Abstract
The mechanisms by which some melanoma cells adapt to BRAF inhibitor therapy are incompletely understood. In the present study, we used mass spectrometry-based phosphoproteomics to determine how BRAF inhibition remodeled the signaling network of melanoma cell lines that were BRAF-mutant and PTEN-null. Short-term BRAF inhibition was associated with marked changes in fibronectin-based adhesion signaling that were PTEN-dependent. These effects were recapitulated through BRAF siRNA knockdown and following treatment with chemotherapeutic drugs. Increased fibronectin expression was also observed in mouse xenograft models as well as specimens from melanoma patients undergoing BRAF inhibitor treatment. Analysis of a melanoma TMA showed loss of PTEN expression to predict for a lower overall survival, with a trend for even lower survival being seen when loss of fibronectin was included in the analysis. Mechanistically, the induction of fibronectin limited the responses of these PTEN-null melanoma cell lines to vemurafenib, with enhanced cytotoxicity observed following the knockdown of either fibronectin or its receptor α5β1 integrin. This in turn abrogated the cytotoxic response to BRAF inhibition via increased AKT signaling, which prevented the induction of cell death by maintaining the expression of the pro-survival protein Mcl-1. The protection conveyed by the induction of fibronectin expression could be overcome through combined treatment with a BRAF and PI3K inhibitor.
INTRODUCTION
BRAF inhibitors are now the mainstay of systemic therapy for the 50% of patients whose melanomas harbor BRAFV600 mutations 1-3. In the clinic, treatment with BRAF inhibitors such as dabrafenib and vemurafenib is associated with impressive responses, with significant levels of tumor shrinkage (as measured by RECIST criteria) being seen in the majority of patients. Most recently, a Phase III randomized study (COMBI-D) has demonstrated an improved progression-free survival and overall response rate for patients treated with a combination of dabrafenib plus the MEK inhibitor trametinib versus dabrafenib alone, and a similar Phase 3 trial has demonstrated improved overall survival in patients treated with the dabrafenib plus trametinib combination compared to vemurafenib monotherapy4, 5. Based on the durable objective responses observed to the combination of dabrafenib and trametinib, the combination gained FDA approval for treatment of unresectable BRAFV600E/K patients. Despite the initial success of BRAF inhibitor therapy, resistance is common in most patients, with the reactivation of the MAPK signaling pathway being observed in >70% of relapsing tumors2. Mechanistically, resistance to BRAF inhibition seems to follow a course of shortterm adaptation in which small populations of cells evade therapy through increased PI3K/AKT and MAPK signaling followed by the acquisition of mutations that sustain growth in the continual presence of drug. Emerging evidence suggests the initial adaptive responses to result in part from the relief of feedback inhibition at the level of Ras that sensitizes the melanoma cells to the pro-survival effects of exogenous growth factors (including IGF-1R, PDGFR, c-MET, HER3 and FGFR)6-10. It has been demonstrated that upregulation of expression and signaling through EGFR can also convey resistance to BRAF inhibition11, 12. Analysis of post-failure patient biopsies has identified multiple resistance mechanisms including mutations in NRAS, MEK1, MEK2 and BRAF-splice mutants7, 13, 14. Melanoma has the highest mutational load of all tumors, and frequently exhibits a complex genetic profile. Among these, melanomas that are both BRAF mutant and PTEN-null (PTEN-) sometimes show a reduced response to vemurafenib and dabrafenib15, 16. A recent analysis of melanoma patients stratified on the basis of their PTEN status, showed aberrant PTEN function not to be associated with the best overall response and these patients to show a trend towards a lower progression-free survival (PFS)17. There is, however, evidence that some BRAF mutant/PTEN- melanoma cell lines still respond to BRAF or MEK inhibitor monotherapy, suggesting that other co-operating factors may also contribute to intrinsic resistance18, 19. At this time, the mechanisms underlying the intrinsic resistance of some BRAF mutant/PTEN- melanomas to vemurafenib therapy remain poorly defined. Most studies have focused upon the genetic changes that are associated with acquired BRAF inhibitor resistance. In the current study, we used a proteomic approach to evaluate modulation of tyrosine phosphorylation in the short-term adaptive responses of BRAFV600E/PTEN-melanoma cell lines to BRAF inhibition. Through these methods, we uncovered a previously uncharacterized fibronectin (FN)-derived protective niche that is induced upon BRAF inhibition that drives therapeutic escape through α5β1 integrinmediated maintenance of pro-survival Mcl-1 expression.
RESULTS
Inhibition of BRAF is associated with adaptive changes in the signaling network
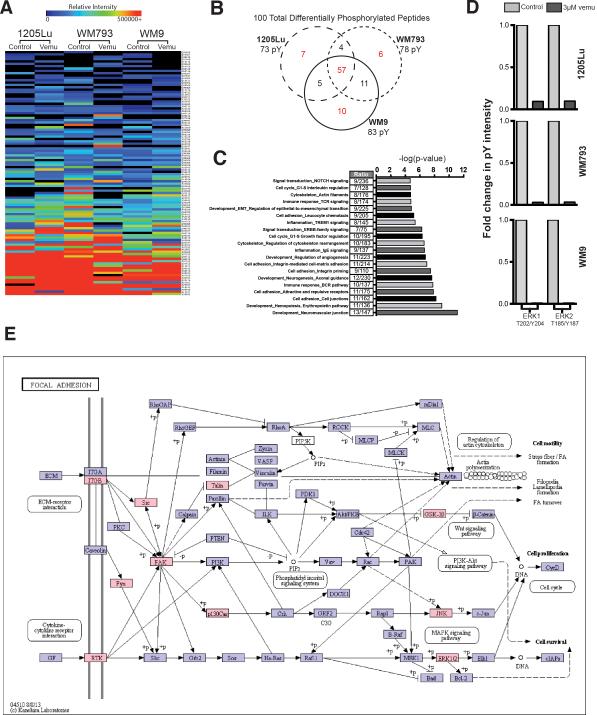
To better understand the global vemurafenib-mediated adaptive changes in signaling, three BRAFV600E/PTEN- mutant melanoma cell lines were treated with either vehicle or vemurafenib (3 μM, 24 hrs), the tyrosine phosphorylated (pTyr) peptides were captured by immunoprecipitation, and changes in the level of phosphorylation were analyzed by mass spectrometry (Supplemental Figure 1)20, 21. Quantification of the tyrosine phosphorylation following BRAF inhibition using label-free MaxQuant revealed changes in 73-83 distinct peptides, depending upon the cell line (Figures 1A-D: see Supplemental Table 1 for peptide list and phosphorylation sites identified)22. Of these, alterations in 57 pTyr peptides were common to all three cell lines (Figure 1A, B). GeneGO process network analysis of proteins exhibiting at minimum of 10% change in phosphorylation highlighted changes in pathways implicated in cytoskeletal remodeling, RTK signaling, integrinmediated adhesion, and epithelial-to-mesenchymal transition (EMT) (Figure 1C). As expected, BRAF inhibitor treatment led to the inhibition of ERK1 and ERK2 phosphorylation in all of the cell lines tested (Figure 1D). Mapping of this data using KEGG software revealed vemurafenib-treatment to drive an adhesion signaling network involving integrin α5β1, Src, Talin and FAK (Figure 1E).
