conceptual developments coupled with new genetic data now offer unprecedented opportunities to characterize the mechanisms of measles vaccine-induced immune responses from a systems-level perspective. However, important scientific questions must still be addressed (Outstanding Questions). The bigger questions are whether the aforementioned advancements and breakthroughs in the field of vaccinology enhance our ability to manipulate specific targets to obtain optimal immune responses and immunological memory. Also, can they lead to the development of prediction algorithms when assessing immune response outcomes after measles vaccination? Can these also be applied to personalized vaccination approaches, and to the evaluation of novel measles (or other pathogen) vaccines to benefit public health?
Trends. Genetic drivers of measles vaccine-induced antibody responses.
2–10% of individuals immunized with two Measles Mump Rubella (MMR) vaccine doses may not have protective measles antibody titers.
Variability in humoral immune responses to measles vaccine is highly heritable (88.5%)
SNP associations in non-HLA genes, together with HLA alleles, may explain ~30% of the inter-individual variability in antibody titers after measles vaccination.
Several class I and class II HLA alleles, as well as the HLA-B7 supertype have been associated with significant differences in antibody titers following two administrated doses of measles-containing vaccine in human subjects.
The measles vaccine (MV) receptor (CD46, SLAM), pattern recognition receptor genes (DDX58, TLR2, TLR4, TLR5, TLR7) and antiviral/signaling (TRIM5, VISA) genes have been associated with inter-individual differences in antibody responses after measles vaccination.
Cytokine (IL12B, IFNA1, IL4, IL6) and cytokine receptor (IL8RA, IL2RB) genes have also been associated with inter-individual differences in antibody titers after measles vaccination.
Acknowledgments
We thank Caroline Vitse for assistance in preparing the manuscript. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R37AI048793 (which recently received a MERIT Award) and AI033144. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of Interest
Dr. Poland is the chair of a Safety Evaluation Committee for novel investigational vaccine trials being conducted by Merck Research Laboratories. Dr. Poland offers consultative advice on vaccine development to Merck & Co. Inc., CSL Biotherapies, Avianax, Dynavax, Novartis Vaccines and Therapeutics, Emergent Biosolutions, Adjuvance, and Microdermis. Drs. Poland and Ovsyannikova hold two patents related to vaccinia and measles peptide research. These activities have been reviewed by the Mayo Clinic Conflict of Interest Review Board and are conducted in compliance with Mayo Clinic Conflict of Interest policies. This research has been reviewed by the Mayo Clinic Conflict of Interest Review Board and was conducted in compliance with Mayo Clinic Conflict of Interest policies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Global progress towards regional measles elimination, worldwide, 2000–2013. Wkly Epidemiol Rec. 2014;89:509–516. [PubMed] [Google Scholar]
- 2.Whitaker JA, Poland GA. Measles and mumps outbreaks in the United States: Think globally, vaccinate locally. Vaccine. 2014;32:4703–4704. doi: 10.1016/j.vaccine.2014.06.088. [DOI] [PubMed] [Google Scholar]
- 3.Haralambieva IH, et al. The genetic basis for interindividual immune response variation to measles vaccine: new understanding and new vaccine approaches. Expert Review of Vaccines. 2013;12:57–70. doi: 10.1586/erv.12.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poland GA, Jacobson RM. The re-emergence of measles in developed countries: time to develop the next-generation measles vaccines? Vaccine. 2012;30:103–104. doi: 10.1016/j.vaccine.2011.11.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clemmons NS, et al. Measles - United States, january 4-april 2, 2015. MMWR. Morbidity and mortality weekly report. 2015;64:373–376. [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Measles Cases and Outbreaks. 2015 http://www.cdc.gov/measles/cases-outbreaks.html.
