Abstract
AIM: To develop a Fok-I nested polymerase chain reaction (PCR)-restriction fragment length polymorphism analysis (PRA) method for the detection of hepatitis B virus X region (HBx) V5M mutation.
METHODS: Nested PCR was applied into DNAs from 198 chronic patients at 2 different stages [121 patients with hepatocellular carcinoma (HCC) and 77 carrier patients]. To identify V5M mutants, digestion of nested PCR amplicons by the restriction enzyme Fok-I (GGA TGN9↓) was done. For size comparison, the enzyme-treated products were analyzed by electrophoresis on 2.5% agarose gels, stained with ethidium bromide, and visualized on a UV transilluminator.
RESULTS: The assay enabled the identification of 69 patients (sensitivity of 34.8%; 46 HCC patients and 23 carrier patients). Our data also showed that V5M prevalence in HCC patients was significantly higher than in carrier patients (47.8%, 22/46 patients vs 0%, 0/23 patients, P < 0.001), suggesting that HBxAg V5M mutation may play a pivotal role in HCC generation in chronic patients with genotype C infections.
CONCLUSION: The Fok-I nested PRA developed in this study is a reliable and cost-effective method to detect HBxAg V5M mutation in chronic patients with genotype C2 infection.
Keywords: Hepatitis B virus, X antigen, Polymerase chain reaction-restriction fragment length polymorphism analysis, V5M mutation, Hepatocellur carcinoma
Core tip: In the present study, we developed a reliable and cost-effective Fok-I nested polymerase chain reaction-restriction fragment length polymorphism analysis (PRA) method for the detection of V5M from chronic patients with genotype C2 infection. In addition, our epidemiological data based on the Fok-I nested PRA method strongly support the previous reports that V5M may play a very pivotal role in hepatocarcinogenesis, at least in chronic patients infected with genotype C2.
INTRODUCTION
Hepatitis B virus (HBV) infection is a global health problem, and more than 350 million people are chronic carriers of the virus[1]. Korea is a recognized endemic area of HBV infection, and an extraordinary prevalence of genotype C2 was also reported in this area, which is known to be more prone to mutations and related to more severe liver diseases and a lower antiviral response compared with genotype B[2,3]. Furthermore, the high prevalence of basal core promoter (BCP) double mutations and the presence of a distinct immune response against HBV proteins in the Korean population could lead to the generation of distinct HBV variants that are rarely encountered in other areas, resulting in distinct clinical manifestations in Korean chronic patients[4-19].
Despite its small size, the HBV genome contains four partially overlapping open reading frames producing at least seven viral gene products[20]. Among these, the HBV X antigen (HBxAg) has been the focus of much attention in recent years because it is implicated strongly in hepatocarcinogenesis. HBxAg is a 154-amino-acid protein with an N-terminal negative regulatory domain and a C-terminal transactivation domain. The HBxAg is multifunctional and affects gene transcription, signaling pathways, genotoxic stress responses, cell-cycle control, and apoptosis, and it also plays an essential role in viral replication[21-24].
During the natural course of HBV infection, naturally occurring mutations may occur, and these could be HBV variants that affect prognosis[25-27]. Several reports have demonstrated that specific point mutations, deletions or insertions in the HBxAg gene were related to severe forms of liver disease, such as cirrhosis of the liver and/or HCC[17,28,29]. In particular, our previous study based on a direct sequencing protocol had introduced a novel N-terminal mutation type, V5M/L, with a mutation in codon 5 of HBxAg from Korean chronic patients with genotype C infections[12]. This mutation type was found significantly more frequently in HCC patients than in patients in other disease groups. This finding suggests that it may play a pivotal role in HCC generation during the natural course of HBV chronic infection. For its monitoring among chronic patients, particularly with genotype C2 infections, the development of a new molecular diagnostic method for its detection is needed. For this purpose, in the present study, we developed a new nested PCR-restriction fragment length polymorphism analysis (PRA) method that can detect V5M and applied it to DNAs from Korean chronic patients.
MATERIALS AND METHODS
Study subjects
Among the patients visiting the Jeju National University Hospital from March to November and the Seoul National University Hospital from January to December in 2005, hepatitis B patients were selected in this study. The patients with a history of anti-viral therapy, an alcoholic liver disease, or hepatitis C involving liver disorders were excluded from this research. For an analysis of the correlation between V5M mutants and hepatocellular carcinoma (HCC), 121 serum samples from HCC patients and 77 samples from carriers were collected and stored at -80 °C. The basic biochemical tests for the serum samples were performed, and hepatitis B e antigen (HBeAg), anti-HBe, HBV-DNA and alpha-fetoprotein (AFP) assays were performed in the case of hepatitis B surface antigen (HBsAg)-positive samples. The detection of HBsAg, HBeAg, anti-HBs, and anti-HBe were performed by a chemiluminescence immunoassay (Abbott ARCHITECT, Abbott, IL, United States), and the determination of the serum HBV DNA was quantified by using a Versant HBV DNA Assay version 3.0 (bDNA, Siemens, NY, United States). This study was approved by the Institutional Review Board of Seoul National University Hospital (IRB No. 1404-070-572).
