Acute myeloid leukemia (AML) with chromosomal rearrangements inv(16)(p13q22) or t(16;16)(p13;q22) (collectively referred to as inv(16)) and t(8;21)(q22;q22) are classified as core binding factor (CBF) AML, which accounts for ~20% of all AML cases (1). The resulting oncogenic fusion genes, CBFB-MYH11 and RUNX1-RUNX1T1, respectively, involve members of the CBF family of transcription factors, RUNX1 and its binding partner CBFβ(1). Overall, patients with CBF AML have a favorable prognosis compared to other AML cytogenetic groups, with the majority of them (>85%) achieving complete remission (CR) with cytarabine/anthracycline-based regimens (2). However, approximately half of the CBF AML patients relapse, with a median time to relapse of 2.5 years after achieving CR (2) and the overall survival at 5 years of only 51% (3). Thus, it is imperative to develop novel and “personalized” treatments for these patients. However, the mechanisms leading to relapse remain elusive. The goal of our study was to elucidate the mechanism of relapse in CBF AML by unbiased genomic approaches. Therefore, we applied whole exome sequencing (WES) and single nucleotide polymorphism (SNP) arrays to analyze the somatic mutational landscape of matched diagnosis and relapse DNA samples from CBF AML patients. Here, we report our data on the identification of putative novel AML driver genes and mechanisms of relapse.
WES and SNP arrays were performed on diagnosis, CR and relapse samples from ten patients and diagnosis and relapse samples from additional three patients on the CALGB 8461 protocol (4). Patient details and study design are described in Supplementary methods, Supplementary Table S1, and Supplementary Figure 1. DNA from CR samples was used as the germline control to identify somatic single nucleotide variants (SNVs) and copy number variants (CNVs). We obtained high quality WES reads with average breadth of coverage of 90.5% of exome (range: 85 – 93%) and average depth of coverage of 82.4X (range: 39.3 – 108.4X) (Supplementary Table S2).
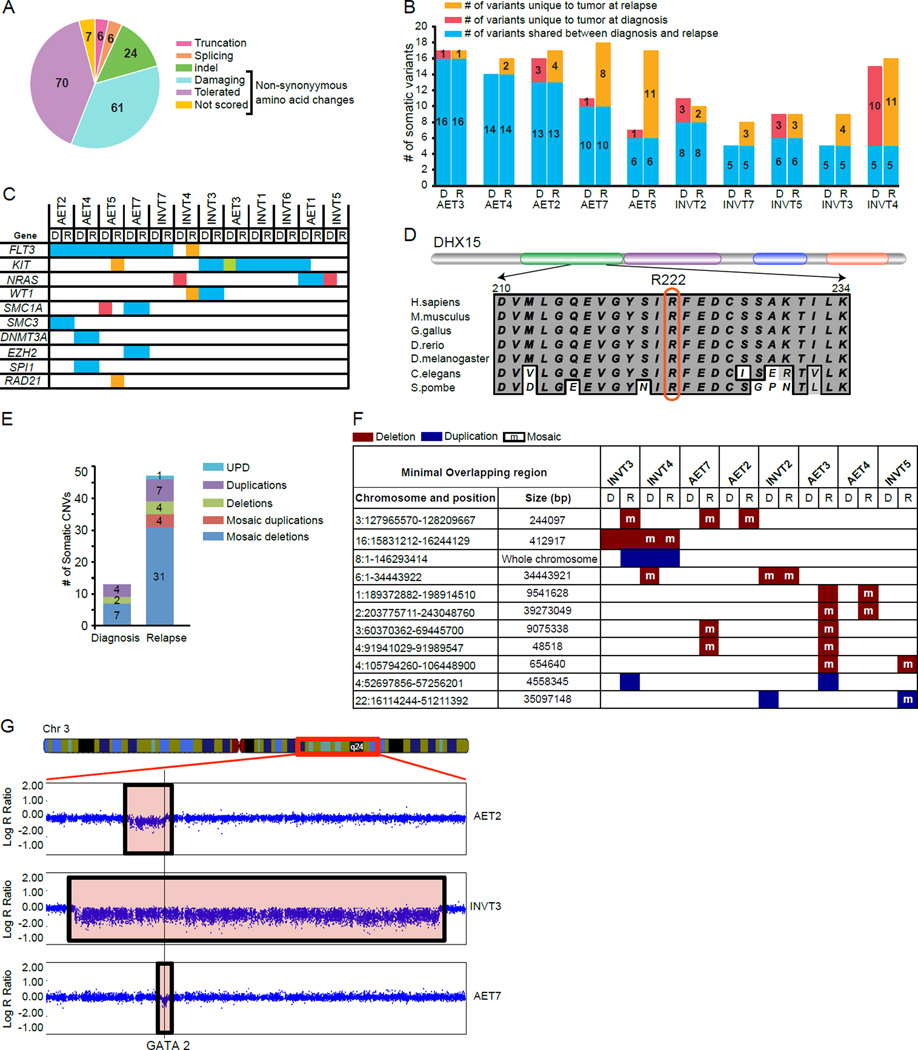
To identify genes involved in CBF AML, we focused on the variants with non-synonymous amino acid changes, splicing changes, and coding region indels (collectively referred to as SNVs). Furthermore, breakpoint analysis identified FLT3-ITD (internal tandem duplications of 35 to 84bp) in 3 patients. A total of 174 unique variants in 163 genes met the criteria of being “somatic” based on their absence in the corresponding germline DNA sample, ruled out as a sequencing artifact and variant allele frequency (VAF) of ≥ 0.10 with high quality reads (Supplementary Tables S3, S4). For the patients lacking WES data on the CR sample, we identified somatic SNVs by manual validation of variants in known AML genes (Supplementary Tables S5 and S6). More than half of the somatic variants were predicted to have deleterious effects on the functions of the corresponding proteins (Figure 1A). However, we did not find any specific