Abstract
Kernel machine based association tests (KAT) have been increasingly used in testing the association between an outcome and a set of biological measurements due to its power to combine multiple weak signals of complex relationship with the outcome through the specification of a relevant kernel. Human genetic and microbiome association studies are two important applications of KAT. However, the classic KAT framework relies on large-sample theory, and conservativeness has been observed for small-sample studies, especially for microbiome association studies. The common approach for addressing the small-sample problem relies on computationally intensive resampling methods. Here, we derive an exact test for KAT with continuous traits, which resolves the small-sample conservatism of KAT without the need for resampling. The exact test has significantly improved power to detect association for microbiome studies. For binary traits, we propose a similar approximate test, and we show that the approximate test is very powerful for a wide range of kernels including common variant- and microbiome-based kernels, and the approximate test produces correct null distribution of association p-values for these kernels. In contrast, the sequence kernel association tests (SKAT) have slightly inflated genomic inflation factors after small-sample adjustment. Extensive simulations and application to a real microbiome association study are used to demonstrate the utility of our method.
Keywords: kernel machine based association tests, small sample problem, exact tests, overdispersion
Introduction
Human genetic association studies, which identify associations between human genetic variation and phenotypic variation, have been instrumental in revealing the genetic architecture of human complex traits and diseases [McCarthy, et al. 2008]. Recently, the human genetic association studies have been extended to the human microbiome, which consists of the collection of the microbial genomes associated with the human body, and have now been seen as the ‘extended’ human genome [Cho and Blaser 2012]. The human microbiome association studies thus aim to establish a link between the human microbiome and a phenotype and can improve our understanding of the non-human genetic component of complex traits and diseases. Although single feature (variant)-based association tests are still being used for both human genetic and microbiome association studies, joint testing of multiple features (e.g. multiple genetic variants from a gene or multiple species in a genus) has become increasingly popular due to its ability to increase the statistical power by reducing the multiple testing burden and by pooling individually weak signals.
One of the most successful joint tests is the kernel association test, wherein the genetic/microbiome effects are specified by a kernel function [Kwee, et al. 2008; Lee, et al. 2012; Liu, et al. 2008; Liu, et al. 2007; Wu, et al. 2011a; Wu, et al. 2010; Wu, et al. 2011b]. The kernel framework allows flexible modeling of complex genetic/microbiome effects through the kernel function, easy adjustment of covariates, and analytic calculation of the p-value without permutation. Many forms of kernel association tests have been proposed, ranging from the sequence kernel association test (SKAT) for human genetic data, to the microbiome regression-based kernel association test (MiRKAT) for human microbiome data [Zhao, et al. 2015]. Kernel-based association tests share a common framework and are based on a score test which compares similarity in the features to similarity in the outcome. However, despite sharing a framework, the approach has evolved particularly with regard to how a p-value for association is computed. The original kernel association tests computed a p-value for association using moment matching, yet this sometimes led to inflated type I error. Thus, the SKAT modified the original kernel association tests to use an asymptotically exact distribution, which represents the current standard. It works sufficiently well for large sample size, for example, over 2,000 samples, but in practice, the sample size of DNA sequence data is still much smaller than array-based genotype data and it is not uncommon to have a sample size less than 200 for an exome sequencing study [Emond, et al. 2012]. This is also the case for microbiome data [Wu, et al. 2011a]. When the sample size is modest, SKAT is often conservative, leading to potential power loss to detect meaningful associations. The conservatism is worse for binary traits and when kernels are more complicated, as in microbiome profiling studies. For binary traits, Lee et al. [Lee, et al. 2012] addressed the small-sample problem by using moment matching with resampling, yet this correction can over-correct and lead to inflated type I error and no correction has been previously published for continuous traits.
To address the problems with current approaches to p-value computation for kernel association tests with modest sample sizes, in this paper, we propose an exact test based on SKAT score statistic for quantitative traits, which resolves the small-sample problem of SKAT for quantitative traits. Then for binary traits, we propose an alternative approximate test based on modified SKAT score statistic to account for overdispersion. We show that the approximate test works sufficiently well for microbiome-based kernels without any resampling. In the following, we first introduce our proposed method and then compare it with SKAT (with/without small-sample adjustment) in both human genetic association studies and microbiome association studies. We also apply our method to a real microbiome data to demonstrate its advantage.
Methods
Exact Test of SKAT for Quantitative Traits
For quantitative traits, we propose an “exact” method for small-sample inference. It does not depend on asymptotics, bootstrapping or permutation. Suppose y is the vector of phenotypes of n individuals, and X is the n × p covariance matrix including the intercept. Then the null model can be expressed as
where I is the n × n identity matrix, σ2I is the variance matrix of error ε. Then the score statistic in SKAT is computed as
where K is the n × n kernel matrix, ŷ0 is the fitted phenotype value under the null model, i.e., ŷ0 = X(XT X)−1 XT y, and P0 = I − X(XT X)−1 XT is a projection matrix. Because P0KP0 is a real symmetric matrix, the eigendecomposition can be written as UΛUT, where U is an orthogonal matrix and Λ is a diagonal matrix. , then z is a vector of independent standard normal variables. The score statistic becomes , where λi is the ith diagonal component of Λ and zi is the ith component of z. Therefore the score statistic follows a mixture of chi-squared distribution and the Davies method [Davies 1980] [Duchesne and De Micheaux 2010] can be used to produce an exact p-value, regardless of the sample size. In reality, σ2 is unknown and we can use a consistent estimate of σ2, such as the maximum likelihood estimate (MLE). In such case, the score statistic will asymptotically have a mixture of chi-squared distribution. Therefore, when the sample size is large, the asymptotic distribution is quite accurate. However, when the sample size is small, the variability of the estimate of σ2, which has a chi-squared distribution, will cause inaccurate approximation of the null distribution of the test statistic, leading to the apparent conservativeness using the Davies method. However, the conservativeness is not due to the Davies method per se but rather not accounting for the variability of the σ2 estimate. If we plug in the MLE estimate σ̂2, the score statistic becomes
which is a ratio of two quadratic forms of normal variables. To derive the p-value, we have
where r is the observed (scaled) score statistic using σ̂2. Therefore, the p-value depends on the distribution of εT (P0KP0 − rP0)ε/σ2, which has an exact mixture of chi-squared distribution because P0KP0 − rP0 is also a real symmetric matrix. Note that the Davies method only requires a linear combination of independent chi-squared random variables, and it can be applied even if some eigenvalues of (P0KP0 − rP0) are negative.
An Alternative Test of SKAT for Binary Traits
For binary traits, we do not have an equivalent exact method. However, we can derive an approximate method that is similar to the exact test for quantitative traits that accounts for trait overdispersion that mitigates the type I error problems associated with kernel association tests with binary traits. Compared to the moment matching method used in [Lee, et al. 2012], the proposed method does not require resampling, and yet it alleviates the conservatism for many kernels including the common variant- and microbiome-based kernels as shown by simulations and real data.
Suppose y is the vector of phenotypes of n individuals, 0 indicates controls and 1 indicates cases. The null model is
where μi = P(yi = 1), (Xγ)i denotes the ith element of Xγ. The original SKAT test statistic for binary traits assumes no overdispersion (dispersion parameter ϕ = 1 for binomial distribution) and has the following form
where K is the n × n kernel matrix, and ŷ0 is the vector of fitted probabilities under the null model. Here we instead propose a modified score statistic
to account for potential overdispersion, which is commonly seen in real data due to unmodelled risk factors in the link function.
We estimate ϕ based on the iteratively reweighted least squares (IRLS) algorithm for generalized linear models. The tth step of IRLS can be expressed as solving the linear model
where ỹt is the working response, σ2 = ϕ the dispersion parameter, and W̃t is the diagonal weight matrix with [McCullagh and Nelder 1989]. We drop the superscript t to denote corresponding quantities at convergence, and let D be a diagonal matrix with the ith diagonal element μ̃i (1 − μ̃i). Our proposed score statistic for binary traits accounting for overdispersion is then equivalent to
where σ̂2 is the dispersion estimate at convergence based on reweighted least squares. If we define ỹ* = W̃1/2ỹ, X* = W̃1/2 X and ε* = W̃1/2ε, the linear model becomes
and
This form is the same as that for the quantitative traits, and we thus have similar results as
where P0 = I − X* (X*T X*)−1 . Assuming the normality of ε*, the p-value can be calculated using Davies method based on
where r is the observed score statistic of R. Because our method is based on the normal approximation of the logistic model at convergence, it is not an exact test. Considering the overdispersion mitigates the conservatism due to small-sample even if no overdispersion really exists as shown by simulations.
Simulation Study
Human Genetic Association
The simulation for human genetic association is similar to that used in Lee et al. [Lee, et al. 2012]. We generated 10,000 chromosomes/haplotypes over 1Mb regions based on the coalescent model using the software cosi [Schaffner, et al. 2005]. Then we randomly sampled a region of length 3 kb for each data set. Genotypes of each individual were generated by randomly sampling two haplotypes.
Type I error Simulations
We simulated both quantitative traits and binary traits for case-control design. For quantitative traits, we used the linear model for individual i:
where yi was the phenotype, x1i and x2i were two covariates, and x1i followed a standard normal distribution, x2i followed a Bernoulli distribution with probability 0.5, ei followed a standard normal distribution. For binary traits, we used the logistic model for individual i:
where P(yi = 1) is the probability of being a case, x1i and x2i had the same distribution as in the quantitative model, γ0 was set to a value so that the prevalence P(yi = 1) was 0.01. The same number of cases and controls were sampled in each data set.
