Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that post-transcriptionally regulate gene expression in organisms. To understand the underlying mechanisms behind the molecular response of the crab to low salt-stress, high-throughput Illumina/Solexa deep sequencing technology was used to investigate the expression profiles of miRNAs under low salinity challenged. Two mixed RNA pool libraries of gill tissues from low salinity challenged (LC) and the control groups (NC) were sequenced on the Illumina platform. A total of 6,166,057 and 7,032,973 high-quality reads were obtained from the NC and LC libraries, respectively. Sixty-seven miRNAs consisting of 16 known and 51 novel ones were identified, among which, 12 miRNAs were differentially expressed in LC compared to NC. Thirty-four of the target genes predicted were differentially expressed in the opposite direction to the miRNAs, which were involved in crucial processes related to osmoregulation by gene ontology (GO) functional enrichment analysis, such as anion transport processes (GO:0006820) and chitin metabolic process (GO:0006030). These results provide a basis for further investigation of the miRNA-modulating networks in osmoregulation of Portunus trituberculatus.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-015-0641-9) contains supplementary material, which is available to authorized users.
Keywords: Portunus trituberculatus, MicroRNA, Salinity, Osmoregulation
Background
Portunus trituberculatus (Crustacea: Decapoda: Brachyura), often known as the swimming crab, is the most important economic crab species and fishery resource in China (Yu et al. 2003). According to China’s Ministry of Agriculture, production of the crab reached 99,580 t in 2012, which was valued at more than AUS$ 2.7 billion.
Salinity is an important abiotic factors that influence the distribution, abundance, physiology, and well-being of crustaceans (Romano and Zeng 2012), which is also an important factor during artificial propagation (Xu and Liu 2011). When the environmental osmolality is lower than that of the hemolymph, it is termed “hypo-osmotic” and crustaceans must compensate for ion loss from their hemolymph via hyper-osmoregulation (Rainbow and Black 2001; Romano and Zeng 2012). Throughout the prolonged culture period, the swimming crab often experiences low salinity stress due to heavy rainfalls, which could have significant impacts on farm productivity. Therefore, the identification and characterization of the genes and the regulatory factors involved in hyper-osmoregulation are now essential for increasing the production and the efficiency of selective breeding programs for this important species. With recent advances in genomic sequencing and transcriptome mapping (microarray and high-throughput sequencing), some salt-related genes and regulatory factors have been identified in P. trituberculatus on a large scale at the genome-wide level (Lv et al. 2013; Xu and Liu 2011). Nevertheless, the molecular basis of tolerance to salinity stress has not yet been identified.
MicroRNAs (miRNAs) are 20–22 nt non-coding RNAs that play an important role in the regulation of many biological processes, such as signal transduction, organ development, cellular proliferation, apoptosis, oncogenesis, and immunity (Ma et al. 2012; Zhou et al. 2014; Nilsen 2007; Yang et al. 2013; Pushpavalli et al. 2014). In animals, miRNAs regulate their target genes through mRNA destabilization and translational inhibition (Bushati and Cohen 2007). So far, miRNAs have been identified in a few aquatic crustacean species, and these were involved in regulation of oocyte meiosis (Song et al. 2014), adaptive immunity (Li et al. 2013; Ou et al. 2012; Zeng et al. 2015), and drug response (Valenzuela-Miranda et al. 2015). Previous reports have shown that some plant miRNAs respond to salinity stress and that some miRNA targets are salt stress-related genes (Xie et al. 2014; Peng et al. 2014; Frazier et al. 2011). This suggests that miRNAs play important roles in plant salt stress responses. However, little is known regarding crustacean miRNA expression profiles in response to salinity stress, and their roles in osmoregulation remain unclear.
High-throughput next generation sequencing technology is an efficient method for discovering genes in an unbiased manner [7–10]. Measurement of miRNA expression levels and elucidation of the regulatory relationship between miRNA and its corresponding mRNA target are important to clarify pathways and biological processes including biotic and abiotic stress responses (Rao et al. 2014). Transcriptome analyses of P. trituberculatus growth (Lv et al. 2015) and responses to salinity challenged (Lv et al. 2013) by high-throughput next-generation sequencing technology have been reported; however, the study or experimental identification of miRNAs has been no performed.
In this study, we examined two miRNA transcriptomes from the gills of P. trituberculatus under normal conditions and low salinity stress using the Illumina/Solexa deep sequencing technology. Then, we elucidated their cellular miRNAs and the genes profiles related to hyper-osmoregulation (Fig. 1). These results provide a basis for further investigation of miRNA-modulating networks underlying the osmoregulation of P. trituberculatus.
Fig. 1.
Schematic of Illumina deep sequencing and analysis
Materials and methods
Ethics statement
P. trituberculatus is not an endangered or protected species. All animal work has been conducted according to the relevant national and international guidelines. No specific permissions are required to work with invertebrates in China. Similarly, no specific permissions are required for the collection of P. trituberculatus from sample sites because they were not collected from protected areas of land.
Salinity challenge experiment and sample preparation
Eighty-day-old swimming crabs were collected from a local farm in Qingdao, China (5.62∼11.66 g in body weight). All the samples were acclimated in the laboratory environment (33 ppt, 18 °C) for 7 days before the experiment. The crabs were divided into two groups (90 crabs per group) and acclimated in non-challenged (NC, 33 ppt) and low salinity challenged (LC, 5 ppt) at 18 °C, respectively. The gills from nine crabs were collected after 10 days, and all samples were stored in RNAlater (Ambion) at 4 °C over night and then at −20 °C.
The construction and deep sequencing of small RNA libraries
Total RNA was isolated from each sample by TRIzol (Invitrogen, CA, USA). RNA degradation and contamination was monitored on 1 % agarose gels. RNA purity was checked using the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA). RNA concentration was measured using a Qubit® RNA Assay Kit in Qubit® 2.0 Flurometer (Life Technologies, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). Then, all samples were pooled in equal amounts in each group to generate two mixed sample. A total of 3 μg pooling samples was used as input material for the small RNA sample preparations. All samples had RIN values above 8. Sequencing libraries were generated using an IlluminaTruSeq™ Small RNA Sample Preparation Kit (Illumina, San Diego, CA, USA) following the manufacturer’s recommendations. The libraries were sequenced on an Illumina Hiseq 2000 platform and 100-bp single-end reads were generated.
miRNA identification
Raw data (raw reads) in the fastq format were first processed through our self-written Perl and Python scripts (Novogene Bioinformatics Institute). In this step, clean data (clean reads) were processed by getting rid of reads with low-quality reads, containing ploy-N, with 5′ primer contaminants, without 3′ primers or the insert tag, and with ploy A or T or G or C from the raw data. At the same time, Q20, Q30, and GC-content of the raw data were calculated. Then, the length of sRNA tags within a certain range from clean reads was remained for all the downstream analysis.