pathways to be significantly enriched among the mutated genes. To understand the relationship between diagnosis and relapse samples, we classified the variants for each patient into those shared by the matched diagnosis and relapse samples and those specific to either time-point (Figure 1B and Supplementary Table S4). Consistent with the previous studies in AML (5), the total number of somatic variants per sample ranged from 5 to 18 (Figure 1B). Relapse samples gained one to eleven additional somatic variants compared with the corresponding diagnosis samples. Two distinct patterns of tumor evolution were evident: 1) diagnosis and relapse leukemias share the majority of the somatic variants indicating origin from a common clone (e.g. AET4 and INVT3); 2) relapse leukemias lost some of the variants detected in the corresponding diagnostic samples, indicating origin from an ancestral pre-leukemic clone (e.g. AET2, INVT2, INVT4 and INVT5).
Figure 1.
Characterization of somatic SNVs and CNVs in CBF leukemia samples. A). Pie chart depicting the distribution of 174 somatic variants based on their effect on protein function. SIFT (sorting intolerant from tolerant) analysis was used to classify non-synonymous amino acid changes into damaging, tolerated or not scored (Supplemental Materials and Methods). B). Histograms showing total number of SNVs/leukemia for ten patients with WES data on matched diagnosis, complete remission and relapse samples. Patients AET1, INVT1 and INVT6 are not shown since total number of shared somatic variants could not be determined due to lack of WES data from their CR samples. For each patient, the numbers of SNVs in diagnosis (D) and relapse (R) leukemia are shown next to each other and are depicted as shared (blue), unique to diagnosis (red) and unique to relapse (orange). Within each cytogenetic abnormality [t(8;21) or inv(16)], the patients are shown in the increasing order of relapse-specific SNVs. C). Relationship of matched diagnosis and relapse leukemia based on the SNVs in known AML driver genes. SNVs shared by matched diagnosis and relapse leukemia are shown in blue, unique to diagnosis in red and unique to relapse in orange. The green color for diagnosis leukemia from AET3 depicts two variants in KIT (D816Y and N822K) of which D816Y was shared with the relapse leukemia. D). A schematic of the DHX15 protein showing its domains: DEXDc domain in green, HELICc domain in purple, HA2 domain in blue and OB-NTP-bind domain in orange (top) and an alignment of a part of the DEXDc domain from human, mouse, chicken, zebrafish, drosophila, worm and yeast DHX15 orthologs (bottom). Amino acid residue R222 is marked by a red rectangle. E). Total number of somatic CNVs detected in the diagnosis and relapse samples. Each histogram is further marked for the number of different types of CNVs: uniparental disomy (UPD), duplications, deletions, mosaic duplications and mosaic deletions. F). Recurrent CNVs arranged by their order of frequency in