Abstract
An important function of glia is the maintenance of the ionic composition and pH of the synaptic microenvironment. In terms of pH regulation, HCO3− buffering has been shown to be important in both glia and neurons. Here, we used in vivo fluorescent pH imaging and RNA sequencing of the amphid sheath glia of Caenorhabditis elegans to reveal a novel mechanism of cellular HCO3− uptake. While the classical mechanism of HCO3− uptake involves Na+/HCO3− cotransporters, here we demonstrate that the C. elegans ClC Cl− channel CLH-1 is highly permeable to HCO3− and mediates HCO3− uptake into amphid sheath glia. CLH-1 has homology and electrophysiological properties similar to the mammalian ClC-2 Cl− channel. Our data suggest that, in addition to maintaining synaptic Cl− concentration, these channels may also be involved in maintenance of synaptic pH via HCO3− flux. These findings provide an exciting new facet of study regarding how pH is regulated in the brain.
SIGNIFICANCE STATEMENT Maintenance of pH is essential for the physiological function of the nervous system. HCO3− is crucial for pH regulation and is transported into the cell via ion transporters, including ion channels, the molecular identity of which remains unclear. In this manuscript, we describe our discovery that the C. elegans amphid sheath glia regulate intracellular pH via HCO3− flux through the voltage-gated ClC channel CLH-1. This represents a novel function for ClC channels, which has implications for their possible role in mammalian glial pH regulation. This discovery may also provide a novel therapeutic target for pathologic conditions, such as ischemic stroke where acidosis leads to widespread death of glia and subsequently neurons.
Keywords: bicarbonate, C. elegans, chloride channels, glia, pH, RNA sequencing
Introduction
Glia are essential for the maintenance and function of synapses in both mammalian and nonmammalian systems. Glia have a diverse array of functions, including regulation of synaptic metabolic supply (Allaman et al., 2011), maintenance of extracellular K+ levels (Sontheimer, 1994; Kressin et al., 1995; Butt and Kalsi, 2006; Han et al., 2013) and glutamate levels (Hansson et al., 1985; Bergles et al., 1997; Coulter and Eid, 2012), and active participation in synaptic function through release of gliotransmitters (Santello et al., 2012). The amphid sheath glia (AMsh) of Caenorhabditis elegans are closely associated with 12 pairs of amphid sensory neurons (Oikonomou and Shaham, 2011). These sensory neurons extend cilia to the nose of C. elegans and function to recognize chemosensory, mechanosensory, and thermosensory information (Bargmann and Horvitz, 1991; Bargmann et al., 1993; Kaplan and Horvitz, 1993; Mori and Ohshima, 1995; Bargmann, 2006). Sheath glia processes also extend to the nose of the animal and help to form a “channel” through which the sensory neurons extend their dendrites before reaching the outer environment. These glia are essential for the proper development and/or function of several of these sensory neurons, at both the cellular and behavioral levels (Bacaj et al., 2008; Procko et al., 2011). Recent work from our laboratory has shown that AMsh glia express the DEG/ENaC channel ACD-1 (Wang et al., 2008). Knock-out of ACD-1, in combination with mutations in neuronally expressed channels DEG-1 and TAX-2, resulted in defects in animal avoidance behavior to acidic conditions, and in attraction to Na+ and the odor isoamyl alcohol (Wang et al., 2008, 2012). Interestingly, ACD-1 currents are strongly inhibited by both extracellular and intracellular acidification (Wang et al., 2008; Wang and Bianchi, 2009). This indicates that regulation of intracellular and extracellular pH in AMsh cells is likely essential for the function of ACD-1, and in turn is essential for sensory behavior in C. elegans. In addition, changes in intracellular and extracellular pH likely influence the function of many other AMsh glia proteins. However, to date, nothing is known regarding the mechanisms that regulate intracellular pH in the AMsh cells of C. elegans. In addition, the greater role that glia play in maintenance of pH of the synaptic environment both in invertebrates and vertebrates is not well understood.
In mammalian systems, several types of transporters are known to mediate regulation of cellular pH. Na+/H+ exchangers, H+-ATPases, Na+/HCO3− cotransporters, and Na+-driven Cl−/HCO3− exchangers are responsible for alkalization of the cytoplasm, whereas Cl−/HCO3− exchangers use Cl− entry to remove HCO3− and acidify the cytoplasm (Boron, 2004). Beyond these transport mechanisms, there is evidence for a substantial role of anion channels in HCO3−-dependent pH regulation in certain cell types. For example, the ATP-binding cassette transporter CFTR has been shown to transport HCO3− in several mammalian cell types (Poulsen et al., 1994; Hug et al., 2003; Chan et al., 2006; Chen et al., 2010). Bestrophin Cl− channels have also been shown to be highly permeable to HCO3− (Qu and Hartzell, 2008), and ionotropic GABA and glycine receptor channels have permeability to HCO3− as well (Bormann et al., 1987; Fatima-Shad and Barry, 1993). Comparatively little is known about the HCO3− permeability of voltage-gated ClC-type Cl− channels. ClC-5, which localizes to endosomes, was shown to be permeable to HCO3− when expressed in Xenopus oocytes (Mo et al., 1999). However, to our knowledge, no studies to date have examined the possible contribution of plasma membrane ClC channels to pH regulation via HCO3− flux, especially not in an in vivo setting.