Figure 1. Phosphoproteomic analysis shows BRAF inhibition to upregulate adhesion signaling in BRAF6000E/PTEN- melanoma cell lines.
A: Heat map showing global changes in tyrosine phosphorylation in 3 melanoma cell lines following vemurafenib treatment (3 μM, 24 hrs, two replicates for each of the 3 cell lines). B: Venn-diagram showing the overlap of tyrosine-phosphorylated peptides following the treatment of 3 melanoma cell lines (1205Lu, WM793, WM9) with vemurafenib (3 μM, 24 hrs.). C: GeneGO network processes analysis of proteins exhibiting a miminum of 10% change in phosphorylation, identifies cell adhesion, regulation of the cytoskeleton and RTK signaling as being the pathways with significant alterations in tyrosine phosphorylation following BRAF inhibition. D: Bar graph showing changes in tyrosine phosphorylation of ERK1 and ERK2 in 3 melanoma cell lines following vemurafenib treatment (3 μM, 24 hrs.). E: KEGG pathway map identifying the activation of integrin-mediated adhesion/RTK 22 signaling following vemurafenib treatment, proteins with changes in phosphorylation (>10%) due to vemurafenib treatment depicted in pink.
BRAF inhibitor-associated integrin/FN signaling
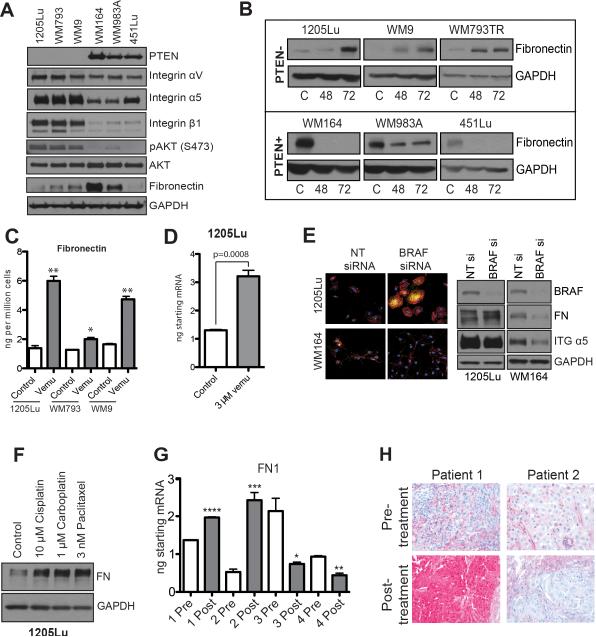
A growth inhibition assay performed on a panel of PTEN- and PTEN+ melanoma cell lines demonstrated that the vemurafenib IC50 was not correlated with PTEN expression, however, we have previously showed that PTEN expression in BRAFV600E cell lines does correlate with the extent of apoptosis induction following BRAF inhibition (Supplemental Figure 2)23. A Western blot analysis of a panel of melanoma cell lines showed the PTEN- cohort to express higher levels of integrin α5β1 and basal pAKT than their PTEN+ counterparts (Figure 2A). As α5β1 integrin signaling is activated through binding to FN, we next asked whether the adhesion phenotype identified using phosphoproteomics was the result of ECM remodeling. BRAFV600E/PTEN- melanoma cell lines showed increased FN expression following vemurafenib treatment, an effect not observed in the BRAFV600E/PTEN+ cohort (Figure 2B, Supplemental Figures 3-5). ELISA assays showed an increase in active secretion of FN into the cell culture media (Figure 2C). BRAF inhibition increased FN expression at the mRNA level as measured by RT-qPCR (Figure 2D). These effects were not restricted to vemurafenib treatment and were recapitulated following siRNA knockdown of BRAF (Figure 2E, Supplemental Figures 6-7) as well as following treatment with cytotoxic drugs such as cisplatin, carboplatin and paclitaxel (Figure 2F, Supplemental Figure 8-9). Vemurafenib-mediated fibronectin upregulation was evident as early as after 2 hours of treatment (Supplemental Figure 10A). The combination of the MEK inhibitor trametinib with vemurafenib showed a similar effect to vemurafenib monotherapy (Supplemental Figure 10B). The upregulation of FN expression is coupled with extracellular deposition of FN as a functional matrix, as demonstrated by the ability of vemurafenib-induced FN to promote melanoma cell adhesion (Supplemental Figure 11). No changes in other ECM proteins that are known interactors of α5β1 integrin (such as vitronectin) were noted (Supplemental Figure 12). Quantitative RT-PCR analysis of tumors from 4 patients failing vemurafenib therapy identified increased intratumoral FN in two cases (Figure 2G). Immunohistochemical (IHC) analysis of an additional group of patient specimens (n=3) identified one tumor with higher FN expression in the post- BRAF inhibitor relapse sample that was absent in the initial specimen (Figure 2H).
Figure 2. Vemurafenib induces FN expression.
A: Western blot showing expression of PTEN, integrin αV, integrin α5, integrin β1, FN pAKT (Ser473) and AKT in three BRAF6000E/PTEN+ melanoma cell lines (WM164, WM983A, 451Lu) and three that are BRAF6000E/PTEN- (1205Lu, WM793, WM9). B: Vemurafenib induces FN expression in BRAF6000E/PTEN- melanoma cell lines. Western blot showing the induction of FN expression following 48- and 72-hour 3μM vemurafenib treatment. C: ELISA showing vemurafenib-mediated induction of fibronectin secretion from 3 BRAF6000E/PTEN+ cell lines. D: qRT-PCR showing vemurafenib-mediated induction of fibronectin mRNA expression in the 1205Lu melanoma cell line. E: siRNA knockdown of BRAF induces FN expression (yellow) in 1205Lu cells (PTEN-) and reduces expression in WM164 cells (PTEN+), phalloidin staining for the cytoskeleton is shown in red. F: Cytotoxic chemotherapy induces FN expression in 1205Lu PTEN- melanoma cells. Cells were treated with 10μM cisplatin, 1μM carboplatin and 3nM paclitaxel for 72 hours before being analyzed by Western blot (right). G: Some melanoma patients failing BRAF inhibitor therapy show increased intratumoral FN mRNA expression. Data shows q-RT-PCR experiments on 4 matched (pre and post treatment) pairs of melanoma patient specimens receiving vemurafenib therapy (960 mg BID), statistics calculated based on technical replicates. H: Increased FN staining in a melanoma specimen from a patient with a tumor resistant to vemurafenib therapy. Data show IHC staining for FN pre- and post-vemurafenib treatment (N=3, 960 mg BID).
