Abstract
Mutations in SETD2, a histone H3 lysine trimethyltransferase, have been identified in clear cell renal cell carcinoma (ccRCC); however it is unclear if loss of SETD2 function alters the genomic distribution of histone 3 lysine 36 trimethylation (H3K36me3) in ccRCC. Furthermore, published epigenomic profiles are not specific to H3K36me3 or metastatic tumors. To determine if progressive SETD2 and H3K36me3 dysregulation occurs in metastatic tumors, H3K36me3, SETD2 copy number (CN) or SETD2 mRNA abundance was assessed in two independent cohorts: metastatic ccRCC (n=71) and the Cancer Genome Atlas Kidney Renal Clear Cell Carcinoma data set (n=413). Although SETD2 CN loss occurs with high frequency (>90%), H3K36me3 is not significantly impacted by monoallelic loss of SETD2. H3K36me3-positive nuclei were reduced an average of ~20% in primary ccRCC (90% positive nuclei in uninvolved vs 70% positive nuclei in ccRCC) and reduced by ~60% in metastases (90% positive in uninvolved kidney vs 30% positive in metastases) (P<0.001). To define a kidney-specific H3K36me3 profile, we generated genome-wide H3K36me3 profiles from four cytoreductive nephrectomies and SETD2 isogenic renal cell carcinoma (RCC) cell lines using chromatin immunoprecipitation coupled with high-throughput DNA sequencing and RNA sequencing. SETD2 loss of methyltransferase activity leads to regional alterations of H3K36me3 associated with aberrant RNA splicing in a SETD2 mutant RCC and SETD2 knockout cell line. These data suggest that during progression of ccRCC, a decline in H3K36me3 is observed in distant metastases, and regional H3K36me3 alterations influence alternative splicing in ccRCC.
Introduction
Cancer of the kidney and renal pelvis affect more than 65 000 patients annually and is 1 of the 10 leading causes of cancer death. The most common histological subtype is clear cell renal cell carcinoma (ccRCC), which accounts for the majority of renal cell carcinoma (RCC)-related deaths. Surgery remains the standard of care for patients with early-stage tumors (stages I–III); however, ~30% of these patients will progress to distant metastases after surgery for localized disease.1 Despite recent advances in systemic therapy, median survival drops to ~2 years after development of metastatic disease.
The von Hippel–Lindau (VHL) tumor-suppressor protein is inactivated in about 90% of ccRCC cases.2 Loss of VHL function occurs early in the development of ccRCC, and it can be detected in the earliest preneoplastic lesions arising in patients with VHL disease. However, Vhl deletion in mice is insufficient for tumorigenesis, which suggests that additional mutations are required.2 Exome sequencing of early-stage ccRCC tumors has identified missense and truncating mutations in those genes that encode for histone-modifying enzymes, such as the genes PBRM1 (polybromo-1) (33–45%),3, 4 BAP1 (15%)4, 5 and SETD2 (3–12%).3, 4, 6 SETD2, a histone H3 methyltransferase, belongs to a superfamily of lysine methyltransferases.3, 6 Histone H3 lysine 36 (H3K36)-specific methyltransferases all contain the catalytic SET domain, with variations in affinity for monomethylation, dimethylation or trimethylation (abbreviated as H3K36me, H3K36me2 and H3K36me3, respectively).7 Homozygous disruption of Setd2 in mice results in embryonic lethality, vascular defects and loss of H3K36me3.8 H3K36me and H3K36me2 were not affected in Setd2-/- mice, which suggests that H3K36 trimethylation is a non-redundant function of Setd2.
We hypothesized that loss of SETD2 histone methyltransferase activity decreases H3K36me3 in human tissue and is associated with genomic alterations of H3K36me3. To determine if there is progressive SETD2 dysregulation, we assessed H3K36me3 staining in primary tumors and in metastatic RCC lesions, and compared these intensities to uninvolved kidney tissue controls. To generate a high-resolution profile of H3K36me3 in ccRCC, we used chromatin immunoprecipitation (ChIP) coupled with high-throughput sequencing to perform a genome-wide analysis of H3K36me3 from four matched ccRCC tumor and uninvolved kidney cases, as well as isogenic SETD2 RCC cell lines and assessed alterations of H3K36me3 at genomic regions with alternative splicing.
Results
SETD2 mutations correlate with decreased H3K36me3 in early-stage ccRCC.
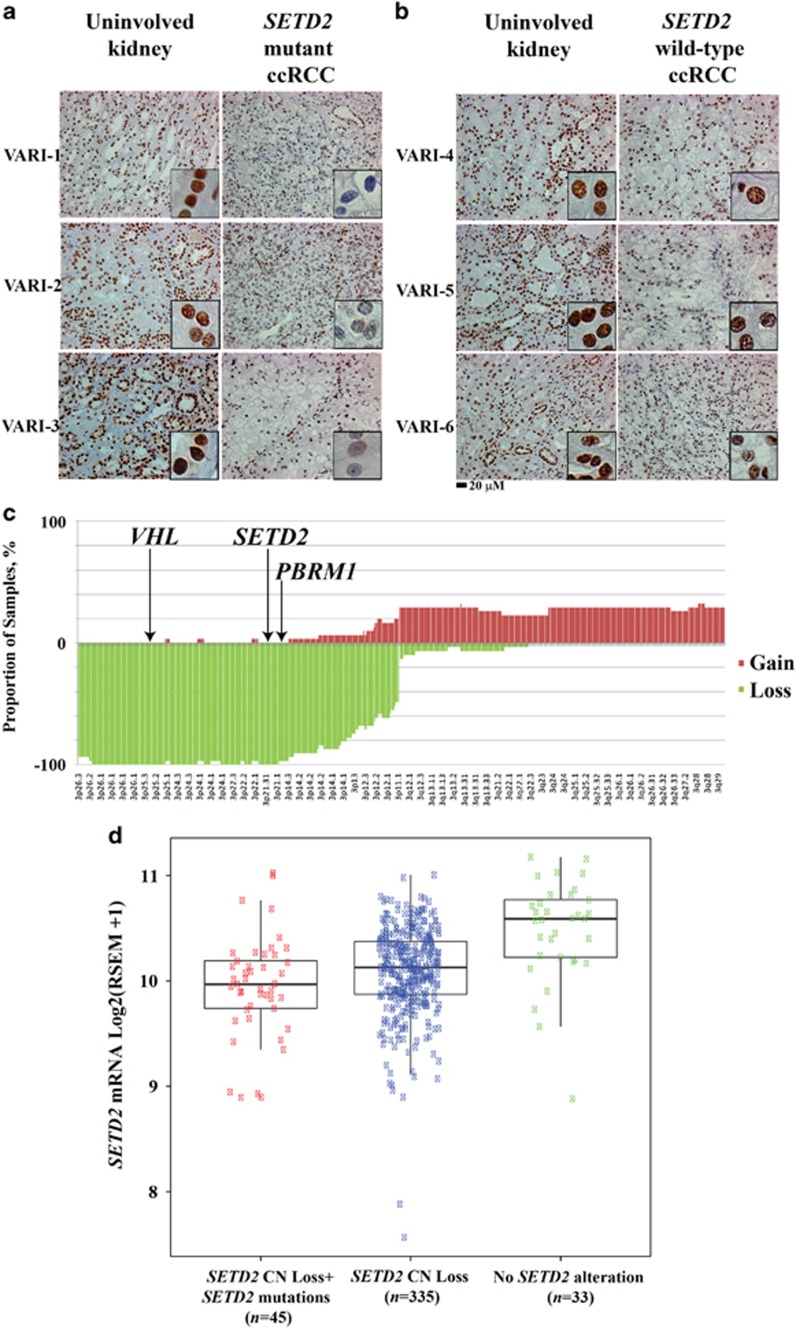
Global levels of H3K36me3 were analyzed in matched uninvolved kidney and ccRCC specimens from patients with early-stage RCC who had concurrent 3p loss and mutations in VHL, PBRM1 and SETD2.6 H3K36me3, as detected using immunohistochemistry (IHC) analysis, was decreased in tumors with mutant SETD2 (28% H3K36me3-positive nuclei; n=6; Figure 1a) compared with tumors with wild-type SETD2 (88% H3K36me3-positive nuclei; n=5, Figure 1b and Supplementary Figure 1C; P<0.05). H3K36me2, as detected using IHC analysis, was unchanged between mutant and wild-type SETD2 (Supplementary Figure 1D; P>0.05), consistent with prior studies of SETD2 protein depletion in cell lines.9 H3K36me3 nuclear staining was preserved in the stromal cells of SETD2 wild-type and SETD2 mutant tumors (Supplementary Figures 1A and B).