We considered both linear and weighted linear kernel for genotype data. For a linear kernel, K = GGT, where Gn×m is the genotype matrix of n individuals and m variants. For linear weighted kernel, K = GWWGT, where W is a diagonal matrix with weights for each variant. Two scenarios were simulated. For the first scenario, both common and rare variants were used with the linear weighted kernel. The weight function used the default beta distribution density function, specifically, wj = Beta(pj; a1, a2), where pj was the estimated minor allele frequency (MAF) for SNP j using all cases and controls, a1 = 1 and a2 = 25. Because common variants were heavily down-weighted, the results were influenced mainly by rare variants. For the second scenario, only common variants with MAF > 0.05 were used with the linear kernel. For each scenario, we considered three different total sample sizes: 50, 100, and 200. To save computing time, as in Lee et al. [Lee, et al. 2012], we simulated 500 phenotype sets for each genotype set. A total of 107 phenotypes were simulated. Type I error was estimated as the proportion of p-values less than the nominal level α. In order to better characterize the inflation and conservativeness, we also provided a 95% confidence interval (CI) of the estimated type I errors. Since by simulation each genotype corresponded to 500 phenotypes, the p-values from the same genotype data were dependent, and the number of rejected hypotheses at a given α will not follow a binomial distribution. We thus used a block bootstrap approach to calculate CI. These 500 p-values were treated as a block (resampling unit) in bootstrap, and 1,000 bootstrap samples were used to estimate the 95% CI (2.5 – 97.5 percentile).
Power Simulations
We added the effects of causal variants into the null model to form the alternative model. Specifically for individual i, for quantitative traits:
and for binary traits:
where (β1, …, βs) were the effect size of the corresponding causal genotypes (gi1, …, gis). Genotypes were additively coded as 0, 1, and 2. The rest were the same as in the null model.
We also considered three scenarios. For the first scenario, all candidate causal variants were rare variants with true MAF < 0.03. We randomly selected 20% of these candidate causal variants as causal variants. Following Lee et al. [Lee, et al. 2012], the effect sizes were specified using |βj| = c|log10(pj)|/2, where pj was the MAF of variant j. For quantitative traits, we set c = log(5); for binary traits, c = log(7). We chose 80% of the causal variants to be deleterious, i.e., positive sign of βj, and the rest were protective, i.e., negative sign of βj. For the second scenario, the causal variants were common variants (true MAF > 0.05). We simulated 1 and 3 common causal variants. For a proper range of power in comparison, for quantitative traits, we set c = log(5) for 1 causal variant and c = log(2.5) for 3 causal variants. For binary traits, we set c = log(10) for 1 causal variant and c = log(7) for 3 causal variants. All common causal variants were deleterious. For the third scenario, both the rare and common variants were causal. The settings for the rare and common causal variants were the same as the first and second scenarios, respectively, except that we only simulated 1 common causal variant. We used the weighted linear kernel for the first scenario and linear kernel for the second. Both kernels were used for the third scenario. We simulated 103 data sets for each setting and the power was estimated as the proportion of p-values less than the nominal level α.
Human Microbiome Association
We simulated microbiome datasets according to the method used in [Chen and Li 2013] and [Zhao, et al. 2015], which has been shown to generate simulated data reflective of real Operational Taxonomic Units (OTU) counts. OTUs are clustered sequence units for 16S rRNA gene targeted microbiome sequencing study [Caporaso, et al. 2010], and are proxies for biological species. Specifically, we simulated datasets with a total sample size of 50, 100, and 200, and generated the OTU counts for each sample from a Dirichlet-multinomial distribution with the parameters estimated from a real upper respiratory tract microbiome dataset, which consists of 856 OTUs measured on each of 60 samples [Charlson, et al. 2010]. To conduct type I error and power analysis, we simulated the outcome using a similar method as the genetic association studies described above. The only difference is that the outcome was related to a cluster of taxa that depend on a phylogenetic tree. For microbiome data, bacterial species exhibit a hierarchical clustering pattern, and the outcome is usually related to the abundance of a cluster of species such as species from the same genus. To reflect this, we partitioned the species into 20 clusters based on the patristic distances (sum of branch lengths on the phylogenetic tree) among the species. We used partitioning around medoid (PAM) for the clustering [Reynolds, et al. 2006], which is a general clustering algorithm based on a distance matrix. Then we let the outcome depend on the abundance of one cluster. Specifically, we chose a moderately abundant OTU cluster that constituted 3.7% of the total OTU reads to be related to the outcome. For continuous traits, we simulated under the model
Where x1i followed a standard normal distribution, x2i followed a Bernoulli distribution with probability 0.5 and standardized to have mean 0 and variance 1, A is the set of OTU indices from the cluster, β is the effect size, and zij is the OTU relative abundance (proportion). Similarly, for binary traits, we have
We considered using the weighted and unweighted UniFrac kernels (Kw and Ku, respectively), the Bray-Curtis kernel (KBC), which were converted from the corresponding distances based on the distance-to-kernel approach in Zhao et al. [Zhao, et al. 2015]. Specifically, let Dist be the pairwise n × n distance matrix, then it can be used to calculate the kernel matrix as:
where I is the n × n identity matrix, 1 is a n × 1 vector with all 1s. Dist2 is the element-wise square of Dist. The three corresponding distances used for kernels are the most used ecological distance metrics for microbiome data and represent a range of different classes of distances [Chen, et al. 2012]. Consider two microbiome communities A and B and suppose that we have a rooted phylogenetic tree with p taxa and m branches. Let bi be the length of the branch i and , are the taxa proportions descending from the branch i for community A and B, respectively. The UniFrac-based distance is defined as
where I(x) = 0 if x = 0, I(x) = 1 elsewhere. The weighted UniFrac distance is defined as
The Bray-Curtis distance is defined on the leaf nodes
The UniFrac-based distance utilizes phylogenetic relationships, whereas the Bray-Curtis distance does not, and the weighted UniFrac and Bray-Curtis distance accounts for abundance information, whereas the unweighted UniFrac distance does not.
To evaluate the type I error, we set the effect size to 0 and generated 105 data sets for each setting. Type I error was estimated as the proportion of p-values less than the nominal level α. We also calculated the 95% CI assuming the type I error is α. Because of the independence among replicated data sets, the number of rejected hypothesis has a binomial distribution, based on which 95% CI was calculated. To evaluate the power, we set different effect sizes so the power varied between 0 and 1. We simulated 103 data sets for each setting and the power was estimated as the proportion of p-values less than the nominal level α.
For quantitative traits, there is no default small-sample adjustment, and the original SKAT on quantitative traits was denoted by uSKAT. For binary traits, the original SKAT without small-sample adjustment was denoted by uSKAT, SKAT with small-sample adjustment was denoted by SKAT, our method was denoted by aSKAT (adjusted SKAT).
Results
Results on Quantitative Traits
Type I Error and Power of Human Genetic Association
Table 1 shows the empirical type I error on both common and rare variants using the linear weighted kernel. Because of the use of weights, the results were influenced mainly by rare variants. The exact test used by aSKAT had type I errors very close to different nominal levels for different sample sizes. Only two 95% CIs were above, but still close to, the nominal level, which was probably due to statistical fluctuation. On the contrary, uSKAT was conservative, which had much smaller type I errors and the upper endpoint of the 95% CI was always lower than the nominal level. Figure 1 shows the power comparison when the causal variants were rare. The power of aSKAT was consistently larger than uSKAT. The increase in power can be 0.04 in absolute terms. Table 2 shows the empirical type I error on common variants using the linear kernel. Again aSKAT controlled the type I errors at nominal levels, and uSKAT was conservative. Figure 2 shows the power comparison when there was one common causal variant. aSKAT had consistently improved power than uSKAT. Figure S1 in supplementary material shows the power comparison when there were three common causal variants and the pattern was similar. Figure S2 shows the power comparison when there were both rare and common causal variants using the weighted linear kernel. Due to downweighting on common variants, the power was mainly influenced by the rare variants and has similar pattern as that in Figure 1. Figure S3 shows the power comparison when there were both rare and common causal variants using the linear kernel. Due to no upweighting on rare variants, the power was mainly influenced by the common variants and has similar pattern as that in Figure 2.
Table 1.