BRAF inhibitor-associated integrin/FN signaling is PTEN dependent
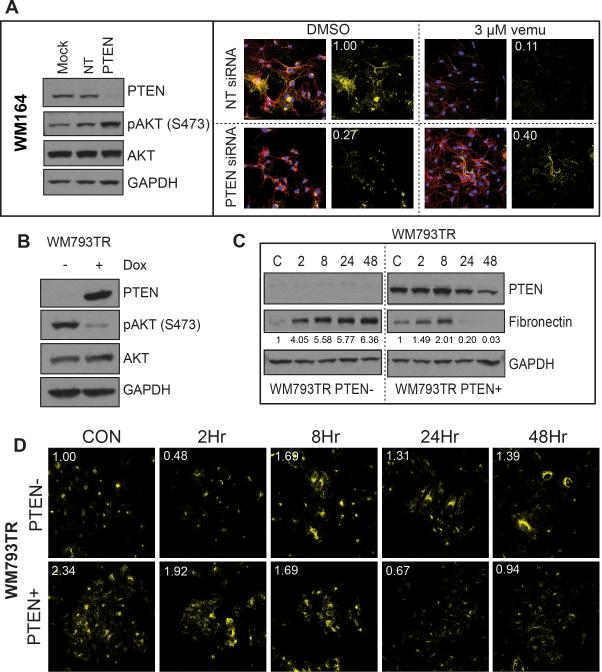
As the induction of FN expression was only observed in the cell lines lacking a functional form of PTEN, we next explored the PTEN dependency of FN induction. As expected, siRNA-mediated knockdown of PTEN in WM164 cells (BRAF6000E/PTEN+) prior to vemurafenib treatment led to increased FN expression upon treatment (Figure 3A, Supplemental Figure 13). Conversely, induction of doxycycline-driven PTEN expression in WM793TR cells (BRAF6000E/PTEN-) blocked the increase of FN expression following 24-hour vemurafenib treatment (Figures 3B-D).
Figure 3. Treatment-associated induction of fibronectin is PTEN-dependent.
A: Knockdown of PTEN leads to FN induction following BRAF inhibition. (left) siRNA knockdown of PTEN in WM164 cells (PTEN+) leads to increased AKT signaling. (right) Immunofluorescence studies showing increased FN staining (yellow) following vemurafenib treatment in cells with PTEN siRNA knockdown (3 μM, 48 hrs.), phalloidin staining for the cytoskeleton is shown in red. B: Doxycyclinemediated induction of PTEN in the WM793 (PTEN-) cell line. Western blot shows the induction of PTEN to reduce AKT phosphorylation (48 hours). C: Induction of PTEN reduces FN expression following vemurafenib treatment. Following either induction or non-induction of PTEN WM793-TR cells were treated with vehicle or vemurafenib (3 μM, 2-48 hrs.) and FN expression was measured by Western blot. D: Immunofluorescence staining showing loss of vemurafenib-associated FN upregulation (yellow) after PTEN induction.
Integrin/FN signaling is crucial for melanoma cell survival on therapy
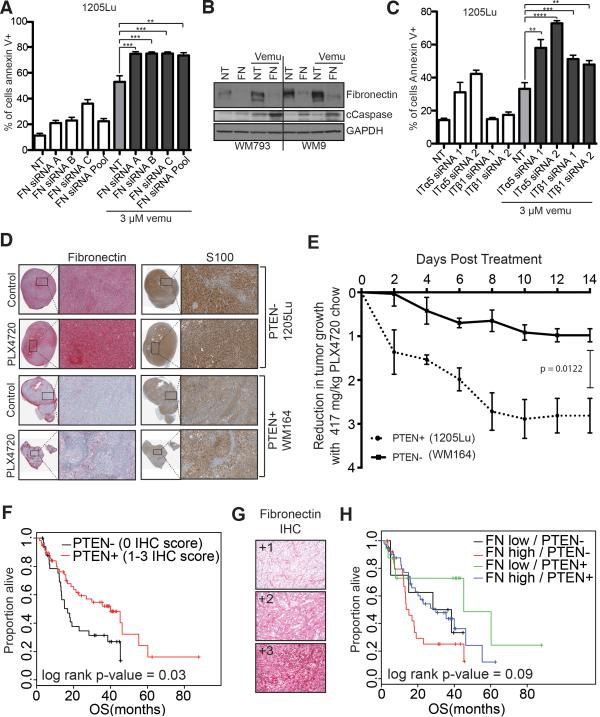
Previous work from our group has shown that melanoma cell lines lacking PTEN expression can show reduced cytotoxicity following BRAF inhibitor treatment15. We next investigated whether increased FN expression secondary to BRAF inhibition could contribute to therapeutic escape. Knockdown of FN using an siRNA pool, as well as by three individual siRNAs, significantly increased vemurafenib-mediated apoptosis (3 μM, 24 hrs) (Figures 4A,B and Supplemental Figure 14A). In these studies, cells were grown in 1% FBS to reduce the survival benefit from FN in the serum. The low serum levels increased basal levels of cell death particularly following treatment with the non-targeting siRNA. The protective effects were mediated through integrin α5β1, with siRNA knockdown of α5 integrin found to significantly increase the level of vemurafenib-induced apoptosis (Figure 4C and Supplemental Figure 14B). Mouse xenograft studies show an induction of FN expression in BRAF/PTEN- melanoma xenografts in response to treatment with the vemurafenib analogue, PLX4720 (Figure 4D). In line with the reduced cytotoxicity seen in vitro due to FN upregulation, the BRAF inhibitor PLX4720 also exhibited significantly weaker anti-tumor activity in BRAF/PTEN- melanoma xenografts compared to BRAF/PTEN+ xenografts (Figure 4E). Unlike the tissue culture experiments, the BRAF6000E/PTEN- tumor xenografts were found to have basal FN expression (Figure 4D). We reasoned that this may have resulted from the increased stress the cells experience under xenograft conditions, and demonstrated that mild acidosis (pH 6.7) - such as that seen in vivo - was sufficient to induce FN expression in BRAF6000E/PTEN- cell lines (Supplemental Figure 14C,D). The clinical significance of PTEN loss and increased FN expression was investigated on an annotated tissue microarray (TMA) of 93 individual stage III/IV melanoma patient specimens (Supplemental Tables 2-4). Here, PTEN loss was associated with significantly reduced (P<0.05) overall survival (OS) (Figure 4F, Supplemental Table 3), with a trend towards reduced OS (median OS 15.1 months vs. 45.2 months for PTEN-/FNHigh and PTEN+/FNLow, respectively) seen in patients with concurrent PTEN-/FNHigh expression (Figure 4G-H, Supplemental Table 4).
Figure 4. Integrin/FN signaling is required for melanoma cell survival on therapy.
A: siRNA knockdown of FN enhances vemurafenib mediated cell death. 1205Lu melanoma cells were transfected with three individual siRNAs and an siRNA pool targeting FN followed by treatment with vemurafenib (3 μM, 72 hours). Annexin V staining was measured by flow cytometry. B: Knockdown of FN enhances vemurafenib-mediated caspase cleavage. WM793 and WM9 cells were treated with either non-targeting or FN siRNA prior to treatment with vemurafenib (3 μM, 24 hours). Western blot shows levels of FN, cleaved caspase-7 (cCaspase) and GAPDH C: Knockdown of α5 and β1 integrin enhance vemurafenib-induced (3 μM, 48 hrs.) apoptosis in 1205Lu cells. Annexin V staining was measured by flow cytometry. D: Vemurafenib induces fibronectin expression, leading to reduced anti-proliferative effects in the 1205Lu BRAF6000E/PTEN- human melanoma xenografts, compared to the WM164 BRAF6000E/PTEN+ xenografts. IHC of representative xenograft tumors show staining for FN (red) and S100 (brown). E: Tumor volume was measured using calipers, and tumor reduction was compared by calculating the fold-decrease in tumor volume (tumor v olume = 1/2 x L x W 2). F: Lack of PTEN and high fibronectin expression predicts for worse overall survival in melanoma patients. Overall survival curve for a cohort of melanoma patients (n=93). G: Representative FN IHC staining on the tissue microarray. H: Lack of PTEN expression and high FN expression predicts for worse overall survival in melanoma patients. Overall survival curve for a cohort of melanoma patients (n=92).