In this study, we used real-time fluorescent pH imaging and sequencing of glial AMsh cells mRNA transcripts to identify the C. elegans ClC-2 channel homolog CLH-1 as a major contributor to HCO3−-dependent intracellular pH regulation in these glial cells. Surprisingly, we found no evidence for the Na+/HCO3− transporters mediating HCO3− uptake in these cells. Electrophysiological characterization of CLH-1 revealed that it shares characteristics of the mammalian ClC-2 channel, such as sensitivity to extracellular pH, and also revealed that it is highly permeable to HCO3−.
Materials and Methods
Molecular biology.
Superecliptic pHluorin in pGEX-27 (designated pGM87) was kindly gifted to us by Dr. Gero Miesenboeck. pHlourin was amplified from pGM87 using platinum TAQ (Invitrogen) using gene specific primers that added kpnI and xhoI restriction sites to the 5′ and 3′ end of the product, respectively. The pHlourin product was then cloned into the pCR2.1-TOPO vector (Invitrogen) for amplification and sequencing. Sequence-verified pHluorin was digested with kpnI and xhoI, and ligated into pPD95.75 downstream from the promoter for T02B11.3, which is expressed exclusively in AMsh cell and a pair of unknown cells in the tail of C. elegans (Bacaj et al., 2008; Wang et al., 2008). The pPD95.75+pHlourin construct was then injected into wild-type Bristol strain N2 to create the strain BLC44 Ex[pT02B11.3::pGM87]. The clh-1 cDNA construct and the clh-1::GFP reporter construct was kindly provided by Dr. Keith Nehrke (University of Rochester, Rochester, NY). For synthesis of clh-1 mRNA, the clh-1 cDNA sequence was subcloned into the pGEM vector using ApaI and KpnI restriction sites to ligate a 2860 bp fragment containing the clh-1 cDNA sequence into the pGEM vector. For the clh-1 rescue construct, the 7884 bp clh-1 genomic DNA sequence was amplified from C. elegans genomic DNA using primers that added BstXI and ApaI restriction sites to the 5′ and 3′ ends, respectively. Clh-1 genomic DNA was then cloned into a vector with the AMsh-specific promoter pT02B11.3. Germline transformation by microinjection was performed as described previously (Mello et al., 1991).
RNAi.
AMsh-specific knockdown of abts-2 and abts-4 expression was achieved using the method described by Esposito et al. (2007). Exon-rich regions (sizes of ∼500 bp) of the abts-2 and abts-4 genes were amplified, whereas the T02B11.3 promoter was also amplified with the addition of a 25 nt region at the 3′ end of the promoter that was complimentary to either the sense or antisense 5′ end of the abts-2 or abts-4 PCR products. The T02B11.3 promoter was then fused to abts-2 or abts-4 sense or antisense by using a forward nested primer for the pT02B11.3 PCR product and a reverse nested primer for either the sense or antisense strands of the abts-2 or abts-4 PCR product. pT02B11.3:abts-2 and pT02B11.3:abts-4 sense and antisense constructs were coinjected into young adult ex[pT02B11.3::pGM87] animals along with an unc-122:GFP transcriptional fusion construct, expressed in seam cells (Loria et al., 2004), as cotransformation marker, to generate the strain ex[pT02B11.3::pGM87;pT02B11.3::abts-2;abts-4RNAi;unc-122::GFP] (BLC289).
C. elegans strains and growth.
Hermaphroditic worms were maintained at 20°C on standard nematode growth media seeded with the OP50 strain of Escherichia coli. BLC292 (abts-4;Ex[pT02B11.3::pGM87]), BLC295 (clh-1;Ex[pT02B11.3::pGM87]), and BLC296 (clh-3;Ex[pT02B11.3::pGM87]) strains were created by first crossing abts-4 (RB1026, R03E9.3(ok953) X), clh-1 (RB833, T27D12.2(ok658) II), or clh-3 (RB900, clh-3(ok763)) strains with BLC44 (ex[pT02B11.3::pGM87]). BLC297 (clh-1;clh-3;Ex[pT02B11.3::pGM87]) was created by crossing BLC295 with BLC296. The knock-out strains were outcrossed twice with male N2 wild-type animals before crossing them with BLC44. All mutations were followed by PCR.
pH imaging.
Transgenic animals expressing the pH sensor pHluorin were immobilized onto a glass coverslip using surgical glue. Using a sharp glass micropipette, an incision perpendicular to the length of the animal was made well posterior to the AMsh glia to relieve internal pressure. A parallel cut was then performed to open the worm to just posterior of the large pharyngeal bulb to allow for perfusion buffers to contact the glial cell. AMsh cells were then imaged using a Lumencor SOLA light engine attached to a Nikon Eclipse E600FN upright microscope. Images were captured using an IMAGO camera. Tillvision v4 software and a polychrome 2 control unit (Till Photonics) were used to control the camera and light engine shutter. Fluorescence values were obtained at a rate of 1 capture/s. For data analysis, raw florescence values were converted to ΔF/F values by calculating the average baseline fluorescence of the glia (F) from the first 30 captures. The rate of recovery from an acid load was defined as the slope of the first 300 s of the recovery in ΔF/F after an acid load. The composition of extracellular buffers used during pH imaging is outlined in Table 1. All solutions were adjusted to pH 7.2. For buffers containing HCO3−, alkalization due to loss of CO2 into the atmosphere was minimized by storing the buffers in airtight bottles at 4°C before use. For Na+- removal experiments in the presence of HCO3−, 20 mm KHCO3 instead of 20 mm NaHCO3 was used in both control and Na+ solutions. The extra K+ in KHCO3 buffers did not cause any measurable difference in pH response compared with solutions buffered with NaHCO3.