All the clean reads were searched against Rfam (http://rfam.sanger.ac.uk/) database for annotation. The reads annotated as tRNA, rRNA, snoRNA and snRNA were discarded. Considering that there was no published genome information of P. trituberculatus available, the remainder small RNA reads were compared to the known miRNA from all metazoan species in the miRBase 20.0 (http://www.mirbase.org/) to identify conserved miRNAs. Only perfect matches were considered as conserved miRNAs. Reads that were not aligned to the miRBase database were used to predict novel miRNAs. The miREvo and mirdeep2 (Friedlander et al. 2012) software were used for novel miRNA prediction by exploring the secondary structure, the Dicer cleavage site, and the minimum free energy of the former unannotated small RNA tags that could be mapped to reference sequences.
Target gene prediction and analyses
As there was no published genome information on P. trituberculatus, we tried to extract the 3′-UTR and 5′-UTR from the P. trituberculatus transcriptome that has been published (Lv et al. 2013). The sequences of transcroptome containing the extracted 3′-UTR and 5′-UTR were considered as candidate gene databases for target gene prediction. MiRanda-3.3 (Enright et al. 2003) software was used to predict the target genes of identified miRNAs. The predicted target genes were aligned by blastx, and then gene ontology (GO) analysis were performed on the target genes by GOEAST (Zheng and Wang 2008).
Differential expression of miRNA
miRNA expression levels were estimated by TPM (transcript per million) through the following criteria (Zhou et al. 2010). Differential expression analysis of two samples was performed using the DEGseq (2010) R package. P-value was adjusted using q value (Storey and Tibshirani 2003). qvalue < 0.01 and |log2 (foldchange) | >1 were set as the threshold for significantly differential expression.
Quantitative real-time PCR assay
MiRNAs expression levels were assayed by quantitative real-time PCR (qPCR) using a SYBR primescriptTM miRNA RT-PCR kit (TaKaRa) (Song et al. 2014) . Total RNAs extracted from the gills were reverse transcribed using a stem-looped antisense primer. The resultant cDNA was submitted to the amplification of mature miRNAs using a miRNA specific primer and a universal primer. U6 snRNA gene was employed as an endogenous control. The primers for miRNAs and U6 snRNA are listed in Table 1. The qPCR was conducted in 15 μl reaction volumes containing 300 nM of each primer and cDNA derived from 0.1 μg of total RNA. Cycling parameters were 95 °C for 1 min, and followed by 40 cycles of 95 °C for 35 s and 60 °C for 30 s. All reactions were run in triplicate. The expression of miRNAs was normalized against U6 snRNA levels. Significant differences were examined by paired t-test and p value < 0.05 was considered to be statistically significant.
Table 1.
The primers for miRNAs real-time RT-PCR
| Primer names | Primer sequences (5′-3′) |
|---|---|
| U6-F | CATATACTAAAATTGGAACGATACAG |
| U6-R | AACGCTTCACGATTTTGCGT |
| miR-2788b | CAATGCCCTTGGAAATCCC |
| novel_1 | AGTTACAGAGATCGTGGAAAGA |
| novel_9 | GCTGTAGGAATATCAAGAGAGT |
| miR-750b | CCAGATCTAACTCTTCCAGCTCA |
| novel_42 | TTGGTCATAGAATTCCTCCGT |
| novel_61 | AGTTCGTTCCTCCCTGGA |
| novel_2 | GTTCCCACACTCCAGGCACTG |
| novel_3 | TCTTTGGTGATCTAGCTGTATGA |
| novel_28 | AGGGGTGTGCAACAAATTATTTT |
| novel_32 | TCGGGCCTCTGTTGTGGTGTGT |
| novel_63 | CCCGAGGCCACACTGTCCTGCAG |
| novel_70 | CGGCACACTCAGTACCAGTA |
| miR-2788a | CAATGCCCTTGGAAATCCCAAAT |
| novel_54 | ACCTGATTGACTCCCCTGGCC |
| novel_67 | GTGGTTTGTCAGGACTCTGAGAGG |
Result
Solexa sequencing of small RNAs
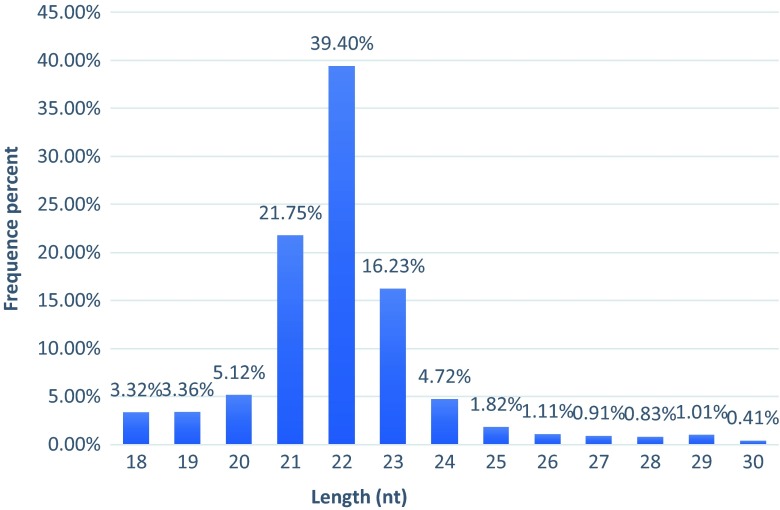
After removal of the 5′ and 3′ adapters, polluted reads and reads smaller than 18 nucleotides (nt), a total of 7,032,973 clean reads, consisting of 153,814,547 nt with an average GC content of 51.18 % were obtained from the LC sample, while 6,166,057 clean reads, consisting of 136,575,094 nt with an average GC content of 51.03 % were obtained from the NC sample (Table 2). 77.39 % of the reads were 21–23 nt in length with a peak at 22 nt (Fig. 2). A total of 2,656,713 of the resulting unique small RNAs were annotated by the RFam 10.1 database in the LC sample while 2,899,003 of the resulting unique small RNAs were annotated in the NC sample. 2,103,956 and 2,501,788 reads of rRNA, tRNA, snoRNA, snRNAs, and other reads were annotated in LC and NC respectively, and then removed from the following analysis (Table 3).
Table 2.
Statistics of microRNA transcriptome output sequencing
| Samples | LC | NC |
|---|---|---|
| Total raw reads | 10,983,455 | 8,819,069 |
| Total clean reads | 7,032,973 | 6,166,057 |
| Total clean nucleotides (nt) | 153,814,547 | 136,575,094 |
| Q20 percentage | 99.28 | 98.63 |
| GC percentage | 51.18 | 51.03 |
Fig. 2.
Length distribution of small RNAs
Table 3.