Empirical type I error of quantitative traits using the linear weighted kernel on all variants in human genetic association.
| α | SKAT | CI_lower | CI_upper | aSKAT | CI_lower | CI_upper |
|---|---|---|---|---|---|---|
| n = 50 | ||||||
| 5.00E−02 | 4.13E−02 | 4.11E−02 | 4.14E−02 | 5.00E−02 | 4.99E−02 | 5.01E−02 |
| 1.00E−02 | 5.94E−03 | 5.89E−03 | 5.99E−03 | 9.97E−03 | 9.91E−03 | 1.00E−02 |
| 1.00E−03 | 2.89E−04 | 2.79E−04 | 3.00E−04 | 1.00E−03 | 9.85E−04 | 1.02E−03 |
| 1.00E−04 | 9.40E−06 | 7.40E−06 | 1.13E−05 | 9.95E−05 | 9.36E−05 | 1.06E−04 |
| 2.50E−06 | 1.00E−07 | 0.00E+00 | 3.00E−07 | 2.50E−06 | 1.60E−06 | 3.50E−06 |
| n = 100 | ||||||
| 5.00E−02 | 4.44E−02 | 4.43E−02 | 4.46E−02 | 5.00E−02 | 4.99E−02 | 5.02E−02 |
| 1.00E−02 | 7.51E−03 | 7.45E−03 | 7.56E−03 | 1.00E−02 | 9.94E−03 | 1.01E−02 |
| 1.00E−03 | 5.29E−04 | 5.14E−04 | 5.42E−04 | 1.02E−03 | 1.00E−03 | 1.04E−03 |
| 1.00E−04 | 3.68E−05 | 3.36E−05 | 4.05E−05 | 1.07E−04 | 9.99E−05 | 1.14E−04 |
| 2.50E−06 | 4.00E−07 | 1.00E−07 | 8.00E−07 | 3.90E−06 | 2.70E−06 | 5.30E−06 |
| n = 200 | ||||||
| 5.00E−02 | 4.69E−02 | 4.67E−02 | 4.70E−02 | 5.01E−02 | 5.00E−02 | 5.03E−02 |
| 1.00E−02 | 8.59E−03 | 8.53E−03 | 8.66E−03 | 1.00E−02 | 9.95E−03 | 1.01E−02 |
| 1.00E−03 | 7.24E−04 | 7.07E−04 | 7.41E−04 | 9.96E−04 | 9.76E−04 | 1.02E−03 |
| 1.00E−04 | 5.75E−05 | 5.25E−05 | 6.20E−05 | 9.78E−05 | 9.18E−05 | 1.04E−04 |
| 2.50E−06 | 6.00E−07 | 2.00E−07 | 1.20E−06 | 2.10E−06 | 1.30E−06 | 3.00E−06 |
The total sample size is denoted by n, α is the nominal significance level. 107 replicates are used. uSKAT does not use small sample adjustment for quantitative trait. aSKAT is the proposed adjusted SKAT. CI_lower and CI_upper are the lower and upper endpoint of the bootstrapped 95% confidence interval of the estimated type I error. Bold face indicates the confidence interval is above α.
Figure 1.
Power comparison of quantitative traits with rare causal variants in human genetic association studies. The linear weighted kernel was used. 20% of the rare variants (MAF < 3%) were causal. 80% of the causal variants were deleterious. Total sample sizes considered were 50, 100 and 200. Significance levels considered were 0.01, 0.001, 10−4 and 2.5×10−6. 103 replicates were used to estimate the power.
Table 2.
Empirical type I error of quantitative traits using the linear kernel on common variants with MAF > 0.05 in human genetic association.
| α | SKAT | CI_lower | CI_upper | aSKAT | CI_lower | CI_upper |
|---|---|---|---|---|---|---|
| n = 50 | ||||||
| 5.00E−02 | 4.76E−02 | 4.75E−02 | 4.77E−02 | 5.02E−02 | 5.00E−02 | 5.03E−02 |
| 1.00E−02 | 8.07E−03 | 8.01E−03 | 8.12E−03 | 1.00E−02 | 9.97E−03 | 1.01E−02 |
| 1.00E−03 | 5.13E−04 | 5.00E−04 | 5.27E−04 | 1.00E−03 | 9.80E−04 | 1.02E−03 |
| 1.00E−04 | 2.38E−05 | 2.07E−05 | 2.69E−05 | 1.04E−04 | 9.78E−05 | 1.11E−04 |
| 2.50E−06 | 3.00E−07 | 0.00E+00 | 7.00E−07 | 2.60E−06 | 1.70E−06 | 3.70E−06 |
| n = 100 | ||||||
| 5.00E−02 | 4.88E−02 | 4.87E−02 | 4.89E−02 | 5.00E−02 | 4.99E−02 | 5.01E−02 |
| 1.00E−02 | 9.08E−03 | 9.03E−03 | 9.14E−03 | 1.00E−02 | 9.96E−03 | 1.01E−02 |
| 1.00E−03 | 7.51E−04 | 7.34E−04 | 7.68E−04 | 1.00E−03 | 9.85E−04 | 1.02E−03 |
| 1.00E−04 | 5.83E−05 | 5.35E−05 | 6.31E−05 | 1.03E−04 | 9.69E−05 | 1.09E−04 |
| 2.50E−06 | 3.00E−07 | 0.00E+00 | 7.00E−07 | 2.40E−06 | 1.50E−06 | 3.40E−06 |
| n = 200 | ||||||
| 5.00E−02 | 4.95E−02 | 4.94E−02 | 4.96E−02 | 5.01E−02 | 4.99E−02 | 5.02E−02 |
| 1.00E−02 | 9.54E−03 | 9.48E−03 | 9.60E−03 | 1.00E−02 | 9.95E−03 | 1.01E−02 |
| 1.00E−03 | 8.68E−04 | 8.49E−04 | 8.86E−04 | 1.01E−03 | 9.87E−04 | 1.03E−03 |
| 1.00E−04 | 7.74E−05 | 7.19E−05 | 8.29E−05 | 1.01E−04 | 9.52E−05 | 1.07E−04 |
| 2.50E−06 | 1.00E−06 | 4.00E−07 | 1.60E−06 | 2.60E−06 | 1.70E−06 | 3.70E−06 |
The total sample size is denoted by n, α is the nominal significance level. 107 replicates are used. uSKAT does not use small sample adjustment for quantitative trait. aSKAT is the proposed adjusted SKAT. CI_lower and CI_upper are the lower and upper endpoint of the bootstrapped 95% confidence interval of the estimated type I error. Bold face indicates the confidence interval is above α.
Figure 2.
Power comparison of quantitative traits with one common causal variant in human genetic association studies. The linear kernel was used. The causal variant was randomly sampled with MAF > 0.05. Total sample sizes considered were 50, 100 and 200. Significance levels considered were 0.01, 0.001, 10−4 and 2.5×10−6. 103 replicates were used to estimate the power.
Type I Error and Power of Human Microbiome Association
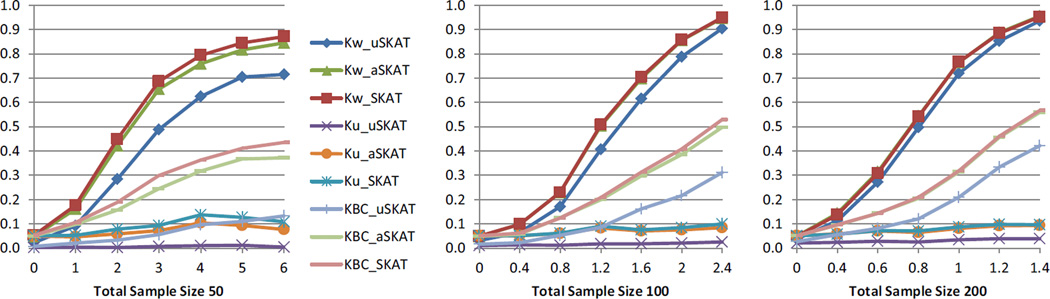
Table 3 shows the empirical type I error using different distance functions. aSKAT maintained the type I error at desired levels, and all empirical type I error fell into the 95% CI assuming the nominal level. uSKAT had conservative type I errors as expected, and in some cases very conservative. For example, when the total sample size was 50 and the nominal level was 10−3, the type I error of uSKAT using Kw distance was 10−5. Therefore we expected the power gain would be much larger in the microbiome genetic association than that in human genetic association. Figure 3 shows the power comparison using significance level 0.05. As expected, the power difference was very dramatic. For example, when the total sample size was 50 and the effect size was 1, the power of uSKAT was 0.382, and the power of aSKAT was 0.626, an increase of 0.244. Even for sample size of 200, the increase of power can be over 0.10 depending on different distance functions.
Table 3.
Empirical type I error of quantitative traits using different distance functions in microbiome genetic association.
| α | CI_lower | CI_upper | Kw_uSKAT | Kw_aSKAT | Ku_uSKAT | Ku_aSKAT | KBC_uSKAT | KBC_aSKAT |
|---|---|---|---|---|---|---|---|---|
| n = 50 | ||||||||
| 5.00E−02 | 4.86E−02 | 5.14E−02 | 1.39E−02 | 4.92E−02 | 6.90E−04 | 5.11E−02 | 2.71E−03 | 5.00E−02 |
| 1.00E−02 | 9.38E−03 | 1.06E−02 | 1.04E−03 | 9.90E−03 | 0.00E+00 | 1.02E−02 | 7.00E−05 | 9.91E−03 |
| 1.00E−03 | 8.04E−04 | 1.20E−03 | 1.00E−05 | 9.60E−04 | 0.00E+00 | 8.10E−04 | 0.00E+00 | 1.00E−03 |
| n = 100 | ||||||||
| 5.00E−02 | 4.86E−02 | 5.14E−02 | 2.53E−02 | 5.05E−02 | 6.09E−03 | 4.96E−02 | 1.18E−02 | 5.03E−02 |
| 1.00E−02 | 9.38E−03 | 1.06E−02 | 3.69E−03 | 1.03E−02 | 2.00E−04 | 9.87E−03 | 6.80E−04 | 1.05E−02 |
| 1.00E−03 | 8.04E−04 | 1.20E−03 | 1.70E−04 | 1.03E−03 | 0.00E+00 | 1.05E−03 | 1.00E−05 | 9.70E−04 |
| n = 200 | ||||||||
| 5.00E−02 | 4.86E−02 | 5.14E−02 | 3.34E−02 | 5.00E−02 | 1.59E−02 | 5.05E−02 | 2.17E−02 | 4.96E−02 |
| 1.00E−02 | 9.38E−03 | 1.06E−02 | 5.33E−03 | 1.01E−02 | 1.04E−03 | 9.69E−03 | 2.30E−03 | 1.03E−02 |
| 1.00E−03 | 8.04E−04 | 1.20E−03 | 5.60E−04 | 1.10E−03 | 3.00E−05 | 9.40E−04 | 1.00E−04 | 1.12E−03 |
The total sample size is denoted by n, α is the nominal significance level. CI_lower and CI_upper are the lower and upper endpoint of the 95% confidence interval assuming the true type I error is α based on 105 replicates. Kw denotes the kernel from the weighted phylogeny-based UniFrac distances, Ku denotes the kernel from the unweighted phylogeny-based UniFrac distances, and KBC denotes the kernel from the nonphylogeny-based Bray-Curtis distance. uSKAT does not use small sample adjustment for quantitative trait. aSKAT is the proposed adjusted SKAT
Figure 3.