Diagnosis of liver disease
A definitive diagnosis of liver disease was made according to the overall findings, including clinical, biochemical, and radiological data. Among patients having positive HBsAg, the subjects with positive HBeAg, anti-HBe negative, HBV DNA positive, and normal serum transaminase were clinically defined as HBeAg-positive healthy carriers if there was no evidence of chronic liver disease in the radiologic findings[30]. Hepatocellular carcinoma was diagnosed by clinical findings such as more than 400 pg/mL in a typical value of the serum alpha-fetoprotein (AFP), typical hyper blood vessels upon computed tomography (CT) or biopsy from the liver[31].
HBV DNA extraction
Viral HBV DNA was extracted from 200 μL of the serum obtained from 121 HCC and 77 carrier patients using the QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany). Briefly, 20 μL of QIAGEN Protease (or proteinase K) was added into the 1.5 mL micro-centrifuge tube containing 200 μL of serum sample. Then, 200 μL of buffer AL was added into the sample and was mixed by pulse-vortexing for 15 s. To ensure efficient lysis, it is essential that the sample and Buffer AL are mixed thoroughly to yield a homogeneous solution. After the sample was incubated at 56 °C for 10 min, 200 μL of ethanol (96%-100%, DukSan, Seoul, Korea) was loaded into the sample and was mixed again by pulse-vortexing for 15 s. After mixing, the mixture was carefully applied to the QIAamp Mini-spin column and was centrifuged at 8000 rpm for 1 min. Then, 500 μL of buffer AW1 was carefully added into the column, and the column was centrifuged at 8000 rpm for 1 min. Next, 500 μL of buffer AW2 was added into the column and was centrifuged at full speed (14000 rpm) for 3 min. After the collection tube containing the filtrate was discarded, 50 μL of buffer AE was added into the column and was incubated at room temperature for 1 min; it was then centrifuged at 8000 rpm for 1 min. The eluted DNA was stored at -20 °C and used for polymerase chain reaction (PCR) mixtures as the template.
PCR amplification and direct sequencing analysis of HBx region
To analyze the mutation patterns in the HBx gene from 69 patients amplified by nested PCR, the PCR product was directly sequenced for sequence analysis. The first round of PCR was carried out using the sense primer X-pro-F1 (5’-CTC TGC CAA GTG TTT GCT GA-3’; GenBank accession number AY641558, positions 1171-1190 nt) and the antisense primer X-pro-R1 (5’-CAA GGC ACA GCT TGG AGG CT-3’; positions 1886-1905 nt), which amplify a HBV X region, while the second round of amplification was performed using the sense primer X-pro-F2 (5’-TTG CTC GCA GCC GGT CTG GA-3’; positions 1295-1314 nt) and the antisense primer X-pro-R2 (5’-TGA ACA GTA GGA CAT GAA CA-3’; antisense positions 1866-1885 nt) (Figure 1A). PCR was initiated using the i-MAX II DNA polymerase (iNtRON Bio, Seoul, Korea) in a 20 μL mixture containing 1X PCR buffer containing 2 mmol/L MgCl2, 2.5 mmol/L dNTP each, 5 pmol primer each, and 5 U of i-MAX II DNA polymerase. The 2 μL of Template DNA extracted from patients’ serum was added into the mixture. The final reaction mixture was subjected to 30 cycles of amplification (60 s at 95 °C, 45 s at 60 °C, and 90 s at 72 °C) followed by a 5 min extension at 72 °C. A 96-well thermocycler (Model 9600 thermocycler, Perkin-Elmer Cetus, Norwalk, United States) was also used. The obtained PCR products were analyzed by electrophoresis on 1% agarose gels, stained with ethidium bromide, and visualized on a UV transilluminator. For direct sequence analysis, an Applied Biosystems model 373A automatic sequencer and a BigDye terminator cycle sequencing kit (Perkin-Elmer Applied Biosystems, Norwalk, United States) were used for the sequencing. For sequencing reactions, 60 ng of PCR-amplified DNA, 5 pmol of reverse primer (X-pro-R2), and 4 μL of BigDye terminator v2.0 100 RR mix (Perkin-Elmer Applied Biosystems, Norwalk, United States) were mixed. Contents were adjusted to a final volume of 10 μL by adding distilled water, and the reaction was run for 30 cycles of 10 s at 96 °C, 5 s at 60 °C, and 4 min at 60 °C. Determined sequences were aligned with the sequences of five HBV references (GenBank accession no. M57663, AB100695, AB074755, AY247032, AY641558, and X02496) using the multiple-alignment algorithm in the MegAlign package (DNASTAR, WS, United States).