Table 1.
Composition of buffers used in pH imaging experimentsa
| NaHCO3 buffer |
KHCO3 buffer |
HCO3−-free buffer |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Normal | NH4+ | Cl−-free |
Normal | NH4+ | Na+-free |
Normal | NH4+ | Na+-free |
||||
| Normal | NH4+ | Normal | NH4+ | Normal | NH4+ | |||||||
| NaCl | 125 | 105 | 130 | 110 | 145 | 125 | ||||||
| KCl | 5 | 5 | 5 | 5 | 5 | 5 | ||||||
| MgCl2 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | ||
| CaCl2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||
| Glucose | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 |
| HEPES | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
| NaHCO3 | 20 | 20 | 20 | |||||||||
| KHCO3 | 20 | 20 | 20 | 20 | ||||||||
| NH4Cl | 20 | 20 | 20 | 20 | 20 | |||||||
| NMDG | 130 | 110 | 145 | 125 | ||||||||
| Na-gluconate | 125 | 125 | ||||||||||
| K-gluconate | 5 | 5 | ||||||||||
| Mg-gluconate | 5 | 5 | ||||||||||
| Ca-gluconate | 1 | 1 | ||||||||||
| NH4HCO3 | 20 | |||||||||||
aAll numbers are in mm concentration.
Cell culture.
Culture of embryonic C. elegans cells was performed as described previously (Sangaletti and Bianchi, 2013). Briefly, ex[pT02B11.3::PGM87] (BLC44) worms were grown on 8P agar plates seeded with the NA22 strain of E. coli. Gravid adults were washed 3 times with sterile H2O; then eggs were released by lysing the worms with a solution containing 5 ml of Fresh Bleach, 1.25 ml of 10 N NaOH and 18.5 ml of sterile H2O. The lysis reaction was stopped by addition of egg buffer (118 mm NaCl, 48 mm KCl, 2 mm CaCl2, 2 mm MgCl2, 25 mm HEPES, pH 7.3, osmolarity 340 mOsm). The eggs were then washed 3 times in egg buffer and isolated from lysed worm debris by centrifugation at 200 × g for 20 min in 30% sucrose in egg buffer. Sucrose was removed by washing the eggs 3 times in egg buffer, and the eggs were pelleted by centrifugation at 200 × g for 10 min. For embryonic cell dissociation, the eggs were then incubated for 10 min in 2 mg/ml chitinase in egg buffer at room temperature. The eggs were then pelleted by centrifugation at 900 × g for 3 min and resuspended in 3 ml of L-15 media. The eggs were then dissociated by aspiration into a sterile syringe with an 18 gauge needle. Isolated cells were passed through a 5 μm filter (Millipore) to remove egg debris and plated at ∼230,000 cells/cm2 in L-15 media overnight at 20°C. The media was replaced the next day, and the cells were cultured for up to 9 d at 20°C.
RNA sequencing.
Adherent C. elegans ex[pT02B11.3::pGM87] cells cultured for 3–5 d were gently washed off culture dishes using egg buffer. GFP-expressing AMsh cells were separated from nonfluorescent cells into different pools by FACS using a FACSAria II cell sorter (BD Biosciences) at the University of Miami Flow Cytometry Core Facility. Cells of both pools were pelleted by centrifugation at 900 × g for 10 min and lysed in Trizol reagent (Invitrogen). RNA from lysed GFP and control pools from three independent cell cultures were isolated and then further processed at Ocean Ridge Biosciences. Libraries were generated using the NuGen Ovation RNA Seq System version 2, and sequenced on a Illumina HiSeq 2500. All samples had a minimum of 59 million passed-filter reads and aligned at 75% efficiency to the reference genome. Tophat 1.4.1 software was used to map reads from input FASTQ files to the Ensembl C. elegans (WS235) reference genome to generate BAM alignment files. Samtools version 0.1.18 was used to generate BAM index files1. The stranded parameter (-library-type fr-unstranded) was provided during the alignment. This indicated that the sequence reads used were from both the strands. The normalized counts of sequence reads (fragments per kilobase of gene per million fragments mapped [FPKM]) mapped to annotated Ensemble genes were determined using cufflinks software. The raw count files were annotated using data from Ensembl C. elegans. The FPKM values were filtered to retain a list of genes with a minimum of ∼50 mapped reads in one or more samples. To avoid reporting large fold changes due to random variation of counts from low abundance mRNA, FPKM values equivalent to a count of ≤10 reads per gene were replaced with the average FPKM value equivalent to 10 reads/gene across all the samples in the experiment. Differences among the two sample groups were assessed by an unpaired, two-tailed t test. False discovery rates were calculated from p values using the method of Benjamini and Hochberg (1995). Fold changes were also calculated between group means.
Electrophysiology.
Capped CLH-1 cRNAs were synthesized using T7 mMESSAGE mMACHINE kit (Ambion) and purified using RNAeasy columns (QIAGEN). cRNA quantification was then performed using spectroscopy. Stage VI defolliculated oocytes from Xenopus laevis were purchased from Ecocyte Bioscience. Oocytes were incubated in ND96 media (96 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, 5 mm HEPES, pH 7.5), supplemented with penicillin streptomycin (0.1 mg/ml) and 2.5 mm Na pyruvate. Oocytes were injected with 69 nl of cRNA mix for a final amount of 20–30 ng/oocyte. Oocytes were incubated in ND96 at 20°C for 1–3 d before recording. Currents were measured using a two-electrode voltage-clamp amplifier (GeneClamp 500B; Molecular Devices) at room temperature. Electrodes (0.3–1 mΩ) were filled with 3 m KCl, and oocytes were perfused with a NaCl solution containing the following (in mm): 100 NaCl, 2 KCl, 1 CaCl2, 2 MgCl2, 10 HEPES, pH 7. When we tested solutions at pH <6, we used 10 mm 2-(N-morpholino)ethanesulphonic acid instead of 10 mm HEPES to buffer the solutions. We used the pCLAMP suite of programs (Molecular Devices) for data acquisition and analysis. Currents were filtered at 200 Hz and sampled at 1 kHz.