independent patient samples. Deletions are marked in red, duplications in blue and mosaic events with an “m” in the corresponding color fill. For each CNV, chromosome number, genomic coordinates and size of the minimal overlapping region in base pairs (bp) are shown in the first two columns. For each patient, diagnosis and relapse leukemia samples are denoted by “D” and “R” respectively. G). Visual representation of deletions (boxed areas) affecting the GATA2 locus (vertical line) in relapse leukemia DNA from AET2, INVT3 and AET7 patients. Shown are Log R ratios, which were ratios between observed normalized intensity of the samples and the expected intensity. A schematic of chromosome 3 with the displayed region marked by a red box (chr3: 117,346,598 – 155,484,300) is shown at the top.
We identified somatic variants in at least one of ten previously known AML driver genes (FLT3, KIT, NRAS, WT1, SMC1A, SMC3, DNMT3A, EZH2, SPI1 and RAD21 , ref. (5, 6) in all patients except INVT2 (Figure 1C and Supplementary Tables S3, S4). FLT3 and KIT were the top two recurrently mutated genes in our patient cohort (Figure 1C, Supplementary Table S7). FLT3 -ITD was detected in diagnosis and relapse samples in five patients, whereas a relapse-specific F594I mutation (in the juxtamembrane domain) was detected in one patient (Supplementary Table S7). KIT mutations in both exons 8 (residues 417–421) and 17 (D816) were detected (Supplementary Table S7). Interestingly, the diagnosis sample in patient AET3 showed two KIT variants, D816Y and N822K (VAF = 0.27 and 0.13 respectively), while the relapse sample contained D816Y variant only (VAF = 0.54; Figure 1C, Supplementary Table S7), indicating at least two clones during diagnosis of which the dominant clone with D816Y is resistant to therapy, leading to relapse.
From the remaining 153 genes (75 shared, 24 diagnosis-specific and 54 relapse -specific), DHX15 was the only gene mutated in more than one patient. We detected an identical non-synonymous variant causing R222G (genomic coordinates: chr 4: G24572314C) in patients AET1 and AET3. Interestingly, the variant was lost in corresponding relapse sample from patient AET1 (Supplementary Figure S2, Supplementary Tables S3 and S4), indicating origin of relapse leukemia from an ancestral clone. The variant is predicted to be damaging by SIFT and is not detected in 938 controls from the Clinseq project (7). The affected amino acid R222 is part of the DEXDc domain, and is evolutionarily conserved from mammals to yeast (Figure 1D). Our data, combined with the previous report of the same variant in a patient with refractory anemia with excess blasts, a form of myelodysplastic syndrome with predisposition to AML (8), suggest DHX15 as a potential new AML driver gene. Functional analyses of Prp43, the yeast homologue of DHX15, have demonstrated its role in the release of the intron lariat during RNA splicing (9), thus adding DHX15 to the growing list of splicing factors involved in AML (5).