- 7.Rosen JB, et al. Outbreak of measles among persons with prior evidence of immunity, New York City, 2011. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2014;58:1205–1210. doi: 10.1093/cid/ciu105. [DOI] [PubMed] [Google Scholar]
- 8.Defay F, et al. Measles in children vaccinated with 2 doses of MMR. Pediatrics. 2013;132:e1126–1133. doi: 10.1542/peds.2012-3975. [DOI] [PubMed] [Google Scholar]
- 9.De Serres G, et al. Higher risk of measles when the first dose of a 2-dose schedule of measles vaccine is given at 12–14 months versus 15 months of age. Clinical Infectious Diseases. 2012;55:394–402. doi: 10.1093/cid/cis439. [DOI] [PubMed] [Google Scholar]
- 10.Zipprich J, et al. Measles outbreak--California, December 2014-February 2015. MMWR. Morbidity and mortality weekly report. 2015;64:153–154. [PMC free article] [PubMed] [Google Scholar]
- 11.Strebel P, et al. Measles Vaccine. In: Plotkin SA, et al., editors. Vaccines. 6. Elsevier; 2013. pp. 352–387. [Google Scholar]
- 12.Haralambieva IH, et al. A large observational study to concurrently assess persistence of measles specific B-cell and T-cell immunity in individuals following two doses of MMR vaccine. Vaccine. 2011;29:4485–4491. doi: 10.1016/j.vaccine.2011.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poland GA, Jacobson RM. Failure to reach the goal of measles elimination. Apparent paradox of measles infections in immunized persons. Archives of Internal Medicine. 1994;154:1815–1820. [PubMed] [Google Scholar]
- 14.Bouche FB, et al. Neutralizing B cell response in measles. Viral Immunol. 2002;15:451–471. doi: 10.1089/088282402760312331. [DOI] [PubMed] [Google Scholar]
- 15.Haralambieva IH, et al. Development of a novel efficient fluorescence-based plaque reduction microneutralization assay for measles immunity. Clin Vaccine Immunol. 2008;15:1054–1059. doi: 10.1128/CVI.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Swart RL, et al. Relative contributions of measles virus hemagglutinin- and fusion protein-specific serum antibodies to virus neutralization. J Virol. 2005;79:11547–11551. doi: 10.1128/JVI.79.17.11547-11551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Swart RL, et al. Depletion of measles virus glycoprotein-specific antibodies from human sera reveals genotype-specific neutralizing antibodies. The Journal of general virology. 2009;90:2982–2989. doi: 10.1099/vir.0.014944-0. [DOI] [PubMed] [Google Scholar]
- 19.Samb B, et al. Serologic status and measles attack rates among vaccinated and unvaccinated children in rural Senegal. Pediatr Infect Dis J. 1995;14:203–209. doi: 10.1097/00006454-199503000-00007. [DOI] [PubMed] [Google Scholar]
- 20.Jacobson RM, et al. Independence of measles-specific humoral and cellular immune responses to vaccination. Human Immunology. 2012;73:474–479. doi: 10.1016/j.humimm.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poland GA, et al. Heterogeneity in vaccine immune response: the role of immunogenetics and the emerging field of vaccinomics. Clin Pharmacol Ther. 2007;82:653–664. doi: 10.1038/sj.clpt.6100415. [DOI] [PubMed] [Google Scholar]
- 22.Poland GA, et al. Vaccinomics and a new paradigm for the development of preventive vaccines against viral infections. Omics. 2011;15:625–636. doi: 10.1089/omi.2011.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poland GA, et al. Vaccinomics and personalized vaccinology: Is science leading us toward a new path of directed vaccine development and discovery? PLoS Pathogens. 2011;7:e1002344. doi: 10.1371/journal.ppat.1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan PL, et al. Twin studies of immunogenicity - determining the genetic contribution to vaccine failure. Vaccine. 2001;19:2434–2439. doi: 10.1016/s0264-410x(00)00468-0. [DOI] [PubMed] [Google Scholar]
- 25.Jacobson RM, et al. Studies of twins in vaccinology. Vaccine. 2007;25:3160–3164. doi: 10.1016/j.vaccine.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 26.Ovsyannikova IG, et al. Replication of rubella vaccine population genetic studies: validation of HLA genotype and humoral response associations. Vaccine. 2009;27:6926–6931. doi: 10.1016/j.vaccine.2009.08.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poland GA, et al. Identification of an association between HLA class II alleles and low antibody levels after measles immunization. Vaccine. 2001;20:430–438. doi: 10.1016/s0264-410x(01)00346-2. [DOI] [PubMed] [Google Scholar]
- 28.Ovsyannikova IG, et al. Associations between human leukocyte antigen (HLA) alleles and very high levels of measles antibody following vaccination. Vaccine. 2004;22:1914–1920. doi: 10.1016/j.vaccine.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 29.Ovsyannikova IG, et al. Human leukocyte antigen haplotypes in the genetic control of immune response to measles-mumps-rubella vaccine. Journal of Infectious Diseases. 2006;193:655–663. doi: 10.1086/500144. [DOI] [PubMed] [Google Scholar]