Annotation of small RNAs
| Types | LC | NC |
|---|---|---|
| known_miRNA | 397,081 | 250,542 |
| rRNA | 299 | 295 |
| tRNA | 5 | 4 |
| snRNA | 27 | 35 |
| snoRNA | 69 | 50 |
| repeat | 2,287 | 2,670 |
| novel_miRNA | 155,676 | 146,673 |
| other | 2,101,269 | 2,498,734 |
| total | 2,656,713 | 2,899,003 |
The discovery of known and novel miRNAs
A total of 16 known miRNAs, belonging to eight families were obtained from LC and NC, which exhibited variable abundance. MiR-750b was the most accumulated miRNA with a total of 349,452 reads detected between the two libraries (220,568 reads in LC and 128,884 in NC), while miR-2788a (20 reads in NC and no reads in LC) were the least accumulated miRNA in the two libraries (Table 4). The results of the family analysis showed that mir-317, mir-2788 and mir-305 were only found in arthropod species (Table S1S1).
Table 4.
miRNA expression in LC and NC
| Type | miRNA | Sequence | LC | NC | log2.Fold_change. | p value | q value | Different expression |
|---|---|---|---|---|---|---|---|---|
| Known miRNA | miR-190 | AGAUAUGUUUGAUAUUCUUGG | 3602 | 1738 | 0.36 | 1.5E−71 | 2.9E−71 | No |
| Known miRNA | miR-2788a | CAAUGCCCUUGGAAAUCCCAAAU | 0 | 20 | −6.66 | 9.0E−13 | 1.2E−12 | Yes |
| Known miRNA | miR-2788b | CAAUGCCCUUGGAAAUCCC | 37 | 5091 | −7.79 | 0.0E+00 | 0.0E+00 | Yes |
| Known miRNA | miR-281a | UGUCAUGGAGUUGCUCUCUUU | 86006 | 56476 | −0.08 | 4.6E−45 | 7.2E−45 | No |
| Known miRNA | miR-281b | AAGAGAGCUAUCCGUCGACAGUACU | 927 | 851 | −0.57 | 2.1E−17 | 3.0E−17 | No |
| Known miRNA | miR-281c | AAGAGAGCUAUCCGUCGACAGU | 30504 | 23961 | −0.34 | 1.5E−105 | 3.2E−105 | No |
| Known miRNA | miR-305a | AUUGUACUUCAUCAGGUGCUCU | 12013 | 7081 | 0.07 | 2.1E−52 | 3.4E−52 | No |
| Known miRNA | miR-305b | AUUGUACUUCAUCAGGUGCUCGG | 20436 | 13941 | −0.14 | 1.3E−01 | 7.3E−02 | No |
| Known miRNA | miR-305c | AUUGUACUUCAUCAGGUGCUC | 12013 | 7081 | 0.07 | 2.1E−52 | 3.4E−52 | No |
| Known miRNA | miR-317a | UGAACACAGCUGGUGGUAUCUCAGU | 33584 | 20252 | 0.04 | 3.8E−110 | 8.5E−110 | No |
| Known miRNA | miR-317b | UGAACACAGCUGGUGGUAUAUCAGU | 79 | 34 | 0.53 | 7.0E−04 | 5.9E−04 | No |
| Known miRNA | miR-750a | CCAGAUCUAACUCUUCCAUAUGACG | 334 | 131 | 0.66 | 3.7E−16 | 4.9E−16 | No |
| Known miRNA | miR-750b | CCAGAUCUAACUCUUCCAGCUCA | 220568 | 128884 | 0.09 | 0.0E+00 | 0.0E+00 | No |
| Known miRNA | miR-7a | UGGAAGACUAGUGAUUUUGUUGU | 10764 | 7053 | −0.08 | 1.1E−06 | 1.2E−06 | No |
| Known miRNA | miR-7b | UGGAAGACUAGUGAUUUUGUUGUUC | 10767 | 7058 | −0.08 | 1.4E−06 | 1.5E−06 | No |
| Known miRNA | miR-9 | UCUUUGGUGAUCUAGCUGUAUGA | 28293 | 15943 | 0.14 | 7.2E−199 | 3.0E−198 | No |
| Novel miRNA | novel_1 | AGUUACAGAGAUCGUGGAAAGA | 9686 | 46459 | −2.95 | 0.0E+00 | 0.0E+00 | Yes |
| Novel miRNA | novel_10 | CUUACGACCGCCUAGCACGGU | 2221 | 782 | 0.82 | 2.4E−131 | 7.9E−131 | No |
| Novel miRNA | novel_11 | CAAGAAAUCACUAAUCCUCCUA | 1773 | 743 | 0.57 | 8.0E−64 | 1.5E−63 | No |
| Novel miRNA | novel_12 | ACUAAGAAUCAAACAUAUUAUCA | 1940 | 1124 | 0.10 | 2.0E−11 | 2.4E−11 | No |
| Novel miRNA | novel_14 | AGUUGGAAGUGGGGAUCUCCGGCA | 983 | 625 | −0.04 | 2.2E−02 | 1.4E−02 | No |
| Novel miRNA | novel_15 | CUGUUGAGCUUGACUCUAGU | 311 | 214 | −0.15 | 9.6E−01 | 4.4E−01 | No |
| Novel miRNA | novel_16 | UUCCCACACUCCAGGCACUG | 256 | 143 | 0.15 | 3.5E−03 | 2.8E−03 | No |
| Novel miRNA | novel_17 | CCAUGGAACCACUAGUGCACC | 207 | 95 | 0.43 | 1.5E−06 | 1.5E−06 | No |
| Novel miRNA | novel_2 | GUUCCCACACUCCAGGCACUG | 35580 | 19762 | 0.16 | 4.3E−287 | 2.1E−286 | No |
| Novel miRNA | novel_20 | CUUUCUCCUCUUCCACUUUCUCG | 25 | 5 | 1.63 | 3.7E−05 | 3.4E−05 | No |
| Novel miRNA | novel_21 | UAAGUGGGAUCGGUAUGCUCAUU | 19 | 6 | 0.97 | 1.1E−02 | 7.8E−03 | No |
| Novel miRNA | novel_26 | UCCAUUUUCACGGAGUCCUGU | 10 | 2 | 1.63 | 9.0E−03 | 6.7E−03 | No |
| Novel miRNA | novel_27 | UUGUAUGUGAAACAAAUACCAUU | 10 | 5 | 0.31 | 3.9E−01 | 2.1E−01 | No |
| Novel miRNA | novel_28 | AGGGGUGUGCAACAAAUUAUUUU | 5 | 0 | 3.97 | 4.5E−03 | 3.4E−03 | Yes |
| Novel miRNA | novel_30 | UGGGGAAAGAUUUGGAGGAACU | 4 | 6 | −1.27 | 8.7E−02 | 5.1E−02 | No |
| Novel miRNA | novel_31 | CAUCCCUUAAUCAGUCUGUCUCU | 4 | 5 | −1.01 | 2.1E−01 | 1.1E−01 | No |
| Novel miRNA | novel_32 | UCGGGCCUCUGUUGUGGUGUGU | 5 | 0 | 3.97 | 4.5E−03 | 3.4E−03 | Yes |
| Novel miRNA | novel_36 | UGAAUCAAAGGAGAUGUAGCA | 3 | 0 | NA | NA | NA | No |
| Novel miRNA | novel_37 | UGACGUGAAGUGAGAGGCUACA | 2 | 2 | −0.69 | 6.0E−01 | 3.0E−01 | No |
| Novel miRNA | novel_39 | UUUAACAAGAUUUCAAGUUGCAU | 3 | 3 | −0.69 | 5.2E−01 | 2.7E−01 | No |