Power comparison of quantitative traits in microbiome genetic association studies. Kw denotes the kernel from the weighted phylogeny-based UniFrac distances, Ku denotes the kernel from the unweighted phylogeny-based UniFrac distances, and KBC denotes the kernel from the nonphylogeny-based Bray-Curtis distance. Total sample sizes considered were 50, 100 and 200. Significance level was set to 0.05. 103 replicates were used to estimate the power.
In summary, for quantitative traits, aSKAT controled the type I error at desired levels due to its exactness. aSKAT had consistently higher power than uSKAT with similar computational cost. The increase of power was huge for microbiome association studies.
Results on Binary Traits
Type I Error and Power of Human Genetic Association
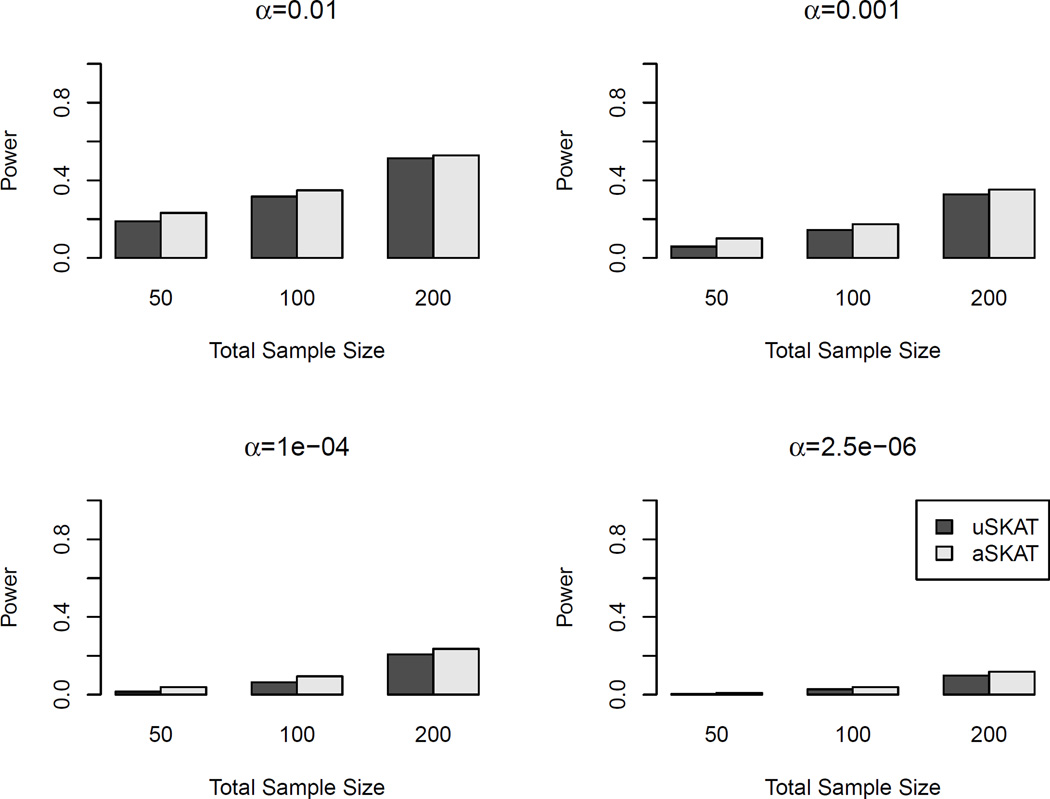
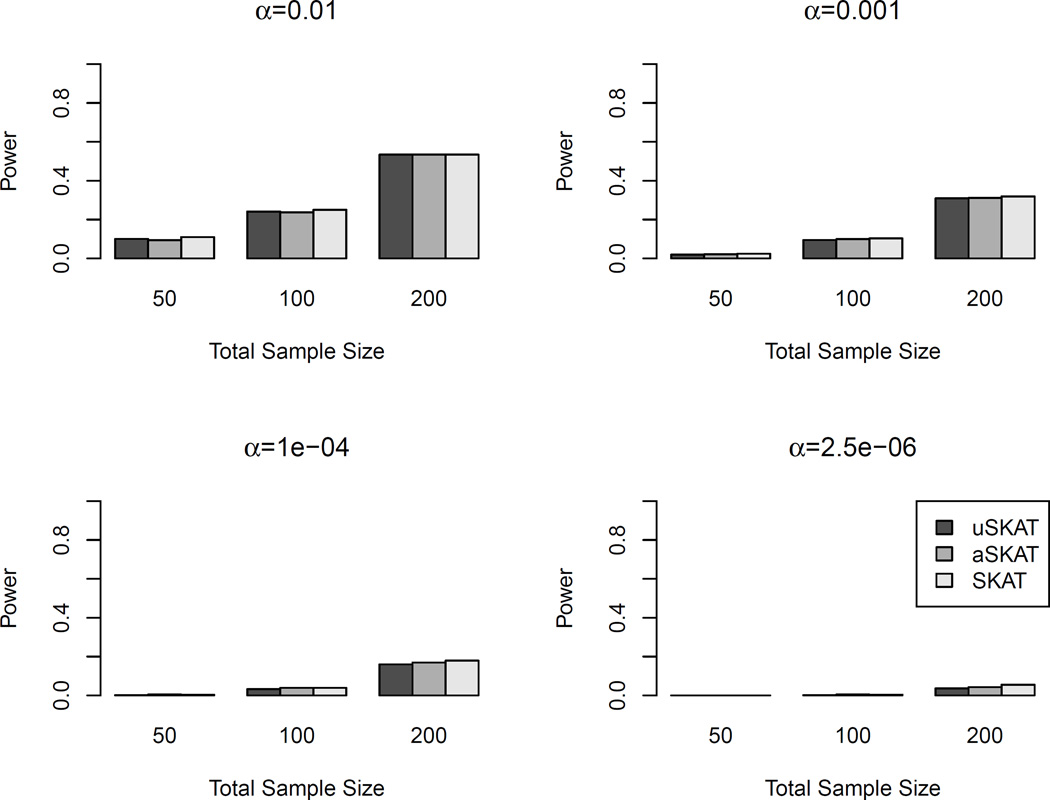
Table 4 shows the empirical type I error for binary traits on both common and rare variants using the linear weighted kernel. Because of the use of weights, the results were influenced mainly by rare variants. uSKAT and aSKAT were both conservative, but aSKAT was less conservative for α < 0.05. Due to the small-sample adjustment, SKAT was much less conservative than uSKAT and aSKAT. However, we found that when the nominal level was relatively large, e.g., 0.05 or 0.01, the empirical type I error of SKAT was slightly anti-conservative. For example, when the total sample size was 50 and the nominal level was 0.05, SKAT had type I error 0.065, estimated from 107 replicates. For α=0.05 and 0.01, all 95% CIs of SKAT were above the nominal level. Figure 4 shows the power comparison when the causal variants were rare. SKAT had the highest power in all scenarios. The increase of power can be over 0.10 than uSKAT and aSKAT. uSKAT and aSKAT had similar power, even though aSKAT usually had slightly higher power than uSKAT.
Table 4.
Empirical type I error of binary traits using the linear weighted kernel on all variants in human genetic association.
