Abstract
Neurofibromatosis type 1 (NF1) patients are predisposed to neurofibromas but the driver(s) that contribute to neurofibroma formation are not fully understood. By cross comparison of microarray gene lists on human neurofibroma-initiating cells and developed neurofibroma Schwann cells (SCs) we identified RUNX1 overexpression in human neurofibroma initiation cells, suggesting RUNX1 might relate to neurofibroma formation. Immunostaining confirmed RUNX1 protein overexpression in human plexiform neurofibromas. Runx1 overexpression was confirmed in mouse Schwann cell progenitors (SCPs) and mouse neurofibromas at the messenger RNA and protein levels. Genetic inhibition of Runx1 expression by small hairpin RNA or pharmacological inhibition of Runx1 function by a Runx1/Cbfβ interaction inhibitor, Ro5-3335, decreased mouse neurofibroma sphere number in vitro. Targeted genetic deletion of Runx1 in SCs and SCPs delayed mouse neurofibroma formation in vivo. Mechanistically, loss of Nf1 increased embryonic day 12.5 Runx1+/Blbp+ progenitors that enable tumor formation. These results suggest that Runx1 has an important role in Nf1 neurofibroma initiation, and inhibition of RUNX1 function might provide a novel potential therapeutic treatment strategy for neurofibroma patients.
INTRODUCTION
Neurofibromatosis type 1 (NF1) is a tumor-suppressor gene that encodes neurofibromin, a Ras-GTPase-activating protein.1,2 NF1 patients are predisposed to neurofibromas, benign Schwann cell tumors.3,4 The molecular mechanisms of tumorigenesis and the molecules that drive neurofibroma formation are incompletely understood. Schwann cells (SCs) are believed to be the primary pathogenic cells in neurofibromas.5 Ablation of Nf1 using Nf1 homozygous (Nf1−/−) embryonic stem cells or ablation of Nf1 in the Schwann cell lineage using Krox20Cre, DhhCre or PLPCre in mice lead to the development of plexiform neurofibromas.6–9 Dermal neurofibromas can develop from skin-derived progenitors through loss of Nf1.10,11 Proliferation of mature non-myelinating SCs is a feature of neurofibroma formation.12 Therefore, Schwann cell progenitors (SCPs) and/or non-myelinating SCs may be cells of origin in neurofibromas.
Runt-related transcription factor-1 (Runx1 or AML1) is a transcription factor that is important for hematopoiesis, angiogenesis, maturation of megakaryocytes and differentiation of T and B cells.13,14 Runx1 is important for neuronal development and glial cell differentiation.15–17 Runx1 gene is expressed in proliferating Mash1+ or NeuroD+ olfactory receptor neuron progenitors on the basal side of the olfactory epithelium.18,19 Exogenous Runx1 expression in cultured olfactory neural progenitors causes an expansion of the mitotic cell population.18
Runx1 has shown paradoxical effects in cancer, in which it can function either as a tumor-suppressor gene or dominant oncogene according to context. Runx1 has been implicated as a tumor suppressor in a variety of solid neoplastic diseases such as breast cancer and gastric cancer.20 Recent evidence shows that Runx1 also acts as an oncogene in skin cancer, endometrial cancer and epithelial cancer.21–23 Here we showed that Runx1 functions as an oncogene in neurofibromas. Specifically, the loss of Nf1 induces Runx1 overexpression in mouse neurofibromas. Genetic inhibition of Runx1 by shRNA or pharmacological inhibition of Runx1 function by a Runx1/Cbfβ interaction inhibitor, Ro5-3335, decreased mouse neurofibroma sphere number in vitro. Targeted genetic deletion of Runx1 in mouse SCs and SCPs delayed neurofibroma formation owing to decreased cell proliferation and increased cell apoptosis. Mechanistically, loss of Nf1 increased embryonic day 12.5 Runx1+/Blbp+ progenitors, which contribute to neurofibroma formation.
RESULTS
Cross comparison of microarray gene lists reveals RUNX1 is only overexpressed in human neurofibroma tumor initiation cells
Previous reports support the notion that SCPs and/or non-myelinating SCs contribute to neurofibroma formation, but beyond Nf1 itself the underlying driving gene(s) are poorly understood. On the basis of the finding that human and mouse neurofibromas contain p75+/EGFR+ SCP-like tumor-initiating cells,24 we sorted p75+/EGFR+ tumor-initiating cells and p75+/EGFR− SCs from four primary human plexiform neurofibromas by using fluorescence-activated cell sorting. We performed microarray on these sorted cells and identified 1140 transcripts that were differentially expressed between p75+/EGFR+ SCP-like tumor-initiating cells and p75+/EGFR− Schwann cell sample classes using a t-test (P <0.05) (Supplementary Figure 1). We hypothesized that genes expressed exclusively in the neurofibroma-initiating cells, but not in the differentiated neurofibromas, might contribute to tumor initiation. By cross comparing this tumor-initiating cell gene list with the previously published differentiated neurofibroma Schwann cell microarray gene list25 and eliminating shared genes, we obtained RUNX1 as a top differentially expressed gene that was overexpressed only in the human neurofibroma-initiating cell microarray gene list (7.6-fold) (Figure 1a).