| Novel miRNA | novel_40 | AGUGCACACGCUGGCUGGGAGU | 3 | 0 | NA | NA | NA | No |
| Novel miRNA | novel_42 | UUGGUCAUAGAAUUCCUCCGU | 17 | 2 | 2.40 | 2.1E−05 | 2.0E−05 | Yes |
| Novel miRNA | novel_46 | UGUGGGACCCGAGUACCGCCGA | 29 | 20 | −0.15 | 1.0E+00 | 4.5E−01 | No |
| Novel miRNA | novel_47 | UGUCCCAGUUGGCUCCAGAGA | 30 | 12 | 0.63 | 1.8E−02 | 1.2E−02 | No |
| Novel miRNA | novel_48 | CUGCUUGCUGAGGGGAGGGA | 516 | 904 | −1.50 | 2.6E−130 | 7.6E−130 | No |
| Novel miRNA | novel_5 | AAAUAUCAGCUGGUAAAUUUGG | 13211 | 7096 | 0.21 | 5.2E−136 | 1.9E−135 | No |
| Novel miRNA | novel_51 | AAACAUCCUUGAGAACUUGGA | 15 | 12 | −0.37 | 5.9E−01 | 2.9E−01 | No |
| Novel miRNA | novel_52 | UAGGCCUUUGGAUCGUCAA | 5 | 4 | −0.37 | 7.6E−01 | 3.6E−01 | No |
| Novel miRNA | novel_53 | GUGGCCAUACUCUAUGCACAUC | 242 | 78 | 0.94 | 5.7E−19 | 8.4E−19 | No |
| Novel miRNA | novel_54 | ACCUGAUUGACUCCCCUGGCC | 0 | 2 | −3.33 | 4.9E−02 | 3.0E−02 | Yes |
| Novel miRNA | novel_55 | GUACAGUGGAACCAUGUGUGCUU | 0 | 1 | NA | NA | NA | No |
| Novel miRNA | novel_56 | AGAGGAGGCCUGGACACCU | 7 | 5 | −0.20 | 9.3E−01 | 4.3E−01 | No |
| Novel miRNA | novel_58 | GCAGCAGUGUGCUCUGCCCU | 17 | 20 | −0.92 | 2.2E−02 | 1.4E−02 | No |
| Novel miRNA | novel_59 | GUUUUAGCGAAUUUUCUGGGU | 4 | 8 | −1.69 | 1.2E−02 | 8.2E−03 | No |
| Novel miRNA | novel_6 | CAUCACAGUGAUAGUACCUUACU | 10329 | 5450 | 0.23 | 1.3E−120 | 3.4E−120 | No |
| Novel miRNA | novel_61 | AGUUCGUUCCUCCCUGGA | 10 | 0 | 4.97 | 2.7E−05 | 2.5E−05 | Yes |
| Novel miRNA | novel_62 | GGUGUAGUGGAAGUUAUUGG | 42 | 38 | −0.54 | 8.8E−02 | 5.1E−02 | No |
| Novel miRNA | novel_63 | CCCGAGGCCACACUGUCCUGCAG | 8 | 0 | 4.64 | 2.0E−04 | 1.7E−04 | Yes |
| Novel miRNA | novel_64 | UGAAUUCUUACACGAUGAGUGC | 28 | 9 | 0.95 | 2.4E−03 | 2.0E−03 | No |
| Novel miRNA | novel_66 | CUUUGAAGGUGUGUUGAACCCC | 3 | 3 | −0.69 | 5.2E−01 | 2.7E−01 | No |
| Novel miRNA | novel_67 | GUGGUUUGUCAGGACUCUGAGAGG | 1 | 8 | −3.69 | 4.0E−05 | 3.6E−05 | Yes |
| Novel miRNA | novel_68 | AUGUAUGGUAUGGAUGUGACUGCA | 11 | 11 | −0.69 | 2.2E−01 | 1.2E−01 | No |
| Novel miRNA | novel_7 | UCCCUGAGACCCUUUCUUGUGA | 7720 | 4957 | −0.05 | 1.3E−08 | 1.5E−08 | No |
| Novel miRNA | novel_70 | CGGCACACUCAGUACCAGUA | 4 | 0 | 3.64 | 1.3E−02 | 9.1E−03 | Yes |
| Novel miRNA | novel_71 | UUGCACCUGAUCCAGGAUUUCU | 5 | 4 | −0.37 | 7.6E−01 | 3.6E−01 | No |
| Novel miRNA | novel_75 | AUCUUGGUCGUUUUGCAAAAUG | 27 | 19 | −0.18 | 9.3E−01 | 4.3E−01 | No |
| Novel miRNA | novel_76 | UGUGACGUCAUCGUGGUACUCUU | 6 | 2 | 0.90 | 1.8E−01 | 9.8E−02 | No |
| Novel miRNA | novel_77 | GUGUUCUGACUGGGGGCUGA | 6 | 1 | 1.90 | 2.7E−02 | 1.7E−02 | No |
| Novel miRNA | novel_78 | CUGGAACGCAUUUUUAUC | 2 | 4 | −1.69 | 7.4E−02 | 4.5E−02 | No |
| Novel miRNA | novel_8 | UUUACGACCGUUUAGCACGGU | 5786 | 2158 | 0.73 | 3.5E−293 | 2.1E−292 | No |
| Novel miRNA | novel_9 | GCUGUAGGAAUAUCAAGAGAGU | 1070 | 4822 | −2.86 | 0.0E+00 | 0.0E+00 | Yes |
Altogether, we obtained 51 predicted novel miRNAs. The novel miRNAs also displayed unequal expression levelsbetween the two libraries (Table 4). Novel_5 was the most accumulated miRNA with a total of 20,307 reads detected between the two libraries (13,211 reads in LC and 7,096 in NC), while novel_40 and novel_36 (three reads in NC and no reads in LC) were the least abundant miRNA in the two libraries.
We matched the miRNAs of P. trituberculatus against the miRNAs of the Chinese mitten crab, Eriocheir sinensis, another important economic aquaculture species in China, and 85.7 % (12 out of 16) of the known miRNAs were matched, whereas none of the novel miRNAs were found among the miRNAs from E. sinensis.
miRNA differential expression analysis
A comparison of miRNA expressions showed that a total of 12 miRNA were differentially expressed after low salinity stress (q_value < 0.005 & |log2 (fold change)| > 1). Six were up-regulated and six were down-regulated between LC and NC (Table 4). Novel_61 (log2.Fold_change = 4.9663) and miR-2788b (log2.Fold_change = -7.7936) were the most up- or down-regulated miRNAs in LC vs. NC respectively. Among the 12 miRNAs, novel_1, novel_9 and miR-2788b were the most abundant miRNAs with 56,145, 5,892, and 5,128 reads detected between the two libraries, respectively (Table 4). Fifteen miRNA (4 known miRNAs and 11 novel miRNAs) were selected to confirm the expression pattern by stem-loop RT-qPCR assays. The results showed that there was a general consistency (86.7 %) between the quantitative assay and deep sequencing analysis for the fifteen miRNAs in terms of directions of regulation and significance (Ou et al. 2012) (Table 5).