Relative permeabilities of anions to chloride were calculated by using the following equation derived from Hille (2001):
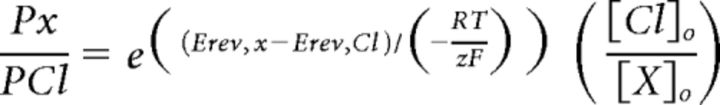 |
Fluorescent microscopy.
Ex[pT02B11.3::pGM87] or Ex[CLH-1::GFP] animals were anesthetized with 20 mm sodium azide on glass slide with an agarose pad. A coverslip was placed on the pad, and fluorescent images were taken at 20–40× using a Leica microscope with a Spot RTslider camera (Diagnostic Instruments) camera and Spot32 acquisition software.
Results
AMsh glia have bicarbonate-dependent pH buffering
To determine the mechanisms by which the AMsh glia regulate intracellular pH, we expressed the GFP-based pH sensor superecliptic pHlourin in AMsh under the control of the AMsh-specific promoter pT02B11.3 (Bacaj et al., 2008; Wang et al., 2008). Transgenic animals were immobilized on glass coverslips and dissected to expose the glia to extracellular solution (Fig. 1A,B). We first wanted to establish whether intracellular pH in AMsh was influenced by extracellular bicarbonate (HCO3−). The composition of C. elegans pseudocoelomic fluid, which bathes the tissues of the animal, has not been determined. However, the pseudocoelomic concentration of HCO3− in the parasitic nematode Ascaris suum was determined to be between 10 and 20 mm (Harpur, 1974). In addition, C. elegans expresses at least two classes of carbonic anhydrase genes (Fasseas et al., 2010, 2011), suggesting that HCO3− has a role in pH buffering in the worm. We thus perfused the AMsh with a solution containing 20 mm HCO3− in addition to 20 mm HEPES. We found that AMsh underwent an initial brief acidification, followed by a slow and sustained alkalization (Fig. 1C). As a further confirmation of the capacity of HCO3− to buffer pH in AMsh, we measured the steady-state baseline pH of AMsh cells in HEPES buffered and HEPES + HCO3−-buffered solutions using the nigericin calibration method (Thomas, 1984). The resting pH was significantly higher in the presence of HCO3− (Fig. 1D; 7.24 ± 0.02, mean ± SEM, n = 3 cells) compared with the resting pH in the absence of HCO3− (Fig. 1D; 7.13 ± 0.03, n = 4 cells, p < 0.05). These results support that mechanisms of HCO3− transport are present in glial AMsh and that these play a key role in intracellular pH regulation, as seen in other cell types (Parker and Boron, 2013).
Figure 1.
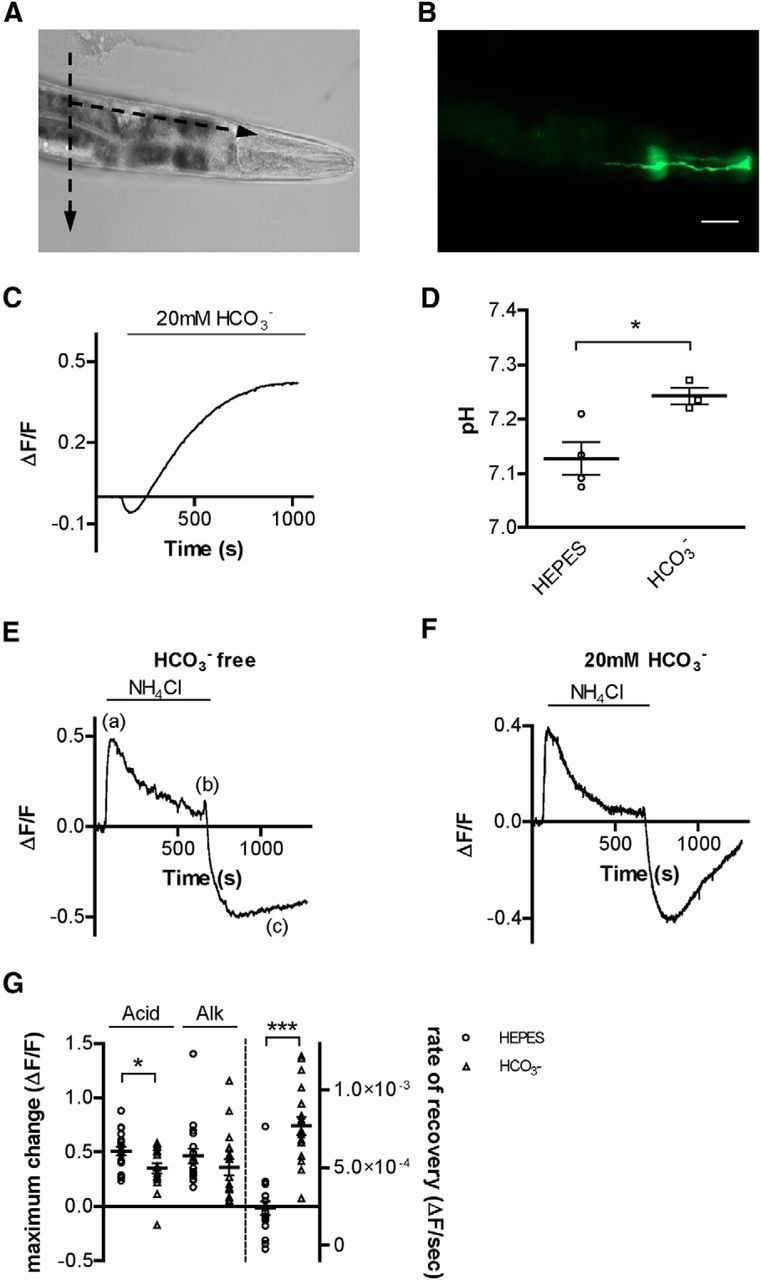
C. elegans AMsh glia buffer intracellular pH via HCO3− transport. A, Bright-field image of C. elegans immobilized on a glass slide. Dashed arrows indicate the direction of the cuts that were made with a glass micropipette before pHlourin pH imaging experiments. B, Fluorescent image of the same animal as in A, showing the pHlourin-mediated fluorescence in an AMsh cell. Scale bars, A, B: 50 μm. C, Representative trace of pHlourin fluorescence in AMsh glia before and after perfusion with a solution containing 20 mm HCO3−. The intracellular pH of glia was alkalized following the perfusion with HCO3−. Data from this trace were used in the analysis shown in Figure 2C. D, Average resting pH of AMsh cells in either the absence (7.13 ± 0.03, n = 4 cells) or presence (7.24 ± 0.02, n = 3 cells) of HCO3−, as determined via nigericin calibration. E, F, Representative traces from NH4Cl acid pulse experiments in AMsh glia in the absence (E) and presence (F) of HCO3−. (a), (b), and (c) indicate the changes of intracellular pH associated with addition (a) and removal (b) of NH4Cl, and the recovery from acid load (c). G, The extent of acidification (Acid, left bars) upon removal of NH4Cl in the AMsh glia cytosol is higher in HEPES buffer alone (ΔF/F = absolute change of 0.51 ± 0.04, n = 18 cells) compared