Figure 1.
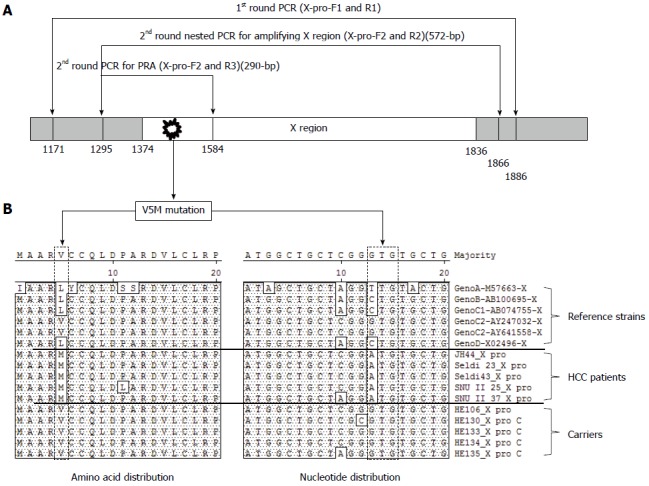
Polymerase chain reaction amplification and direct sequencing analysis of hepatitis B virus X region. A: Location of primer set for the nested PCR direct sequencing (1st primer set; X-pro-F1 and X-pro-R1, 2nd primer set; X-pro-F2 and X-pro-R2) and Fok-I based nested PRA (1st primer set; X-pro-F1 and X-pro-R1, 2nd primer set; X-pro-F2 and X-pro-R3) for the detection of HBxAg V5M mutations and the 3 polymorphisms [V5(GTG), M5(ATG), and L5(CTG or TTG)] of the 5th codon of HBxAg; B: The amino acid and nucleotide sequences of the X region from six reference strains (Genotype A, B, C1, C2, and D) and 10 chronic hepatitis patients (five carriers and five HCC patients) were aligned.
Detection of V5M mutation by Fok-I PCR-restriction analysis
To identify V5M mutants conveniently from PCR product, the restriction enzyme Fok-I (GGA TGN9↓) was used in this study. Amplification of the targeted X region was performed under the same conditions in the first round of the nested PCR using the same primer set (X-pro-F1 and X-pro-R1). The second round of PCR was conducted using the sense primer X-pro-F2 (5’-TTG CTC GCA GCC GGT CTG GA-3’; positions 1295-1314 nt) and the antisense primer X-pro-R3 (5’-CGT GCA GAG GTG AAG CGA AG-3’; antisense 1584-1603 nt), which could amplify the size of the 290-bp product (Figure 1A). After amplification, 5 units of Fok-I (New England Biolabs, MA, United States) restriction enzyme were treated with 10x buffer and PCR product at 37 °C for 1 h. For size comparison, the enzyme-treated products were analyzed by electrophoresis on 2.5% agarose gels, stained with ethidium bromide, and visualized on a UV transilluminator (Figure 2).
Figure 2.
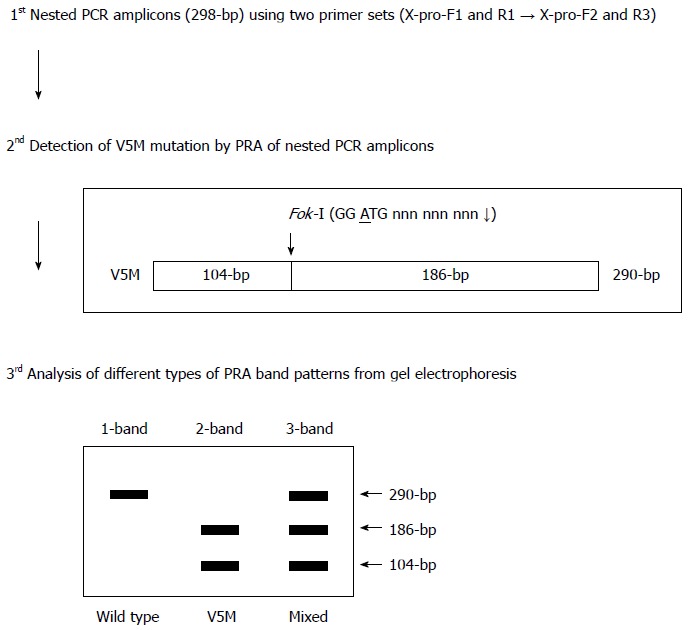
Algorithm of Fok-I nested polymerase chain reaction-restriction fragment length polymorphism analysis method for the detection of V5M mutation in the HBV X antigen region used in the present study.