- 30.Ovsyannikova IG, et al. HLA supertypes and immune responses to measles-mumps-rubella viral vaccine: Findings and implications for vaccine design. Vaccine. 2007;25:3090–3100. doi: 10.1016/j.vaccine.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 31.Ovsyannikova IG, et al. Consistency of HLA associations between two independent measles vaccine cohorts: a replication study. Vaccine. 2012;30:2146–2152. doi: 10.1016/j.vaccine.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobson RM, et al. Human leukocyte antigen associations with humoral and cellular immunity following a second dose of measles-containing vaccine: Persistence, dampening, and extinction of associations found after a first dose. Vaccine. 2011;29:7982–7991. doi: 10.1016/j.vaccine.2011.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ovsyannikova IG, et al. Human leukocyte antigen haplotypes in the genetic control of immune response to measles-mumps-rubella vaccine. J Infect Dis. 2006;193:655–663. doi: 10.1086/500144. [DOI] [PubMed] [Google Scholar]
- 34.Ovsyannikova IG, et al. Extended LTA, TNF, LST1 and HLA gene haplotypes and their association with rubella vaccine-induced immunity. PLoS ONE. 2010;5:e11806. doi: 10.1371/journal.pone.0011806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ovsyannikova IG, et al. Naturally processed measles virus peptide eluted from class II HLA-DRB1*03 recognized by T lymphocytes from human blood. Virology. 2003;312:495–506. doi: 10.1016/s0042-6822(03)00281-2. [DOI] [PubMed] [Google Scholar]
- 36.Ovsyannikova IG, et al. Identification and characterization of novel, naturally processed measles virus class II HLA-DRB1 peptides. Journal of Virology. 2004;78:42–51. doi: 10.1128/JVI.78.1.42-51.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poland GA, et al. Application of pharmacogenomics to vaccines. Pharmacogenomics. 2009;10:837–852. doi: 10.2217/PGS.09.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kennedy RB, et al. Multigenic control of measles vaccine immunity mediated by polymorphisms in measles receptor, innate pathway, and cytokine genes. Vaccine. 2012;30:2159–2167. doi: 10.1016/j.vaccine.2012.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dhiman N, et al. Associations between measles vaccine immunity and single nucleotide polymorphisms in cytokine and cytokine receptor genes. J Infect Dis. 2007;195:21–29. doi: 10.1086/510596. [DOI] [PubMed] [Google Scholar]
- 40.Dhiman N, et al. Variations in measles vaccine-specific humoral immunity by polymorphisms in SLAM and CD46 measles virus receptors. Journal of Allergy and Clinical Immunology. 2007;120:666–672. doi: 10.1016/j.jaci.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 41.Ovsyannikova IG, et al. Effects of vitamin A and D receptor gene polymophisms/haplotypes on immune responses to measles vaccine. Pharmacogenetics and Genomics. 2012;22:20–31. doi: 10.1097/FPC.0b013e32834df186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ovsyannikova IG, et al. The association of CD46, SLAM, and CD209 cellular receptor gene SNPs with variations in measles vaccine-induced immune responses--a replication study and examination of novel polymorphisms. Human Heredity. 2011;72:206–223. doi: 10.1159/000331585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haralambieva IH, et al. Genetic polymorphisms in host antiviral genes: associations with humoral and cellular immunity to measles vaccine. Vaccine. 2011;29:8988–8997. doi: 10.1016/j.vaccine.2011.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haralambieva IH, et al. Associations between single nucleotide polymorphisms and haplotypes in cytokine and cytokine receptor genes and immunity to measles vaccination. Vaccine. 2011;29:7883–7895. doi: 10.1016/j.vaccine.2011.08.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoo KH, et al. Assessment of humoral and cell-mediated immune response to measles-mumps-rubella vaccine viruses among patients with asthma. Allergy Asthma Proc. 2010;31:499–506. doi: 10.2500/aap.2010.31.3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dhiman N, et al. Associations between SNPs in toll-like receptors and related intracellular signaling molecules and immune responses to measles vaccine: preliminary results. Vaccine. 2008;26:1731–1736. doi: 10.1016/j.vaccine.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poland GA, et al. Genetics and immune response to vaccines. In: Kaslow RA, et al., editors. Genetic Susceptibility to Infectious Diseases. Oxford University Press; 2008. pp. 1–447. [Google Scholar]