Treatment-associated integrin/FN signaling protects melanoma cells from vemurafenib-mediated cytotoxicity through increased PI3K/AKT signaling
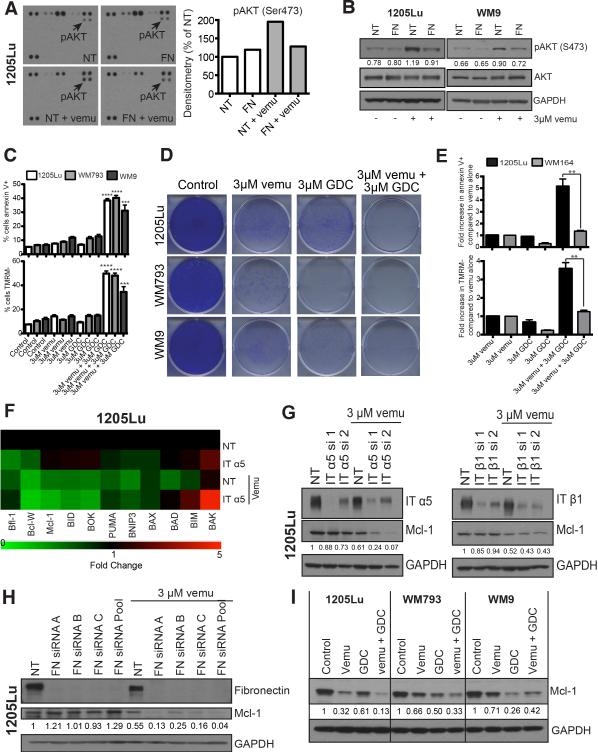
Previous work from our group and others has demonstrated BRAF/MEK inhibition to be associated with adaptive PI3K/AKT signaling15, 19. We next utilized kinome arrays and western blot studies to show that siRNA knockdown of FN abrogated the vemurafenib-mediated increases in AKT phosphorylation (Figures 5A,B). The protective role of PI3K/AKT signaling was confirmed by the ability of the PI3K inhibitor, GDC-0941, to enhance vemurafenib-mediated apoptosis and reduce survival in long-term colony formation assays (Figures 5C,D). The enhanced proapoptotic effects of combined BRAF/PI3K inhibition were PTEN dependent and were not observed in WM164 cells that are BRAF6000E/PTEN+ (Figure 5E). Liquid chromatography multiple reaction monitoring mass spectrometry (LC-MRM) was next used to identify the BH3 family proteins required for FN-mediated survival (Figure 5F, Supplemental Figure 15A) 15, 24. These studies showed Mcl-1 expression to be dependent upon signaling through α5 integrin and FN, with siRNA knockdown of either of these proteins decreasing Mcl-1 expression when BRAF was inhibited (Figures 5F-H, Supplemental Figure 15A). In line with FN being a stress-induced response in PTEN- cells, the expression of FN was upregulated in serum-starvation transfection conditions and in the presence of lipofectamine (Figure 5H, Supplemental Figure 15B). The adhesion-mediated Mcl-1 expression was also dependent upon PI3K/AKT signaling with expression being reduced following treatment with the combination of vemurafenib and GDC0941 (Figure 5I).
Figure 5. Treatment-associated integrin/FN signaling protects melanoma cells from vemurafenib-mediated cytotoxicity through increased PI3K/AKT signaling and control of Mcl-1 expression.
A: (left) Kinome array demonstrating that FN knockdown attenuates vemurafenib mediated AKT signaling (3 μM, 8 hrs.). (right) Quantification of pAKT densitometry on the kinome array B: Western blot confirmation of the kinome array from A demonstrating that FN knockdown prevents adaptive AKT signaling following BRAF inhibition (3 μM, 8 hrs.). pAKT was quantified using densitometry. C: Inhibition of PI3K/AKT signaling in combination with BRAF inhibition enhances cytotoxicity. Treatment with vemurafenib (3 μM) + GDC-0941 (3 μM) induces significantly more apoptosis compared to either inhibitor 25 alone. Cells were treated with the stated drug combinations for 48 hrs followed by Annexin-V/TMRM staining and flow cytometry. D: Combined BRAF/PI3K inhibition is associated with prolonged suppression of growth of BRAF6000E/PTEN- melanoma cell lines. Cells were treated with 3 μM vemurafenib and 3 μM GDC-0941 for 2 weeks. Colonies were visualized following staining with crystal violet. E: Inhibition of PI3K/AKT signaling in combination with BRAF inhibition does not enhance cytotoxicity in PTEN+ WM164 cells compared to PTEN- 1205Lu. Cells were treated with the stated drug combinations for 48 hrs. followed by Annexin-V/TMRM staining and flow cytometry. F: α5 integrin is required for maintenance of Mcl-1 expression following BRAF inhibition. LC-MRM experiment showing the relative fold changes in 11 BH3 only family proteins following siRNA knockdown of α5 integrin +/− 3 μM vemurafenib treatment in 1205Lu cell line, green indicating a decrease in expression and red indicating an increase in expression. G: Western blot showing siRNA knockdown of integrin α5 and β1 to be critical for the regulation of Mcl-1 expression in 1205Lu cell line. H: Induction of fibronectin regulates Mcl-1 expression. siRNA knockdown of FN is associated with decreased Mcl-1 expression following 3 μM vemurafenib treatment (72 hours) in 1205Lu cell line. I: Combined treatment with a BRAF and PI3K inhibitor downregulates Mcl-1 expression. Cells were treated with either 3 μM vemurafenib or 3 μM PI3K inhibitor (GDC-0941) for 72 hours followed by Western blotting for Mcl-1.