with experiments done in the presence of HCO3− (ΔF/F = 0.35 ± 0.05, n = 18 cells). However, the extent of alkalization (Alk, middle bars) upon NH4Cl application was similar in HEPES buffer alone (ΔF/F = 0.36 ± 0.07, n = 18 cells) and 20 mm HCO3− (ΔF/F = 0.47 ± 0.07, n = 18 cells). The rate of recovery from acid load (Acid recovery, right bars) is significantly higher in the presence of 20 mm HCO3− (ΔF/F/s = 7.68 × 10−4 ± 5.92 × 10−5) compared with HEPES buffer alone (ΔF/F/s = 2.40 × 10−4 ± 4.31 × 10−5). Data are mean ± SEM. *p < 0.05 (Student's t test). ***p < 0.0001 (Student's t test).
AMsh cells are exposed to the outside environment and have been implicated in the animal response to acidic solutions through the activity of the acid-sensitive DEG/ENaC channel ACD-1 (Wang et al., 2008). Therefore, we next wanted to establish to what extent HCO3− transport mechanisms are involved in pH regulation after an acid load. Thus, in the next set of experiments, we induced an ammonium chloride (NH4Cl)-mediated acid load in the glia in either the presence or absence of 20 mm HCO3−. After dissociation of the NH4Cl salt, both the protonated and unprotonated forms of ammonium (NH4+ and NH3) enter the cell, with NH3 permeating more rapidly and abundantly. This causes alkalization of the cytosol due to sequestration of cytosolic H+ ions by NH3 to form NH4+, followed by a slow acidification due to extracellular NH4+ entry through an unknown channel (Thomas, 1984) (Fig. 1E, (a)). Removal of NH4Cl at this point results in the exit of only NH3. The remaining NH4+ must shed its H+ ion to leave the cell, resulting in acidification beyond the initial baseline pH (Thomas, 1984) (Fig. 1E, (b)). The rate of recovery from acid load (Fig. 1E, (c)) is dependent on the cell pH recovery mechanisms (Thomas, 1984).
Because of the nonratiometric nature of superecliptic pHlourin and the cytotoxic nature of nigericin calibration, we could not calibrate our fluorescence values to absolute values of pH for all experiments. However, the average change in ΔF/F when switching from HEPES to HCO3− buffer was 0.30 ± 0.05 (Fig. 1C), and the average difference in resting pH in HEPES versus HCO3− buffer was 0.11 pH units (Fig. 1D). Thus, we can assume that the changes in fluorescence we are observing in our pH imaging experiments represent physiologically relevant changes in absolute pH of the AMsh glia, as our ΔF/F values for other experiments in this study were generally between 0.10 and 0.40 units. In addition, the sigmoidal curve describing the dynamic range of changes in fluorescence of superecliptic pHlourin at different pH values is linear between pH 6.5 and pH 7.5 (Sankaranarayanan et al., 2000). Intracellular alkalization due to perfusion with NH4Cl is independent from HCO3− (Boron and De Weer, 1976). Thus, as expected, we did not find difference in the extent of alkalization in the presence and absence of HCO3− (Fig. 1E–G; ΔF/F = 0.47 ± 0.07 and 0.36 ± 0.07, respectively, p = 0.2989, n = 18 cells each). However, when we analyzed the extent of acid load in the absence and presence of HCO3−, we found that it was larger in the absence of HCO3− (Fig. 1E–G; ΔF/F = absolute change of 0.51 ± 0.04 and 0.35 ± 0.05, respectively, p = 0.0154, n = 18 cells each). This result indicates that glial AMsh use HCO3− as a pH buffer and suggests that HCO3− is transported into the cell to serve this function. Furthermore, when we looked at the recovery from acid load, we found that it was significantly faster in the presence of HCO3− (Fig. 1F,G; ΔF/F/s = 7.68 × 10−4 ± 5.92 × 10−5, n = 18 cells) compared with HCO3−-free conditions (Fig. 1E,G; 2.40 × 10−4 ± 4.31 × 10−5, p < 0.0001, n = 18 cells).