Table 5.
Gene expression levels of 15 selected miRNAs for the comparison of non-challenged and low salinity-challenged groups according to Illumina sequence and qPCR analysis
| Gene | qPCR | Illumina sequence |
|---|---|---|
| LC vs. NC log2 (fold change) | LC vs. NC log2 (fold change) | |
| novel_54 | −1.22* | −3.3337* |
| novel_67 | −0.49 | −3.6893* |
| novel_9 | −1.86* | −2.8613* |
| novel_1 | −1.81* | −2.9513* |
| miR-2788a | −3.78* | −6.6556* |
| miR-2788b | −6.64* | −7.7936* |
| novel_32 | 1.50* | 3.9663* |
| novel_70 | 1.38* | 3.6444* |
| novel_28 | 1.32* | 3.9663* |
| novel_63 | 1.78* | 4.6444* |
| novel_42 | 1.64* | 2.3982* |
| novel_61 | 0.15 | 4.9663* |
| miR-750b | 0.55 | 0.08585 |
| novel_2 | 0.42 | 0.15904 |
| miR-9 | 0.16 | 0.13823 |
*Significant difference between salinity-challenged group and control group (p < 0.05)
Targets prediction and go enrichment analysis
A total of 94,511 transcriptome unigene of P. trituberculatus (Lv et al. 2013) were used to identify miRNA targets. The results show that 67 miRNA had 4702 affiliated target genes (70 target genes / miRNA), among which 12 differentially expressed miRNAs had 2051 affiliated targets (171 target genes / miRNA). GO enrichment analysis demonstrated that these targets were involved in many biological processes including regulation of cellular process (GO:0050794), cellular response to stimulus (GO:0051716), transport (GO:0006810), cell communication (GO:0007154), protein modification (GO:0036211), signal transduction (GO:0007165), phosphorylation (GO:0016310), etc. (Table S2).
To increase efficiency to discover true target genes, we filtered the targets showing differential expression in the opposite direction compared to miRNA. In total, 34 of the target genes predicted by miRanda had an opposite differential expression to the miRNAs, which included sodium-independent sulfate anion transporter, sodium-dependent phosphate transporter 1-B, S-adenosylmethionine synthase, RNA-directed DNA polymerase from mobile element jockey, ribonuclease H1, protein henna, polypeptide N-acetylgalactosaminyltransferase 2, phosphoethanolamine N-methyltransferase 1, chorion peroxidase, carbohydrate sulfotransferase 3, etc. (Table 6). Biological processes of the 34 genes were further analyzed according to GO functional enrichment analysis, which were involved in six biological processes. The main processes involving unigenes were anion transport (GO:0006820, two unigenes), chitin metabolic process (GO:0006030, two unigenes), alpha-amino acid metabolic process (GO:1901605, two unigenes), intracellular protein transport (GO:0006886, three unigenes), organic substance transport (GO:0071702, four unigenes), and cytoskeleton organization (GO:0007010, two unigenes) (Fig 3).
Table 6.
The predicted targets of the differentially expressed miRNAs
| Type | miRNA ID | Putative target | Annotation |
|---|---|---|---|
| Down | novel_1 | comp72508_c0 | Phenylalanine hydroxylase |
| comp73104_c0 | Chitin-binding peritrophin-A | ||
| comp77164_c0 | Zinc finger protein ZPR1 | ||
| Down | novel_25 | comp44245_c0 | S-Adenosylmethionine synthase |
| Down | novel_54 | comp71288_c0 | Calcineurin B homologous protein 1 |
| Down | novel_67 | comp70423_c0 | Anaphylotoxin-like domain |
| comp71288_c0 | Calcineurin B homologous protein 1 | ||
| comp77164_c0 | Zinc finger protein ZPR1 | ||
| Down | novel_9 | comp55772_c0 | Sodium-independent sulfate anion transporter |
| comp72508_c0 | Phenylalanine hydroxylase | ||
| Down | miR-2788b | comp44245_c0 | S-Adenosylmethionine synthase |
| Up | novel_28 | comp38756_c0 | Collagen alpha-1(IV) chain |
| comp43033_c0 | – | ||
| comp67982_c0 | Alcohol acetyltransferase | ||
| comp70501_c0 | Transmembrane protein 70 homolog, mitochondrial | ||
| comp72677_c0 | Outer dense fiber protein 3 | ||
| comp74429_c0 | Microtubule-associated protein TAU | ||
| comp76458_c0 | Polypeptide N-acetylgalactosaminyltransferase 2 | ||
| comp76540_c0 | Egl nine homolog 1 | ||
| Up | novel_32 | comp70501_c0 | Transmembrane protein 70 homolog, mitochondrial |
| comp75735_c1 | – | ||
| Up | novel_42 | comp66615_c0 | Sodium-dependent phosphate transporter 1-B |
| comp70501_c0 | Transmembrane protein 70 homolog, mitochondrial | ||
| comp72399_c0 | RNA polymerase II, large subunit/ameloblastin precursor | ||
| comp74211_c0 | Carbohydrate sulfotransferase 3 | ||
| comp77007_c0 | 2-Hydroxyacylsphingosine 1-beta-galactosyltransferase | ||
| Up | novel_61 | comp55240_c0 | Phosphoethanolamine N-methyltransferase 1 |
| comp76538_c0 | Compound eye opsin BCRH2 | ||
| Up | novel_63 | comp44799_c0 | Arrestin homolog |
| comp66615_c0 | Sodium-dependent phosphate transporter 1-B | ||
| comp68146_c0 | Cuticlin-1 | ||
| comp70501_c0 | Transmembrane protein 70 homolog, mitochondrial | ||
| comp74011_c0 | Helix-loop-helix protein delilah | ||
| comp74806_c0 | Splicing coactivator SRm160/300 | ||
| Up | novel_70 | comp44799_c0 | Arrestin homolog |
| comp55240_c0 | Phosphoethanolamine N-methyltransferase 1 | ||
| comp65138_c0 | Ribonuclease H1 | ||
| comp66513_c0 | – | ||
| comp75339_c0 | Filamin-A | ||
| comp76267_c0 | Chorion peroxidase | ||
| comp76440_c0 | Villin headpiece domain | ||
| comp76744_c0 | Chitin-binding peritrophin-A /cytochrome oxidase c subunit | ||
| comp76758_c2 | Apolipophorin-III precursor (apoLp-III) | ||
| comp77517_c0 | RNA-directed DNA polymerase from mobile element jockey |
Fig. 3.