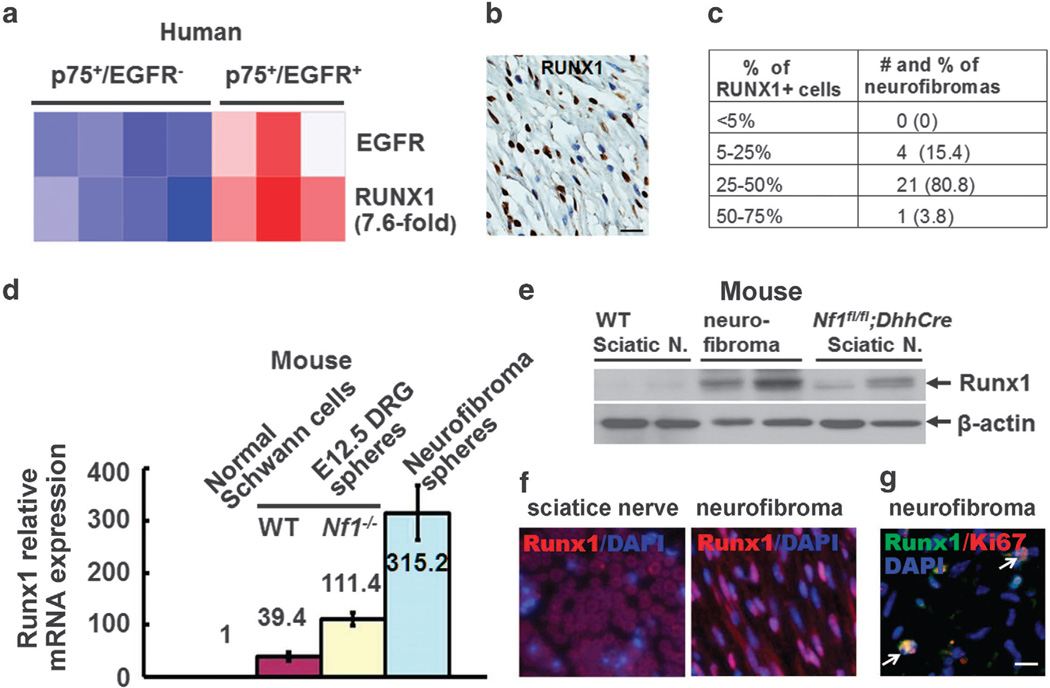
Figure 1.
Runx1 is overexpressed in Schwann cell progenitors and neurofibroma Schwann cells. (a) Microarray heat map shows Runx1 expression in human P75+/EGFR− Schwann cells and P75+/EGFR+ SCP-like tumor-initiating cells. (b) Representative Immunohistochemistry staining showing RUNX1+ cells (brown staining) in a human plexiform neurofibroma. Tissues embedded in paraffin were cut into six-μm sections. Sections were incubated overnight at 4 °C with anti-RUNX1 antibody (Abcam, Cambridge, MA, USA) and then incubated in appropriate biotinylated secondary antibody and visualized with DAB (brown). Blue was hematoxylin counterstaining. (c) Quantification of distribution of RUNX1+ cells in NF1 human plexiform neurofibromas (n=26). (d) Loss of Nf1 increases Runx1 expression on E12.5 WT, E12.5 Nf1−/− DRG and Nf1fl/fl;DhhCre mouse neurofibroma spheres detected by qRT–PCR. E12.5 WT DRG, E12.5 Nf1−/− DRG or mouse neurofibromas from the Nf1fl/fl;DhhCre mouse was enzymatically dissociated and cells were cultured on low binding plates in a serum-free medium to generate SCPs as described.24 Medium was added every 3 days. We extracted mRNA from wild-type Schwann cells (WT SCs), E12.5 WT, E12.5 Nf1−/− and mouse neurofibroma spheres using the RNeasy mini kit (Qiagen, Valencia, CA, USA). We reverse transcribed mRNA using the Superscript System (ABI, Grand Island, NY, USA). We amplified Tubulin as a control for each sample. We carried out quantitative real-time PCR experiments in the presence of SYBR green using the Runx1 primers. We pre-formed replicate reactions in an ABI Prism 7500 Sequence Detection System Cycler according to manufacturer's instructions. We confirmed all PCR products on 2% agarose gels. We calculated ΔCt values relative to Tubulin expression. We calculated the fold changes in spheres normalized to WT Schwann cells using the equation 2−ΔΔCt, where Ct is the cycle number at the chosen amplification threshold as determined by PE Biosystems software, ΔCt = Ct(Runx1)–Ct(tubulin), and ΔΔCt =ΔCt(sphere) –ΔCt(WT SCs). Statistics were performed by ordinary one way-ANOVA. (e) Western blots of Runx1 on Nf1fl/fl;DhhCre mouse neurofibromas and sciatic nerves. Western blots were performed as described9 using antibodies recognizing Runx1 (Abcam), and β-actin (Cell Signaling, Danvers, MA, USA). At least three different tumor/cell lysates were analyzed per antigen. Wild-type (WT) mouse sciatic nerves were used as controls. (f) Representative pictures of immunofluorescence staining of Runx1(red) on mouse WT sciatic nerve (left) and mouse neurofibroma (right). Nuclei were stained with DAPI. (g) Representative pictures of immunofluorescence staining of Runx1 (Abcam, green) and Ki67 (Leica Biosystems, Buffalo Grove, IL, USA) on mouse neurofibromas. Nuclei were stained with DAPI. Bar= 25μM.
We labeled human plexiform neurofibroma sections with an anti-RUNX1 antibody. Staining was detected in all human plexiform neurofibromas (n = 26). Three to sixty percent of human neurofibroma cells expressed RUNX1 (Figures 1b and c).
Runx1 is overexpressed in mouse SCPs and mouse neurofibromas We used neurofibroma sphere culture, a system to determine the proliferation of SCPs, to characterize Runx1 gene expression in embryonic day 12.5 (E12.5) wild-type (WT) spheres, E12.5 Nf1−/− mouse dorsal root ganglia (DRG) spheres, and mouse neurofibroma spheres by quantitative reverse transcription PCR (qRT– PCR). The overexpression of Runx1 was detected in all progenitors when normalized to WT SCs, but was much higher in E12.5 Nf1−/− mouse DRG spheres and mouse neurofibroma spheres compared with WT spheres (Figure 1d). There was a significant difference in Runx1 expression between Nf1−/− and WT spheres (P <0.001) or neurofibroma spheres vs WT (P <0.001). QRT–PCR showed that Cbf-β messenger RNA (mRNA) relative expressions were within twofold range in three different neurofibromas when we normalized to age-matched WT mouse sciatic nerve mRNA expressions, and there was no significant difference in mRNA relative expression levels between these two groups (P = 0.15, not shown). Runx1 protein expression was detected by western blot and by immunofluorescence in mouse neurofibromas but rarely in WT mouse sciatic nerves (Figures 1e and f). We performed Runx1 and Ki67 double-labeling on mouse neurofibromas to further characterize these Runx1−expressing cells. We detected Runx1/Ki67 double-positive cells (25.5% of all Runx1−positive cells or 5.9% of total cells) within these tumors (Figure 1g), suggesting some of the Runx1−expressing cells in neurofibromas were mitotic.