Figure 1.
Analysis of SETD2 and H3K36me3 in clear cell renal cell carcinoma (ccRCC) specimens. H3K36me3 immunohistochemical staining of uninvolved kidney and tumor specimens with mutant SETD2 (a) and with wild-type SETD2 (b). SETD2 mutations correlate with decreased H3K36me3 in early-stage ccRCC. Scale bar, 20 μm. (c) Virtual karyotypes formalin-fixed paraffin-embedded metastatic ccRCC specimens (n=51) were generated using Affymetrix single-nucleotide polymorphism arrays. Copy number (CN) loss of the SETD2 gene locus was observed in >90% of tested ccRCC tumors. SETD2 is located at 3p21.31; VHL, 3p25.3; BAP1, 3p21.31-p21.2 (not shown); PBRM1, 3p21.1. (d) Scatter plot and box plot overlay of SETD2 mRNA abundance estimates by RNA-Seq by Expectation-Maximization (RSEM) in the Cancer Genome Atlas Kidney Renal Clear Cell Carcinoma data set (n=413). Boxes represent the 25th, median and 75th interquartile ranges.
In a prior analysis of intratumor heterogeneity, distinct SETD2 loss-of-function mutations that disrupt the SET domain were noted in matched primary tumor and metastasis specimens.10 We assessed primary RCC from patients with advanced RCC for VHL, PBRM1 and SETD2 copy number (CN) loss by generating virtual karyotypes from 51 formalin-fixed paraffin-embedded ccRCC nephrectomy specimens using Affymetrix GeneChip Mapping 250 K Nsp single-nucleotide polymorphism arrays (Figure 1c).11, 12, 13 Tumor DNA was isolated from microdissected paraffin sections. CN loss was observed at the VHL, PBRM1 and SETD2 loci in more than 90% of cases. Further analysis demonstrated that loss of heterozygosity at these loci occurred as part of a generalized loss of 3p in these tumor samples. To evaluate mRNA expression after CN loss, SETD2 mRNA abundance was analyzed from the Cancer Genome Atlas (TCGA) KIRC data set. Of the 413 samples analyzed, 81% (335/413), 11% (45/413) and 8% (33/413) had SETD2 CN loss alone, SETD2 CN loss with a concurrent SETD2 mutation and no SETD2 alterations, respectively. SETD2 mRNA abundance was lowest in samples with SETD2 CN loss and concurrent SETD2 mutations (Figure 1d) when compared with samples with SETD2 CN loss alone (P<0.05) or no SETD2 alterations (P<0.05), suggesting that biallelic inactivation leads to decreased mRNA expression.
H3K36me3 was progressively dysregulated in metastases
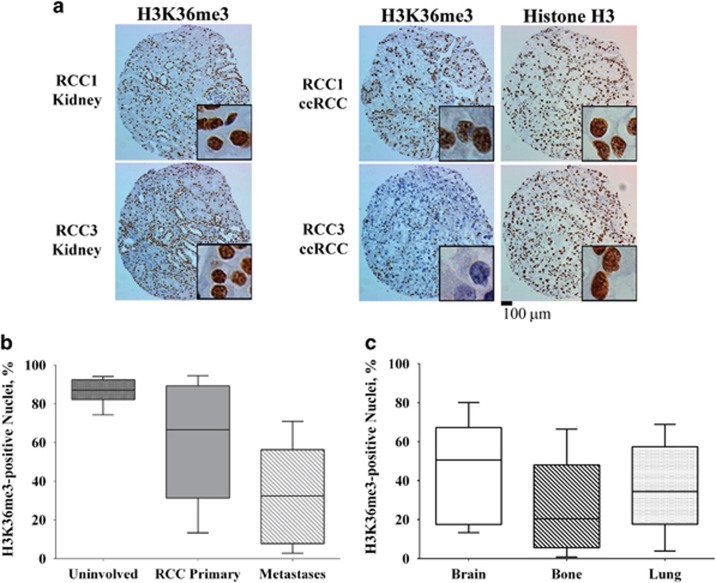
Since more than 90% of tumors had SETD2 CN losses in both our tumors (n=51) and the TCGA data set (n=413), we examined H3K36me3 levels by IHC as a surrogate of SETD2 H3K36 methyltransferase activity to identify tumors with loss of H3K36me3 (Figure 2a, bottom panel, 7% H3K36me3-positive nuclei). In a cohort of unmatched uninvolved kidney, primary RCC and resected metastases (bone, brain and lungs), H3K36me3-positive nuclei were reduced an average of ~20% in primary RCC (90% positive nuclei in uninvolved vs 70% positive nuclei in RCC) and were reduced by ~60% in metastases (90% positive in uninvolved kidney vs 30% positive in metastases; P<0.001; Figure 2b). H3K36me3 nuclear staining was analyzed across metastatic sites: positive staining for H3K36me3 was reduced in the brain (46%), bone (28%) and lung (37%) metastases relative to uninvolved kidney tissue (P=0.007; Figure 2c and Supplementary Figure 2). Together, these data indicate that although SETD2 CN losses occur with high frequency (>90%) in primary RCC, H3K36 trimethyltransferase activity is not impacted by monoallelic loss of SETD2, decreases during progression of RCC and is lowest in metastatic lesions. In patients presenting with metastatic disease, overall levels of H3K36me3 in the primary tumors (n=71), as assessed by the percentage of nuclear staining for H3K36me3 by IHC (expressed as 0–100%), was not predictive of clinical site of metastases or of the number of distant metastases at the time of diagnosis (Table 1). These data suggest that complete loss of H3K36 trimethyltransferase function is not required for the development of the initial cancer phenotype or metastases, but that a progressive decrease in H3K36me3 may result from adaptation processes or cooperate with other mechanisms in distant metastases.