Statistical analysis
Statistical analysis of the data in this study was conducted using the SPSS version 21.0 software program (Professional Statistic, Chicago, IL), and the results are expressed as percentages. Frequency tables were analyzed using the χ2 tests or Fisher’s exact test. Student’s t test was used when the data showed a normal distribution. In all tests, P values < 0.05 were considered to be statistically significant.
RESULTS
Design of a novel Fok-I nested PRA for the detection V5M mutation in HBxAg
Previously, 3 polymorphisms, V5(GTG), L5(CTG) and M5(ATG), were reported to be found in the 5th codon of HBxAg in Korean chronic patients[12]. The V5(GTG) type is wild type for genotype C2. Although L5(CTG) is rare in genotype C2, it is also found in the wild types of genotype A, B, C1 and D (Figure 1B). We therefore sought to develop the diagnostic method of Fok-I-based nested PRA to detect the M5(ATG) type (hereafter designated V5M), a genuine mutation among 3 types of polymorphisms in the 5th codon of HBxAg in this study. First, for the nested PCR, the total sequences of four HBV genotype C2 strains retrieved from GenBank were aligned using the SeqManTM II software (DNASTAR) (data not shown), and two primer pairs (outer and inner for nested PCR) for the amplification of an HBxAg fragment were designed using Oligo V 6.5 (Molecular Biology Insights), producing a 290-bp nested PCR amplicon (Figure 1A and Figure 2). Second, to enable the simple separation between the V5M type and 2 wild types, V(GTG) and L(CTG), we developed a novel Fok-I (GGATGN9↓) PRA algorithm using MapDraw (version 3.14; DNASTAR, Madison, Wis.). The Fok-I restriction enzyme can recognize the V5M mutation but not the 2 wild types, V(GTG) and L(CTG). It produced 3 types of PRA pattern. In the presence of wild type [V(GTG) or L(CTG)] alone in blood sample DNA, it can produce one band of undigested 290-bp in an agarose gel. In the presence of V5M mutant type alone having the Fok-I recognition site in blood sample DNA, it can produce two completely digested bands, 186-bp and 104-bp, in an agarose gel. Finally, with the coexistence of both wild type and V5M type, it can produce three bands, an undigested one (290-bp) from the wild type and 2 digested bands, 186-bp and 104-bp, from the X5M type in an agarose gel (Figures 2 and 3).
Figure 3.
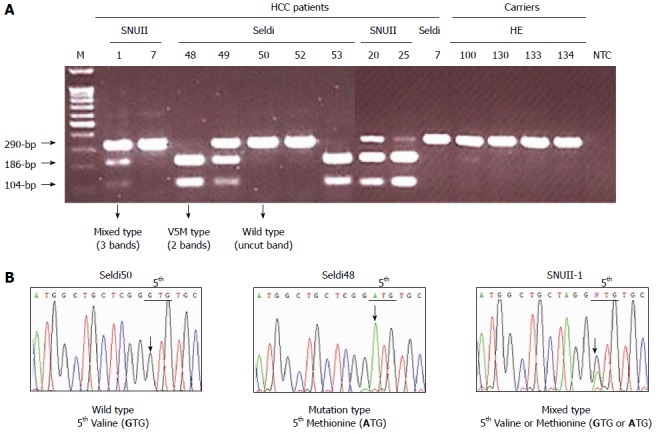
Application of Fok-I nested polymorphism analysis method (A) and direct sequencing analysis (B) into serum DNAs. A: The Fok-I PRA application of nested PCR products could produce three types of PRA pattern on an agarose gel (2.5%): an undigested 290-bp from the wild type, V5(GTG); two bands of complete digestion, 186-bp and 104-bp, from the V5M mutant and three bands, an undigested one of 290-bp from the wild type and 2 digested bands, 186-bp and 104-bp, from V5M mutant. B: Direct sequencing could also produce three types: wild type only [V5(GTG)], mutation only [M5(ATG)], and mixed types with the wild type V5(GTG) and M5(ATG). The mixed types showed the mixed peaks (G and A) at the first nucleotide of the 5th codon of HBxAg (NTT). The results of the Fok-I nested PRA method were completely concordant with those obtained by direct sequencing analysis. Lane M, 100-bp DNA ladder marker.