Next we analyzed the SNP array data from >900,000 polymorphic markers to identify recurrently occurring CNVs with putative roles in the relapse of CBF AML. We identified 52 somatic CNVs ranging in size from 20kb to an entire chromosome (Supplementary Table S8). Consistent with previous data (10, 11), relapse samples showed an increase in the number of CNVs compared to the diagnosis samples (Figure 1E, Supplementary Table S8). In particular, relapsed leukemia cells from patient AET3 harbored 17 somatic CNVs compared to only one CNV in its diagnosis sample (Supplementary Table S8). Importantly, even removing AET3 the rest of the relapse samples still showed increased CNVs (Supplementary Figure S3). Of the eleven genomic regions with recurrent CNVs (Figure 1F, Supplementary Figures S4, S5), trisomy 8 is known to be involved in AML and has been detected in 16% of AML patients with inv(16) (12). Among the CNVs affecting sub-chromosomal regions, deletion of overlapping regions on chromosome 3 was detected in relapse samples from three patients (Figure 1G, Supplementary Figure S4). The minimal overlapping region of deletions in these relapse samples (chr3: 127965570–128209667) contains EEFSEC, DNAJB8 and GATA2 . Of these, GATA2 encodes a master regulator of hematopoiesis. Recent studies have shown that both mutations and expression level changes in GATA2 play a causative role in AML (13–15). Our observation is the first indication that GATA2 haploinsufficiency may be important for CBF leukemia relapse.
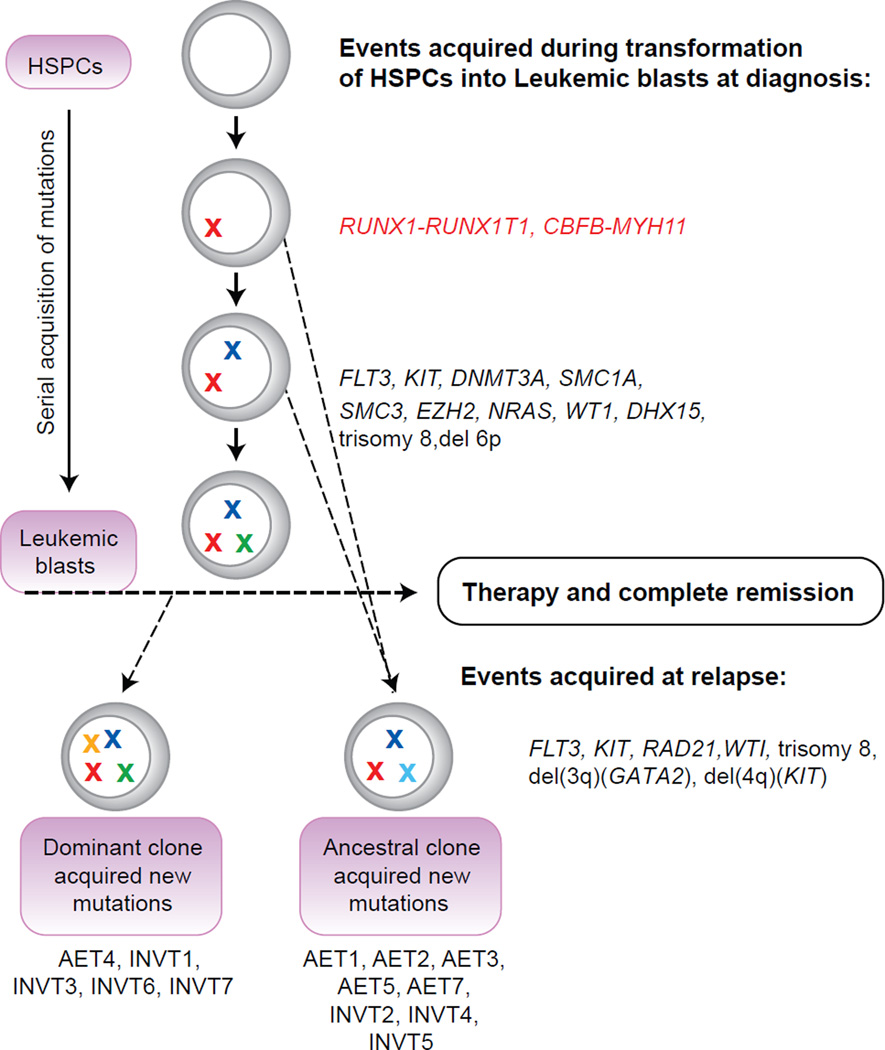
We deduced clonal evolution during relapse by integrated analysis of the SNVs and CNVs in matched diagnosis and relapse samples (Figure 2). Our data suggested two potential mechanisms of relapse: 1. A subclone of the diagnosis leukemia survived therapy and reemerged after accumulating additional mutations; 2. A pre-leukemia clone survived therapy, acquired additional mutations during remission, and gave rise to relapse leukemia (Figure 2). Common to both mechanisms is the existence of a clone with potential to evolve into leukemia that was not completely eliminated by the treatment after diagnosis. These findings are consistent with other recent studies involving analysis of matched diagnosis and relapse AML samples (11, 16, 17). It is worth noting that the RUNX1-RUNX1T1 and CBFB-MYH11 fusion genes were present at both diagnosis and relapse in all patients and mutations in one or more of the known AML driver genes tend to be present at both stages as well. On the other hand, mutations that emerged at relapse are mostly in the genes that have not been previously linked to leukemia. Additional CBF AML samples need to be screened for mutations in these genes and functional studies need to be carried out to determine if they contribute to relapse.