Figure 2.
H3K36me3 immunohistochemical analysis of uninvolved kidney, clear cell renal cell carcinoma (ccRCC) and metastases. (a) Compared with matched uninvolved kidney parenchyma, a subset of ccRCC tumors had decreased H3K36me3. Top panel, >60% positive nuclei; bottom panel, <10% positive nuclei. Scale bar, 100 μm. (b) Box-and-whiskers plots of H3K36me3 in tissue microarrays, representing uninvolved kidney tissue (n=30), primary RCC tumors (n=71) and metastases (n=45). Boxes represent the 25th, median and 75th percentiles. Whiskers represent the 10th and 90th percentiles. (c) H3K36me3 in brain (n=12), bone (n=27) and lung (n=6) metastases.
Table 1. Association of histone H3 lysine 36 trimethylation (H3K36me3) immunohistochemical staining in nephrectomy specimens with clinical findings at the time of diagnosis for patients with metastatic RCC.
| Number of metastatic sitesa | n | Mean (H3K36me3%) | s.d. (%) | Median (%) | Minimum (%) | Maximum (%) | P-value |
|---|---|---|---|---|---|---|---|
| 1 | 18 | 64.43 | 29.40 | 70.75 | 6.70 | 95.66 | |
| 2 | 33 | 61.89 | 29.50 | 71.41 | 6.87 | 95.95 | |
| 3 | 10 | 48.16 | 33.16 | 34.96 | 10.66 | 95.15 | |
| 4 | 9 | 59.74 | 30.54 | 64.78 | 11.12 | 94.71 | 0.56b |
| Lung | |||||||
| No | 14 | 63.90 | 30.58 | 74.04 | 6.97 | 95.26 | |
| Yes | 57 | 58.64 | 30.25 | 63.62 | 6.70 | 95.95 | 0.56c |
| Bone | |||||||
| No | 54 | 59.87 | 29.81 | 64.42 | 6.70 | 95.95 | |
| Yes | 17 | 59.06 | 32.22 | 71.96 | 6.97 | 95.15 | 0.92c |
| Lymph nodes | |||||||
| No | 37 | 62.12 | 31.22 | 71.50 | 6.70 | 95.66 | |
| Yes | 34 | 57.02 | 29.22 | 64.20 | 6.87 | 95.95 | 0.48c |
| Liver | |||||||
| No | 64 | 58.33 | 30.49 | 64.29 | 6.70 | 95.95 | |
| Yes | 7 | 71.97 | 25.80 | 84.61 | 20.11 | 95.26 | 0.26c |
Abbreviation: RCC, renal cell carcinoma.
One patient with missing number of metastatic sites.
F test by analysis of variance.
t-Test.
ChIP sequencing identifies the genome-wide loss of H3K36me3 in human nephrectomy specimens
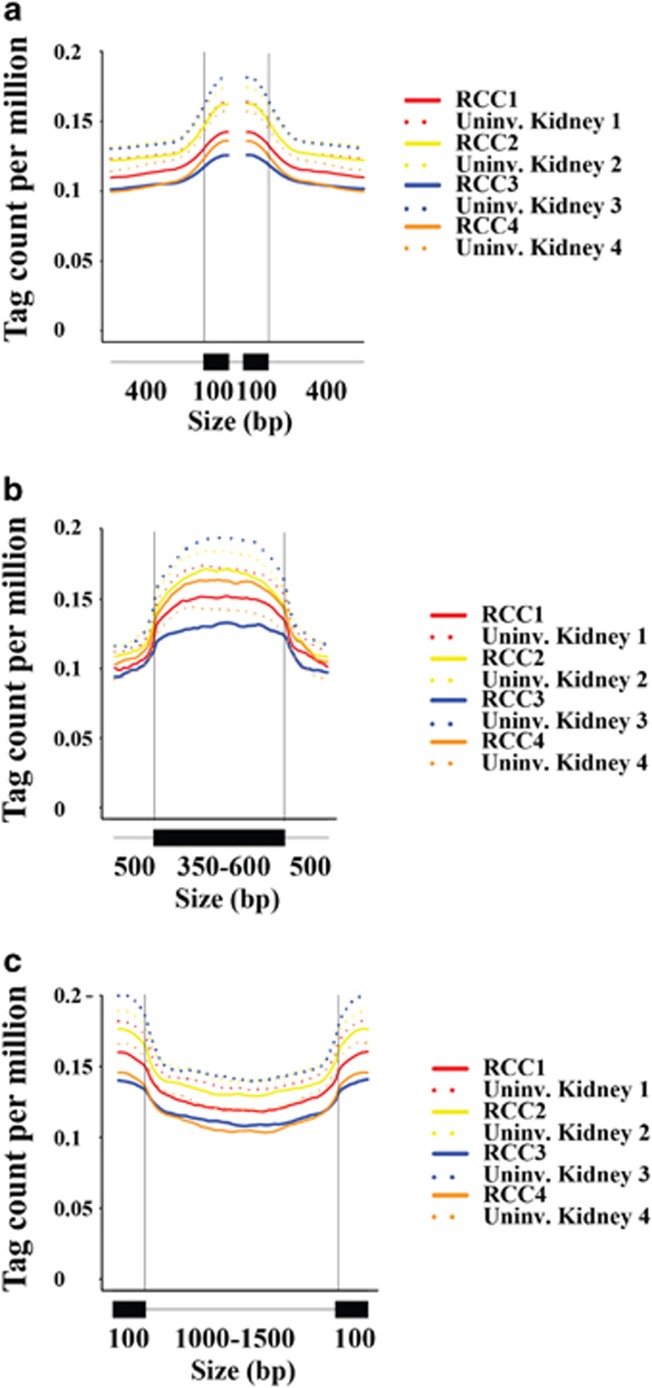
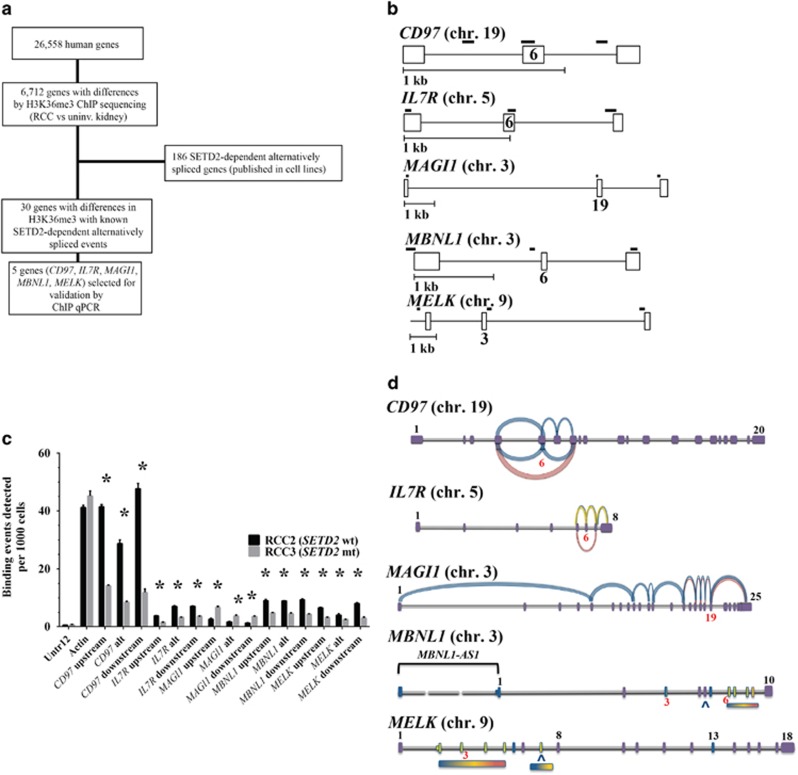
Formaldehyde-assisted chromatin capture of regulatory elements identified altered chromatin accessibility in RCC specimens from patients with localized disease, but these epigenomic profiles were not specific to H3K36me3 or metastatic tumors.14, 15 We wanted to assess the effect of SETD2 mutations from patient tumors on H3K36me3 distribution profiles and the impact of this altered distribution profile on alternative splicing of candidate genes. To define the H3K36me3 profile in cytoreductive nephrectomy tissue from patients with advanced RCC, we examined the genome-wide distribution of H3K36me3 by ChIP sequencing (Supplementary Table 1). Since the detection of SETD2 mutations may vary because of tumor heterogeneity, IHC of H3K36me3 was used to phenotype the tumors into either a high-H3K36me3 group (more than 60% of the tumor staining for H3K36me3) or a low-H3K36me3 group (less than 10% staining; Figure 2a). Targeted exome sequencing confirmed SETD2 mutations in one sample (RCC3) with decreased H3K36me3 (Supplementary Table 2). In Caenorhabditis elegans and mouse chromatin, H3K36me3 is known to be predominantly associated with gene bodies, with much higher coverage for exons than introns.16 To evaluate the human genome-wide distribution of H3K36me3, the H3K36me3 was analyzed across intron–exon and exon–intron boundaries. Consistent with prior studies in other species, we observed enrichment of H3K36me3 signals in exon regions compared with introns for both uninvolved kidney and RCC (Figure 3). Of all eight ChIP sequencing