Pharmacological and genetic inhibition of Runx1 decreases mouse neurofibroma sphere number in vitro
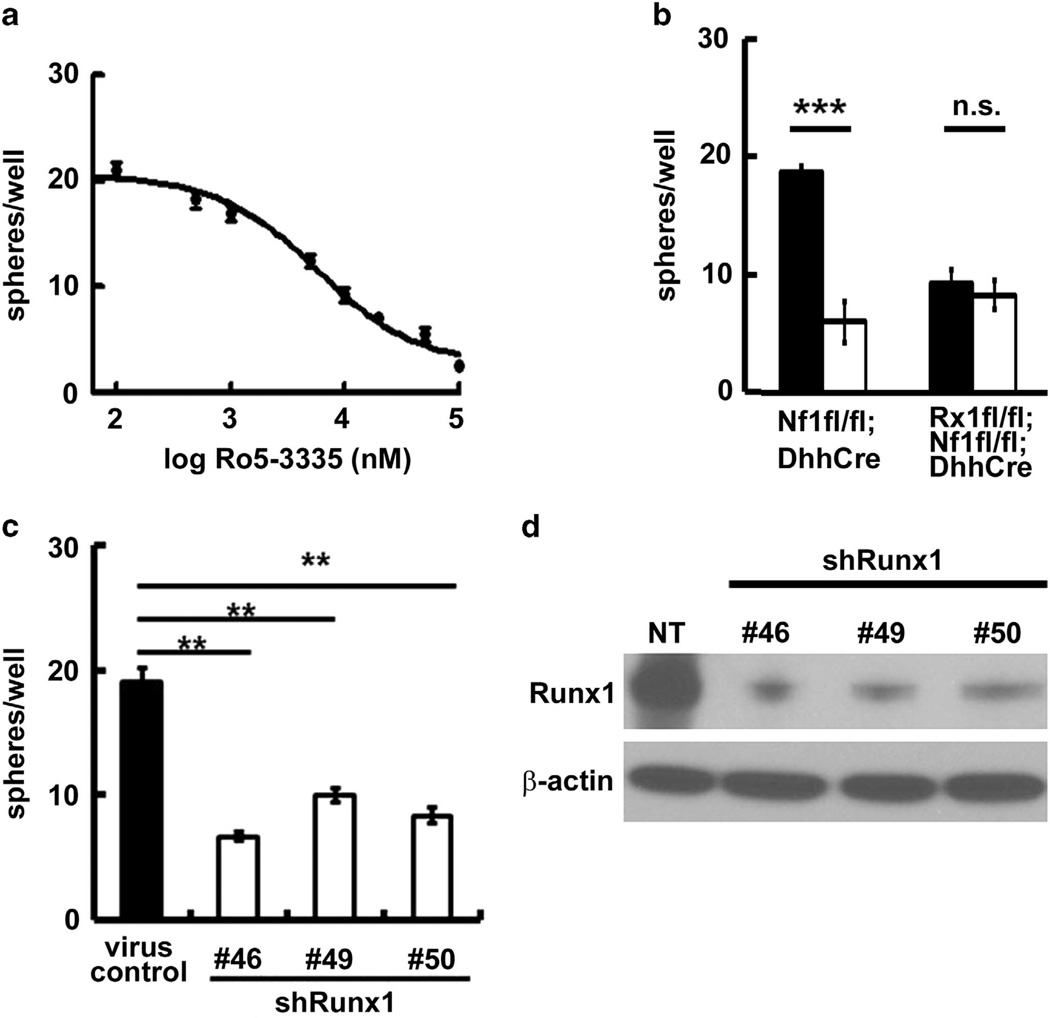
To determine whether Runx1 relates to SCP number, we treated secondary Nf1fl/fl;DhhCre mouse neurofibroma SCP-like sphere cells with a novel potent Runx1/Cbfβ interaction inhibitor, Ro5-3335,26 at a range of doses. Ro5-3335 inhibited sphere formation in a dose-dependent manner (Figure 2a). To test if the inhibitory effect was specifically caused by Runx1, we treated the 2-month-old Nf1fl/fl;DhhCre (Runx1 intact) and Runx1fl/fl;Nf1fl/fl; DhhCre (Runx1 deleted) mouse DRG/tumor spheres with Ro5-3335. Ro5-3335 decreased sphere numbers significantly in the Runx1 intact Nf1fl/fl;DhhCre DRG/tumor spheres but had little effect in Runx1 deleted Runx1fl/fl;Nf1fl/fl;DhhCre DRG/tumor spheres (Figure 2b), confirming that Runx1 contributes to sphere formation. In both cases we did not detect a Runx1 protein change because Ro5-3335 inhibits Runx1/Cbfβ interaction rather than the Runx1 protein (not shown).26 We further tested the effects of Runx1 on sphere number using small hairpin RNAs. We infected mouse neurofibroma sphere cells with three shRunx1 lentiviruses. All three shRunx1 clones inhibited mouse neurofibroma progenitor cell number as evidenced by sphere numbers compared with non-target control (Figure 2c). Western blots showed the three clones knocked down 470% of Runx1 proteins compared with nontarget control (Figure 2d).
Figure 2.