Together, these results show that AMsh use HCO3− to buffer intracellular protons and that they have mechanisms to transport HCO3− into the cell from the extracellular milieu. Moreover, these data support that these HCO3−-mediated buffering mechanisms are engaged not only at baseline (Fig. 1C,D) but also following an acid load (Fig. 1F).
Na+ dependent HCO3− transporters are not involved in HCO3− uptake at baseline pH in C. elegans
The most common mechanism of HCO3− transport into cells is through Na+/HCO3− cotransporters and/or Na+-driven Cl−/HCO3− exchangers encoded by the SLC4 family in mammals (Romero et al., 2013). In C. elegans, the ABTS family of transporters shares the highest degree of homology with the mammalian SLC4 family (Sherman et al., 2005). ABTS-1 in particular is homologous to the mammalian Na+-dependent Cl−/HCO3− exchangers SLC4A8 and SLC4A10 (51% and 48% identity, respectively) (Sherman et al., 2005) and was shown to mediate Na+-driven Cl−/HCO3− exchange when heterologously expressed in Xenopus oocytes (Bellemer et al., 2011). ABTS-2 and ABTS-3 share the highest homology with the mammalian Na+-coupled borate transporter SLC4A11 (35% and 43% identity, respectively). Thus, the activity of all these transporters is dependent on extracellular Na+. Surprisingly however, removal of extracellular Na+ had no effect on AMsh cell HCO3− entry at baseline after switching from HCO3−-free to 20 mm HCO3− buffer (Fig. 2A,C; ΔF/F for control = 0.30 ± 0.05, n = 21 cells and for Na+-free = 0.22 ± 0.05, n = 8 cells), in contrast to what has been observed previously in mammalian cells (Buckler et al., 1991; Bevensee et al., 1997). Similarly, the concentration of extracellular and intracellular Cl− influences the activity of Na+-dependent Cl−/HCO3− exchangers, as well as Na+-independent Cl−/HCO3− exchangers, such as the mammalian Cl−/HCO3− exchanger anion exchange protein 3, to which ABTS-4 from C. elegans is most homologous (28% identity). However, Cl− removal caused no change in HCO3− entry after switching from HCO3−-free to 20 mm HCO3− buffer at baseline pH (Fig. 2C; 0.35 ± 0.06, n = 7 cells vs 0.30 ± 0.05). Moreover, disodium 4,4′-diisothiocyanatostilbene-2,2′-disulfonate (DIDS), which blocks Na+/HCO3− transporters, Cl−/HCO3− exchangers, and Cl− channels (Wulff, 2008; Romero et al., 2013), had no effect on HCO3− entry at baseline conditions (Fig. 2C; 0.32 ± 0.08, n = 9 cells vs 0.30 ± 0.05). Together, these results argue against the involvement of Na+/HCO3− cotransporters and/or Cl−/HCO3− exchangers in bicarbonate-dependent pH buffering in C. elegans glial AMsh cells.
Figure 2.
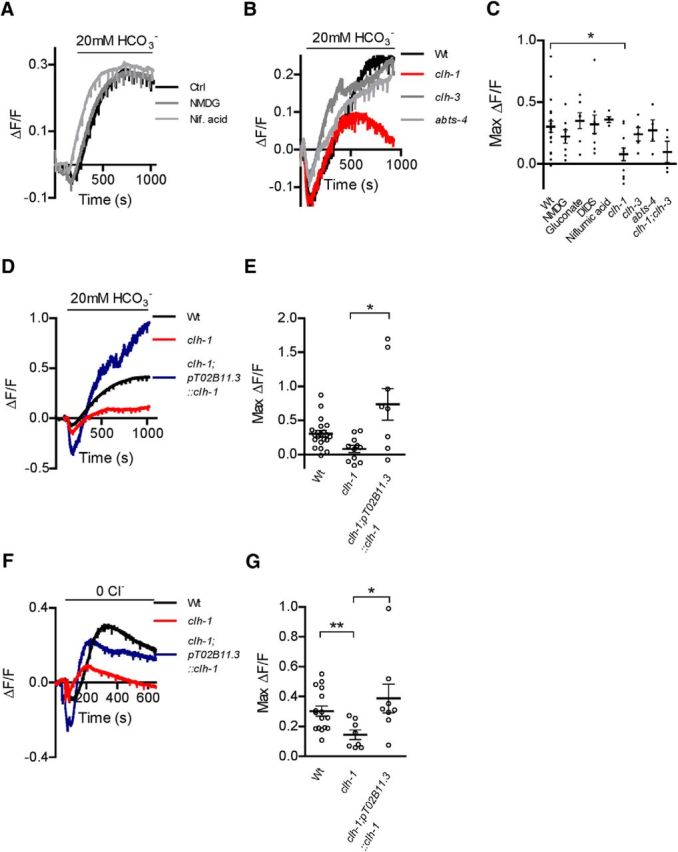
AMsh from clh-1 knock-out animals have impaired HCO3− entry. A, Representative traces from AMsh cells switched from HCO3−-free buffer to 20 mm HCO3−, under control conditions, Na+-free conditions (NMDG, dark gray trace), and solution containing niflumic acid (Nif. acid, 100 μm, light gray trace). B, Same as in A, in wild-type (Wt, black trace), clh-1 (red trace), clh-3 (dark gray trace), and abts-4 (light gray trace) animals. C, Knock-out of clh-1 significantly reduced alkalization in AMsh glia upon HCO3− addition compared with wild-type animals (Wt) (ΔF/F = 0.08 ± 0.05, n = 11 cells vs 0.30 ± 0.05, n = 21 cells, respectively). In contrast, removal of Na+ (NMDG, ΔF/F = 0.22 ± 0.05, n = 8 cells) or Cl− (gluconate, ΔF/F = 0.35 ± 0.06, n = 7 cells), the presence of DIDS (250 μm, ΔF/F = 0.32 ± 0.08, n = 9 cells) and niflumic acid (100 μm, ΔF/F = 0.36 ± 0.02, n = 5 cells), and knock-out of clh-3 (ΔF/F = 0.24 ± 0.05, n = 5 cells) and abts-4 (ΔF/F = 0.26 ± 0.07, n = 4 