| α | uSKAT | CI_lower | CI_upper | aSKAT | CI_lower | CI_upper | SKAT | CI_lower | CI_upper |
|---|---|---|---|---|---|---|---|---|---|
| n = 50 | |||||||||
| 5.00E−02 | 1.61E−02 | 1.54E−02 | 1.66E−02 | 1.47E−02 | 1.42E−02 | 1.52E−02 | 6.46E−02 | 6.33E−02 | 6.59E−02 |
| 1.00E−02 | 1.08E−03 | 9.94E−04 | 1.16E−03 | 1.28E−03 | 1.19E−03 | 1.38E−03 | 1.26E−02 | 1.20E−02 | 1.31E−02 |
| 1.00E−03 | 5.42E−05 | 4.92E−05 | 5.90E−05 | 7.95E−05 | 7.22E−05 | 8.63E−05 | 9.38E−04 | 8.24E−04 | 1.06E−03 |
| 1.00E−04 | 9.90E−06 | 8.00E−06 | 1.18E−05 | 1.21E−05 | 9.90E−06 | 1.43E−05 | 7.90E−05 | 5.33E−05 | 1.15E−04 |
| 2.50E−06 | 2.60E−06 | 1.60E−06 | 3.70E−06 | 2.50E−06 | 1.60E−06 | 3.50E−06 | 1.10E−06 | 5.00E−07 | 1.80E−06 |
| n = 100 | |||||||||
| 5.00E−02 | 2.22E−02 | 2.12E−02 | 2.31E−02 | 2.16E−02 | 2.06E−02 | 2.25E−02 | 5.97E−02 | 5.79E−02 | 6.13E−02 |
| 1.00E−02 | 1.83E−03 | 1.61E−03 | 2.07E−03 | 2.07E−03 | 1.83E−03 | 2.32E−03 | 1.22E−02 | 1.15E−02 | 1.28E−02 |
| 1.00E−03 | 4.49E−05 | 2.48E−05 | 7.72E−05 | 6.85E−05 | 4.21E−05 | 1.11E−04 | 1.01E−03 | 8.63E−04 | 1.17E−03 |
| 1.00E−04 | 1.30E−06 | 7.00E−07 | 2.00E−06 | 1.70E−06 | 9.00E−07 | 2.80E−06 | 7.79E−05 | 5.07E−05 | 1.14E−04 |
| 2.50E−06 | 1.00E−07 | 0.00E+00 | 3.00E−07 | 1.00E−07 | 0.00E+00 | 3.00E−07 | 3.40E−06 | 1.00E−07 | 9.40E−06 |
| n = 200 | |||||||||
| 5.00E−02 | 3.31E−02 | 3.18E−02 | 3.46E−02 | 3.29E−02 | 3.16E−02 | 3.44E−02 | 5.59E−02 | 5.41E−02 | 5.78E−02 |
| 1.00E−02 | 3.83E−03 | 3.40E−03 | 4.25E−03 | 4.09E−03 | 3.64E−03 | 4.51E−03 | 1.13E−02 | 1.05E−02 | 1.20E−02 |
| 1.00E−03 | 1.09E−04 | 7.63E−05 | 1.51E−04 | 1.47E−04 | 1.06E−04 | 2.02E−04 | 1.03E−03 | 8.67E−04 | 1.21E−03 |
| 1.00E−04 | 1.30E−06 | 5.00E−07 | 2.40E−06 | 2.20E−06 | 1.10E−06 | 3.70E−06 | 7.50E−05 | 4.74E−05 | 1.13E−04 |
| 2.50E−06 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 2.70E−06 | 3.00E−07 | 7.00E−06 |
The total sample size is denoted by n, α is the nominal significance level. 107 replicates are used. uSKAT is the original SKAT without small sample size adjustment. aSKAT is the proposed adjusted SKAT. SKAT is the SKAT with small sample size adjustment using bootstrapping. CI_lower and CI_upper are the lower and upper endpoint of the bootstrapped 95% confidence interval of the estimated type I error. Bold face indicates the confidence interval is above α.
Figure 4.
Power comparison of binary traits with rare causal variants in human genetic association studies. The linear weighted kernel was used. 20% of the rare variants (MAF < 3%) were causal. 80% of the causal variants were deleterious. Total sample sizes considered were 50, 100 and 200. Significance levels considered were 0.01, 0.001, 10−4 and 2.5×10−6. 103 replicates were used to estimate the power.
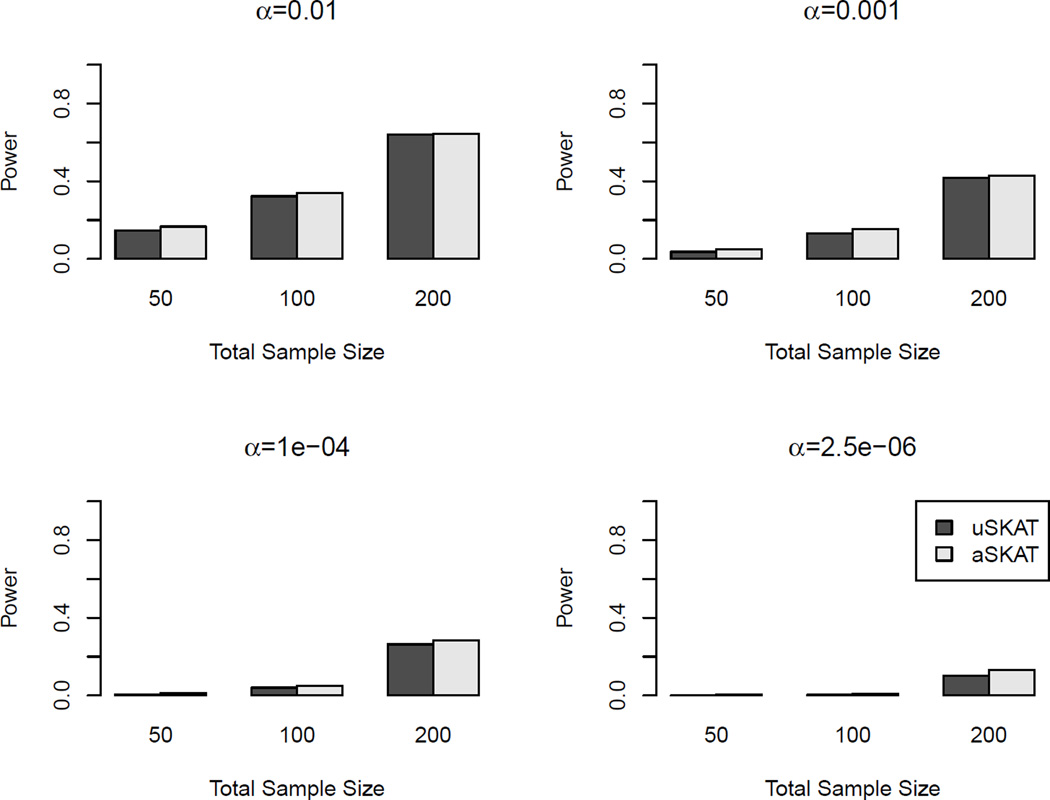
Table 5 shows the empirical type I error for common variants. The pattern was different from that of rare variants. When α ≥ 10−3, all methods showed no conservativeness, however, the 95% CIs for SKAT were all above the nominal level 0.05 for all three different sample sizes, which indicates inflated type I error. When α was 10−4 or lower, uSKAT was the most conservative, aSKAT was less conservative and usually had type I error close to that of SKAT. However, the differences of all three methods were not as large as in the scenario of rare variants. Figure 5 shows the power comparison when there was one common causal variant. All three methods had similar performance. SKAT had slightly higher power than aSKAT, and aSKAT usually had slightly higher power than uSKAT. Figure S4 in supplementary material shows the power comparison when there were three common causal variants and the pattern was similar. Figure S5 and S6 shows the power comparison when there were both rare and common causal variants using the weighted linear kernel and the linear kernel, respectively. Due to specific weighting on rare and common variants, the power pattern in Figure S5 is similar as in Figure 4, and the power pattern in Figure S6 is similar as in Figure 5.
Table 5.
Empirical type I error of binary traits using the linear kernel on common variants with MAF > 0.05 in human genetic association.
| α | uSKAT | CI_lower | CI_upper | aSKAT | CI_lower | CI_upper | SKAT | CI_lower | CI_upper |
|---|---|---|---|---|---|---|---|---|---|
| n = 50 | |||||||||
| 5.00E−02 | 5.71E−02 | 5.50E−02 | 5.91E−02 | 5.14E−02 | 4.95E−02 | 5.33E−02 | 5.90E−02 | 5.69E−02 | 6.11E−02 |
| 1.00E−02 | 1.12E−02 | 1.04E−02 | 1.22E−02 | 1.08E−02 | 1.00E−02 | 1.17E−02 | 1.34E−02 | 1.24E−02 | 1.43E−02 |
| 1.00E−03 | 9.14E−04 | 7.21E−04 | 1.15E−03 | 1.20E−03 | 9.79E−04 | 1.47E−03 | 1.53E−03 | 1.27E−03 | 1.82E−03 |
| 1.00E−04 | 3.17E−05 | 1.57E−05 | 5.51E−05 | 9.88E−05 | 5.86E−05 | 1.57E−04 | 9.93E−05 | 5.82E−05 | 1.58E−04 |
| 2.50E−06 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 1.30E−06 | 3.00E−07 | 2.70E−06 | 5.00E−07 | 1.00E−07 | 1.10E−06 |
| n = 100 | |||||||||
| 5.00E−02 | 5.18E−02 | 5.00E−02 | 5.37E−02 | 4.90E−02 | 4.71E−02 | 5.08E−02 | 5.22E−02 | 5.03E−02 | 5.41E−02 |
| 1.00E−02 | 9.68E−03 | 8.91E−03 | 1.05E−02 | 9.43E−03 | 8.68E−03 | 1.02E−02 | 1.06E−02 | 9.76E−03 | 1.14E−02 |
| 1.00E−03 | 8.40E−04 | 6.21E−04 | 1.08E−03 | 9.26E−04 | 7.01E−04 | 1.18E−03 | 1.09E−03 | 8.47E−04 | 1.36E−03 |
| 1.00E−04 | 7.15E−05 | 3.92E−05 | 1.13E−04 | 1.14E−04 | 6.39E−05 | 1.78E−04 | 1.39E−04 | 8.05E−05 | 2.16E−04 |
| 2.50E−06 | 1.00E−07 | 0.00E+00 | 4.00E−07 | 6.00E−07 | 2.00E−07 | 1.20E−06 | 6.00E−07 | 1.00E−07 | 1.40E−06 |
| n = 200 | |||||||||
| 5.00E−02 | 5.25E−02 | 5.05E−02 | 5.46E−02 | 5.11E−02 | 4.91E−02 | 5.31E−02 | 5.21E−02 | 5.01E−02 | 5.42E−02 |
| 1.00E−02 | 1.09E−02 | 1.01E−02 | 1.18E−02 | 1.07E−02 | 9.93E−03 | 1.16E−02 | 1.13E−02 | 1.05E−02 | 1.22E−02 |
| 1.00E−03 | 8.42E−04 | 6.63E−04 | 1.06E−03 | 8.91E−04 | 7.06E−04 | 1.11E−03 | 1.03E−03 | 8.28E−04 | 1.27E−03 |
| 1.00E−04 | 3.85E−05 | 1.94E−05 | 6.60E−05 | 4.84E−05 | 2.48E−05 | 8.23E−05 | 6.17E−05 | 3.33E−05 | 1.01E−04 |
| 2.50E−06 | 1.00E−07 | 0.00E+00 | 3.00E−07 | 2.00E−07 | 0.00E+00 | 6.00E−07 | 3.00E−07 | 0.00E+00 | 1.00E−06 |
The total sample size is denoted by n, α is the nominal significance level. 107 replicates are used. uSKAT is the original SKAT without small sample size adjustment. aSKAT is the proposed adjusted SKAT. SKAT is the SKAT with small sample size adjustment using bootstrapping. CI_lower and CI_upper are the lower and upper endpoint of the bootstrapped 95% confidence interval of the estimated type I error. Bold face indicates the confidence interval is above α.