libraries, the SETD2 mutant (RCC3) had the largest depletion of H3K36me3 at exons. To make comparisons across eight ChIP libraries, we determined the average fragment densities for 26 558 human genes. The calculated average fragment densities for all human gene bodies were used for further analysis to screen samples for differences in H3K36me3 marked genes. Analysis of variance comparing the average fragment densities of genes from RCC to those of uninvolved kidney identified 6712 genes (false discovery rate <0.1, corresponding P<0.05, Figure 4a and Supplementary Tables 3-5). To further refine intragenic H3K36me3 alterations, we examined the overlap between the genes identified in tissue by RCC ChIP sequencing and published SETD2-regulated alternative splicing events in cell lines.17 In a comparison of tumor and uninvolved kidney specimens, 30 SETD2-regulated alternatively spliced genes had statistically significant differences in average H3K36me3 fragment densities (Figure 4a and Supplementary Table 3). Our data suggest that for these alternative splicing events, there is a complex interplay between loss of SETD2 histone methyltransferase activity, preferential depletion of H3K36me3 at exons, and alternative exon usage.
Figure 3.
Whole-genome H3K36me3 chromatin immunoprecipitation of four paired nephrectomy samples (uninvolved kidney and renal cell carcinoma (RCC). (a–c) H3K36me3 signal intensity across intron–exon and exon–intron boundaries. Only genes with at least three exons were selected. (a) For each gene, the first and last exons, as well as exons of <100 bp and introns of <400 bp were excluded from the plots. Each colored line represents the H3K36me3-binding profile over 400 bp intron and 100 bp exon sequence, with the two vertical lines denoting intron–exon and exon–intron junction sites, respectively. (b) H3K36me3 signal intensity across exons of 350–600 bp and 500-bp flanking intronic sequence. Only exons of 350–600 bp flanked by introns of ⩾500 bp at both sides were selected. (c) H3K36me3 signal intensity across introns of 1000–1500 bp and 100-bp flanking exonic sequence. Only introns of 1000–1500 bp flanked by exons of ⩾100 bp at both sides were selected.
Figure 4.
Analysis of SETD2-genotyped clear cell renal cell carcinoma (ccRCC). (a) Selection of 30 candidate genes for further analysis of H3K36me3. The average fragment densities across gene bodies were determined for 26 558 human genes. Analysis of variance comparing the average fragment densities of genes from ccRCC to those of uninvolved kidney identified 6712 genes with differences in H3K36me3 (P<0.05). One hundred eighty-six genes were previously reported to undergo SETD2-dependent alternative splicing in human mesenchymal stem cells.17 (b) Intragenic regions selected for ChIP qPCR analysis. The alternative exons are numbered and the solid bold lines represent the regions examined by three PCR primer pairs (upstream, alternative exon and downstream). (c) ChIP qPCR assay of DNA isolated from SETD2-wild-type (wt) and SETD2 mutant (mt) ccRCCs. Error bars represent s.e.m. Asterisk (*) indicates P<0.05 by t-test. (d) Relative abundance of splicing isoforms identified by RNA sequencing of SETD2 mt ccRCC. RNA sequencing predictions identify specific exons, or 'hotspot' regions containing several exons, which are involved in alternative splicing events. RNA abundances are based on fragments per kilobase of exon per million fragments mapped values binned into high (red), medium (gold) and low (blue) transcripts; exons are numerically labeled. Exons examined by ChIP qPCR are labeled in red.