cells) showed no significant differences. In addition, clh-1;clh-3 double knock-out (ΔF/F = 0.10 ± 0.09, n = 4 cells) was not significantly different from clh-1 knock-out alone. *p < 0.05 (one-way ANOVA, Tukey's multiple-comparison test). D, E, Rescue of clh-1 expression in AMsh cells of clh-1 animals (clh-1;pT02B11.3:clh-1 restored HCO3−-mediated alkalization; ΔF/F = 0.73 ± 0.23, n = 8 cells). Rescue animals had a difference in variance compared with the data from A–C (Bartlett's test, p < 0.0001); thus, values were compared with t test. *p < 0.05. F, Representative traces of removal of extracellular Cl− at baseline pH in buffer containing HCO3−, in the AMsh cells of wild-type (Wt, black trace), clh-1 (red trace), and clh-1 rescue (clh-1;pT02B11.3::clh-1, blue trace) animals. G, Alkalization in AMsh cells after Cl− removal is significantly smaller in clh-1 animals (ΔF/F = 0.15 ± 0.03, n = 8 cells) compared with wild-type animals (Wt, ΔF/F = 0.30 ± 0.03, n = 16 cells), and restored to wild-type levels in clh-1 rescue animals (clh-1;pT02B11.3::clh-1, ΔF/F = 0.39 ± 0.10, n = 8 cells). As in F, clh-1;pT02B11.3::clh-1 animals had unequal variance (Bartlett's test, p = 0.0114), so t test was used. Data are mean ± SEM. **p < 0.01 (t test).
We also tested the effect of other pharmacological agents that are known to block channels that may be involved in transport of HCO3−. For example, some Cl− channels, including bestrophins, are permeable to HCO3− (Qu and Hartzell, 2008). Thus, we tested the effect of the chloride channel blocker niflumic acid (100 μm) (White and Aylwin, 1990). However, we did not find that it caused a difference in the extent of alkalization mediated by HCO3− from a baseline pH (Fig. 2A,C; 0.36 ± 0.02, n = 5 cells vs 0.30 ± 0.05). Many Cl− channels are also blocked by DIDS (Wulff, 2008), which we also showed has no effect on the rate of alkalization caused by HCO3− from baseline pH.
Bicarbonate-independent acid removal does not depend on Na+/H+ exchange
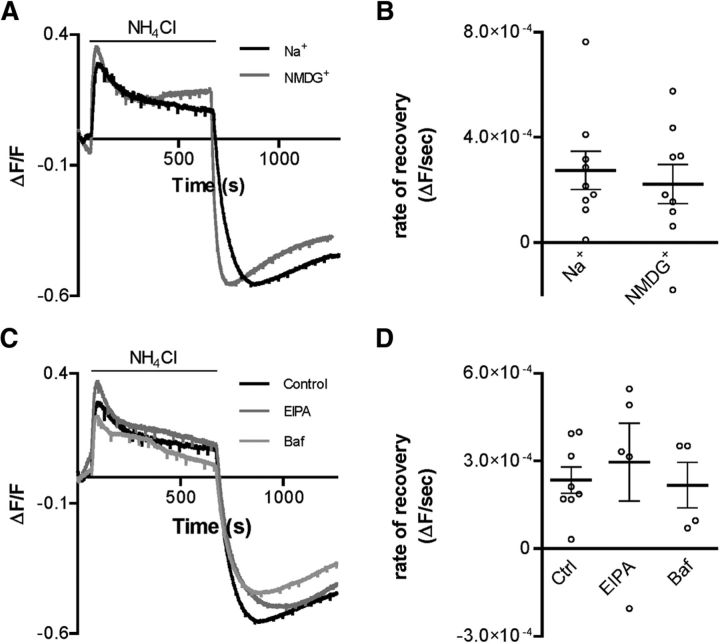
Even in the absence of HCO3−, AMsh can still recover from acid load, albeit more slowly (Fig. 1E,G). This suggests that these glial cells have other mechanisms of proton buffering or transport. One known mechanism of proton transport from the cytoplasm to the extracellular environment is through the action of Na+/H+ exchangers, which use sodium entry down the electrochemical gradient to remove protons from the cytoplasm (Boron, 2004). To determine whether AMsh glia use Na+/H+ exchangers to remove protons from the cytoplasm, we first examined recovery from an NH4Cl acid load in HCO3−-free media with or without extracellular Na+. The rate of recovery from acid load was similar in both the presence (ΔF/F/s = 2.7 × 10−4 ± 7.2 × 10−5, n = 9 cells) and absence (2.2 × 10−4 ± 7.4 × 10−5, n = 9 cells) of extracellular Na+ (Fig. 3A,B; p = 0.6244). These data argue against the Na+/H+ exchangers being involved in recovery from an acid load in AMsh cells. Furthermore, there was no difference in the extent of acidification after removal of NH4Cl, between the two conditions (ΔF/F = −0.58 ± 0.06, n = 9 and −0.45 ± 0.10, n = 9 in the presence and absence of Na+, respectively). To further test the involvement of the Na+/H+ exchangers, we perfused the AMsh with the Na+/H+ exchanger inhibitor 5-(N-ethyl-N-isopropyl) amiloride (EIPA, 20 μm). But we found no effect of this inhibitor on recovery from an acid load in HCO3−-free buffer in the absence or presence of EIPA (Fig. 3C,D; control ΔF/F/s = 3.0 × 10−4 ± 1.3 × 10−4, n = 5 cells and EIPA ΔF/F/s = 2.4 × 10−4 ± 4.5 × 10−5, n = 8 cells, p = 0.8066). To conclude, these results provide no evidence that that the Na+/H+ exchanger is involved in pH regulation in AMsh glia under our experimental conditions.