Application of Fok-I nested PRA into DNAs from 198 chronic patients
Our Fok-I nested PRA protocol designed in this study was applied to the DNA from 198 chronic patients of 2 different stages (121 patients with hepatocellular carcinoma and 77 carrier patients). We produced the nested PCR amplicons from 69 patients (sensitivity of 34.8%; 46 HCC patients and 23 carrier patients) (Table 1). In 46 HCC patients with PCR-positive amplicons, 24 (52.2%), 17 (37.0%) and 5 (10.8%) were identified as wild-type infection alone, V5M mutant type infection alone, and coinfection with both types, wild and mutant, respectively (Figure 3). A total of 22 (47.8 %) from 46 HCC patients with positive PCR amplicons were identified as infection with the V5M mutant. All 23 carriers with PCR positive amplicons were identified as wild-type infection alone. No patients with V5M mutant infection were found in the carriers. This result was completely concordant with that obtained from a direct sequencing protocol (specificity of 100%). The prevalence of infection of the V5M mutant type was significantly higher in HCC compared with carriers [47.8% (22/46 patients) vs 0 % (0/23 patients), P < 0.001] (Table 2).
Table 1.
Sensitivity of nested polymerase chain reaction assay for the amplification of the 572-bp HBV X antigen fragment n (%)
|
No. of samples (n = 198) |
Sensitivity | ||
| HCC (n =121) | Carrier (n =77) | ||
| PCR amplification | 46 (38.0) | 23 (29.9) | 34.8% |
PCR: Polymerase chain reaction.
Table 2.
Evaluation of Fok-I nested polymorphism analysis assay for the detection of V5M mutations from 69 samples amplified by nested PCR by comparing with the results of the direct sequencing method n (%)
| Type of HBxAg 5th codon |
Result for |
|||
|
Fok-I nested PRA |
Direct sequencing |
|||
| HCC (n =46) | Carrier (n =23) | HCC (n =46) | Carrier (n =23) | |
| Wild type | 24 (52.2) | 23 (100) | 24 (52.2) | 23 (100) |
| V5M | 17 (37.0) | 0 (0) | 17 (37.0) | 0 (0) |
| Wild type + V5M | 5 (10.8) | 0 (0) | 5 (10.8) | 0 (0) |
PRA: Polymorphism analysis; HCC: Hepatocellular carcinoma; HBxAg: HBV X antigen.
DISCUSSION
Previously, we introduced a total of 5 types of mutations in the X gene (V5M/L, P38S, H94Y, I127T/N, and K130M and V131I) that were significantly related to the clinical severity of chronic patients infected with genotype C2 via a molecular epidemiologic study of Korean chronic patients[12]. Of these, the V5M/L type was first introduced by us and has some properties that are distinct from other mutations. First, this is a genuine HCC-specific mutation as its prevalence in HCC patients was significantly higher than even that of cirrhosis patients or in chronic hepatitis or carriers. Recently, the combination of both BCP double mutations and both types of the V5M mutation, V5M and V5L, has also been reported to increase the risk of HCC by 5.34 times compared with wild type, suggesting that V5M with HBV genotype C2 is a risk factor for the development of HCC and can be used to predict the clinical impact of chronic HBV infection[32]. Second, it is more prevalent in HBeAg-negative patients than in HBeAg-positive patients, suggesting that it may be generated from host immune pressure[12]. Third, of note, it has a positive correlation with the BCP mutations K130M and V131I[12]. Taken together, it may play a very pivotal role in hepatocarcinogenesis during the HBeAg-negative immune active stage in the natural course of genotype C2 HBV infection. The development of molecular diagnosis for the detection of V5M must therefore be accomplished in genotype C2 endemic areas such as South Korea and China.
For the molecular detection of V5M mutants of the HBV X region, we developed a molecular-based approach, Fok-I nested PRA, which allowed for the rapid detection of the V5M mutant type without sequence analysis. There are some noteworthy advantages in our Fok-I nested PRA method. First, the 5-bp consecutive HBV sequences (GGATG) corresponding to the Fok-I (GGATGN9↓) recognition site of the V5M mutant type are highly conserved among the HBV genotype C2 strains having the V5M mutation, as shown in Figure 1B. When we analyzed the HBxAg sequences from 45 independent chronic patients infected with genotype C2 who had been identified as having the V5M mutation by direct sequencing analysis[12], no mutations were found in the 5-bp consecutive HBV sequences (GGATG) (data not shown). This result could guarantee the enhanced specificity of our Fok-I nested PRA method. Actually, our Fok-I PRA method can detect all of the V5M mutations with 100% specificity compared with the direct sequencing protocol (Table 2). Second, this method can also detect mixed type infections consisting of both the V5M mutant and the wild type in addition to being able to detect V5M mutation infections alone and wild type infections alone. However, some concerns regarding this assay should also be noted. First, this method may underestimate the frequency of the V5M/L mutation because it is blind to the L(CTG) genotype starting from the CTG in the 5th codon of the X region, which is found in wild-type genotype A, B, C1 or D, but very rare in genotype C2 as shown in Figure 1B. However, given that the V5L genotype is the wild type in genotype A, B or D, it seems more likely and reasonable that unlike the V5M type, it may be one of the wild types in genotype C2 rather than the mutation generated by host immune pressure. Second, despite the nested PCR protocol, our assay had a low level of sensitivity (34.8 %, 69/198 patients) (Table 1). This low sensitivity may be due to the absence of the target X region rather than a defect of our assay as previously reported[33] because in 129 samples not amplified by our assay, our repeated attempts to amplify a partial X gene also failed, but the nested PCR protocol targeting the partial S gene could amplify most of them (data not shown). Our epidemiological data based on our Fok-I nested PRA method showed that V5M mutations are significantly more prevalent in HCC patients than in carrier stage patients [47.8% (22/46 patients) vs 0% (0/23 patients), P < 0.001], strongly supporting previous reports that this mutation may play a very pivotal role in hepatocarcinogenesis, at least in chronic patients infected with genotype C2[12,32].