Loss of SETD2 methyltransferase activity is associated with alterations in mRNA splicing in human ccRCC
H3K36me3 readers, such as MRG15, function as adaptor proteins for alternative splicing factors.7 To further assess the impact of SETD2 loss on splicing in RCC, we designed ChIP quantitative PCR primers to bind the upstream and downstream genomic sequences flanking the alternative exon for selected genes: CD97, IL7R, MAGI1, MBNL1 and MELK (Figure 4b). These genes were both identified by our ChIP sequencing profiles and previously identified in a high-throughput cDNA screen of cell lines with alternative splicing events sensitive to SETD2 downregulation.17 Actin was used as a positive control, and a “gene desert” on chromosome 12 was used as a negative control. We detected differences in H3K36me3-binding events at the regions flanking the alternative cassette exons in a SETD2-wild-type RCC (RCC2) compared with a SETD2 mutant RCC (RCC3; P<0.05; Figure 4c and Supplementary Table 6). No significant differences were observed in the number of binding events at chromosome 12 (negative control) or at actin (positive control) in the SETD2-wild-type (RCC2) when compared with those of the SETD2 mutant (RCC3) tumor. The ChIP PCR results were also concordant with the calculated average H3K36me3 fragment densities from ChIP sequencing; smaller average H3K36me3 fragment densities identified by ChIP sequencing were associated with decreased H3K36me3-binding events detected by ChIP PCR. Interestingly, ChIP sequencing identified genes such as MAGI1 with paradoxically higher H3K36me3 enrichment in SETD2 mutant RCC and global loss of H3K36me3 at the ChIP qPCR-amplified sites was not observed (Figure 4c).
To assess the diversity of RNA splicing isoforms, we performed in parallel, ChIP and RNA sequencing, of the SETD2 mutant tumor (RCC3) and matched uninvolved kidney specimen. Consistent with published SETD2-depleted cell lines, we identified multiple splicing alterations at genes (CD97, IL7R, MAGI1, MBNL1 and MELK) with alterations in H3K36me3 validated by ChIP quantitative PCR (qPCR; Figure 4d). For all of the genes, exon–intron islands were identified where differential splicing occurs more frequently in association with loss of H3K36me3 than in other areas of the gene. For example, for IL7R, RNA sequencing identified splicing events involving exons 5, 6, 7 and 8 in SETD2 mutant tumor (RCC3). The most abundant RNA splicing isoform (highlighted in red) is a splice event where exon 6 is excluded. Our data indicate that H3K36me3 redistribution occurs at discrete intragenic regions in SETD2 mutant RCC (RCC3) and is associated with diverse RNA splicing isoforms.
Loss of SETD2 methyltransferase activity is associated with genome-wide alterations of H3K36me3 and diverse RNA splicing isoforms in isogenic RCC cell lines
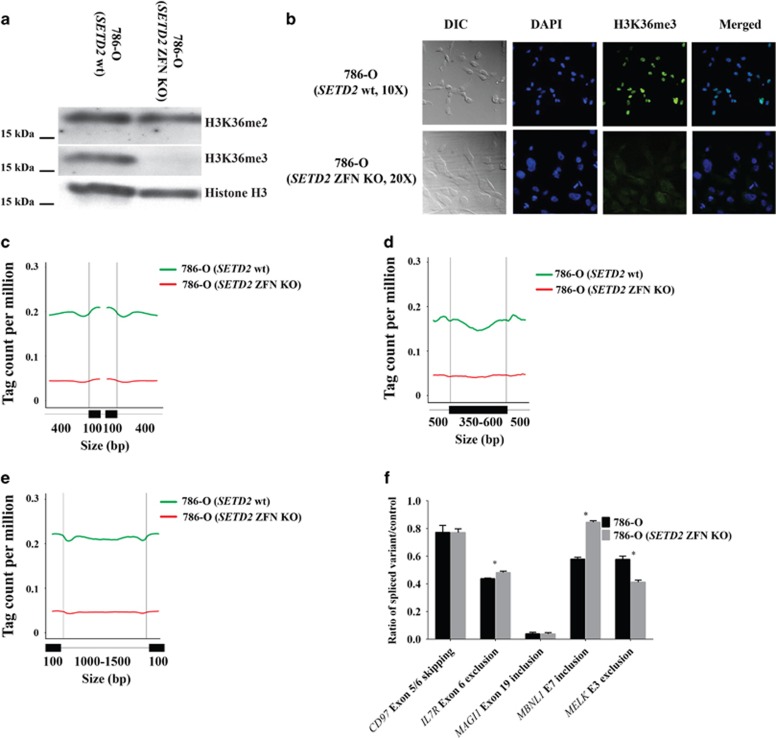
To further study SETD2-regulated events in vitro, we generated isogenic SETD2 cell lines to analyze both alterations in H3K36me3 and RNA splicing.10, 18 We used zinc finger nucleases (ZFNs) to target the SETD2 gene to generate homozygous deletions in VHL-deficient (786-O) RCC cell lines (Supplementary Figure 3). Depletion of SETD2 methyltransferase activity was confirmed by decreased H3K36me3 by western blot and immunofluorescence (Figures 5a and b). Consistent with our analysis of SETD2 mutant RCC and published Setd2-deficient mice, H3K36me2 levels by western blot were not affected in the SETD2 ZFN knockout (KO) cell line.8 To assess the diversity of RNA splicing isoforms, we performed in parallel, ChIP and RNA sequencing, of SETD2 ZFN KO and parental 786-O cell lines. To evaluate the genome-wide distribution of H3K36me3, the H3K36me3 was analyzed across intron–exon and exon–intron boundaries. Consistent with our nephrectomy samples, we observed enrichment of H3K36me3 signals in exon regions compared with introns. Genome-wide depletion of H3K36me3 was observed in the SETD2 ZFN KO when compared with the parental cell line 786-O (Figures 5c and e). Our analysis of RNA sequencing identified both differentially spliced exons and exon regions between the SETD2 wt and KO cell lines (Supplementary Figure 4). To further analyze the alternative splicing events, two sets of reverse transcription–PCR primers were designed to quantitate mRNA splicing isoforms with cassette exon skipping or inclusion (Figure 5f and Supplementary Table 7). Results are expressed as a normalized ratio of splicing isoforms to control. Loss of H3K36me3 in cell lines was associated with an increase in IL7R exon 6 exclusion, MBNL1 exon 7 inclusion and MELK exon 3 exclusion (P<0.05 by t-test). We did not detect statistically significant changes in CD97 exon 5/6 skipping or MAGI1 exon 19 inclusion (P>0.05 by t-test). Our data in cell lines indicate that SETD2 deletion leads to genomic redistribution of H3K36me3 associated with splicing isoforms.