Pharmacological and genetic inhibition of Runx1 decrease mouse neurofibroma sphere number in vitro. (a) Ro5-3335 inhibited mouse neurofibroma sphere number in a dose-dependent manner. (b) Specificity of Ro5-3335 showed that it has little effect on sphere number when Runx1 is knocked out (Runx1fl/fl;Nf1fl/fl;DhhCre). (c) Three clones of shRunx1 decrease mouse neurofibroma sphere number compared with non-target virus control. We infected secondary Nf1fl/fl;DhhCre mouse neurofibroma spheres with shRunx1 and non-target control (Sigma, St Louis, MO, USA). We incubated lentiviral particles with neurofibroma spheres for 4 days and counted spheres. (d) Western blot confirmed Runx1 knockdown. **P <0.01, ***P <0.001, ns =no significant difference. Rx1 =Runx1. All statistical analyses were performed by unpaired, two-tailed student t-test.
Targeted genetic deletion of Runx1 in SCs and SCPs delays mouse neurofibroma formation in vivo
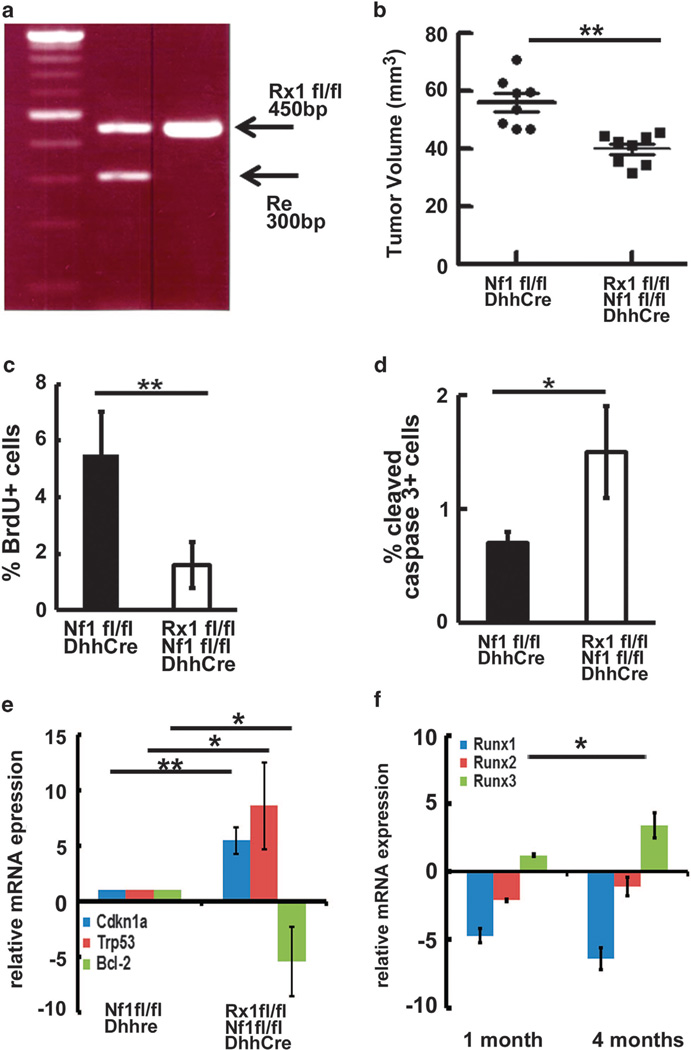
To test if Runx1 contributes to neurofibroma initiation, we targeted genetically deleted Runx1 in SCs and SCPs using a specific SCP/Schwann cell driver, Desert hedgehog (Dhh).27 The Runx1 knockout mouse dies at E11–E12, requiring use of a conditional knock out of Runx1. We obtained the Runx1fl/fl;Nf1fl/fl; DhhCre mouse after two generations at regular Mendel ratios. Mouse genotypes were confirmed by PCR (Figure 3a). We performed magnetic resonance imaging scanning at 4 months of age on Runx1fl/fl;Nf1fl/fl;DhhCre (n = 8) and Nf1fl/fl; DhhCre (n = 8) mice. Volumetric measurements were performed to obtain tumor size. An unpaired t-test showed that neurofibroma volumes decreased significantly in the Runx1fl/fl;Nf1fl/fl;DhhCre mice compared with the Nf1fl/fl;DhhCre mice at 4 months of age (54.5 ± 3.9mm2 vs 34.2 ± 1.8mm2, P <0.01) (Figure 3b).
Figure 3.
Targeted genetic deletion of Runx1 in SCs and SCPs delays mouse neurofibroma formation in vivo. (a) The animal care and use committees of Cincinnati Children’s Hospital Medical Center approved all animal procedures. Mice were housed in temperature- and humidity-controlled facilities on 12-h dark–light cycles with free access to food and water. We bred the Runx1fl/flmice onto the Nf1fl/fl background to obtain F1 generation (Runx1fl/+;Nf1fl/+). We also bred the Runx1fl/fl mice with Nf1fl/+;DhhCre+ to obtain Runx1fl/+;Nf1fl/+;DhhCre mice. We then bred Runx1fl/+;Nf1fl/+with Runx1fl/+;Nf1fl/+;DhhCre mice to obtain Runx1fl/fl;Nf1fl/fl;DhhCre mice. Littermates, Nf1fl/fl;DhhCre mice or Runx1fl/+;Nf1fl/fl;DhhCre, served as controls. PCR genotyping of Runx1fl/fl mouse. (b) Volumetric measurements on 4-month-old Runx1fl/fl;Nf1fl/fl; DhhCre (n = 8) and Nf1fl/fl;DhhCre mice (n =8). (c) BrdU analysis on Nf1fl/fl;DhhCre (black) and Runx1fl/fl;Nf1fl/fl;DhhCre (white). (d) Cleaved caspase 3 analysis on Nf1fl/fl;DhhCre (black) and Runx1fl/fl;Nf1fl/fl;DhhCre (white). (e) qRT–PCR of the indicated genes. mRNAs were extracted from the DRG/tumors of Nf1fl/fl;DhhCre (n =3) or Runx1fl/fl;Nf1fl/fl;DhhCre (n =3) mice at 4 months of age. qRT–PCRs were performed as above. (f) qRT–PCR of Runx1, Runx2 and Runx3 from DRG/tumors of Runx1fl/fl;Nf1fl/fl;DhhCre (n =3) mice at 1 month and 4 months of age and age-matched wild-type mouse sciatic nerves. Relative mRNA expression levels were normalized to wild-type mouse sciatic nerves gene expressions. **P <0.01, *P <0.05.