Figure 5.
Power comparison of binary traits with one common causal variant in human genetic association studies. The linear kernel was used. The causal variant was randomly sampled with MAF > 0.05. Total sample sizes considered were 50, 100 and 200. Significance levels considered were 0.01, 0.001, 10−4 and 2.5×10−6. 103 replicates were used to estimate the power.
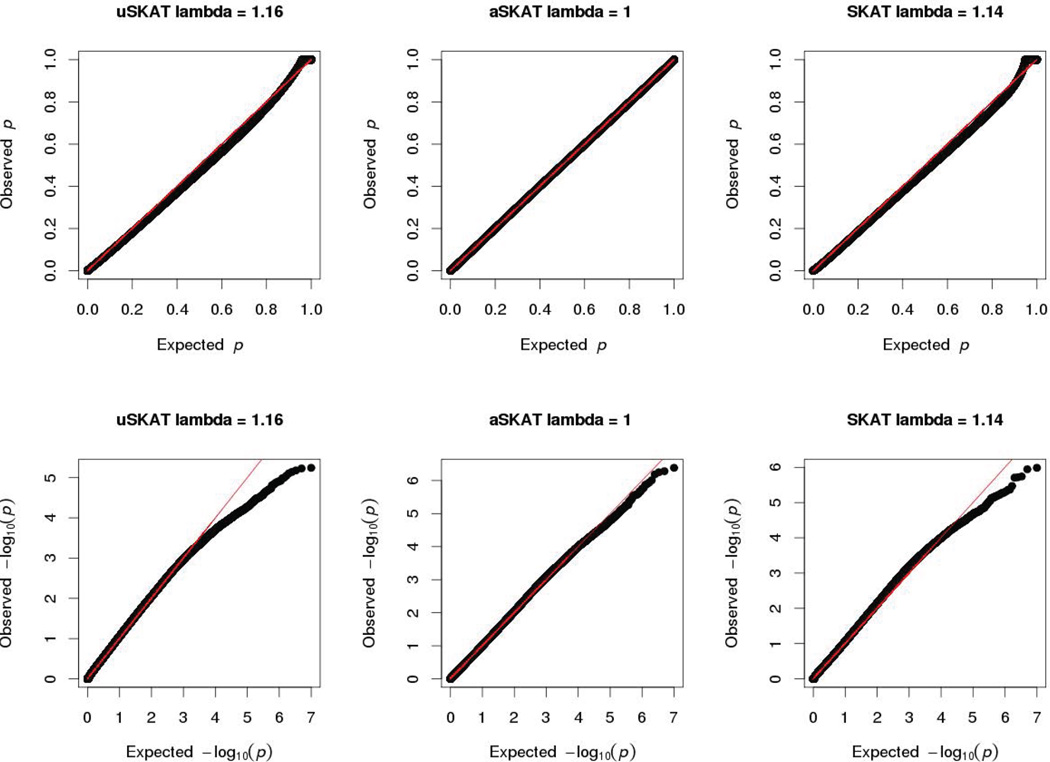
For common variants association, though the power difference is modest between methods, an advantage of using aSKAT is that under the null hypothesis, its QQ-plot behaves better than uSKAT and SKAT when the sample size is small. Figure 6 shows the QQ-plot where the total sample size is 50 for binary traits. On the original scale, when the p-values were large, uSKAT and SKAT had smaller than expected p-values. On the log10 scale, SKAT had larger deviation from the expected values in the middle than aSKAT. To assess the deviation, we adapted the concept of inflation indicator. Specifically, we first converted these p-values to test statistics assuming a χ2(1) distribution, then we took the median test statistic and divided it by 0.455, which is the theoretic median of χ2(1). If the median of these p-values is close to 0.5, the inflation indicator should be close to 1. The inflation indicator λ was 1.16 for uSKAT, 1.14 for SKAT and 1 for aSKAT. Therefore for common variants, when the sample size is small, QQ-plot of aSKAT behaves as expected, while uSKAT and SKAT can produce misleading QQ-plots.
Figure 6.
QQ-plots of different methods on binary traits and common variants. The total sample size was 50. The x- axis and y-axis of the upper 3 plots were on original scale. The x- axis and y-axis of the lower 3 plots were on log10 scale. The inflation indicator lambda was calculated for each method.
Type I Error and Power of Human Microbiome Association
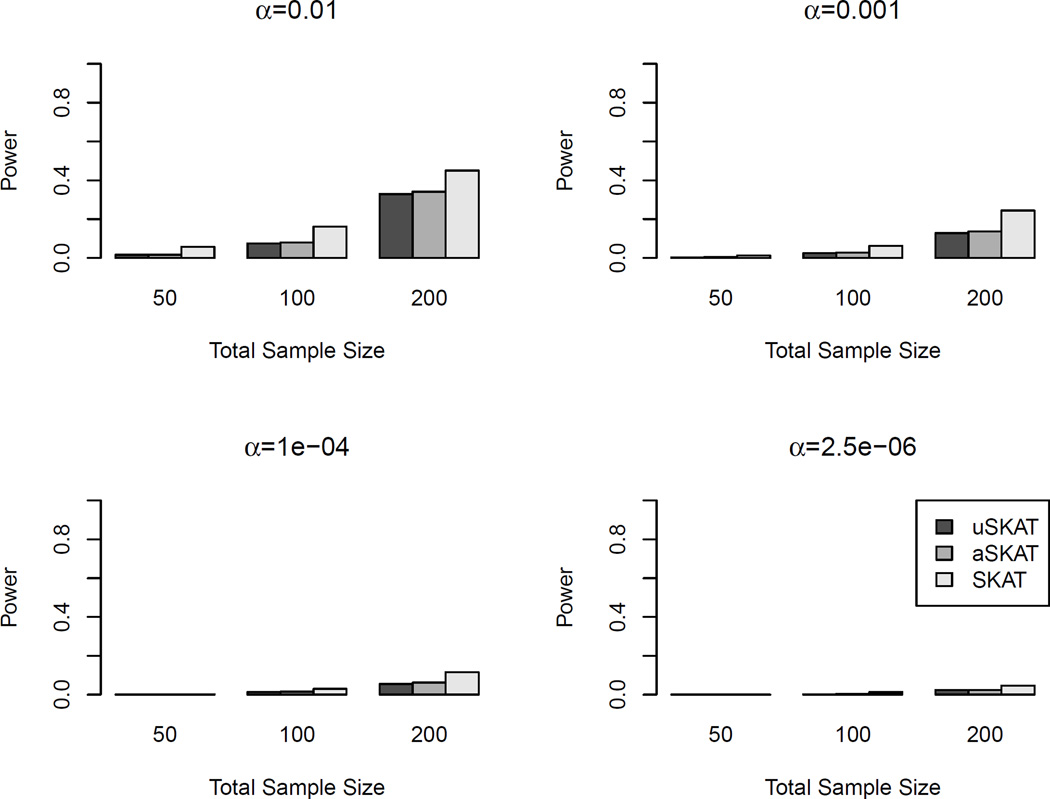
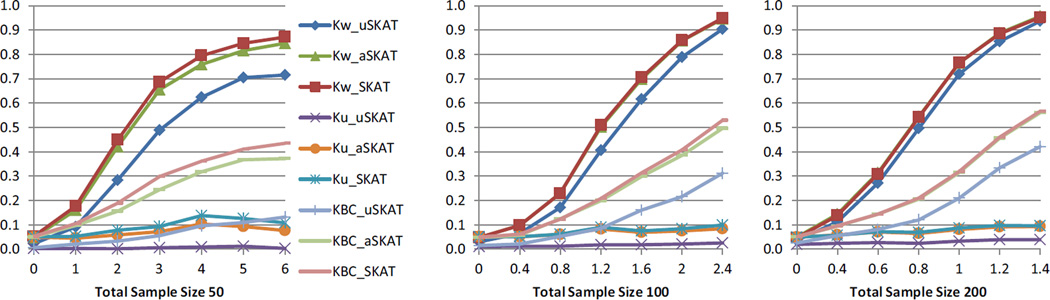
Table 6 shows the empirical type I error using different distance functions. The pattern was more similar to the scenario of common variants in human genetic association. aSKAT had type I errors close to the nominal levels, though sometimes slightly conservative. For SKAT, there were several empirical type I errors above the desired 95% CIs. uSKAT was more conservative than aSKAT and SKAT. Figure 7 shows the power comparison. When the total sample size was 100 and 200, aSKAT and SKAT almost overlapped with each other. Both showed much higher power than uSKAT. When the total sample size was 50, SKAT had slightly higher power than aSKAT when the effect size was large. The increase of power was very modest (0.03 or 0.04). Both SKAT and aSKAT had much larger power than uSKAT, where the increase of power can be over 0.20.