Figure 5.
Analysis of Isogenic SETD2 renal cell carcinoma cell lines. The VHL-deficient, SETD2 wild-type 786-O cell line was transfected with zinc finger constructs that generate a deletion in the SETD2 catalytic domain. Single cell-derived clones in a 96-well plate were analyzed by fragment length analysis to identify those with frameshifts (11 bp deletion) and then expanded for additional analyses. (a) Western blot confirming depletion of H3K36me3, but not H3K36me2. Total levels of histone H3 was used as a loading control. (b) Immunofluorescence confirming depletion of H3K36me3 from an expanded single-cell clone. (c) For each gene, the first and last exons, as well as exons of <100 bp and introns of <400 bp were excluded from the plots. Each colored line represents the H3K36me3-binding profile over 400 bp intron and 100 bp exon sequence, with the two vertical lines denoting intron–exon and exon–intron junction sites, respectively. (d) H3K36me3 signal intensity across exons of 350–600 bp and 500 bp flanking intronic sequence. Only exons of 350–600 bp flanked by introns of ⩾500 bp at both sides were selected. (e) H3K36me3 signal intensity across introns of 1000–1500 bp and 100 bp flanking exonic sequence. Only introns of 1000–1500 bp flanked by exons of ⩾100 bp at both sides were selected. (f) Analysis of alternative splicing. Two sets of reverse transcription–PCR primers were designed to quantitate alternative splicing isoforms. Results are expressed as a normalized ratio of splicing isoforms to control with error bars representing standard error. DAPI, 4',6-diamidino-2-phenylindole; DIC, differential interference contrast; ZFN, zinc finger nuclease.
Discussion
The discovery that mutations in histone-modifying genes occur in a significant percentage of RCC has led to a search for the functional significance of these alterations. To our knowledge, our data represent the first comparison of H3K36me3-specific profiles of metastatic RCC tumors with a defined SETD2 genotype. Some of the first questions we asked were whether CN loss of SETD2 is linked to H3K36me3 levels, and whether there is progressive loss of H3K36me3 as a function of tumor progression. Our data from nephrectomy specimens indicate that that chromosome 3p loss leads to SETD2 CN loss in more than 90% of tested specimens and SETD2 mutations decrease H3K36me3, which is consistent with prior observations.4 However, SETD2 CN loss alone may not have a proportional effect on loss of H3K36me3, with a relatively high level of H3K36me3 levels remaining in tumors with 3p loss. This observation suggests that decreased SETD2 mRNA expression after monoallelic loss is not sufficient to alter SETD2 methyltransferase activity. It is nevertheless possible that subtle changes in SETD2 protein levels may have a cooperative biological effect in these early tumors through the monoallelic loss of other chromosome 3p tumor suppressors.
Biallelic SETD2 inactivation may be a later event during tumor progression. We observed a substantial decrease of nuclear staining for H3K36me3 in bone, brain and lung metastases compared with primary lesions (P<0.05). These findings suggest a link between loss of H3K36me3 and RCC tumor progression or adaptation. The predicted mutational frequency of SETD2 in early-stage primary RCC tumors is 12% however, our data indicated a considerably higher loss of H3K36me3 in more advanced lesions, which suggests either a higher prevalence of SETD2 mutations in metastatic RCC or the presence of additional post-translational mechanisms that downregulate H3K36me3. Indeed, multiregion sequencing of ccRCC has identified parallel evolution of distinct SETD2 mutations that phenotypically converge on loss of SETD2 catalytic activity.10 In addition to RCC, SETD2 mutations (12–15%) have been characterized in bladder cancers, gliomas and sarcomas, and we hypothesize cancers with loss of SETD2 methyltransferase function share similar genomic regions with H3K36me3 alterations.19, 20, 21 Sequencing of matched diagnosis-remission-relapse samples in pediatric leukemias identified acquisition of SETD2 mutations further supporting a link between decreased H3K36me3 and disease progression or adaptation.19 High-resolution H3K36me3 profiling of other SETD2 mutant cancers may identify these regions with aberrant chromatin accessibility.
To better characterize the effects of SETD2 dysregulation on genome-wide H3K36me3 levels and on mRNA splicing, we performed H3K36me3 profiling in four paired RCC tumors and uninvolved kidneys through ChIP sequencing and interrogation of H3K36me3 at genomic sites flanking the alternatively spliced cassette exons. The impact of SETD2 loss of function on H3K36me3 profiles at SETD2-dependent alternatively spliced exons in RCC has not been previously examined. We selected five genes with SETD2-dependent alternatively spliced exons for further study by ChIP PCR and identified dysregulation of H3K36me3 in samples with biallelic SETD2 loss. Our DNA and RNA sequencing in SETD2 mutated tumors and cell lines suggest that a key result is the manifestation of H3K36me3 redistribution and aberrant splicing isoforms. We confirmed by quantitative reverse transcription–PCR changes in alternative splicing isoforms with H3K36me3 depletion. At this time, the clinical and functional significance of these alternate splice variants and their impact on RCC tumor biology are not known, but form the basis of future work on elucidating their effect on RCC biology.