- 48.Clifford HD, et al. TLR3 and RIG-I gene variants: Associations with functional effects on receptor expression and responses to measles virus and vaccine in vaccinated infants. Human Immunology. 2012;73:677–685. doi: 10.1016/j.humimm.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 49.Clifford HD, et al. CD46 measles virus receptor polymorphisms influence receptor protein expression and primary measles vaccine responses in naive Australian children. ClinVaccine Immunol. 2012;19:704–710. doi: 10.1128/CVI.05652-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clifford HD, et al. Toll-like receptor 7 and 8 polymorphisms: associations with functional effects and cellular and antibody responses to measles virus and vaccine. Immunogenetics. 2012;64:219–228. doi: 10.1007/s00251-011-0574-0. [DOI] [PubMed] [Google Scholar]
- 51.Clifford HD, et al. SLAM and DC-SIGN measles receptor polymorphisms and their impact on antibody and cytokine responses to measles vaccine. Vaccine. 2011;29:5407–5413. doi: 10.1016/j.vaccine.2011.05.068. [DOI] [PubMed] [Google Scholar]
- 52.Newport MJ. The genetic regulation of infant immune responses to vaccination. Front Immunol. 2015;6:18. doi: 10.3389/fimmu.2015.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clifford HD, et al. Polymorphisms in key innate immune genes and their effects on measles vaccine responses and vaccine failure in children from Mozambique. Vaccine. 2012;30:6180–6185. doi: 10.1016/j.vaccine.2012.07.063. [DOI] [PubMed] [Google Scholar]
- 54.Feenstra B, et al. Common variants associated with general and MMR vaccine-related febrile seizures. Nature Genetics. 2014;46:1274–1282. doi: 10.1038/ng.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muhlebach MD, et al. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature. 2011;480:530–533. doi: 10.1038/nature10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noyce RS, et al. Tumor Cell Marker PVRL4 (Nectin 4) Is an Epithelial Cell Receptor for Measles Virus. PLoS Pathog. 2011;7:e1002240. doi: 10.1371/journal.ppat.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seegers D, et al. A TaqI polymorphism in the 3′UTR of the IL-12 p40 gene correlates with increased IL-12 secretion. Genes Immun. 2002;3:419–423. doi: 10.1038/sj.gene.6363919. [DOI] [PubMed] [Google Scholar]
- 59.Yucesoy B, et al. Influence of cytokine gene variations on immunization to childhood vaccines. Vaccine. 2009;27:6991–6997. doi: 10.1016/j.vaccine.2009.09.076. [DOI] [PubMed] [Google Scholar]
- 60.Ovsyannikova IG, et al. Single-nucleotide polymorphism associations in common with immune responses to measles and rubella vaccines. Immunogenetics. 2014;66:663–669. doi: 10.1007/s00251-014-0796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lambert ND, et al. Polymorphisms in HLA-DPB1 are associated with differences in rubella-specific humoral immunity after vaccination. Journal of Infectious Diseases. 2015;211:898–905. doi: 10.1093/infdis/jiu553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kennedy RB, et al. Genome-wide genetic associations with IFNgamma response to smallpox vaccine. Human Genetics. 2012;131:1433–1451. doi: 10.1007/s00439-012-1179-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ovsyannikova IG, et al. Genome-wide association study of antibody response to smallpox vaccine. Vaccine. 2012;30:4182–4189. doi: 10.1016/j.vaccine.2012.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oberg AL, et al. Systems biology approaches to new vaccine development. Current Opinion in Immunology. 2011;23:436–443. doi: 10.1016/j.coi.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kennedy RB, Poland GA. The Top Five “Game Changers” in Vaccinology: Toward Rational and Directed Vaccine Development. Omics. 2011;15:533–537. doi: 10.1089/omi.2011.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oberg AL, et al. Lessons learned in the analysis of high-dimensional data in vaccinomics. Vaccine. 2015 doi: 10.1016/j.vaccine.2015.04.088. S0264-410X, 00574-00575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharma D, et al. Unraveling the web of viroinformatics: computational tools and databases in virus research. Journal of Virology. 2015;89:1489–1501. doi: 10.1128/JVI.02027-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He L, Zhu J. Computational tools for epitope vaccine design and evaluation. Curr Opin Virol. 2015;11:103–112. doi: 10.1016/j.coviro.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paquet E, Viktor HL. Molecular dynamics, monte carlo simulations, and langevin dynamics: a computational review. Biomed Res Int. 2015;2015:183918. doi: 10.1155/2015/183918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee PS, Wilson IA. Structural characterization of viral epitopes recognized by broadly cross-reactive antibodies. Current Topics in Microbiology and Immunology. 2015;386:323–341. doi: 10.1007/82_2014_413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Correia BE, et al. Proof of principle for epitope-focused vaccine design. Nature. 2014;507:201–206. doi: 10.1038/nature12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Newell EW, et al. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36:142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Newell EW, et al. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nature Biotechnology. 2013;31:623–629. doi: 10.1038/nbt.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Han A, et al. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nature Biotechnology. 2014;32:684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shalek AK, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shalek AK, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poland GA, et al. Vaccinomics, adversomics, and the immune response network theory: Individualized vaccinology in the 21st century. Seminars in Immunology. 2013;25:89–103. doi: 10.1016/j.smim.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pulendran B, et al. Systems vaccinology. Immunity. 2010;33:516–529. doi: 10.1016/j.immuni.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Querec TD, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nakaya HI, et al. Systems biology of seasonal influenza vaccination in humans. Nat Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Obermoser G, et al. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013;38:831–844. doi: 10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Teunissen MB, et al. Insight into the immunobiology of human skin and functional specialization of skin dendritic cell subsets to innovate intradermal vaccination design. Current Topics in Microbiology and Immunology. 2012;351:25–76. doi: 10.1007/82_2011_169. [DOI] [PubMed] [Google Scholar]
- 83.Low N, et al. A randomized, controlled trial of an aerosolized vaccine against measles. The New England Journal of Medicine. 2015;372:1519–1529. doi: 10.1056/NEJMoa1407417. [DOI] [PubMed] [Google Scholar]
- 84.Mayor S. Inhaled measles vaccine is less protective than injected vaccine, study shows. Bmj. 2015;350:h2006. doi: 10.1136/bmj.h2006. [DOI] [PubMed] [Google Scholar]
- 85.Edens C, et al. A microneedle patch containing measles vaccine is immunogenic in non-human primates. Vaccine. 2015 doi: 10.1016/j.vaccine.2015.02.074. S0264-410X, 00285-00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pe’er I, et al. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genetic Epidemiology. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 87.Wacholder S, et al. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J NatlCancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jacobsen M, et al. Candidate biomarkers for discrimination between infection and disease caused by Mycobacterium tuberculosis. J Mol Med (Berl) 2007;85:613–621. doi: 10.1007/s00109-007-0157-6. [DOI] [PubMed] [Google Scholar]
- 90.Lambert ND, et al. Understanding the immune response to seasonal influenza vaccination in older adults: a systems biology approach. Expert Rev Vaccines. 2012;11:985–994. doi: 10.1586/erv.12.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Haralambieva IH, et al. Profiling of Measles-Specific Humoral Immunity in Individuals Following Two Doses of MMR Vaccine Using Proteome Microarrays. Viruses. 2015;7:1113–1133. doi: 10.3390/v7031113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bian X, et al. Antiviral antibody profiling by high-density protein arrays. Proteomics. 2015;15:2136–2145. doi: 10.1002/pmic.201400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.de Vries RD, et al. Measles immune suppression: lessons from the macaque model. PLoS Pathog. 2012;8:e1002885. doi: 10.1371/journal.ppat.1002885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gans H, et al. Immune responses to measles and mumps vaccination of infants at 6, 9, and 12 months. J Infect Dis. 2001;184:817–826. doi: 10.1086/323346. [DOI] [PubMed] [Google Scholar]
- 95.Gans HA, et al. Humoral and cell-mediated immune responses to an early 2-dose measles vaccination regimen in the United States. J Infect Dis. 2004;190:83–90. doi: 10.1086/421032. [DOI] [PubMed] [Google Scholar]
- 96.LeBaron CW, et al. Persistence of measles antibodies after 2 doses of measles vaccine in a postelimination environment. Arch Pediatr Adolesc Med. 2007;161:294–301. doi: 10.1001/archpedi.161.3.294. [DOI] [PubMed] [Google Scholar]
- 97.Davidkin I, et al. Persistence of measles, mumps, and rubella antibodies in an MMR-vaccinated cohort: a 20-year follow-up. J Infect Dis. 2008;197:950–956. doi: 10.1086/528993. [DOI] [PubMed] [Google Scholar]
- 98.Kakoulidou M, et al. Kinetics of antibody and memory B cell responses after MMR immunization in children and young adults. Vaccine. 2013;31:711–717. doi: 10.1016/j.vaccine.2012.11.031. [DOI] [PubMed] [Google Scholar]
- 99.Ovsyannikova IG, et al. Associations between polymorphisms in the antiviral TRIM genes and measles vaccine immunity. Human Immunology. 2013;74:768–774. doi: 10.1016/j.humimm.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]