Abstract
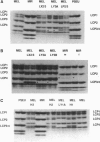
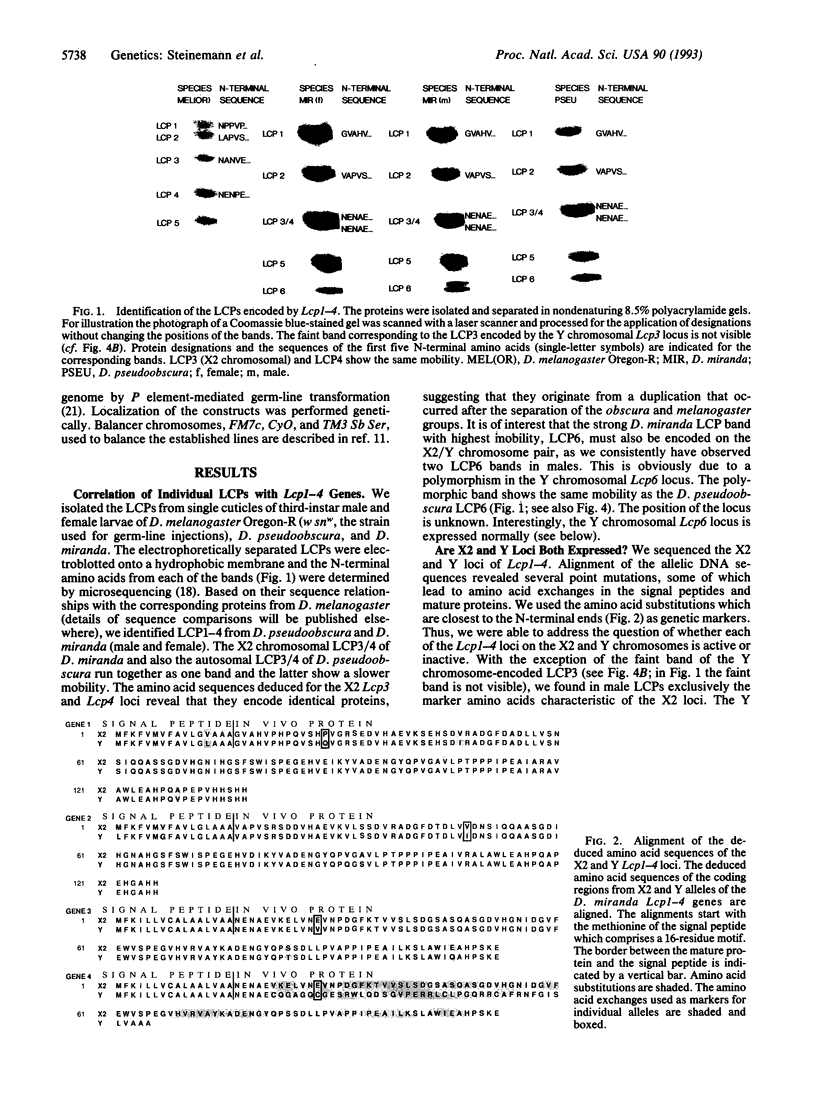
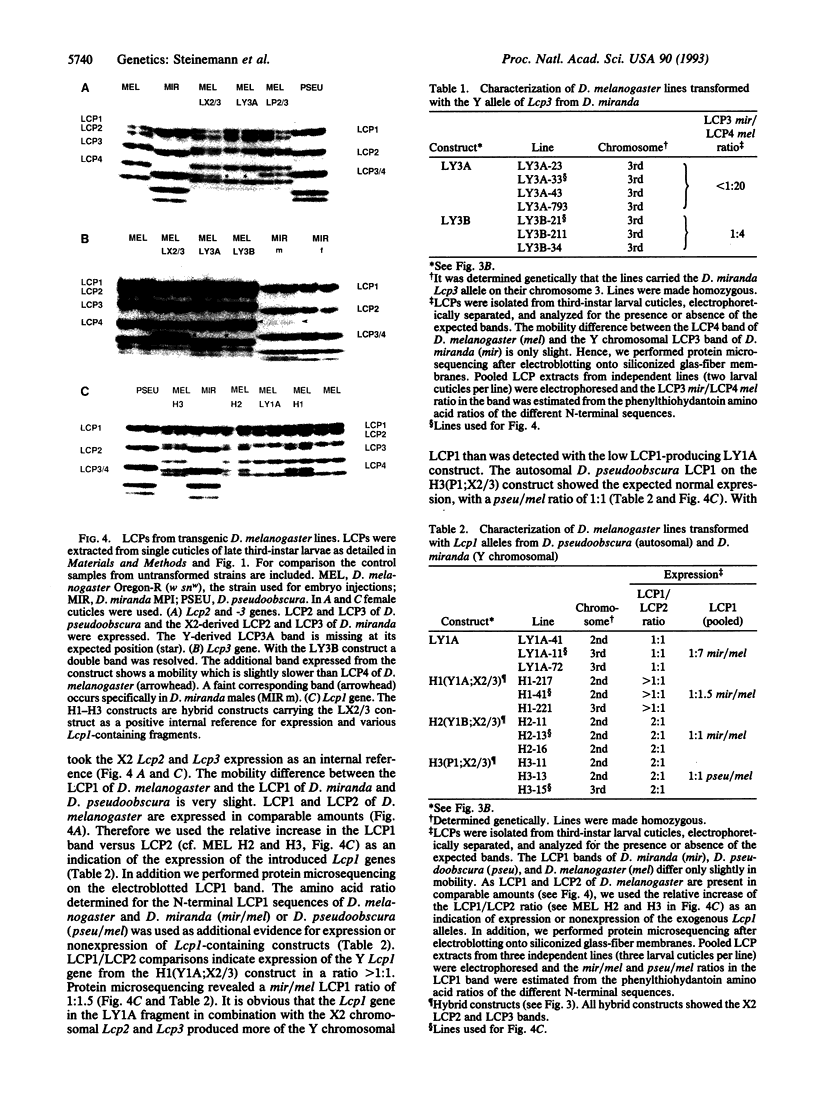
We have investigated the mechanistic aspects of inactivation of the major larval cuticle protein genes (Lcp1-4) in Drosophila miranda during Y chromosome evolution. The Lcp genes are located on the X2 and neo-Y chromosomes in D. miranda but are autosomally inherited in all other Drosophila species investigated so far. In the neo-Y chromosome all four Lcp loci are embedded within a dense cluster of transposable elements. The X2 Lcp1-4 loci are expressed, while the Y chromosomal Lcp3 locus shows only reduced activity and the Lcp1, Lcp2, and Lcp4 are completely inactive. Our results suggest that Lcp1 and Lcp3 loci on the degenerating Y chromosome of D. miranda are silenced by neighboring transposable elements. These observations support our assumption that the first step in Y chromosome degeneration is the successive silencing of Y chromosomal loci caused by trapping and accumulation of transposons.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cavener D. R. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987 Feb 25;15(4):1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. The evolution of sex chromosomes. Science. 1991 Mar 1;251(4997):1030–1033. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Drosophila Miranda, a New Species. Genetics. 1935 Jul;20(4):377–391. doi: 10.1093/genetics/20.4.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerskorn C., Mewes W., Goretzki H., Lottspeich F. A new siliconized-glass fiber as support for protein-chemical analysis of electroblotted proteins. Eur J Biochem. 1988 Oct 1;176(3):509–519. doi: 10.1111/j.1432-1033.1988.tb14308.x. [DOI] [PubMed] [Google Scholar]
- Fristrom J. W., Hill R. J., Watt F. The procuticle of Drosophila: heterogeneity of urea-soluble proteins. Biochemistry. 1978 Sep 19;17(19):3917–3930. doi: 10.1021/bi00612a005. [DOI] [PubMed] [Google Scholar]