In the present study, we developed a reliable and cost-effective Fok-I nested PRA method for the detection of V5M from chronic patients with genotype C2 infection. In addition, our epidemiological data based on the Fok-I nested PRA method strongly support the previous reports that V5M may play a very pivotal role in hepatocarcinogenesis, at least in chronic patients infected with genotype C2.
COMMENTS
Background
The HBxAg V5M mutation proved to be associated with liver disease progression in chronic subjects with genotype C2 using our previous molecular epidemiologic study based on direct sequencing protocol. So, for its monitoring among genotype C2 infected chronic patients, the development of a new molecular diagnostic method for its detection is needed.
Research frontiers
For molecular epidemiologic study for detection of V5M mutation, in this study, we developed the novel nested polymerase chain reaction (PCR)-restriction fragment length polymorphism analysis (PRA) method that can detect V5M and applied it to DNAs from Korean chronic patients.
Innovations and breakthroughs
The present epidemiological data based on the Fok-I nested PRA method strongly support the previous reports that V5M may play a very pivotal role in hepatocarcinogenesis, at least in chronic patients infected with genotype C2.
Applications
The FRET-based real-time PCR for detection of the preS1 deletion developed in this study might have potential for early prediction for risk of liver disease progression in chronic subjects.
Terminology
The nested PRA method that can detect V5M could be used for molecular epidemiologic purposes. It can permit not only the simultaneous identification of coexisting quasispecies of both wild type and variant, but also the direct identification of target mutations from primary specimens such as serum.
Peer-review
The authors developed a reliable and cost-effective Fok-I nested PRA method for the detection of V5M from chronic patients with genotype C2 infection, which could be effectively used for its screening instead of direct sequencing protocols. Furthermore, they also confirmed that V5M may play a very pivotal role in hepatocarcinogenesis, at least in chronic patients infected with genotype C2.
Footnotes
Supported by a National Research Foundation (NRF) of Korea grant funded by the Korean government (Ministry of Education, Science, and Technology, MEST), Grant No. 2013-005810.
Institutional review board statement: The study was reviewed and approved by the Institutional Review Board of Seoul National University Hospital (Seoul) (IRB No. 1404-070-572).
Informed consent statement: All study participants, or their legal guardian, provided informed written consent prior to study enrollment.
Conflict-of-interest statement: There are no conflicts of interest to report.