Table 6.
Empirical type I error of binary traits using different distance functions in microbiome genetic association.
| α | CI_lower | CI_upper | Kw_uSKAT | Kw_aSKAT | Kw_SKAT | Ku_uSKAT | Ku_aSKAT | Ku_SKAT | KBC_uSKAT | KBC_aSKAT | KBC_SKAT |
|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 50 | |||||||||||
| 5.00E−02 | 4.86E−02 | 5.14E−02 | 2.25E−02 | 4.86E−02 | 5.29E−02 | 3.01E−03 | 4.98E−02 | 5.11E−02 | 6.94E−03 | 5.02E−02 | 5.31E−02 |
| 1.00E−02 | 9.38E−03 | 1.06E−02 | 2.00E−03 | 9.97E−03 | 9.91E−03 | 1.00E−05 | 1.03E−02 | 8.29E−03 | 1.90E−04 | 9.73E−03 | 8.48E−03 |
| 1.00E−03 | 8.04E−04 | 1.20E−03 | 1.00E−04 | 1.02E−03 | 1.02E−03 | 0.00E+00 | 1.17E−03 | 8.30E−04 | 0.00E+00 | 1.03E−03 | 6.90E−04 |
| n = 100 | |||||||||||
| 5.00E−02 | 4.86E−02 | 5.14E−02 | 3.09E−02 | 4.86E−02 | 4.90E−02 | 1.07E−02 | 5.13E−02 | 5.18E−02 | 1.62E−02 | 4.90E−02 | 5.04E−02 |
| 1.00E−02 | 9.38E−03 | 1.06E−02 | 4.03E−03 | 9.12E−03 | 8.97E−03 | 4.60E−04 | 1.04E−02 | 8.84E−03 | 1.33E−03 | 9.60E−03 | 8.77E−03 |
| 1.00E−03 | 8.04E−04 | 1.20E−03 | 2.00E−04 | 8.50E−04 | 9.60E−04 | 0.00E+00 | 9.10E−04 | 7.80E−04 | 3.00E−05 | 1.07E−03 | 8.90E−04 |
| n = 200 | |||||||||||
| 5.00E−02 | 4.86E−02 | 5.14E−02 | 3.71E−02 | 4.94E−02 | 4.74E−02 | 2.07E−02 | 5.05E−02 | 5.06E−02 | 2.62E−02 | 5.07E−02 | 5.07E−02 |
| 1.00E−02 | 9.38E−03 | 1.06E−02 | 6.04E−03 | 9.70E−03 | 9.00E−03 | 1.50E−03 | 1.00E−02 | 9.18E−03 | 2.86E−03 | 1.04E−02 | 9.61E−03 |
| 1.00E−03 | 8.04E−04 | 1.20E−03 | 5.50E−04 | 1.04E−03 | 1.13E−03 | 0.00E+00 | 9.40E−04 | 7.80E−04 | 1.10E−04 | 1.00E−03 | 9.40E−04 |
The total sample size is denoted by n, α is the nominal significance level. CI_lower and CI_upper are the lower and upper endpoint of the 95% confidence interval assuming the true type I error is α based on 105 replicates. Kw denotes the kernel from the weighted phylogeny-based UniFrac distances, Ku denotes the kernel from the unweighted phylogeny-based UniFrac distances, and KBC denotes the kernel from the nonphylogeny-based Bray-Curtis distance. uSKAT is the original SKAT without small sample size adjustment. aSKAT is the proposed adjusted SKAT. SKAT is the SKAT with small sample size adjustment using bootstrapping.
Figure 7.
Power comparison of binary traits in microbiome genetic association studies. Kw denotes the kernel from the weighted phylogeny-based UniFrac distances, Ku denotes the kernel from the unweighted phylogeny-based UniFrac distances, and KBC denotes the kernel from the nonphylogeny-based Bray-Curtis distance. Total sample sizes considered were 50, 100 and 200. Significance level was set to 0.05. 103 replicates were used to estimate the power.
In summary, for rare variants using the linear weighted kernel, SKAT had much higher power than both uSKAT and aSKAT. However, for common variants in human genetic association, as well as microbiome association, aSKAT had power similar or close to SKAT with a huge reduction in computational cost. Compared to uSKAT, being similar in the computational cost, aSKAT had the power increase most obvious in the application of microbiome genetic association.
Type I Error and Power when the model is misspecified
We studied the influence on type I error and power when the model assumptions were violated. All simulations were the same as in the microbiome association with quantitative traits except the following three types of misspecifications. Firstly, we replaced the normal error term with a heavy-tailed t distribution with degree of freedom (df) 1 and 5. The type I errors were reported in Table S1 (supplementary information). All methods showed no inflation on type I error even though all of them assume a normal distribution for the error term. However, the power was influenced by the misspecification as shown in Figure S7 (supplementary information). When the true distribution (t distribution with 1 df) was far from normal, all methods had very low power. When the true distribution (t distribution with 5 df) was closer to normal, the power pattern was similar to that using a normal error term, and aSKAT achieved higher power than uSKAT.
Secondly, we simulated nonlinear covariate effect. Specifically, the true model used was
while only the linear terms were modeled in testing. The type I errors were reported in Table S2 (supplementary information), where all methods showed no inflation of type I error. The power was shown in Figure S8 (supplementary information), where aSKAT still achieved higher power than uSKAT. If there are strong nonlinear effects from covariates, we anticipate that all methods will have low power, similar as that in the first type of misspecification.
Thirdly, we simulated an interaction effect between the covariate and the genetic components. Specifically, the true model used was
where the interaction term was not included in the model for testing. The power was shown in Figure S9, where aSKAT achieved higher power than uSKAT.
In summary, when the model assumptions were slightly violated as shown above, aSKAT achieved higher power than uSKAT while controlling the type I error; when the true model was very different from the assumption, all methods had low power. This illustrates the importance of careful model selection and diagnostics.
Results on Real Data
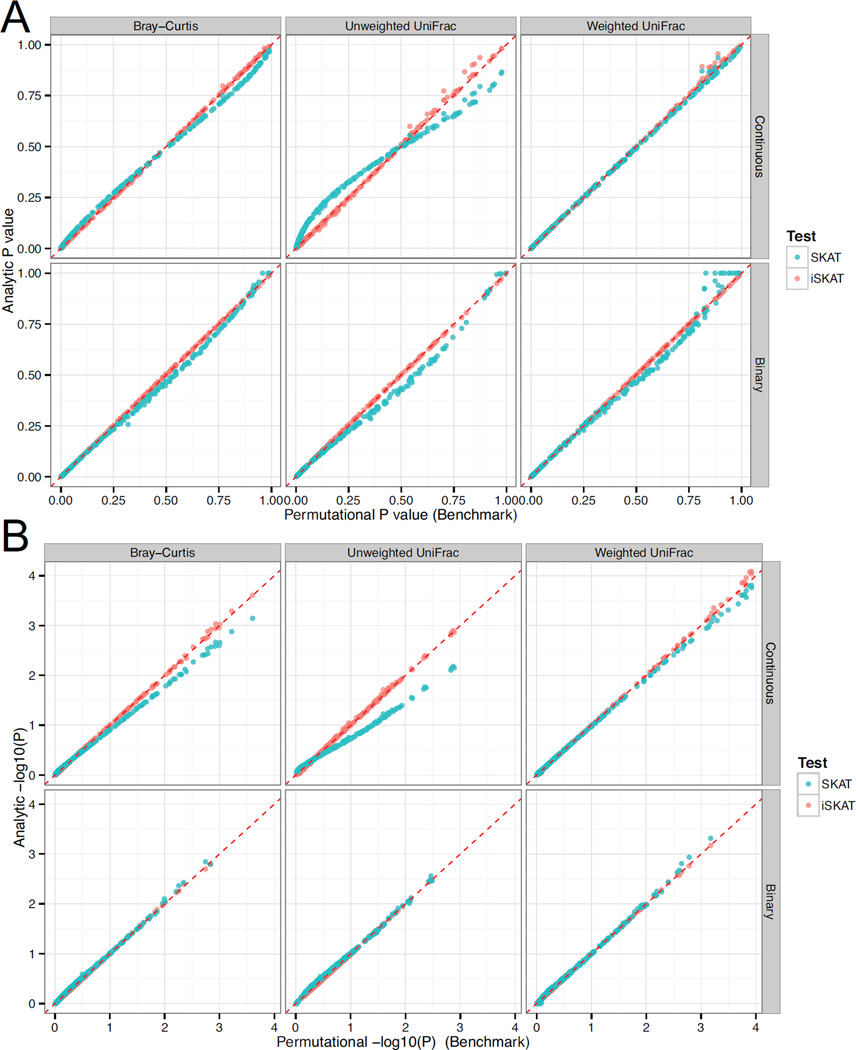
To demonstrate the small-sample performance of our proposed method, we applied our method to a microbiome data set from a study of long-term dietary effects on the human gut microbiome [Wu, et al. 2011a]. One goal of the study was to identify nutrients associated with the gut microbiome composition. In this study, both gut microbiome data and nutrient intake data were available for 98 healthy subjects. The gut microbiome data was produced by sequencing the V1–V2 region of the bacterial 16S rRNA gene using 454/Roche Titanium technology after the bacterial DNA was extracted from the stool and PCR amplified. Multiplexed 454 pyrosequencing generated about 900,000 high quality, partial (~ 370 bp) 16S rRNA gene sequences. These sequences were analyzed using the Qiime pipeline [Caporaso, et al. 2010], where the sequences were clustered at 97% sequence identity into OTUs. Each OTU was represented by a DNA sequence, which was used to build a phylogenetic tree. Based on the OTU abundance data and the phylogenetic tree, we calculated the three most used distance metrics for microbiome data: the nonphylogeny-based Bray-Curtis distance and the phylogeny-based UniFrac distances (unweighted and weighted). The study also measured the intake amounts of 214 nutrients, calculated based on a carefully designed food frequency questionnaire. The nutrient data was further standardized against total caloric intake by regressing the nutrient intake on the total caloric intake and taking residuals. We converted the distance matrices into kernel matrices using the same method from [Zhao, et al. 2015] and conducted kernel-based association tests for these 214 nutrients separately. We compared aSKAT and SKAT to a permutation method (105 permutations), where we permuted the rows (and corresponding columns) of the kernel matrix to get a permutation p-value based on the score statistic. The permutation p-values were used as benchmarks. The original nutrient intake values were continuous and, to demonstrate the performance of aSKAT on binary traits, we also discretized the nutrient intake data into “Low” and “High” intake using the median value as the cutoff. The p-values from aSKAT agreed perfectly with the permutation p-values in both large and small p-value regions across the three distance-based kernels, see Figure 8. For the continuous traits, aSKAT produced “exact” p-values and we had expected perfect correlation with the permutation p-values under any type of kernel. For binary traits, however, the aSKAT p-value is approximate and the performance depends on the type of kernel. Consistent with the simulation results, binary-case aSKAT performed adequately for microbiome-based kernels in this application. In contrast, the continuous-case SKAT, which does not take into account the estimation uncertainty of the error variance, was slightly anti-conservative in large p-value region and conservative in small p-value region. The problem was most serious for unweighted UniFrac-based kernel, indicating potential power loss for SKAT under certain scenarios. For the binary-case SKAT, which uses a small-sample correction strategy, the SKAT p-values agreed well with the permutation-based p-values in the tail region while a small inflation was observed in the non-tail region. In computational aspect, aSKAT was much faster and completed the analysis in less than 1 second on a desktop computer since it does not reply on permutation or bootstrapping. On the other hand, SKAT, which employs computationally intensive bootstrapping to estimate the moments, took around 300 seconds to complete the analysis since it needed to fit the null model for each nutrient. In summary, for the continuous traits, aSKAT produced “exact” p-values and was more powerful than SKAT for small-sample association studies. For the binary traits, aSKAT was more computationally efficient and had comparable performance to SKAT for microbiome association studies.