DISCUSSION
Melanoma cells readily adapt to nearly every treatment intervention and possess multiple routes to therapeutic escape. Understanding of the process of adaptation and resistance has been complicated by the diversity of signaling in melanoma and the ability of subtle alterations in levels of signaling molecules and RTKs to reduce sensitivity to BRAF and MEK inhibitors (e.g. cyclin D1, COT, BRAF, CRAF, MEK1 mutations, PDGFR, IGF1R) (reviewed in25, 26). In the current study, we used massspectrometry based phosphoproteomic analysis to identify a novel mechanism by which BRAF inhibition remodels the extracellular environment of melanoma cells and stimulating signaling through integrins, allowing for therapeutic escape. Proteomic analysis of phosphotyrosine signaling a panel of BRAF6000E/PTEN-melanoma cell lines demonstrated vemurafenib to induce signaling through proteins implicated in EMT, including α5β1 integrin, FN, focal adhesion kinase (FAK), Src, paxillin, and talin 27. Previous mass spectrometry work, in which one BRAF mutant melanoma cell line was treated with the MEK inhibitor U0126, has already demonstrated a role for oncogenic BRAF in cytoskeletal regulation 28. EMT has been best characterized in the oncogenic transformation of epithelial cells, where it is typified by increased motility, upregulated expression of ECM (such as FN), apoptosis resistance and drug resistance 27, 29. Although melanocytes are neural crest-derived and not epithelial in origin, the introduction of mutant BRAF can lead to an EMT-like state 30-32. We here provide evidence that inhibition of BRAF also induces an EMT-like phenotype in melanoma cells that lack PTEN expression. Loss of PTEN/increased AKT signaling mediates a mesenchymal switch in multiple epithelial tumor types including prostate cancer and nasopharyngeal tumors 33-35. In BRAF6000E/PTEN- melanoma cells, the vemurafenib-induced EMT was characterized by increased FN/α5β1 integrin signaling. These observations were not restricted to vemurafenib and could be recapitulated through cytotoxic chemotherapeutic drugs. Other cancer-relevant forms of stress, such as mild acidosis (pH 6.7) also increased FN expression in this system. These findings mirror those seen in breast cancer patients, where resistance to multiple drugs; including chemotherapeutic agents (paclitaxel, cisplatin), signal transduction inhibitors (PI3K, MEK and PI3K/mTOR inhibitors) and RTK inhibitors (EGFR, HER3 inhibitors) was associated with a switch to a more mesenchymal phenotype 29. A link between loss of PTEN and increased FN deposition has been observed in other systems and is characteristic of pathological fibrotic states such as pulmonary fibrosis, where PTEN downregulation in infiltrating fibroblasts leads to increased FN deposition 36, 37. Evaluation of a melanoma TMA demonstrated the role of PTEN loss as a negative prognostic factor with a significant correlation observed between decreased PTEN expression and worse overall survival. When FN expression was included in the survival analysis, patients whose tumors lacked PTEN and expressed high FN showed a poorer survival than those with tumors that retained PTEN and lacked FN expression (15.1 vs. 45.2 months, median OS).
Attachment to extracellular matrix has been identified as an important mechanism of drug resistance in a number of tumor systems. In myeloma, β1-integrin-mediated adhesion serves to increase STAT3 signaling leading to increased Bcl-XL and decreased BIM expression 38. Protection from cytotoxic drugs has also been observed when lung cancer and ovarian cancer cells are adhered onto laminin and collagen 39, 40. In a 3D cell culture model of ovarian cancer, adhesion to reconstituted basement membrane mediated resistance to PI3K/mTOR inhibition 41. Use of a reverse phase protein array (RPPA) system revealed this matrix-driven protection to be mediated through increased EGFR, IGFR1 and c-KIT activity, leading to the suppression of apoptosis. The increase in cell survival was dependent upon enhanced Bcl-2 protein expression and could be overcome through the combination of a PI3K/mTOR inhibitor and a Bcl-2 protein inhibitor or a PI3K/mTOR inhibitor combined with an EGFR or IGF1R inhibitor 41.
In tissue culture studies, the induction of integrin/FN signaling limited the cytotoxic effects of vemurafenib. Mechanistically, integrin α5 signaling was required for the maintenance of Mcl-1 expression, a key anti-apoptotic protein that exerts its effects through binding to BIM-EL and inhibition of pro-apoptotic Bak/Bax 42, 43. In melanoma cells, adhesion to fibronectin prevents the induction of anoikis through increased PI3K signaling 44. The expression of Mcl-1 is regulated at the transcriptional level by PI3K/AKT and post-translationally by MEK-mediated phosphorylation events that prevents its proteasomal degradation 42, 45. A role for the PI3K/AKT pathway in the integrin-mediated maintenance of Mcl-1 levels was suggested by the decreased AKT signaling seen following the siRNA knockdown of FN and the ability of the BRAF/PI3K inhibitor combination to downregulate Mcl-1 expression. The role of PI3K/AKT signaling in the adaptive responses that drive therapeutic escape was suggested by the finding that the BRAF/PI3K inhibitor combination led to increased cytotoxicity.
The observation that anti-cancer therapies induce the establishment of a pro-survival niche that allows for therapeutic escape is not without precedent. There is evidence from mouse xenograft studies that although radiation treatment induces massive cell death in tumors, it also generates signals that promote the survival of small numbers of tumor cells that can ultimately repopulate the tumor. In this instance, radiation-mediated apoptotic cell death led to caspase-3 dependent expression of prostaglandin E2 (PGE2) which limited cytotoxicity 46. The importance of caspase-3 in this process was demonstrated by studies showing that animals deficient in this enzyme showed enhanced responses to radiation therapy. Supporting studies in human cancer patients further demonstrated that high levels of intratumoral caspase-3 predicted for an increased rate of cancer recurrence 46.
The ability of the tumor cells to adapt to therapy and provide a sanctuary for some of its own cells is likely to be of major clinical significance given that even one melanoma cell can establish new tumors in vivo 47. Together, our data demonstrate a role for the PI3K/AKT signaling pathway as the major mediator of adhesionmediated protection. A number of clinical trials are now ongoing to determine whether the dual targeting of BRAF and PI3K/AKT can abrogate or delay the onset of resistance in BRAF mutant melanoma patients (NCT01902173, NCT01820364, NCT01616199).
MATERIALS AND METHODS
Cell culture and reagents
The 1205Lu, WM9, WM793, WM164, WM983A and 451Lu melanoma cells lines were a generous gift from Dr. Meenhard Herlyn (The Wistar Institute, Philadelphia, PA) and were genotyped as being BRAFV600E mutant. WM793TR (tet repressor) cell line, engineered to inducibly express PTEN, was a generous gift from Dr. Andrew Aplin (Kimmel Cancer Center, Philadelphia, PA). Inducible expression of PTEN was achieved by treatment of cultures with 100 ng/mL doxycycline. The identities of all cell lines were confirmed by Biosynthesis Inc. (Lewisville, TX) through STR validation analysis. Cell lines were maintained in 5% FBS/RPMI-1640, and routinely tested negative for mycoplasma contamination. Acidic media experiments were carried out using DMEM/F12 containing 25mM Pipes, 25mM HEPES, and 10% FBS, then pH was adjusted using NaOH.
Phosphoproteomics sample preparation, LC-MS/MS and analysis
Briefly, cells were lysed in denaturing buffer followed by protein reduction, alkylation and trypsin digestion. The tryptic peptides were then desalted. Following lyophilization, phosphotyrosine-containing peptides were enriched by immunoprecipitation with immobilized anti-phosphotyrosine antibody p-Tyr-100 (Cell Signaling Technology) 20, 21. A nanoflow ultra high performance liquid chromatograph (RSLC, Dionex, Sunnyvale, CA) coupled to an electrospray ion trap mass spectrometer (LTQ-Orbitrap, Thermo, San Jose, CA) was used for tandem mass spectrometry peptide sequencing experiments. Sequest (Thermo, San Jose, CA) and Mascot 48 searches were performed and the results were summarized in Scaffold 3.0 (www.proteomesoftware.com). The integrated peak areas for pY peptide quantification were calculated from extracted ion chromatograms (EIC) using QuanBrowser from Xcalibur 2.0 (Thermo, San Jose, CA). Label-Free quantification was performed using MaxQuant (v. 1.2.2.5) 22. Heat maps of pY ion signals were generated using MultiExperiment Viewer (version 4.8.1)49. Process network enrichment analysis was performed using GeneGO Pathway Maps in Metacore (Thomson Reuters). Additional pathway analysis was performed using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database 50. Both GeneGo and KEGG analyses were performed on proteins with a change in phosphorylation levels greater than 10%.
