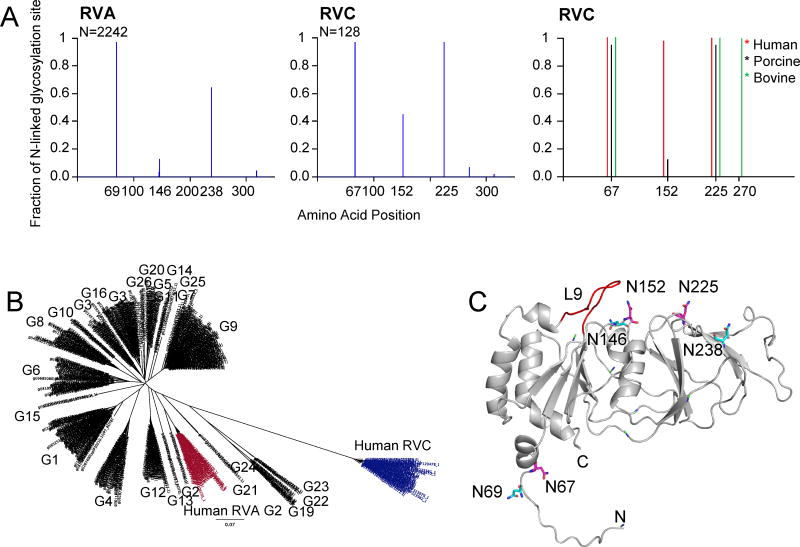
Figure 6.
Potential N-linked glycosylation sites of VP7 proteins. (A) Graphical representation of predicted glycosylation sites for RVA (left) and RVC (middle) VP7 proteins. Number of sequences used for the prediction (N) is shown on top left corner of the graphs. Distribution of most conserved N-linked glycosylation sites in human (red), porcine (blue) and bovine (green) RVC VP7 groups are also shown (left). (B) Phylogenetic tree of RVA and human RVC VP7 proteins. Human RVA G2-type VP7 sequences with 3 N-linked glycosylation sites are shown in red and human RVC VP7s are shown in blue. Corresponding G-types based on Matthijnssens et al., (2012) are also shown. (C) Mapping of potential conserved N-linked glycosylation sites on human RVC VP7 model. RVC conserved N-linked glycosylation sites are labeled and shown in magenta. RVA conserved N-linked glycosylation site locations are projected on the RVC model and are shown in cyan.