Figure 8.
Comparison of SKAT and aSKAT on a real microbiome association study of nutrient intake and the gut microbiome composition. The nutrient data were discretized into binary data to illustrate the performance on binary traits. The original scale p-values were plotted in (A) and the log scale p-values were plotted in (B). In each subfigure, the upper panel shows the result on continuous traits, and the lower panel shows the result on binary traits.
Discussion
We proposed two improved kernel association tests based on SKAT. For quantitative traits, it is an exact test and completely solves the small-sample problem of SKAT. The increase of power was more dramatic for microbiome association studies, though the increase of power in human genetic association studies was limited. The exact test works on any type of kernel and thus is recommended for all kernel-based association tests for quantitative traits.
For binary traits, the proposed test is approximate and the association p-values are calculated analytically without using any resampling method. It worked much better than the original SKAT without small-sample adjustment in terms of both type I error control and power for all scenarios considered in the simulations. Interestingly, it also had similar results as SKAT with adjustment for common variant- or microbiome-based kernels, though no resampling was performed to adjust for small sample sizes. The computation speed was therefore much faster than SKAT with small-sample adjustment. For rare variants, when the sample size was small, SKAT with adjustment still had the best results, even though it was more time-consuming. However, in our simulations we found that the result of SKAT with adjustment could be anti-conservative when the nominal significance level was relatively large, e.g., 0.05. This might be due to the fact that the small-sample adjustment was focused on the tail region at the slight sacrifice of the accuracy in the non-tail region. The consequence was the slight inflation of the genomic inflation factor, which is typically used to assess the potential confounding in genetic association studies such as population stratification. Therefore, users have to be aware of this artifact when performing sequence-based genetic association studies using SKAT. Further research on binary traits and rare variants in the small-sample setting may be needed.
In principle, the proposed method can be extended to other SKAT-based tests since it does not require any specific kernels. For example, we can apply the proposed method to SKAT-O, where the only change is the kernel matrix [Lee, et al. 2012]. Another example is the combined test of both rare and common variants as implemented in SKAT-C, which is based on a different type of kernel matrix [Ionita-Laza, et al. 2013]. The method can also be extended to testing the global gene/environment effect in presence of GxE interaction. For example, we can use a sum of three kernels, one for the genotypes, one for the specific environment covariates and one for the GxE effects [Wang, et al. 2015]. The null hypothesis for the global test is that there is no effect for the genetic component, no effect for the specific environment covariates and no effect for the interaction between the genetic component and the specific environment covariates. Because the sum of the three kernels can be viewed as a new kernel, the extension of our method to this global test is straightforward. However, extension to testing GxE effects directly conditioning on genetic and environmental effects may need further investigation.
We anticipate many other types of kernel association tests may benefit from our method. Association tests between a phenotype and gene expression from a gene pathway or CpG methylation in a gene region are both potential applications of our methods. These data are more like the common variants scenario in human genetic association and we expect our method will work well, even for binary traits.
Supplementary Material
Acknowledgements
This research was supported by the Mayo Clinic Gerstner Family Career Development Award in Individualized Medicine, and the U.S. Public Health Service, National Institutes of Health, contract grant number GM065450.
References
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlson ES, Chen J, Custers-Allen R, Bittinger K, Li H, Sinha R, Hwang J, Bushman FD, Collman RG. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PloS one. 2010;5(12):e15216. doi: 10.1371/journal.pone.0015216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bittinger K, Charlson ES, Hoffmann C, Lewis J, Wu GD, Collman RG, Bushman FD, Li H. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics. 2012;28(16):2106–2113. doi: 10.1093/bioinformatics/bts342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li H. Kernel Methods for Regression Analysis of Microbiome Compositional Data. In: Hu M, Liu Y, Lin J, editors. Topics in Applied Statistics. New York: Springer; 2013. pp. 191–201. [Google Scholar]
- Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nature reviews. Genetics. 2012;13(4):260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies RB. Algorithm AS 155: The Distribution of a Linear Combination of χ2 Random Variables. Journal of the Royal Statistical Society. Series C (Applied Statistics) 1980;29(3):323–333. [Google Scholar]
- Duchesne P, De Micheaux PL. Computing the distribution of quadratic forms: Further comparisons between the Liu-Tang-Zhang approximation and exact methods. Computational Statistics & Data Analysis. 2010;54(4):858–862. [Google Scholar]
- Emond MJ, Louie T, Emerson J, Zhao W, Mathias RA, Knowles MR, Wright FA, Rieder MJ, Tabor HK, Nickerson DA, et al. Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nature genetics. 2012;44(8):886–889. doi: 10.1038/ng.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, Lin X. Sequence kernel association tests for the combined effect of rare and common variants. American journal of human genetics. 2013;92(6):841–853. doi: 10.1016/j.ajhg.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwee LC, Liu D, Lin X, Ghosh D, Epstein MP. A powerful and flexible multilocus association test for quantitative traits. American journal of human genetics. 2008;82(2):386–397. doi: 10.1016/j.ajhg.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, Christiani DC, Wurfel MM, Lin X. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. American journal of human genetics. 2012;91(2):224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Ghosh D, Lin X. Estimation and testing for the effect of a genetic pathway on a disease outcome using logistic kernel machine regression via logistic mixed models. BMC bioinformatics. 2008;9:292. doi: 10.1186/1471-2105-9-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Lin X, Ghosh D. Semiparametric regression of multidimensional genetic pathway data: least-squares kernel machines and linear mixed models. Biometrics. 2007;63(4):1079–1088. doi: 10.1111/j.1541-0420.2007.00799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nature reviews. Genetics. 2008;9(5):356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- McCullagh P, Nelder JA. Generalized Linear Models. Chapman and Hall/CRC; 1989. [Google Scholar]
- Reynolds AP, Richards G, de la Iglesia B, Rayward-Smith VJ. Clustering Rules: A Comparison of Partitioning and Hierarchical Clustering Algorithms. Journal of Mathematical Modelling and Algorithms. 2006;5(4):475–504. [Google Scholar]
- Schaffner SF, Foo C, Gabriel S, Reich D, Daly MJ, Altshuler D. Calibrating a coalescent simulation of human genome sequence variation. Genome research. 2005;15(11):1576–1583. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Maity A, Luo Y, Neely ML, Tzeng JY. Complete effect-profile assessment in association studies with multiple genetic and multiple environmental factors. Genetic epidemiology. 2015;39(2):122–133. doi: 10.1002/gepi.21877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011a;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Kraft P, Epstein MP, Taylor DM, Chanock SJ, Hunter DJ, Lin X. Powerful SNP-set analysis for case-control genome-wide association studies. American journal of human genetics. 2010;86(6):929–942. doi: 10.1016/j.ajhg.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. American journal of human genetics. 2011b;89(1):82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Chen J, Carroll IM, Ringel-Kulka T, Epstein MP, Zhou H, Zhou JJ, Ringel Y, Li H, Wu MC. Testing in Microbiome-Profiling Studies with MiRKAT, the Microbiome Regression-Based Kernel Association Test. American journal of human genetics. 2015;96(5):797–807. doi: 10.1016/j.ajhg.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.