GO enrichment analysis of the differentially expressed targets genes
Discussion
Although miRNAs have been researched extensively in recent years, no systematic study has been reported for P. trituberculatus, which is one of the most economically important mariculture species in China. In this study, we combined high-throughput Illumina sequencing technology with bioinformatic methods to efficiently identify and systematically analyze the miRNA response to low salinity stress in P. trituberculatus. The miRNA sequencing pipeline yielded 6,166,057 and 7,032,973 clean reads in the NC and LC libraries, respectively. The peak size was 22 nt, which was consistent with the results for Triops cancriformis (Ikeda et al. 2015), E. sinensis (Song et al. 2014), and Scylla paramamosain (Li et al. 2013). This suggests that 22 nt may be a typical size for crustacean miRNAs.
A total of 67 miRNAs were identified in this study, among which, 76.1 % (51 out of 67) were novel miRNAs. A similar result was found in E. sinensis (71.2 % were novel miRNA) (Song et al. 2014). The results suggest that the basis for the study of miRNA is weak in aquatic crustacean animals. So far, only 52 crustacean miRNAs have been listed in the miRBase, which accounted for 0.21 % of the total (24,521) (Kozomara and Griffiths-Jones 2013). We matched P. trituberculatus miRNAs against E. sinensis miRNAs (Song et al. 2014; Ou et al. 2012), another important economic aquaculture species in China. A total of 85.7 % (12 out of 16) of the known miRNAs could be matched with the miRNAs from E. sinensis, whereas none of the novel miRNAs matched the miRNAs from E. sinensis. The results indicate that P. trituberculatus miRNAs have high homologies to miRNAs from E. sinensis, but there are also many P. trituberculatus-specific miRNAs.
Many miRNAs are highly conserved among organisms, and they probably have relatively conservative and important roles in regulating basic cellular pathways in both lower and higher organisms (Glazov et al. 2008). In this study, mir-317, mir-2788, and mir-305 of P. trituberculatus specifically existed in the arthropod. The mir-317 influences reproductive responses by females to male sex peptide (Fricke et al. 2014) and was involved in cell cycle control in association with G2/M cyclin B in Drosophila (Pushpavalli et al. 2014). miR-305 regulates the Notch and insulin pathways in the intestinal stem cells, which are required for adaptive homeostasis in the Drosophila gut (Foronda et al. 2014). mir-2788a plays a potential role in butterfly wing development in Heliconius (Surridge et al. 2011). Previous research indicated that mir-317, mir-2788, and mir-305 may have similar roles in P. trituberculatus, but this needs further verification.
Comparison of gene expression among the different treatment groups in the current experiment was helpful for identification of candidate miRNAs underlying responses to salinity stress in P. trituberculatus. In this study, we detected 12 miRNAs differentially expressed between the comparison of LC vs NC (six upregulated and six downregulated). Identification of targets improves our understanding of the physiological functions of these differentially expressed miRNAs. As miRNA binding to 3′-UTR of target gene generally caused destabilization and degradation of mRNA (Guo et al. 2010), for increasing the strength to discover true target genes, we chose to narrow down potential targets to those showing differential expression in the opposite direction compared to miRNA. In total, 34 target genes were identified, among which, S-adenosylmethionine synthetase (SAM) was the only predicted target of the only known differentially expressed miRNA mir-2788. SAM is also known as methionine adenosyltransferase (MAT), catalyzes the formation of S-adenosylmethionine by joining methionine and ATP (Horikawa et al. 1990), and plays an important role in plant responses to salt, drought, alkali, and flood stresses (Kamal et al. 2012; Chen et al. 2014). SAM is also a methyl donor, which controls gene expression by DNA methylation (Reytor et al. 2009). The mir-2788 miRNA was the only potential regulatory miRNA of SAM and was differentially expressed between LC and NC in this study, which indicated that it may play an important role in hyper-osmoregulation by regulating the expression of SAM.
Interestingly, many other genes encoding biological enzymes other than SAM were predicted as targets of differentially expressed miRNAs. These accounted for 38.2 % of the total predicted targets (13 out of 34). Enzymes are macromolecular biological catalysts that can increase the reaction rate by lowering the activation energy. They are able to catalyze more than 5000 biochemical reactions (Schomburg et al. 2013), and some can make reactions of substrates to products occur millions of times faster. Orotidine 5′-phosphate decarboxylase is an extreme example, which catalyzes a reaction that would otherwise take millions of years to occur in milliseconds (Radzicka and Wolfenden 1995; Callahan and Miller 2007). Four out of 13 genes encoding biological enzymes have been reported to play an important role in salinity adaptation. These are SAM [29–32], phosphoethanolamine N-methyltransferase 1 (Wu et al. 2007), cytochrome oxidase c (Zikos et al. 2014; Tine et al. 2011), and calcineurin B (Chen et al. 2015; Gu et al. 2008). Other enzyme genes have functions in anoxic response (alcohol acetyltransferase) (Kwast et al. 2002), disease responses (phenylalanine hydroxylase and polypeptide N-acetylgalactosaminyltransferase 2) (Kaufman 1999) and stress responses (RNA polymerase II) (Bourbonnais et al. 2001). These results indicate that miRNA play important roles in responding to low salinity stress through a series of enzymatic reactions that are highly efficient and have a low energy consumption. However, the hypothesis needs further confirmation.
Six GO biological processes, which help P. trituberculatus to adapt to low salinity environments, were significantly enriched. The top two processes based on the enrichment level (p value), were anion transport (GO:0006820) and chitin metabolic process (GO:0006030). It is worth noting that the two processes were also shown to be enriched in the transcriptome analysis of P. trituberculatus when responding to salinity stress (Lv et al. 2013). Anion transport (GO:0006820) is the directed movement of anions within or between cells by transporters or pores. It is important in “compensatory process” adopted by crustaceans for osmoregulation and predominately occurs in gills (Péqueux 1995; Rainbow and Black 2001). The sodium-independent sulfate anion transporter and sodium-dependent phosphate transporter 1-B were involved in this process. Previous study showed that the two genes were also important osmoregulation factors in E. sinensis (Hui et al. 2014). Peritrophins, which are tightly associated with the peritrophic membrane (PM) or chitin fibrils, were first isolated from insect PMs. They protect insects from invasion microorganisms and stimulate digestion (Loongyai et al. 2007). Peritrophins are also involved in anti-bacterial innate immunity in crabs (Huang et al. 2015); however, their role of osmoregulation has not been reported. In this study, two peritrophin-like genes containing chitin binding peritrophin-A domains were the predicted targets of two differentially expressed miRNAs (novel_1 and novel_70) enriched in the chitin metabolic process (GO:0006030). These results indicated that anion transport and the chitin metabolic process play important roles in osmoregulation; moreover, these processes were controlled by miRNAs according to regulate specific target genes.
Conclusion
This study is the first systematic analysis of miRNAs in P. trituberculatus by high-throughput Illumina sequencing technology. Sixty-seven miRNAs were identified, among which, 12 miRNAs were related to low salinity stress. Thirty-four of the target genes predicted were differentially expressed in the opposite way to the miRNAs, which were enriched in six important GO biological processes related to osmoregulation, such as anion transport and the chitin metabolic process. The results provide a basis for further investigation of miRNA-modulated osmoregulation networks in P. trituberculatus.