Figure 2.
Model of clonal evolution leading to relapse in each patient inferred from the pattern of somatic SNVs and CNVs. Mutations in driver genes are depicted as “X” of different colors to indicate mutations in different driver genes. The initiating event of inv(16) or t(8;21) is depicted as X in red color. HSPCs: hematopoietic stem and progenitor cells. Patient id’s are given below their corresponding mechanism of relapse out of the two possible scenarios.
In summary, we have systematically surveyed the genomic landscape of CBF AML at both diagnosis and relapse, and deduced two potential mechanisms of relapse. This study can serve as a starting point for future investigations into the mechanism of CBF AML relapse. Genes and chromosomal regions identified here, specifically DHX15 and GATA2 , can be screened by targeted approaches in additional CBF AML patients to determine their contribution to CBF AML and its relapse. Combining our data with findings from previous studies on the clonal evolution of relapse leukemias suggest that targeting the leukemia initiating events, such as the CBF fusion proteins and mutations in known driver genes should be explored further to lower the incidence of relapse.
Supplementary Material
Acknowledgements
We thank Ms. Julia Fekecs for her help with illustrations and Ms. Ursula Harper for performing DNA matching using a panel of polymorphic markers randomly distributed throughout the genome.
Financial support
Supported by the Intramural Research Program of National Human Genome Research Institute, NIH; CA101140, CA16058, CA140158, CA180861 and the Leukemia Clinical Research Foundation
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary information is available at Leukemia's website.
References
- 1.Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. [Review] 2002 Jul;2(7):502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 2.Marcucci G, Mrozek K, Ruppert AS, Maharry K, Kolitz JE, Moore JO, et al. Prognostic factors and outcome of core binding factor acute myeloid leukemia patients with t(8;21) differ from those of patients with inv(16): a Cancer and Leukemia Group B study. J Clin Oncol. [Research Support N.I.H., Extramural Research Support, Non-U.S. Gov't Research Support U.S. Gov't, P.H.S.] 2005 Aug 20;23(24):5705–5717. doi: 10.1200/JCO.2005.15.610. [DOI] [PubMed] [Google Scholar]
- 3.Hospital MA, Prebet T, Bertoli S, Thomas X, Tavernier E, Braun T, et al. Core-binding factor acute myeloid leukemia in first relapse: a retrospective study from the French AML Intergroup. Blood. [Clinical Trial Multicenter Study] 2014 Aug 21;124(8):1312–1319. doi: 10.1182/blood-2014-01-549212. [DOI] [PubMed] [Google Scholar]
- 4.Bloomfield CD, Mrozek K, Caligiuri MA. Cancer and leukemia group B leukemia correlative science committee: major accomplishments and future directions. Clin Cancer Res. [Review] 2006 Jun 1;12(11 Pt 2):3564s–3571s. doi: 10.1158/1078-0432.CCR-06-9002. [DOI] [PubMed] [Google Scholar]
- 5.Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. [Research Support, N.I.H., Extramural] 2013 May 30;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mueller BU, Pabst T, Osato M, Asou N, Johansen LM, Minden MD, et al. Heterozygous PU.1 mutations are associated with acute myeloid leukemia. Blood. [Research Support, Non-U.S. Gov't Research Support, U.S. Gov't, P.H.S.] 2002 Aug 1;100(3):998–1007. doi: 10.1182/blood.v100.3.998. [DOI] [PubMed] [Google Scholar]