Growth inhibition assay
Cells were plated in a 96-well plate and left to attach overnight before being treated with vehicle or increasing doses of vemurafenib for 72 hours. Cells were then incubated with MTT reagent (Sigma Aldrich, St. Louis, MO) for three hours, then crystals were solubilized in DMSO followed by a measurement of absorbance at 570 nm.
Western blotting
Proteins were extracted using RIPA lysis buffer (0.5M Tris, Triton X-100, Na-deoxycholate, 10% SDS, NaCl, 0.5M EDTA) containing Complete Mini protease inhibitor cocktail tablet (Roche, Indianapolis, IN). Total protein was quantified using Pierce BCA Protein Assay Kit (Thermo Scientific, Rockford, IL). Protein extracts were resolved on Novex 8-16% Tris-glycine gels (Life Technologies, Carlsbad, CA) and were electrophoretically transferred onto PVDF membranes (Millipore, Bedford, MA). Membranes were blocked using 5% non-fat dry milk/TBST and incubated with primary antibody overnight at 4°C. Membranes were then incubation with isotype-specific HRP-conjugated secondary antibody diluted in 5% milk/TBST (Amersham Pharmacia, Little Chalfont, UK). Immunocomplexes were visualized using the enhanced chemiluminescence substrate (PerkinElmer, Waltham, MA) and detected on autoradiography film (Denville Scientific, Metuchen, NJ). Uniform protein loading was confirmed by blotting for GAPDH. The antibodies to PTEN (9188), Integrin α5 (4705), Integrin β1 (4706), BRAF (9433), phospho-AKT S473 (4058), total AKT (9272), Caspase 7 (9492), phospho-ERK (9101), total ERK (9102), and MCL-1 (4572) were from Cell Signaling Technology (Beverly, MA). Laminin 5 (ab14509) antibody was purchased from Abcam (Cambridge, MA) while the antibody against Vitronectin (sc-28929) was from Santa Cruz Biotechnology (Santa Cruz, CA). Fibronectin (610077) antibody was purchased from BD (San Jose, CA), GAPDH (G8795) was from Sigma (St. Luis, MO), and Phalloidin (A22287) was from Invitrogen (Life Technologies, Carlsbad, CA).
Immunofluorescent staining
Melanoma cells were plated on glass coverslips in six-well plates and incubated overnight prior to treatment. Cells were then fixed using 4% paraformaldehyde solution (Electron Microscopy Sciences, Hatfield, PA) and permeabilized with 0.2% Triton X-100 prior to being blocked in 1% BSA/PBS. Coverslips were incubated with primary antibodies overnight at 4°C. Coverslips were then washed in PBS, incubated with secondary antibodies for 1 hour at RT, and washed again in PBS and sterile water. Coverslips were mounted with ProLong Gold antifade reagent with DAPI (Life Technologies, Carlsbad, CA) and imaged using confocal microscopy.
RNA interference
Cells were plated and left to grow overnight in 5% FBS/RPMI. 5%FBS/RPMI media was replaced with Opti-MEM (Invitrogen). Fibronectin pool, Fibronectin siRNA A GACUGGUGGUUACAUGUUAtt, Fibronectin siRNA B CGCAUCACUUGCACUUCUAtt, Fibronectin siRNA C GAUCCUGUCUACUUCACAAtt, Integrin α5 siRNA A GUCAGAAUUUCGAGACAAAtt, Integrin α5 siRNA B CCACUGACCAGAACUAGAAtt, Integrin β1 siRNA A GAGAUGAGGUUCAAUUUGAtt, Integrin β1 siRNA B GAUGAGGUUCAAUUUGAAAtt, (25nM, Santa Cruz), Integrin α5, Integrin β1, and PTEN (all 25nM, Cell Signaling Technologies), and BRAF (25nM, Thermo Scientific) siRNAs in complex with Lipofectamine 2000 (Invitrogen) were added. Scrambled, non-targeting siRNAs were used as controls. A final concentration of 5% FBS in RPMI was added the next day. For fibronectin knockdowns, final concentration 1% FBS in RPMI was added next day. Cells were transfected for 24-72 hours prior to treatment.
Kinase Arrays
Phosphorylation levels of forty-three phosphorylation sites on human kinases were determined using the Proteome Profiler Human Phospho-Kinase Array Kit (R&D Systems), following manufacturer's instructions.
Animal Studies
Seven week old female BALB SCID mice (The Jackson Laboratory, Bar Harbor, ME) were subcutaneously injected with 2.5 ×106 cells per mouse. Tumors were allowed to grow to until palpable. Mice were randomly separated into treatment cohorts of approximately equal average initial tumor volumes, three mice per treatment group. Mice were administered D10001 control chow or AIN-76A 417 mg/kg PLX4720- formulated chow (Research Diets, New Brunswick, NJ) daily. Mouse weights and tumor volumes (½ x L x W2) were measured every 48 hours. All animal experiments were carried out in compliance with ethical regulations and protocols approved by the University of South Florida Institutional Animal Care and Use Committee.
ELISA Assays
Fibronectin ELISA Kit was obtained from Millipore (Billerica, MA), and utilized abiding by manufacturer's instructions.
qRT-PCR
Cells were treated for 72 hours then total RNA was isolated using Qiagen's RNeasy mini kit. The following TaqMan® Gene Expression Assays primer/probes were used: Hs00365052_m1 (Fibronectin), P/N 4319413E (18S) and Hs99999905_m1 (GAPDH). The 18S + GAPDH data were utilized to normalize Fibronectin. qRT-PCR reactions were performed in duplicate as previously described 15. Analyzed cDNA from patient specimens was collected under approved protocols by the Institutional Review Board at Massachusetts General Hospital.
Flow cytometry
Cells were grown in 6 well tissue culture plates overnight, then treated with vehicle (DMSO) 3 μM vemurafenib, 3 μM GDC-0941, or the two drugs in combination for 72 hours. In some studies, cells were transfected with non-targeting siRNA or siRNAs targeting fibronectin, integrin α5, or integrin β1 for 24 hours prior to treatment and harvested after 48 or 72 hours of treatment. Cells were then washed with 1x Annexin V Binding Buffer (BD, San Jose, CA), resuspended in 100 μl of binding buffer containing 2 μl APC-conjugated Annexin V (550474, R&D Systems, Minneapolis, MN) and 25 nmol/L tetramethylrhodamine, methyl ester, perchlorate (TMRM; Molecular Probes, Eugene, OR). Cells were incubated at 37°C for 15 min before being analyzed for Annexin V fluorescence and TMRM retention using flow cytometry. Gates were delineated based on regions of distinct populations.