Loss of SETD2 function can affect tumor biology in a number of different ways, including a direct impact on p53 transcriptional regulation, and an indirect effect via loss of H3K36me3.9, 22 H3K36me3 regulates DNA mismatch repair, DNA double-strand break repair, alternative splicing and chromatin remodeling, and these functions are linked to chromatin readers with proline–tryptophan–tryptophan–proline domains that interact with methylated lysine residues.17, 23, 24, 25, 26 Treatment of SETD2 protein-deficient cell lines with DNA-damaging agents fail to activate DNA repair mechanisms and leads to increased genomic deletions.25, 26As SETD2 is a non-redundant histone H3 methyltransferase, it is likely that loss of SETD2 will change gene marking by H3K36me3, thus altering the recruitment of chromatin readers. H3K36me3 is recognized by chromatin readers, and regional alterations of H3K36me3 caused by SETD2 dysregulation may lead to repositioning of other proline–tryptophan–tryptophan–proline domain-containing proteins. Misregulated splicing can also affect diverse cellular processes including embryonic development, cellular signaling, apoptosis, angiogenesis and epithelial-to-mesenchymal transition re-programming of cells to cause human disease.27, 28, 29, 30
Similar to published genome-wide analyses of patient-derived biopsy tissue,3, 6, 31 ChIP sequencing in whole tissue presents logistical challenges. Although the tumors were selected from cytoreductive nephrectomies and underwent macrodissection, our H3K36me3 profile represents the aggregate epigenetic environment of the kidney, which includes stroma and infiltrating immune cells such as lymphocytes. We used both input DNA and matched uninvolved kidney as controls, but we did not disaggregate the tumor from the stroma, given the large amount of total DNA required for ChIP sequencing. Our paradoxical finding of increased H3K36me3 in specific genomic regions may also be influenced by large-scale recruitment of specific non-tumor cell populations that possess these histone methylation patterns. Additional filters, such as using a minimal fragment density threshold, could reduce the number of false-positives. Nonetheless, we believe these data provide an accurate representation of H3K36me3 marked genes in tissue because ChIP PCR confirmed differences in H3K36me3 binding to specific genomic sites among PBRM1 mutant, SETD2 mutant, PBRM1 mutant and SETD2 wild-type specimens. As RCC can display intratumoral heterogeneity, we attempted to control for macroscopic heterogeneity by using H3K36me3 IHC of whole-tumor specimens as a surrogate for SETD2 methyltransferase activity to stratify the cytoreductive nephrectomies.10, 18
In conclusion, there was relatively high level of H3K36me3 in primary RCC despite the near universal loss of one SETD2 allele in clear cell RCC. We observed a significant decrease of H3K36me3 activity in distant metastases relative to primary tumor, however, H3K36me3 levels were not predictive of clinical site of metastases or of the number of distant metastases. In tumors and cell lines with SETD2 mutations, we observed significant intragenic redistribution of H3K36me3, and alterations in splice isoforms when compared with non-SETD2 mutated controls. The phenotypic impact of these epigenetic alterations is challenging to define, because recruitment of chromatin readers may have an immediate effect on the cellular phenotype, or may be part of a more complex process that evolves over time in conjunction with other genomic or phenotypic alterations in the cancer cell. Nonetheless, as we develop a mechanistic understanding of the role SETD2 loss and H3K36me3 play in RCC tumors, we will be able to tailor drug strategies that target the downstream consequences of SETD2 loss and H3K36me3 dysregulation more precisely, and alter the course of RCC and other SETD2 mutated cancers.32, 33, 34, 35, 36
Materials and methods
Patient samples
Early-stage SETD2 wild-type and SETD2 mutant RCC samples were identified by the Van Andel Research Institute, Grand Rapids, Michigan, as previously described.6 For metastatic renal cell carcinoma (RCC), tissue samples were collected from patients enrolled in two phase II trials at the University of Texas MD Anderson Cancer Center, Houston, Texas, while undergoing either radical or partial nephrectomy for sporadic clear cell RCC (ccRCC). In a prospective phase II trial, untreated patients with metastatic ccRCC were randomly allocated to receive sorafenib 400 mg orally twice daily with or without subcutaneous interferon (0.5 mU twice daily). Participants were enrolled from 25 June 2005 to 18 June 2007.37 The second trial was a prospective single-arm phase II trial, for untreated patients with metastatic ccRCC whose primary tumors were considered resectable.38 Patients received bevacizumab with or without erlotinib. Participants were enrolled between March 2005 and March 2008 and included in the analysis. All patients had signed an informed consent approved by an institutional review board under protocols 2003-0982, 2004-0526 and 08-0186 (ClinicalTrials.gov Identifiers NCT00126594 and NCT00113217). End points for both trials included progression-free survival and overall survival. Continuous variables were compared between groups using the two-sample t-test or, if there were more than two groups, by an analysis of variance.
ChIP sequencing
ChIP and input DNAs were prepared at ActiveMotif (Carlsbad, CA, USA) by converting overhangs into phosphorylated blunt ends, the addition of an adenine to the 3' ends and the addition of Illumina adaptors (Illumina, San Diego, CA, USA). The library was size-selected (175–225 bp) and amplified for 18 cycles. The resulting amplified DNAs (DNA libraries) were purified, quantified and tested by qPCR at the same specific genomic regions as the original ChIP DNA to assess the quality of the amplification reactions. Amplified DNAs were sequenced on the Illumina Genome Analyzer II. The obtained raw FASTQ sequences (36–50 nt reads, single end) were aligned to the human genome (hg19) using the Burrows-Wheeler Alignment.39 To illustrate the changes of H3K36me3 signals across exon–intron and intron–exon junctions, we generated three subsets of junctions from the hg19 refGene annotation. The first subset represents junctions between exons of ⩾100 bp and introns of ⩾400 bp from genes with at least three exons; the first and last exons were excluded from the analysis. The second subset represents junctions with exons of 350–600 bp flanked by introns of ⩾500 bp at both sides. The third subset represents junctions with introns of 1000–1500 bp flanked by exons of ⩾100 bp at both sides. H3K36me3 signal was calculated as number of reads per million uniquely mapped reads and plotted using the ngsplot software (http://code.google.com/p/ngsplot/).40
ChIP qPCR
Nephrectomy samples underwent gross macrodissection with mirror hematoxylin–eosin staining to ensure that they contained >60–70% tumor then shipped to ActiveMotif. Matched uninvolved kidney and tumor were fixed with 1% formaldehyde for 15 min and quenched with 0.125 M glycine. After the addition of lysis buffer and disruption with a Dounce homegenizer, the DNA was sheared with sonication to an average length of 300–500 bp. As a control, input genomic DNA was prepared by treating chromatin with RNase, proteinase K and heat.
An aliquot of chromatin (25–30 μg) was pre-cleared with protein A agarose beads (Invitrogen, Carlsbad, CA, USA). Genomic DNA regions of interest were isolated using 4 μg antibody to H3K36me3 (#61101, ActiveMotif). After overnight incubation at 4 °C, protein complexes were eluted from the beads with SDS buffer, then treated with RNase and proteinase K treatment. Reversal of crosslinks was performed at 65 °C overnight. ChIP qPCR was carried out in triplicate with a standard curve on specific genomic regions using SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA). Negative control primers were purchased from ActiveMotif (#71001, ActiveMotif). Cycling conditions were 95 °C for 15 s, 58 °C for 20 s and 72 °C for 20 s. The resulting signals were normalized for primer efficiency by carrying out qPCR for each primer pair using input DNA. Primer efficiency ratios were determined for each primer used in the experiment. The primer efficiency ratio was calculated by dividing the average input value (value given by primers when using 12.5 ng of unprecipitated genomic DNA) by the expected CN of the input. Data were normalized using the formula: ([{average raw data} × {resuspension volume/5}] × [1000/cell equivalents in IP])/(primer efficiency ratio).
mRNA splicing analysis in tissue
Frozen nephrectomy tissue was thawed in the presence of RNAlater solution (Qiagen, Valencia, CA, USA), and total RNA was extracted from ~30 mg using the RNease Mini Kit (Qiagen, cat# 74104). Residual DNA was removed by treatment with DNAse I. Libraries were prepared with Illumina's TruSeq RNA sample preparation V2 kit with final amplification using TruSeq bar-coded primers. Libraries were sequenced on an Illumina HiSeq 2500 to generate paired-end 100 nucleotide reads by ActiveMotif. Analysis of RNA sequencing data sets using the cufflinks41 and ChIP-RNAseq-PRO42 software packages was used to generate prediction models of transcript splice isoforms in H3K36me3-deficient regions. RNA abundance was predicted by calculating the fragments per kilobase of exon per million fragments mapped (FPKM) for each transcript prediction. The most abundant isoform for each gene was assigned a score of 1000 and minor isoforms are scored by examining the ratio of minor FPKM to major FPKM. Transcript abundance was defined as low (abundance score (as) <500); intermediate abundance (as >500, <1000) and the most abundant transcript (as=1000).