To determine the potential mechanisms underlying tumor growth, we performed BrdU and cleaved caspase 3 staining on 2-month-old Nf1fl/fl;DhhCre and Runx1fl/fl;Nf1fl/fl;DhhCre mouse DRGs. BrdU staining showed that cell proliferation decreased significantly in Runx1fl/fl;Nf1fl/fl;DhhCre mice compared with Nf1fl/fl; DhhCre (P <0.01, Figure 3c), and the cleaved caspase 3-positive cell (apoptotic cells) increased significantly (P <0.05, Figure 3d). To determine how loss of Runx1 decreased proliferation and induced apoptosis, we performed qRT–PCR to check the mRNA expression levels on Bcl-2, BMI-1, Cdkn1a (p21), Cdkn2a (p16), c-Myc, Mcl-1 and Trp53 in the Nf1fl/fl;DhhCre and Runx1fl/fl;Nf1fl/fl; DhhCre mouse DRG/tumors at 4 months of age. We found significantly increased Cdkn1a (p21) and Trp53 relative mRNA expression and significantly decreased Bcl-2 relative mRNA expression in Runx1fl/fl;Nf1fl/fl;DhhCre DRG/tumors when we normalized the expression levels to Nf1fl/fl;DhhCre mice (Figure 3e).
We show that targeted genetic deletion of Runx1 in mouse SCs and SCPs delayed neurofibroma growth significantly at 4 months in the genetically engineered Nf1fl/fl;DhhCre neurofibroma mouse model, but we could not eliminate neurofibroma formation in the Runx1fl/fl;Nf1fl/fl;DhhCre mouse. It is possible that phenotypes observed on conditional inactivation of Runx1 were attenuated by compensation of other Runx family members including Runx2 and/or Runx3, because all three Runx proteins bind to the same consensus sequence.28 To determine the possibility, we performed qRT–PCR to check the relative mRNA expression levels of Runx1, Runx2 and Runx3 in DRG/tumors of Runx1fl/fl;Nf1fl/fl;DhhCre mice at 1 month and 4 months of age. We normalized the relative mRNA expression levels to age-matched WT mouse sciatic nerve gene mRNA expressions. Runx1 relative mRNA expressions were maintained at low levels. Runx2 relative mRNA expression levels were within two-fold change. Runx3 relative mRNA expression increased significantly at 4 months (P <0.05, Figure 3f), suggesting that Runx3 compensates for the conditional loss of Runx1 and possibly contributes to the delayed neurofibroma formation.
Loss of Nf1 leads to increase in Runx1+/Blbp+ progenitors in E12.5 DRGs
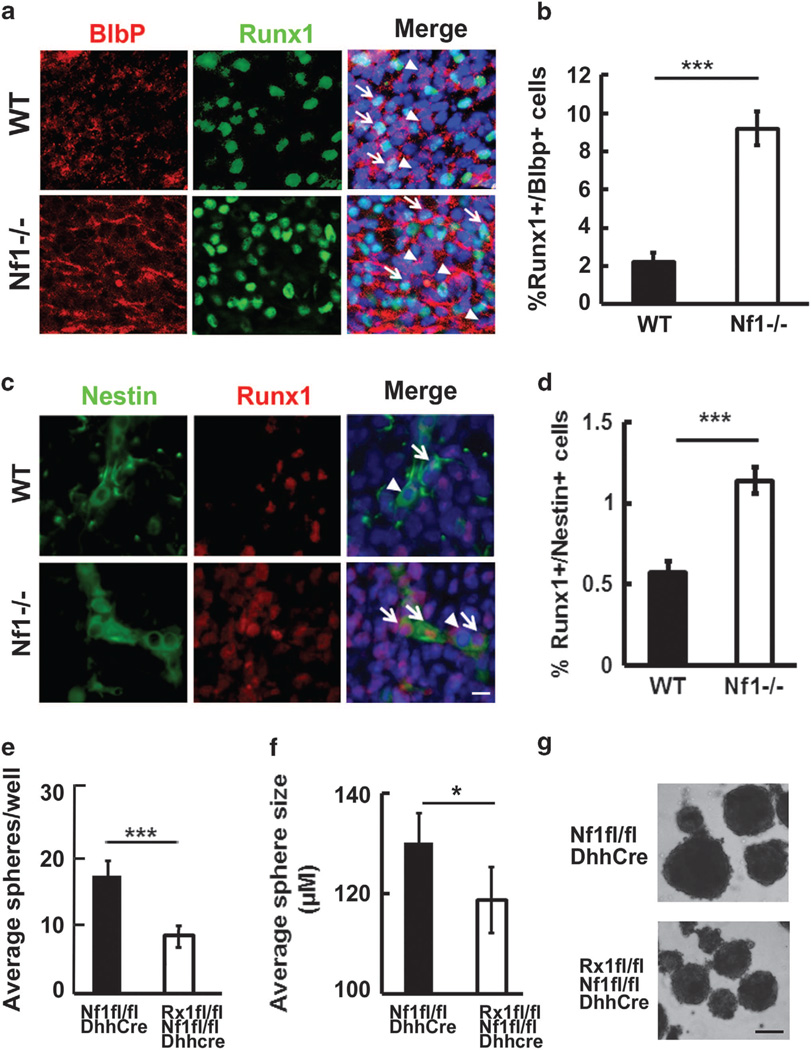
Runx1 expression in mouse embryos has been reported.17 We have shown that neurofibromas develop at E12.5 in the Nf1fl/fl;DhhCre mice.9 To determine the potential underlying mechanism of Runx1 in contributing to neurofibroma formation, we performed immunofluorescence staining of Runx1 on E12.5 WT and E12.5 Nf1−/− mouse DRG containing SCPs, prior to embryonic death caused by loss of Nf1. We detected increased numbers of Runx1+/Blbp+ progenitors in Nf1−/− mouse DRGs (n = 5) compared with WT controls (n =3, P <0.001) directly after removal from mice (Figures 4a and b), supporting the increased Runx1 expression in SCPs on loss of Nf1. Similar results were obtained by using a different SCP marker P0 (Supplementary Figure 2).