- 7.Biesecker LG, Mullikin JC, Facio FM, Turner C, Cherukuri PF, Blakesley RW, et al. The ClinSeq Project: piloting large-scale genome sequencing for research in genomic medicine. Genome Res. [Research Support, N.I.H., Intramural] 2009 Sep;19(9):1665–1674. doi: 10.1101/gr.092841.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taskesen E, Havermans M, van Lom K, Sanders MA, van Norden Y, Bindels E, et al. Two splice-factor mutant leukemia subgroups uncovered at the boundaries of MDS and AML using combined gene expression and DNA-methylation profiling. Blood. [Research Support, Non-U.S. Gov't] 2014 May 22;123(21):3327–3335. doi: 10.1182/blood-2013-07-512855. [DOI] [PubMed] [Google Scholar]
- 9.Walbott H, Mouffok S, Capeyrou R, Lebaron S, Humbert O, van Tilbeurgh H, et al. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. Embo J. [Research Support, Non-U.S. Gov't] 2010 Jul 7;29(13):2194–2204. doi: 10.1038/emboj.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhn MW, Radtke I, Bullinger L, Goorha S, Cheng J, Edelmann J, et al. High-resolution genomic profiling of adult and pediatric core-binding factor acute myeloid leukemia reveals new recurrent genomic alterations. Blood. [Research Support N.I.H., Extramural Research Support, Non-U.S. Gov't] 2012 Mar 8;119(10):e67–e75. doi: 10.1182/blood-2011-09-380444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kronke J, Bullinger L, Teleanu V, Tschurtz F, Gaidzik VI, Kuhn MW, et al. Clonal evolution in relapsed NPM1-mutated acute myeloid leukemia. Blood. [Research Support, Non-U.S. Gov't] 2013 Jul 4;122(1):100–108. doi: 10.1182/blood-2013-01-479188. [DOI] [PubMed] [Google Scholar]
- 12.Paschka P, Du J, Schlenk RF, Gaidzik VI, Bullinger L, Corbacioglu A, et al. Secondary genetic lesions in acute myeloid leukemia with inv(16) or t(16;16): a study of the German-Austrian AML Study Group (AMLSG) Blood. [Research Support, Non-U.S. Gov't] 2013 Jan 3;121(1):170–177. doi: 10.1182/blood-2012-05-431486. [DOI] [PubMed] [Google Scholar]
- 13.Hyde RK, Liu PP. GATA2 mutations lead to MDS and AML. Nat Genet. [Comment News] 2011 Oct;43(10):926–927. doi: 10.1038/ng.949. [DOI] [PubMed] [Google Scholar]
- 14.Bresnick EH, Katsumura KR, Lee HY, Johnson KD, Perkins AS. Master regulatory GATA transcription factors: mechanistic principles and emerging links to hematologic malignancies. Nucleic Acids Res. [Research Support N.I.H., Extramural Research Support, Non-U.S. Gov't Review] 2012 Jul;40(13):5819–5831. doi: 10.1093/nar/gks281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BA, Erpelinck C, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. [Research Support, Non-U.S. Gov't] 2014 Apr 10;157(2):369–381. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 16.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. [Research Support N.I.H., Extramural Research Support, Non-U.S. Gov't] 2012 Jan 26;481(7382):506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkin B, Ouillette P, Li Y, Keller J, Lam C, Roulston D, et al. Clonal evolution and devolution after chemotherapy in adult acute myelogenous leukemia. Blood. [Research Support N.I.H., Extramural Research Support, Non-U.S. Gov't] 2013 Jan 10;121(2):369–377. doi: 10.1182/blood-2012-04-427039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.