- Ganguly R., Swanson K. D., Ray K., Krishnan R. A BamHI repeat element is predominantly associated with the degenerating neo-Y chromosome of Drosophila miranda but absent in the Drosophila melanogaster genome. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1340–1344. doi: 10.1073/pnas.89.4.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. M. Transposable elements in Drosophila and other Diptera. Annu Rev Genet. 1980;14:109–120. doi: 10.1146/annurev.ge.14.120180.000545. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Position-effect variegation after 60 years. Trends Genet. 1990 Dec;6(12):422–426. doi: 10.1016/0168-9525(90)90304-o. [DOI] [PubMed] [Google Scholar]
- James T. C., Elgin S. C. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986 Nov;6(11):3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R., Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991 Mar 8;64(5):941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- Kostriken R., Strathern J. N., Klar A. J., Hicks J. B., Heffron F. A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell. 1983 Nov;35(1):167–174. doi: 10.1016/0092-8674(83)90219-2. [DOI] [PubMed] [Google Scholar]
- Langley C. H., Montgomery E., Hudson R., Kaplan N., Charlesworth B. On the role of unequal exchange in the containment of transposable element copy number. Genet Res. 1988 Dec;52(3):223–235. doi: 10.1017/s0016672300027695. [DOI] [PubMed] [Google Scholar]
- Laski F. A., Rio D. C., Rubin G. M. Tissue specificity of Drosophila P element transposition is regulated at the level of mRNA splicing. Cell. 1986 Jan 17;44(1):7–19. doi: 10.1016/0092-8674(86)90480-0. [DOI] [PubMed] [Google Scholar]