Data sharing statement: No additional data are available.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: June 11, 2015
First decision: July 10, 2015
Article in press: September 30, 2015
P- Reviewer: Haruki K S- Editor: Yu J L- Editor: A E- Editor: Liu XM
References
- 1.Orito E, Mizokami M, Sakugawa H, Michitaka K, Ishikawa K, Ichida T, Okanoue T, Yotsuyanagi H, Iino S. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Japan HBV Genotype Research Group. Hepatology. 2001;33:218–223. doi: 10.1053/jhep.2001.20532. [DOI] [PubMed] [Google Scholar]
- 2.Kao JH, Chen PJ, Lai MY, Chen DS. Genotypes and clinical phenotypes of hepatitis B virus in patients with chronic hepatitis B virus infection. J Clin Microbiol. 2002;40:1207–1209. doi: 10.1128/JCM.40.4.1207-1209.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lavanchy D. Worldwide epidemiology of HBV infection, disease burden, and vaccine prevention. J Clin Virol. 2005;34 Suppl 1:S1–S3. doi: 10.1016/s1386-6532(05)00384-7. [DOI] [PubMed] [Google Scholar]
- 4.Kim BJ. Hepatitis B virus mutations related to liver disease progression of Korean patients. World J Gastroenterol. 2014;20:460–467. doi: 10.3748/wjg.v20.i2.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim DW, Lee SA, Hwang ES, Kook YH, Kim BJ. Naturally occurring precore/core region mutations of hepatitis B virus genotype C related to hepatocellular carcinoma. PLoS One. 2012;7:e47372. doi: 10.1371/journal.pone.0047372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim DW, Lee SA, Kim H, Won YS, Kim BJ. Naturally occurring mutations in the nonstructural region 5B of hepatitis C virus (HCV) from treatment-naïve Korean patients chronically infected with HCV genotype 1b. PLoS One. 2014;9:e87773. doi: 10.1371/journal.pone.0087773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim H, Jee Y, Mun HS, Park JH, Yoon JH, Kim YJ, Lee HS, Hyun JW, Hwang ES, Cha CY, et al. Characterization of two hepatitis B virus populations in a single Korean hepatocellular carcinoma patient with an HBeAg-negative serostatus: a novel X-Gene-deleted strain with inverted duplication sequences of upstream enhancer site II. Intervirology. 2007;50:273–280. doi: 10.1159/000103915. [DOI] [PubMed] [Google Scholar]
- 8.Kim H, Jee Y, Mun HS, Song BC, Park JH, Hyun JW, Hwang ES, Cha CY, Kook YH, Kim BJ. Comparison of full genome sequences between two hepatitis B virus strains with or without preC mutation (A1896) from a single Korean hepatocellular carcinoma patient. J Microbiol Biotechnol. 2007;17:701–704. [PubMed] [Google Scholar]
- 9.Kim H, Jee YM, Song BC, Hyun JW, Mun HS, Kim HJ, Oh EJ, Yoon JH, Kim YJ, Lee HS, et al. Analysis of hepatitis B virus quasispecies distribution in a Korean chronic patient based on the full genome sequences. J Med Virol. 2007;79:212–219. doi: 10.1002/jmv.20789. [DOI] [PubMed] [Google Scholar]
- 10.Kim H, Jee YM, Song BC, Shin JW, Yang SH, Mun HS, Kim HJ, Oh EJ, Yoon JH, Kim YJ, et al. Molecular epidemiology of hepatitis B virus (HBV) genotypes and serotypes in patients with chronic HBV infection in Korea. Intervirology. 2007;50:52–57. doi: 10.1159/000096313. [DOI] [PubMed] [Google Scholar]
- 11.Kim H, Lee SA, Kim DW, Lee SH, Kim BJ. Naturally occurring mutations in large surface genes related to occult infection of hepatitis B virus genotype C. PLoS One. 2013;8:e54486. doi: 10.1371/journal.pone.0054486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim HJ, Park JH, Jee Y, Lee SA, Kim H, Song BC, Yang S, Lee M, Yoon JH, Kim YJ, et al. Hepatitis B virus X mutations occurring naturally associated with clinical severity of liver disease among Korean patients with chronic genotype C infection. J Med Virol. 2008;80:1337–1343. doi: 10.1002/jmv.21219. [DOI] [PubMed] [Google Scholar]
- 13.Lee SA, Cho YK, Lee KH, Hwang ES, Kook YH, Kim BJ. Gender disparity in distribution of the major hydrophilic region variants of hepatitis B virus genotype C according to hepatitis B e antigen serostatus. J Med Virol. 2011;83:405–411. doi: 10.1002/jmv.21988. [DOI] [PubMed] [Google Scholar]