Figure 4.
Loss of Nf1 leads to increase in Runx1+/Blbp+ progenitors and Runx1+/Nestin+ neural crests in E12.5 DRGs. (a) Immunofluorescence staining of Runx1/Blbp on E12.5 DRGs. Tissues were embedded in OCT and cut into 12-μm sections. Sections were incubated overnight at 4 °C with antibodies: anti- Runx1 (Abcam) and anti-Blbp (Millipore, Billerica, MA, USA) and then incubated with FITC-anti-goat and TRITC-anti-rabbit secondary antibody at room temperature for 1 h. Nuclei were stained with DAPI. Top: WT, Bottom: Nf1−/−, Red: Blbp, Green: Runx1, Blue: nuclei. White arrow: Runx1+/Blbp+ progenitors. Arrow head: Runx1−/Blbp+ progenitors. (b) Quantification on percentages of Runx1+/Blbp+ progenitors in E12.5 WT (black bar) and E12.5 Nf1−/− DRGs (white bar). (c) Immunofluorescence staining of Runx1/Nestin on E12.5 DRGs. Tissues were prepared as above. Sections were incubated overnight at 4 °C with antibodies: anti- Runx1 (Abcam) and anti-Nestin (Millipore) and then incubated with FITC-anti-mouse and TRITC-anti-rabbit secondary antibody at room temperature for 1 h. Nuclei were stained with DAPI. Top: WT, Bottom: Nf1−/−, Red: Runx1, Green: Nestin, Blue: nuclei. White arrow: Runx1+/Nestin+ neural crests. Arrow head: Runx1−/Nestin+ neural crests. (d) Quantification on percentages of Runx1+/Nestin+ neural crests in E12.5 WT (black bar) and E12.5 Nf1−/− DRGs (white bar). Bar=50 μm. (e) Average sphere counting in 2 months Nf1fl/fl;DhhCre (black) and Runx1fl/fl;Nf1fl/fl;DhhCre (white) mouse DRG/tumors spheres. Statistical analyses were performed by unpaired, two-tailed student t-test. (f) Average DRG/tumor sphere size in Nf1fl/fl;DhhCre (black) and Runx1fl/fl;Nf1fl/fl;DhhCre (white) mice at 2 months of age. (g) Representative mouse DRG/tumor spheres of Nf1fl/fl;DhhCre (top) and Runx1fl/fl;Nf1fl/fl; DhhCre (bottom) mice at 2 months of age. Bar=100 μm. ***P <0.001, *P <0.05.
It has been shown that the E12.5 spheres express neural crest markers TWIST, Nestin, P75, Wint5a and Sox9 by qRT–PCR.24 To further determine whether there are poorly differentiated Runx1−expressing SCPs on loss of Nf1, we double labeled E12.5 Nf1−/− and WT DRGs using a neural crest marker, Nestin, with Runx1. There were increased numbers of Runx1+/Nestin+ neural crest progenitors in Nf1−/− mouse DRGs (n = 3) compared with WT controls (n=3, P <0.001) (Figures 4c and d). We also performed Runx1 staining on Nf1fl/fl;DhhCre;EGFP mouse neurofibromas, in which EGFP serves as a surrogate marker of SCs and SCPs. We detected Runx1+ cells in both EGFP+ and EGFP− SCs/SCPs within the tumors. Quantitatively, 21.5% of the EGFP+ cells were EGFP+/Runx1+ SCs/SCPs. This represented 56.5% of total Runx1+ cells. Consistent with the idea that some of the Runx1+ cells were progenitors, there were much fewer spheres in Runx1fl/fl;Nf1fl/fl; DhhCre mouse DRG/tumor spheres compared with WT Runx1 Nf1fl/fl;DhhCre DRG/tumor spheres (P <0.001) (Figure 4e). To determine whether Runx1 contributes to cell growth, we randomly measured 30 spheres in each group. Sphere size varied, but the overall average sphere size was significantly smaller in the Runx1fl/fl;Nf1fl/fl;DhhCre mouse DRG/tumor spheres compared with Nf1fl/fl;DhhCre DRG/tumor spheres (P <0.05, Figures 4f and g). Together, these results proved that Runx1 contributes to the number of stem-like cells as well as cell growth in the Nf1fl/fl; DhhCre tumors.
DISCUSSION
Our data indicate that Runx1 acted as an oncogene in the context of NF1. We identified that Runx1 might contribute to neurofibroma initiation by cross comparison of two distinct microarray data. Targeted genetic deletion of Runx1 in mouse SCs and SCPs delayed neurofibroma growth significantly at 4 months in the genetically engineered Nf1fl/fl;DhhCre neurofibroma mouse model, confirming the oncogenic function of Runx1. There was sphere formation in the Runx1fl/fl;Nf1fl/fl;DhhCre DRG/tumors, even though the sphere number was significantly less than age-matched Nf1fl/fl; DhhCre mouse neurofibroma spheres (Figure 2b). This suggests that the Runx3 compensation process might start before 2 months, but we could not determine when this process started. We could not eliminate the possibility that neurofibromas will form in the Runx1fl/fl;Nf1fl/fl;DhhCre mouse because Runx3 compensates for the signaling on conditional inactivation of Runx1. It will be interesting to test if knockdown of Runx1 and Runx3 simultaneously in Nf1fl/fl; DhhCre mice will eliminate neurofibroma sphere formation.