Acknowledgments
We thank Ian Davis, W Kimryn Rathmell, Kathryn E Hacker and Jeremy M Simon for helpful discussions and assistance with the genotyping of tissue. THH is supported by funding from the ASCO Young Investigator Award from the Kidney Cancer Association, the Action to Cure Kidney Cancer Organization, the MD Anderson Hematology-Oncology Fellowship, NIH grant K12 CA90628, a Mayo Clinic CR5 grant, the Mayo Clinic Center for Individualized Medicine Epigenomics Translational Program and a Kathryn H and Roger Penske Career Development Award to Support Medical Research. MDC is supported by the Mayo Clinic Center for Individualized Medicine.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med 2005; 353: 2477–2490. [DOI] [PubMed] [Google Scholar]
- Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: a metabolic disease. Nat Rev Urol 2010; 7: 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 2011; 469: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013; 499: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, Leng N, Pavia-Jimenez A, Wang S et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet 2012; 44: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 2010; 463: 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 2012; 13: 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Sun XJ, Zhang YL, Kuang Y, Hu CQ, Wu WL et al. Histone H3 lysine 36 methyltransferase Hypb/Setd2 is required for embryonic vascular remodeling. Proc Natl Acad Sci USA 2010; 107: 2956–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008; 27: 406–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012; 366: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Shen SS, Ayala AG, Ro JY, Truong LD, Alvarez K et al. Virtual-karyotyping with SNP microarrays in morphologically challenging renal cell neoplasms: a practical and useful diagnostic modality. Am J Surg Pathol 2009; 33: 1276–1286. [DOI] [PubMed] [Google Scholar]
- Alvarez K, Kash SF, Lyons-Weiler MA, Kim HJ, Peterson LE, Mathai B et al. Reproducibility and performance of virtual karyotyping with SNP microarrays for the detection of chromosomal imbalances in formalin-fixed paraffin-embedded tissues. Diagn Mol Pathol 2010; 19: 127–134. [DOI] [PubMed] [Google Scholar]
- Monzon FA, Alvarez K, Peterson L, Truong L, Amato RJ, Hernandez-McClain J et al. Chromosome 14q loss defines a molecular subtype of clear-cell renal cell carcinoma associated with poor prognosis. Mod Pathol 2011; 24: 1470–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JM, Hacker KE, Singh D, Brannon AR, Parker JS, Weiser M et al. Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res 2013; 24: 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MJ, Raaijmakers LM, Ramakrishnan S, Wang D, Valiyaparambil S, Liu S et al. Alterations in chromatin accessibility and DNA methylation in clear cell renal cell carcinoma. Oncogene 2013. [DOI] [PubMed]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet 2009; 41: 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science 2010; 327: 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 2014; 46: 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mar BG, Bullinger LB, McLean KM, Grauman PV, Harris MH, Stevenson K et al. Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat Commun 2014; 5: 3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontebasso AM, Schwartzentruber J, Khuong-Quang DA, Liu XY, Sturm D, Korshunov A et al. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol 2013; 125: 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph CG, Hwang H, Jiao Y, Wood LD, Kinde I, Wu J et al. Exomic analysis of myxoid liposarcomas, synovial sarcomas, and osteosarcomas. Genes Chromosomes Cancer 2014; 53: 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie P, Tian C, An L, Nie J, Lu K, Xing G et al. Histone methyltransferase protein SETD2 interacts with p53 and selectively regulates its downstream genes. Cell Signal 2008; 20: 1671–1678. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 2010; 142: 967–980. [DOI] [PubMed] [Google Scholar]
- Li F, Mao G, Tong D, Huang J, Gu L, Yang W et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell 2013; 153: 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho S, Vitor AC, Sridhara SC, Martins FB, Raposo AC, Desterro JM et al. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. Elife 2014; 3: e02482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister SX, Ahrabi S, Zalmas LP, Sarkar S, Aymard F, Bachrati CZ et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombintion repair and genome stability. Cell Rep 2014; 7: 2006–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J 2004; 23: 3103–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnal S, Vigevani L, Valcarcel J. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov 2012; 11: 847–859. [DOI] [PubMed] [Google Scholar]
- Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med 2012; 18: 472–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 2013; 13: 97–110. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network.. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008; 455: 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai A, Meetze K, Vo NY, Kollipara S, Mazsa EK, Winston WM et al. GP369, ;an FGFR2-IIIb-specific antibody, exhibits potent antitumor activity against human cancers driven by activated FGFR2 signaling. Cancer Res 2010; 70: 7630–7639. [DOI] [PubMed] [Google Scholar]
- Finley SD, Popel AS. Predicting the effects of anti-angiogenic agents targeting specific VEGF isoforms. AAPS J 2012; 14: 500–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivas-Mejia PE, Rodriguez-Aguayo C, Han HD, Shahzad MM, Valiyeva F, Shibayama M et al. Silencing survivin splice variant 2B leads to antitumor activity in taxane—resistant ovarian cancer. Clin Cancer Res 2011; 17: 3716–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James LI, Barsyte-Lovejoy D, Zhong N, Krichevsky L, Korboukh VK, Herold JM et al. Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain. Nat Chem Biol 2013; 9: 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011; 146: 904–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonasch E, Corn P, Pagliaro LC, Warneke CL, Johnson MM, Tamboli P et al. Upfront, randomized, phase 2 trial of sorafenib versus sorafenib and low-dose interferon alfa in patients with advanced renal cell carcinoma: clinical and biomarker analysis. Cancer 2010; 116: 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonasch E, Wood CG, Matin SF, Tu SM, Pagliaro LC, Corn PG et al. Phase II presurgical feasibility study of bevacizumab in untreated patients with metastatic renal cell carcinoma. J Clin Oncol 2009; 27: 4076–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 2014; 15: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010; 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion MD. ChIP-RNA-seqPRO: A strategy for identifying regions of epigenetic deregulation associated with aberrant transcript splicing and RNA-editing sites 2014. Available from: http://sourceforge.net/projects/chiprnaseqpro/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.