- 14.Lee SA, Kim H, Won YS, Seok SH, Na Y, Shin HB, Inn KS, Kim BJ. Male-specific hepatitis B virus large surface protein variant W4P potentiates tumorigenicity and induces gender disparity. Mol Cancer. 2015;14:23. doi: 10.1186/s12943-015-0303-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SA, Kim K, Kim H, Kim BJ. Nucleotide change of codon 182 in the surface gene of hepatitis B virus genotype C leading to truncated surface protein is associated with progression of liver diseases. J Hepatol. 2012;56:63–69. doi: 10.1016/j.jhep.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 16.Lee SA, Kim KJ, Kim DW, Kim BJ. Male-specific W4P/R mutation in the pre-S1 region of hepatitis B virus, increasing the risk of progression of liver diseases in chronic patients. J Clin Microbiol. 2013;51:3928–3936. doi: 10.1128/JCM.01505-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SA, Mun HS, Kim H, Lee HK, Kim BJ, Hwang ES, Kook YH, Kim BJ. Naturally occurring hepatitis B virus X deletions and insertions among Korean chronic patients. J Med Virol. 2011;83:65–70. doi: 10.1002/jmv.21938. [DOI] [PubMed] [Google Scholar]
- 18.Mun HS, Lee SA, Jee Y, Kim H, Park JH, Song BC, Yoon JH, Kim YJ, Lee HS, Hyun JW, et al. The prevalence of hepatitis B virus preS deletions occurring naturally in Korean patients infected chronically with genotype C. J Med Virol. 2008;80:1189–1194. doi: 10.1002/jmv.21208. [DOI] [PubMed] [Google Scholar]
- 19.Mun HS, Lee SA, Kim H, Hwang ES, Kook YH, Kim BJ. Novel F141L pre-S2 mutation in hepatitis B virus increases the risk of hepatocellular carcinoma in patients with chronic genotype C infections. J Virol. 2011;85:123–132. doi: 10.1128/JVI.01524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Summers J, Mason WS. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 21.Murakami S. [Expression and function of hepatitis B virus (HBV) X protein] Seikagaku. 1999;71:1309–1326. [PubMed] [Google Scholar]
- 22.Murakami S. Hepatitis B virus X protein: a multifunctional viral regulator. J Gastroenterol. 2001;36:651–660. doi: 10.1007/s005350170027. [DOI] [PubMed] [Google Scholar]
- 23.Tang SG, Yang BC. [Clone of hepatitis B virus X gene and its protein expression] Zhongnandaxue Xuebao Yixueban. 2005;30:525–528. [PubMed] [Google Scholar]
- 24.Yang Y, Ma Y, Zhen L, Chen Y, Ma W, Murakami S. HBV X protein (HBX) interacts with general transcription factor TFIIB both in vitro and in vivo. Chin Med Sci J. 1999;14:152–157. [PubMed] [Google Scholar]
- 25.Fattovich G, Bortolotti F, Donato F. Natural history of chronic hepatitis B: special emphasis on disease progression and prognostic factors. J Hepatol. 2008;48:335–352. doi: 10.1016/j.jhep.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 26.Lapiński TW, Pogorzelska J, Flisiak R. HBV mutations and their clinical significance. Adv Med Sci. 2012;57:18–22. doi: 10.2478/v10039-012-0006-x. [DOI] [PubMed] [Google Scholar]
- 27.Xia L, Huang W, Tian D, Zhu H, Zhang Y, Hu H, Fan D, Nie Y, Wu K. Upregulated FoxM1 expression induced by hepatitis B virus X protein promotes tumor metastasis and indicates poor prognosis in hepatitis B virus-related hepatocellular carcinoma. J Hepatol. 2012;57:600–612. doi: 10.1016/j.jhep.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 28.Kim S, Park SY, Yong H, Famulski JK, Chae S, Lee JH, Kang CM, Saya H, Chan GK, Cho H. HBV X protein targets hBubR1, which induces dysregulation of the mitotic checkpoint. Oncogene. 2008;27:3457–3464. doi: 10.1038/sj.onc.1210998. [DOI] [PubMed] [Google Scholar]
- 29.Park EH, Koh SS, Srisuttee R, Cho IR, Min HJ, Jhun BH, Lee YS, Jang KL, Kim CH, Johnston RN, et al. Expression of HBX, an oncoprotein of hepatitis B virus, blocks reoviral oncolysis of hepatocellular carcinoma cells. Cancer Gene Ther. 2009;16:453–461. doi: 10.1038/cgt.2008.95. [DOI] [PubMed] [Google Scholar]
- 30.Lok AS, Heathcote EJ, Hoofnagle JH. Management of hepatitis B: 2000--summary of a workshop. Gastroenterology. 2001;120:1828–1853. doi: 10.1053/gast.2001.24839. [DOI] [PubMed] [Google Scholar]
- 31.Bruix J, Sherman M, Llovet JM, Beaugrand M, Lencioni R, Burroughs AK, Christensen E, Pagliaro L, Colombo M, Rodés J. Clinical Management of Hepatocellular Carcinoma. Conclusions of the Barcelona-2000 EASL Conference. J Hepatol. 2001;35:421–430. doi: 10.1016/s0168-8278(01)00130-1. [DOI] [PubMed] [Google Scholar]
- 32.Lee JH, Han KH, Lee JM, Park JH, Kim HS. Impact of hepatitis B virus (HBV) x gene mutations on hepatocellular carcinoma development in chronic HBV infection. Clin Vaccine Immunol. 2011;18:914–921. doi: 10.1128/CVI.00474-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Datta S, Banerjee A, Chandra PK, Biswas A, Panigrahi R, Mahapatra PK, Panda CK, Chakrabarti S, Bhattacharya SK, Chakravarty R. Analysis of hepatitis B virus X gene phylogeny, genetic variability and its impact on pathogenesis: implications in Eastern Indian HBV carriers. Virology. 2008;382:190–198. doi: 10.1016/j.virol.2008.09.007. [DOI] [PubMed] [Google Scholar]