Contrary to the role of Runx1 in leukemia,29 our results show that loss of Runx1 in SCs decreased cell proliferation by activating Trp53-p21, and/or increased cell apoptosis by activating Trp53 or inhibiting antiapoptotic gene Bcl-2 in the context of Nf1−/− Schwann cell environment. It is possible that the level of Runx1 within the neurofibromas determines cell fate through Trp53. Further experiments are needed to determine how p21 and Trp53 were activated, or Bcl-2 was inactivated in the Runx1fl/fl;Nf1fl/fl; Dhhre tumors. We cannot exclude the possibility that other genes might also be involved in neurofibroma formation.
We showed that Runx1 mRNA expression was significantly higher in mouse E12.5 Nf1−/− DRG and mouse neurofibroma progenitors by qRT–PCR. Furthermore, at the protein level, we showed Runx1+/Nestin+ and Runx1+/Blbp+ progenitors increased significantly on loss of Nf1, and we observed Runx1+ SCs/SCPs within neurofibromas. Reports show that Runx1 has a key role in brain neuronal progenitor function. There is an increased subpopulation of brain progenitor cells that are Nestin+/Runx1+, GFAP+/Runx1+ or Sox2+/Runx1+ in the sub-ventricular zone or dentate gyrus after brain injury.30 It has also been reported that Runx1 is expressed in hair follicle, oral epithelium and intestine. Runx1 is required for the development of malignant skin squamous cell carcinoma and papilloma (benign squamous, progenitors of squamous cell carcinoma) that originate from Runx1− expressing tumor initiation cells in the hair follicle stem cells.21 Consistent with these reports, the increased Runx1+/Blbp+ progenitors might contribute to neurofibroma formation.
Targeting transcription factors, which have traditionally been considered untargetable, is becoming a realistic option with increased understanding of transcription factor biology and technological advances. We showed that the Runx1/Cbfβ interaction inhibitor Ro5-3335 decreased mouse neurofibroma sphere number significantly in vitro. The Runx1 protein binds to a consensus sequence with Cbfβ. The protein Cbfβ lacks a DNA-binding domain, but when bound to Runx1, it increases the DNA-binding affinity of the Runt domain several folds, substantially enhancing Runx1 transcriptional activity.28 Ro5-3335 exerts its inhibitory effect on sphere formation by disruption of Runx1/Cbfβ interaction. Ro5-3335 and its analog, Ro24-7429, were initially developed as anti-HIV drugs to inhibit Tat-mediated transactivation. Ro24-7429 entered phase II clinical trial, and the drug was tolerated well by patients. It is possible that both Ro5-3335 and Ro24-7429 might show efficacy in NF1 by inhibiting Runx1/Cbfβ interaction rather than Tat. In vivo Ro5-3335 preclinical therapeutic trial using the neurofibroma mouse model will provide more information on this compound.
In summary, loss of Nf1 leads to an increase in Runx1+/Blbp+ progenitors, which enable tumor initiation. The in vitro efficacy of Ro5-3335 in reducing mouse neurofibroma sphere number suggests inhibiting Runx/Cbf-β interaction might provide a novel therapeutic target for neurofibroma treatment.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Nancy Ratner for providing the microarray data (supported by NS28840 to NR) and critical review and helpful discussions of the manuscript. This work was supported by a Cincinnati Children’s Hospital Trustee Grant to JW.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
REFERENCES
- 1.McCormick F. Ras signaling and NF1. Curr Opin Genet Dev. 1995;5:51–55. doi: 10.1016/s0959-437x(95)90053-5. [DOI] [PubMed] [Google Scholar]
- 2.Le LQ, Parada LF. Tumor microenvironment and neurofibromatosis type I: connecting the GAPs. Oncogene. 2007;26:4609–4616. doi: 10.1038/sj.onc.1210261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blatt J, Jaffe R, Deutsch M, Adkins J. Neurofibromatosis and childhood tumors. Cancer. 1986;57:1225–1229. doi: 10.1002/1097-0142(19860315)57:6<1225::aid-cncr2820570627>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 4.Friedman J. Epidemiology of neurofibromatosis type 1. Am J Med Genet. 1999;89:1–6. [PubMed] [Google Scholar]
- 5.Kim HA, Ling B, Ratner N. Nf1-deficient mouse Schwann cells are angiogenic and invasive and can be induced to hyperproliferate: reversion of some phenotypes by an inhibitor of farnesyl protein transferase. Mol Cell Biol. 1997;17:862–872. doi: 10.1128/mcb.17.2.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cichowski K, Shih TS, Schmitt E, Santiago S, Reilly K, McLaughlin ME, et al. Mouse models of tumor development in neurofibromatosis type 1. Science. 1999;286:2172–2176. doi: 10.1126/science.286.5447.2172. [DOI] [PubMed] [Google Scholar]