Electronic supplementary material
(XLSX 15 kb)
(XLSX 50 kb)
Acknowledgments
This research was supported by the National High Technology Research and Development Program of China (Project 2012AA10A409) and the National Natural Science Foundation of China (Grant No. 41576147 and 41306177).
References
- Bourbonnais Y, Faucher N, Pallotta D, Larouche C. Multiple cellular processes affected by the absence of the Rpb4 subunit of RNA polymerase II contribute to the deficiency in the stress response of the yeast rpb4 delta mutant. Mol Gen Genet. 2001;264:763–772. doi: 10.1007/s004380000365. [DOI] [PubMed] [Google Scholar]
- Cohen SM, Bushati N. MicroRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Callahan BP, Miller BG. OMP decarboxylase—an enigma persists. Bioorg Chem. 2007;35:465–469. doi: 10.1016/j.bioorg.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Chen L, Ren J, Shi H, Zhang Y, You Y, Fan J, Chen K, Liu S, Nevo E, Fu J, Peng J. TdCBL6, a calcineurin B-like gene from wild emmer wheat (Triticum dicoccoides), is involved in response to salt and low-K+ stresses. Mol Breed. 2015;35:1. doi: 10.1007/s11032-015-0202-z. [DOI] [Google Scholar]
- Chen Y, Chen X, Wang H, Bao Y, Zhang W. Examination of the leaf proteome during flooding stress and the induction of programmed cell death in maize. Proteome Sci. 2014;12:33. doi: 10.1186/1477-5956-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A.J, John B, Gaul U, Tuschl T, Chris S, Marks, DS (2003). 2003 Enright Et Volume Al. 5, Issue 1, Article R1 Open Access Research Microrna Targets In Drosophila. Genome Biol, 5:R1: R1. [DOI] [PMC free article] [PubMed]
- Foronda D, Weng R, Verma P, Chen Y-W, Cohen SM. Coordination of insulin and notch pathway activities by microRNA miR-305 mediates adaptive homeostasis in the intestinal stem cells of the Drosophila gut. Genes Dev. 2014;28:2421–2431. doi: 10.1101/gad.241588.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier TP, Sun G, Burklew CE, Zhang B. Salt and drought stresses induce the aberrant expression of microRNA genes in tobacco. Mol Biotechnol. 2011;49:159–165. doi: 10.1007/s12033-011-9387-5. [DOI] [PubMed] [Google Scholar]
- Fricke C, Green D, Smith D, Dalmay T, Chapman T. MicroRNAs influence reproductive responses by females to male sex peptide in Drosophila melanogaster. Genetics. 2014;198:1603. doi: 10.1534/genetics.114.167320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res. 2008;18:957–964. doi: 10.1101/gr.074740.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Ma B, Jiang Y, Chen Z, Su X, Zhang H. Expression analysis of the calcineurin B-like gene family in rice (Oryza sativa L.) under environmental stresses. Gene. 2008;415:1–12. doi: 10.1016/j.gene.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa S, Sasuga J, Shimizu K, Ozasa H, Tsukada K. Molecular cloning and nucleotide sequence of cDNA encoding the rat kidney S-adenosylmethionine synthetase. J Biol Chem. 1990;265:13683–13686. [PubMed] [Google Scholar]
- Huang Y, Ma F, Wang W, Ren Q. Identification and molecular characterization of a peritrophin-like gene, involved in the antibacterial response in Chinese mitten crab, Eriocheir sinensis. Dev Comp Immunol. 2015;50:129–138. doi: 10.1016/j.dci.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Hui M, Liu Y, Song C, Li Y, Shi G, Cui Z. Transcriptome changes in Eriocheir sinensis megalopae after desalination provide insights into osmoregulation and stress adaption in larvae. PLoS ONE. 2014;9:e114187. doi: 10.1371/journal.pone.0114187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda KT, Hirose Y, Hiraoka K, Noro E, Fujishima K, Tomita M, Kanai A. Identification, expression, and molecular evolution of microRNAs in the “living fossil” Triops cancriformis (tadpole shrimp) Rna Publ Rna Soc. 2015;21:230–242. doi: 10.1261/rna.045799.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal AHM, Cho K, Kim D-E, Uozumi N, Chung K-Y, Lee SY, Choi J-S, Cho S-W, Shin C-S, Woo SH. Changes in physiology and protein abundance in salt-stressed wheat chloroplasts. Mol Biol Rep. 2012;39:9059–9074. doi: 10.1007/s11033-012-1777-7. [DOI] [PubMed] [Google Scholar]
- Kaufman S. A model of human phenylalanine metabolism in normal subjects and in phenylketonuric patients. Proc Natl Acad Sci. 1999;96:3160–3164. doi: 10.1073/pnas.96.6.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S, (2013). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res: gkt1181. [DOI] [PMC free article] [PubMed]
- Kwast KE, Lai LC, Menda N, James DT, Aref S, Burke PV. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J Bacteriol. 2002;184:250–265. doi: 10.1128/JB.184.1.250-265.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SK, Zhu S, Li CB, Zhang Z, Zhou LZ, Wang SJ, Wang SQ, Zhang YL, Wen XB. Characterization of microRNAs in mud crab Scylla paramamosain under Vibrio parahaemolyticus infection. PLoS ONE. 2013;8:e73392. doi: 10.1371/journal.pone.0073392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loongyai W, Avarre J-C, Cerutti M, Lubzens E, Chotigeat W. Isolation and functional characterization of a new shrimp ovarian peritrophin with antimicrobial activity from Fenneropenaeus merguiensis. Mar Biotechnol. 2007;9:624–637. doi: 10.1007/s10126-007-9019-z. [DOI] [PubMed] [Google Scholar]
- Lv J, Liu P, Wang Y, Gao B, Chen P, Li J. Transcriptome analysis of Portunus trituberculatus in response to salinity stress provides insights into the molecular basis of osmoregulation. PLoS ONE. 2013;8:e82155. doi: 10.1371/journal.pone.0082155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J, Zhang D, Gao B, Liu P, Li J. Transcriptome and MassARRAY analysis for identification of transcripts and SNPs for growth traits of the swimming crab Portunus trituberculatus. Gene. 2015;566:229–235. doi: 10.1016/j.gene.2015.04.050. [DOI] [PubMed] [Google Scholar]
- Ma H, Hostuttler M, Wei HR, Rexroad CE, Yao JB. Characterization of the rainbow trout egg MicroRNA transcriptome. PLoS ONE. 2012;7:E39649. doi: 10.1371/journal.pone.0039649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Ou J, Meng Q, Li Y, Xiu Y, Du J, Gu W, Wu T, Li W, Ding Z, Wang W. Identification and comparative analysis of the Eriocheir sinensis microRNA transcriptome response to Spiroplasma eriocheiris infection using a deep sequencing approach. Fish Shellfish Immunol. 2012;32:345–352. doi: 10.1016/j.fsi.2011.11.027. [DOI] [PubMed] [Google Scholar]