- Macknight R H. The Sex-Determining Mechanism of Drosophila Miranda. Genetics. 1939 Mar;24(2):180–201. doi: 10.1093/genetics/24.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A. The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell. 1982 Sep;30(2):567–578. doi: 10.1016/0092-8674(82)90253-7. [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C. Genetic transformation of Drosophila with transposable element vectors. Science. 1982 Oct 22;218(4570):348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Snyder M., Hirsh J., Davidson N. The cuticle genes of drosophila: a developmentally regulated gene cluster. Cell. 1981 Jul;25(1):165–177. doi: 10.1016/0092-8674(81)90241-5. [DOI] [PubMed] [Google Scholar]
- Snyder M., Hunkapiller M., Yuen D., Silvert D., Fristrom J., Davidson N. Cuticle protein genes of Drosophila: structure, organization and evolution of four clustered genes. Cell. 1982 Jul;29(3):1027–1040. doi: 10.1016/0092-8674(82)90466-4. [DOI] [PubMed] [Google Scholar]
- Steinemann M. Multiple sex chromosomes in Drosophila miranda: a system to study the degeneration of a chromosome. Chromosoma. 1982;86(1):59–76. doi: 10.1007/BF00330730. [DOI] [PubMed] [Google Scholar]
- Steinemann M., Steinemann S. Degenerating Y chromosome of Drosophila miranda: a trap for retrotransposons. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7591–7595. doi: 10.1073/pnas.89.16.7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinemann M., Steinemann S. Preferential Y chromosomal location of TRIM, a novel transposable element of Drosophila miranda, obscura group. Chromosoma. 1991 Dec;101(3):169–179. doi: 10.1007/BF00355366. [DOI] [PubMed] [Google Scholar]
- Strathern J. N., Klar A. J., Hicks J. B., Abraham J. A., Ivy J. M., Nasmyth K. A., McGill C. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell. 1982 Nov;31(1):183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- Tartof K. D., Hobbs C., Jones M. A structural basis for variegating position effects. Cell. 1984 Jul;37(3):869–878. doi: 10.1016/0092-8674(84)90422-7. [DOI] [PubMed] [Google Scholar]