- 7.Zhu Y, Ghosh P, Charnay P, Burns D, Parada L. Neurofibromas in NF1: Schwann cell origin and role of tumor environment. Science. 2002;296:920–922. doi: 10.1126/science.1068452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayes DA, Rizvi TA, Cancelas JA, Kolasinski NT, Ciraolo GM, Stemmer-Rachamimov AO, et al. Perinatal or adult Nf1 inactivation using tamoxifen-inducible PlpCre each cause neurofibroma formation. Cancer Res. 2011;71:4675–4685. doi: 10.1158/0008-5472.CAN-10-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu J, Williams JP, Rizvi TA, Kordich JJ, Witte D, Meijer D, et al. Plexiform and dermal neurofibromas and pigmentation are caused by Nf1 loss in desert hedgehog-expressing cells. Cancer Cell. 2008;13:105–116. doi: 10.1016/j.ccr.2007.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le LQ, Shipman T, Burns DK, Parada LF. Cell of origin and microenvironment contribution for NF1-associated dermal neurofibromas. Cell Stem Cell. 2009;4:453–463. doi: 10.1016/j.stem.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jouhilahti EM, Peltonen S, Callens T, Jokinen E, Heape AM, Messiaen L, et al. The development of cutaneous neurofibromas. Am J Pathol. 2011;178:500–505. doi: 10.1016/j.ajpath.2010.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng H, Chang L, Patel N, Yang J, Lowe L, Burns DK, et al. Induction of abnormal proliferation by nonmyelinating schwann cells triggers neurofibroma formation. Cancer Cell. 2008;13:117–128. doi: 10.1016/j.ccr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Nagamachi A, Htun PW, Ma F, Miyazaki K, Yamasaki N, Kanno M, et al. A 5' untranslated region containing the IRES element in the Runx1 gene is required for angiogenesis, hematopoiesis and leukemogenesis in a knock-in mouse model. Dev Biol. 2010;345:226–236. doi: 10.1016/j.ydbio.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 14.Ichikawa M, Asai T, Saito T, Seo S, Yamazaki I, Yamagata T, et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 15.Inoue K, Shiga T, Ito Y. Runx transcription factors in neuronal development. Neural Dev. 2008;3:20. doi: 10.1186/1749-8104-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aldskogius H, Berens C, Kanaykina N, Liakhovitskaia A, Medvinsky A, Sandelin M, et al. Regulation of boundary cap neural crest stem cell differentiation after transplantation. Stem Cells. 2009;27:1592–1603. doi: 10.1002/stem.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simeone A, Daga A, Calabi F. Expression of runt in the mouse embryo. Dev Dyn. 1995;203:61–70003C. doi: 10.1002/aja.1002030107. [DOI] [PubMed] [Google Scholar]
- 18.Theriault FM, Nuthall HN, Dong Z, Lo R, Barnabe-Heider F, Miller FD, et al. Role for Runx1 in the proliferation and neuronal differentiation of selected progenitor cells in the mammalian nervous system. J Neurosci. 2005;25:2050–2061. doi: 10.1523/JNEUROSCI.5108-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murthy M, Bocking S, Verginelli F, Stifani S. Transcription factor Runx1 inhibits proliferation and promotes developmental maturation in a selected population of inner olfactory nerve layer olfactory ensheathing cells. Gene. 2014;540:191–200. doi: 10.1016/j.gene.2014.02.038. [DOI] [PubMed] [Google Scholar]
- 20.Chimge NO, Frenkel B. The RUNX family in breast cancer: relationships with estrogen signaling. Oncogene. 2012;32:2121–2130. doi: 10.1038/onc.2012.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheitz CJ, Lee TS, McDermitt DJ, Tumbar T. Defining a tissue stem cell-driven Runx1/Stat3 signalling axis in epithelial cancer. EMBO J. 2012;31:4124–4139. doi: 10.1038/emboj.2012.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Planaguma J, Diaz-Fuertes M, Gil-Moreno A, Abal M, Monge M, Garcia A, et al. A differential gene expression profile reveals overexpression of RUNX1/AML1 in invasive endometrioid carcinoma. Cancer Res. 2004;64:8846–8853. doi: 10.1158/0008-5472.CAN-04-2066. [DOI] [PubMed] [Google Scholar]
- 23.Scheitz CJ, Tumbar T. New insights into the role of Runx1 in epithelial stem cell biology and pathology. J Cell Biochem. 2013;114:985–993. doi: 10.1002/jcb.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams JP, Wu J, Johansson G, Rizvi TA, Miller SC, Geiger H, et al. Nf1 mutation expands an EGFR-dependent peripheral nerve progenitor that confers neurofibroma tumorigenic potential. Cell Stem Cell. 2008;3:658–669. doi: 10.1016/j.stem.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller SJ, Jessen WJ, Mehta T, Hardiman A, Sites E, Kaiser S, et al. Integrative genomic analyses of neurofibromatosis tumours identify SOX9 as a biomarker and survival gene. EMBO Mol Med. 2009;1:236–248. doi: 10.1002/emmm.200900027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cunningham L, Finckbeiner S, Hyde RK, Southall N, Marugan J, Yedavalli VR, et al. Identification of benzodiazepine Ro5-3335 as an inhibitor of CBF leukemia through quantitative high throughput screen against RUNX1-CBFbeta interaction. Proc Natl Acad Sci USA. 2012;109:14592–14597. doi: 10.1073/pnas.1200037109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaegle M, Ghazvini M, Mandemakers W, Piirsoo M, Driegen S, Levavasseur F, et al. The POU proteins Brn-2 and Oct-6 share important functions in Schwann cell development. Genes Dev. 2003;17:1380–1391. doi: 10.1101/gad.258203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bushweller JH. CBF--a biophysical perspective. Semin Cell Dev Biol. 2000;11:377–382. doi: 10.1006/scdb.2000.0182. [DOI] [PubMed] [Google Scholar]
- 29.Motoda L, Osato M, Yamashita N, Jacob B, Chen LQ, Yanagida M, et al. Runx1 protects hematopoietic stem/progenitor cells from oncogenic insult. Stem Cells. 2007;25:2976–2986. doi: 10.1634/stemcells.2007-0061. [DOI] [PubMed] [Google Scholar]
- 30.Logan TT, Villapol S, Symes AJ. TGF-beta superfamily gene expression and induction of the Runx1 transcription factor in adult neurogenic regions after brain injury. PloS One. 2013;8:e59250. doi: 10.1371/journal.pone.0059250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.