- Péqueux A. Osmotic regulation in crustaceans. J Crust Biol. 1995;15:1–60. doi: 10.2307/1549010. [DOI] [Google Scholar]
- Peng Z, He S, Gong W, Sun J, Pan Z, Xu F, Lu Y, Du X. Comprehensive analysis of differentially expressed genes and transcriptional regulation induced by salt stress in two contrasting cotton genotypes. BMC Genomics. 2014;15:760. doi: 10.1186/1471-2164-15-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushpavalli SNCVL, Sarkar A, Bag I, Hunt CR, Ramaiah MJ, Pandita TK, Bhadra U, Pal-Bhadra M. Argonaute-1 functions as a mitotic regulator by controlling cyclin B during Drosophila early embryogenesis. FASEB J. 2014;28:655–666. doi: 10.1096/fj.13-231167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzicka A, Wolfenden R. A proficient enzyme. Science. 1995;267:90–93. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- Rainbow P, Black W. Effects of changes in salinity on the apparent water permeability of three crab species: Carcinus maenas eriocheir sinensis and Necora puber. J Exp Mar Biol Ecol. 2001;264:1–13. doi: 10.1016/S0022-0981(01)00289-1. [DOI] [Google Scholar]
- Rao G, Sui J, Zeng Y, He C, Duan A, Zhang J. De novo transcriptome and small RNA analysis of two Chinese willow cultivars reveals stress response genes in Salix matsudana. PLoS ONE. 2014;9:e109122. doi: 10.1371/journal.pone.0109122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reytor E, Pérez-Miguelsanz J, Alvarez L, Pérez-Sala D, Pajares MA. Conformational signals in the C-terminal domain of methionine adenosyltransferase I/III determine its nucleocytoplasmic distribution. FASEB J. 2009;23:3347–3360. doi: 10.1096/fj.09-130187. [DOI] [PubMed] [Google Scholar]
- Romano N, Zeng CS. Osmoregulation in decapod crustaceans: implications to aquaculture productivity, methods for potential improvement and interactions with elevated ammonia exposure. Aquaculture. 2012;334:12–23. doi: 10.1016/j.aquaculture.2011.12.035. [DOI] [Google Scholar]
- Schomburg I, Chang A, Placzek S, Söhngen C, Rother M, Lang M, Munaretto C, Ulas S, Stelzer M, Grote A. BRENDA in 2013: integrated reactions, kinetic data, enzyme function data, improved disease classification: new options and contents in BRENDA. Nucleic Acids Res. 2013;41(D1):D764–D772. doi: 10.1093/nar/gks1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y-N, Shi L-L, Liu Z-Q, Qiu G-F. Global analysis of the ovarian microRNA transcriptome: implication for miR-2 and miR-133 regulation of oocyte meiosis in the Chinese mitten crab, Eriocheir sinensis (Crustacea: Decapoda) BMC Genomics. 2014;15:547. doi: 10.1186/1471-2164-15-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surridge AK, Lopez-Gomollon S, Moxon S, Maroja LS, Rathjen T, Nadeau NJ, Dalmay T, Jiggins CD. Characterisation and expression of microRNAs in developing wings of the neotropical butterfly Heliconius melpomene. BMC Genomics. 2011;12:62. doi: 10.1186/1471-2164-12-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tine M, Mckenzie DJ, Bonhomme F, Durand J-D. Salinity-related variation in gene expression in wild populations of the black-chinned tilapia from various west African coastal marine, estuarine and freshwater habitats. Estuar Coast Shelf Sci. 2011;91:102–109. doi: 10.1016/j.ecss.2010.10.015. [DOI] [Google Scholar]
- Valenzuela-Miranda D, Nunez-Acuna G, Valenzuela-Munoz V, Asgari S, Gallardo-Escarate C. MicroRNA biogenesis pathway from the salmon louse (Caligus rogercresseyi): emerging role in delousing drug response. Gene. 2015;555:231–241. doi: 10.1016/j.gene.2014.11.008. [DOI] [PubMed] [Google Scholar]
- Wu S, Yu Z, Wang F, Li W, Ye C, Li J, Tang J, Ding J, Zhao J, Wang B. Cloning, characterization, and transformation of the phosphoethanolamine N-methyltransferase gene (ZmPEAMT1) in maize (Zea mays L.) Mol Biotechnol. 2007;36:102–112. doi: 10.1007/s12033-007-0009-1. [DOI] [PubMed] [Google Scholar]
- Xie F, Stewart CN, Jr, Taki FA, He Q, Liu H, Zhang B. High-throughput deep sequencing shows that microRNAs play important roles in switchgrass responses to drought and salinity stress. Plant Biotechnol J. 2014;12:354–366. doi: 10.1111/pbi.12142. [DOI] [PubMed] [Google Scholar]
- Xu QH, Liu Y. Gene expression profiles of the swimming crab Portunus trituberculatus exposed to salinity stress. Mar Biol. 2011;158:2161–2172. doi: 10.1007/s00227-011-1721-8. [DOI] [Google Scholar]
- Yang H, Cho ME, Li TW, Peng H, Ko KS, Mato JM, Lu SC. MicroRNAs regulate methionine adenosyltransferase 1A expression in hepatocellular carcinoma. J Clin Invest. 2013;123:285. doi: 10.1172/JCI63861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Song H, Yao G. Geographical distribution and faunal analysis of crab resources in the East China Sea. J Zhejiang Ocean Univ. 2003;22:108–113. [Google Scholar]
- Zeng D, Chen X, Xie D, Zhao Y, Yang Q, Wang H, Li Y, Chen X. Identification of highly expressed host microRNAs that respond to white spot syndrome virus infection in the Pacific white shrimp Litopenaeus vannamei (Penaeidae) Genet Mol Res. 2015;14:4818. doi: 10.4238/2015.May.11.14. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Wang X-J. GOEAST: a web-based software toolkit for gene ontology enrichment analysis. Nucleic Acids Res. 2008;36:W358–W363. doi: 10.1093/nar/gkn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Chen J, Li Z, Li X, Hu X, Huang Y, Zhao X, Liang C, Wang Y, Sun L. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27. 3 associate with clear cell renal cell carcinoma. PLoS ONE. 2010;5:e15224. doi: 10.1371/journal.pone.0015224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Wang LL, Song LS, Liu R, Zhang H, Huang MM, Chen H. The identification and characteristics of immune-related MicroRNAs in haemocytes of oyster Crassostrea gigas. PLoS ONE. 2014;9:e88397. doi: 10.1371/journal.pone.0088397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zikos A, Seale AP, Lerner DT, Grau EG, Korsmeyer KE. Effects of salinity on metabolic rate and branchial expression of genes involved in ion transport and metabolism in Mozambique tilapia (Oreochromis mossambicus) Comp Biochem Physiol Mol Int Physiol. 2014;178:121–131. doi: 10.1016/j.cbpa.2014.08.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 15 kb)
(XLSX 50 kb)