LC-MRM
LC-MRM was performed as described previously 24, 51. Protein expression was determined using the ratio of peak area of the endogenous peptide to corresponding internal standard; normalization of results was performed using GAPDH to accommodate differences in total protein loading. Data were then normalized to the non-targeting siRNA controls and graphed to show the changes in expression after transfection and drug treatment.
Colony Formation
Cells were seeded out into six-well plates at 1 × 104 per well and grown overnight before being treated with vehicle, 3 μM vemurafenib, 3 μM GDC-0941, or the two drugs in combination. Cells were left to grow for 2 weeks with new drug added twice per week. Wells were washed with PBS and stained with crystal violet solution (50% methanol + 50% H2O + 0.5% crystal violet).
TMA and patient specimens
Immunohistochemical staining of melanoma specimens and a tissue microarray (TMA) containing a spectrum of melanoma specimens was constructed and analyzed under approved protocols by the Institutional Review Board at the University of South Florida (including informed consent). It contained samples from four groups: primary cutaneous melanoma, lymph node metastases, dermal/subcutaneous metastases, and visceral metastases. After identification of tumor tissue, two 1.5 mm cores were obtained from each donor block. For the TMA, a single core from each block was positioned on a grid of 100 samples processed with the Beecher Microarray System. Targeted gene sequencing was performed on extracted DNA from the second core, and analyzed via the Sequenom platform (MassARRAY Analyzer 4 and Melacarta™ Panel). Clinical data was collected on the patients. Medical records and the social security death index were searched for demographic, pathology, staging and recurrence/survival data. Analyses were performed using SPSS and SAS software. TMA sections were evaluated for total FN and PTEN expression via IHC with optimized anti-FN (ab2413, Abcam, Cambridge, MA) and anti-PTEN (E4250, Spring Bioscience, Pleasanton, CA) antibodies in the Tissue Core at Moffitt Cancer Center. TMA was stained using a Ventana Discovery XT automated system (Ventana Medical Systems, Tucson, AZ) following manufacturer's protocol with proprietary reagents. Detection was carried out using the Ventana ChromoMap Red kit with Hematoxylin counterstain. Staining intensity was scored by the dermatopathologist as follows: absent staining=0, weak staining=1, moderate staining=2, strong staining =3. For both markers, all tumor cells in the infiltrate showed diffuse, uniform staining. Dichotomous high and low FN levels were assigned based on IHC scores of 0-1 and 2-3, respectively. PTEN dichotomous scoring was assigned as absent (IHC=0) versus present (IHC=1-3).
Cell Adhesion Assay
1205Lu melanoma cells were plated onto 6-well plates and allowed to attach overnight. They were then treated for 72 hours with 3μM vemurafenib. Following the treatment, the cells were detached using 10mM EDTA in PBS. The plates were incubated overnight with 5% acetic acid at 4°C. Plates were then washed three times with PBS. These vemurefenib-induced FN plates were compared to regular plastic cell culture plates, and commercially available FN-coated plates from BD (San Jose, CA ). 1205Lu cells were plated onto these plates and monitored for cell attachment after a 15-minute incubation period. The cells were fixed with 4% paraformaldehyde and stained with crystal violet. Cells were imaged, and 9 frames from each surface was used for cell counts
Densitometry
The mean pixel intensity for each band and the background was determined. The background was subtracted from each sample and the resulting value was divided by the appropriate control.
Statistical Analysis
Results are reported as mean values, error bars indicating ± SEM, calculated from three biological replicates unless otherwise indicated. Sample sizes are calculated to allow significance to be reached. GraphPad Prism 6 software was used to calculate statistical significance of magnitude of changes between different conditions was calculated using the parametric paired t-test with p-values depicted as follows: *p≤0.05, ** p≤0.01, *** p≤0.001, **** p≤0.0001.
Supplementary Material
ACKNOWLEDGEMENT
We would like to thank Gideon Bollag (Plexxikon) for providing vemurafenib and the PLX4720 chow, Noel Clark, Ashley Troutman, Kelli Noyd, Jayme O'Neal, and Holly Crandell for assistance with IHC staining, Marta Perez and Monica Torres for assistance with xenograft studies, Agnieszka Kasprazak and Joseph Johnson for assistance with microscopy and Laura Hall for assistance with qRT PCR.
Grant support: Work in the Smalley lab is supported by R01 CA161107 from the National Institutes of Health
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. The New England journal of medicine. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 3.McArthur GA, Chapman PB, Robert C, Larkin J, Haanen JB, Dummer R, et al. Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation-positive melanoma (BRIM-3): extended follow-up of a phase 3, randomised, open-label study. Lancet Oncol. 2014;15:323–332. doi: 10.1016/S1470-2045(14)70012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med. 2014;371:1877–1888. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- 5.Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med. 2015;372:30–39. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 6.Lito P, Pratilas CA, Joseph EW, Tadi M, Halilovic E, Zubrowski M, et al. Relief of Profound Feedback Inhibition of Mitogenic Signaling by RAF Inhibitors Attenuates Their Activity in BRAFV600E Melanomas. Cancer Cell. 2012;22:668–682. doi: 10.1016/j.ccr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du JY, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–U118. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abel EV, Basile KJ, Kugel CH, 3rd, Witkiewicz AK, Le K, Amaravadi RK, et al. Melanoma adapts to RAF/MEK inhibitors through FOXD3-mediated upregulation of ERBB3. The Journal of clinical investigation. 2013;123:2155–2168. doi: 10.1172/JCI65780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun C, Wang L, Huang S, Heynen GJ, Prahallad A, Robert C, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508:118–122. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- 12.Girotti MR, Pedersen M, Sanchez-Laorden B, Viros A, Turajlic S, Niculescu-Duvaz D, et al. Inhibiting EGF receptor or SRC family kinase signaling overcomes BRAF inhibitor resistance in melanoma. Cancer Discov. 2013;3:158–167. doi: 10.1158/2159-8290.CD-12-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poulikakos PI, Persaud Y, Janakiraman M, Kong XJ, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature. 2011;480:387–U144. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paraiso KH, Xiang Y, Rebecca VW, Abel EV, Chen YA, Munko AC, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Research. 2011;71:2750–2760. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing F, Persaud Y, Pratilas CA, Taylor BS, Janakiraman M, She QB, et al. Concurrent loss of the PTEN and RB1 tumor suppressors attenuates RAF dependence in melanomas harboring (V600E)BRAF. Oncogene. 2011 doi: 10.1038/onc.2011.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nathanson KL, Martin AM, Wubbenhorst B, Greshock J, Letrero R, D'Andrea K, et al. Tumor Genetic Analyses of Patients with Metastatic Melanoma Treated with the BRAF Inhibitor Dabrafenib (GSK2118436). Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:4868–4878. doi: 10.1158/1078-0432.CCR-13-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sondergaard JN, Nazarian R, Wang Q, Guo D, Hsueh T, Mok S, et al. Differential sensitivity of melanoma cell lines with BRAFV600E mutation to the specific Raf inhibitor PLX4032. J Transl Med. 2010;8:39. doi: 10.1186/1479-5876-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gopal YN, Deng W, Woodman SE, Komurov K, Ram P, Smith PD, et al. Basal and treatment-induced activation of AKT mediates resistance to cell death by AZD6244 (ARRY-142886) in Braf-mutant human cutaneous melanoma cells. Cancer Res. 2010;70:8736–8747. doi: 10.1158/0008-5472.CAN-10-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Rix U, Fang B, Bai Y, Edwards A, Colinge J, et al. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat Chem Biol. 2010;6:291–299. doi: 10.1038/nchembio.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nature Biotechnology. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 22.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 23.Paraiso KH, Xiang Y, Rebecca VW, Abel EV, Chen YA, Munko AC, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res. 2011;71:2750–2760. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang Y, Remily-Wood ER, Oliveira V, Yarde D, He L, Cheng JQ, et al. Monitoring a nuclear factor-kappaB signature of drug resistance in multiple myeloma. Molecular & cellular proteomics : MCP. 2011;10:M110 005520. doi: 10.1074/mcp.M110.005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fedorenko IV, Paraiso KH, Smalley KS. Acquired and intrinsic BRAF inhibitor resistance in BRAF V600E mutant melanoma. Biochem Pharmacol. 2011;82:201–209. doi: 10.1016/j.bcp.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solit DB, Rosen N. Resistance to BRAF Inhibition in Melanomas. New Engl J Med. 2011;364:772–774. doi: 10.1056/NEJMcibr1013704. [DOI] [PubMed] [Google Scholar]
- 27.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. Journal of Clinical Investigation. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Old WM, Shabb JB, Houel S, Wang H, Couts KL, Yen CY, et al. Functional proteomics identifies targets of phosphorylation by B-Raf signaling in melanoma. Molecular cell. 2009;34:115–131. doi: 10.1016/j.molcel.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu MY, Meier FE, Nesbit M, Hsu JY, Van Belle P, Elder DE, et al. E-cadherin expression in melanoma cells restores keratinocyte-mediated growth control and down-regulates expression of invasion-related adhesion receptors. The American journal of pathology. 2000;156:1515–1525. doi: 10.1016/S0002-9440(10)65023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li G, Schaider H, Satyamoorthy K, Hanakawa Y, Hashimoto K, Herlyn M. Downregulation of E-cadherin and Desmoglein 1 by autocrine hepatocyte growth factor during melanoma development. Molecular Biology of the Cell. 2001;12:45A–45A. doi: 10.1038/sj.onc.1205034. [DOI] [PubMed] [Google Scholar]
- 32.Boyd SC, Mijatov B, Pupo GM, Tran SL, Gowrishankar K, Shaw HM, et al. Oncogenic B-RAF(V600E) Signaling Induces the T-Box3 Transcriptional Repressor to Repress E-Cadherin and Enhance Melanoma Cell Invasion. The Journal of investigative dermatology. 2013;133:1269–1277. doi: 10.1038/jid.2012.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, Quah SY, Dong JM, Manser E, Tang JP, Zeng Q. PRL-3 down-regulates PTEN expression and signals through PI3K to promote epithelialmesenchymal transition. Cancer Research. 2007;67:2922–2926. doi: 10.1158/0008-5472.CAN-06-3598. [DOI] [PubMed] [Google Scholar]
- 34.Song LB, Li J, Liao WT, Feng Y, Yu CP, Hu LJ, et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. The Journal of clinical investigation. 2009;119:3626–3636. doi: 10.1172/JCI39374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mulholland DJ, Kobayashi N, Ruscetti M, Zhi A, Tran LM, Huang J, et al. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Research. 2012;72:1878–1889. doi: 10.1158/0008-5472.CAN-11-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muro AF, Moretti FA, Moore BB, Yan M, Atrasz RG, Wilke CA, et al. An essential role for fibronectin extra type III domain A in pulmonary fibrosis. American journal of respiratory and critical care medicine. 2008;177:638–645. doi: 10.1164/rccm.200708-1291OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White ES, Atrasz RG, Hu B, Phan SH, Stambolic V, Mak TW, et al. Negative regulation of myofibroblast differentiation by PTEN (Phosphatase and Tensin Homolog Deleted on chromosome 10). American journal of respiratory and critical care medicine. 2006;173:112–121. doi: 10.1164/rccm.200507-1058OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meads MB, Gatenby RA, Dalton WS. Environment-mediated drug resistance: a major contributor to minimal residual disease. Nat Rev Cancer. 2009;9:665–674. doi: 10.1038/nrc2714. [DOI] [PubMed] [Google Scholar]
- 39.Sethi T, Rintoul RC, Moore SM, MacKinnon AC, Salter D, Choo C, et al. Extracellular matrix proteins protect small cell lung cancer cells against apoptosis: A mechanism for small cell lung cancer growth and drug resistance in vivo. Nature Medicine. 1999;5:662–668. doi: 10.1038/9511. [DOI] [PubMed] [Google Scholar]
- 40.Sherman-Baust CA, Weeraratna AT, Rangel LBA, Pizer ES, Cho KR, Schwartz DR, et al. Remodeling of the extracellular matrix through overexpression of collagen VI contributes to cisplatin resistance in ovarian cancer cells. Cancer Cell. 2003;3:377–386. doi: 10.1016/s1535-6108(03)00058-8. [DOI] [PubMed] [Google Scholar]
- 41.Muranen T, Selfors LM, Worster DT, Iwanicki MP, Song L, Morales FC, et al. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell. 2012;21:227–239. doi: 10.1016/j.ccr.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang JM, Chao JR, Chen W, Kuo ML, Yen JJ, Yang-Yen HF. The antiapoptotic gene mcl-1 is up-regulated by the phosphatidylinositol 3-kinase/Akt signaling pathway through a transcription factor complex containing CREB. Molecular and Cellular Biology. 1999;19:6195–6206. doi: 10.1128/mcb.19.9.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kozopas KM, Yang T, Buchan HL, Zhou P, Craig RW. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:3516–3520. doi: 10.1073/pnas.90.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boisvert-Adamo K, Aplin AE. B-RAF and PI-3 kinase signaling protect melanoma cells from anoikis. Oncogene. 2006;25:4848–4856. doi: 10.1038/sj.onc.1209493. [DOI] [PubMed] [Google Scholar]
- 45.Domina AM, Vrana JA, Gregory MA, Hann SR, Craig RW. MCL1 is phosphorylated in the PEST region and stabilized upon ERK activation in viable cells, and at additional sites with cytotoxic okadaic acid or taxol. Oncogene. 2004;23:5301–5315. doi: 10.1038/sj.onc.1207692. [DOI] [PubMed] [Google Scholar]
- 46.Huang Q, Li F, Liu X, Li W, Shi W, Liu FF, et al. Caspase 3-mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat Med. 2011;17:860–866. doi: 10.1038/nm.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 49.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, opensource system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 50.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Research. 2012;40:D109–114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Remily-Wood ER, Liu RZ, Xiang Y, Chen Y, Thomas CE, Rajyaguru N, et al. A database of reaction monitoring mass spectrometry assays for elucidating therapeutic response in cancer. Proteomics Clinical applications. 2011;5:383–396. doi: 10.1002/prca.201000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.