Figure 3.
HCO3−-independent alkalization in AMsh is not affected by blockers of Na+/H+ or H+-ATPase transporters. A, Representative traces of NH4Cl acid pulse experiment in HCO3−-free buffer, in either the presence (Na+, black trace) or absence (NMDG+, gray trace) of extracellular Na+. B, The rate of alkalization after an NH4Cl acid pulse in HCO3−-free conditions is not significantly different in the presence (Na+, ΔF/F/s = 2.7 × 10−4 ± 7.2 × 10−5, n = 9 cells) versus absence (NMDG+, ΔF/F/s = 2.2 × 10−4 ± 7.4 × 10−5, n = 9 cells) of Na+ in HCO3−-free conditions. C, Representative traces of NH4Cl acid pulse experiments in HCO3−-free control buffer (control) and in the presence of EIPA (20 μm, EIPA, dark gray trace), or bafilomycin (100 nm, Baf, light gray trace). D, The rate of recovery after an NH4Cl-mediated acid load in HCO3−-free conditions is not significantly different in the presence of EIPA (20 μm, EIPA, ΔF/F/s = 3.0 × 10−4 ± 1.3 × 10−4, n = 5 cells) or bafilomycin (100 nm Baf, ΔF/F/s = 2.2 × 10−4 ± 7.8 × 10−5, n = 4 cells), compared with control conditions (control, ΔF/F/s = 2.4 × 10−4 ± 4.5 × 10−5, n = 8 cells). Data are mean ± SEM. Statistical analysis: C, Student's t test; D, ANOVA.
Another known mechanism of cytosolic alkalization is via ATP-driven proton transporters, such as the H+-ATPase (Boron, 2004). To test the possibility that HCO3−-independent recovery from acid load occurs via this mechanism, we perfused AMsh glia with the H+-ATPase blocker bafilomycin (100 nm). However, bafilomycin had no effect on the rate of recovery from an acid load in HCO3−-free conditions (Fig. 3C,D; control ΔF/F/s = 3.0 × 10−4 ± 1.3 × 10−4, n = 5 cells and bafilomycin ΔF/F/s = 2.2 × 10−4 ± 7.8 × 10−5, n = 4 cells, p = 0.8066). Thus, these data provide no evidence that AMsh cells use H+-ATPases to remove protons from the cytoplasm after an acid load. Because these experiments did not indicate any obvious mechanism for HCO3−-independent pH regulation, we turned our focus to the HCO3−-dependent mechanisms.
RNA-seq analysis of embryonic AMsh cells revealed multiple potential functions of these cells
Given that our experiments failed to identify any obvious mechanisms of HCO3− entry in AMsh cells, we surmised that these cells may have a unique mechanism of HCO3− entry, perhaps an anion channel or transporter that is not sensitive to the altered conditions or drugs we tested. Expression analysis on AMsh cells had previously been performed using microarray technology (Bacaj et al., 2008); however, these data did not provide any obvious candidate gene (Bacaj et al., 2008; S. Shaham, personal communication). Given the inherent bias of microarray analysis of gene expression, which is highly dependent on the quality of the microarray chip, we decided to use another less biased method of analysis of transcripts expressed in AMsh cells, namely, RNA sequencing (RNA-seq) (Wang et al., 2009). We thus isolated cultured AMsh cells expressing GFP by cell sorting, extracted RNA, and performed RNA sequencing. We then compared transcript expression in AMsh cells with transcripts expressed in all the other C. elegans embryonic cells.
The results from the RNA-seq of AMsh cells revealed 1135 different transcripts that were significantly enriched in AMsh cells (p < 0.05, n = 3 independent AMsh and control pools), 239 of which were found to be below the false discovery rate threshold as calculated by using the statistical method of Benjamini and Hochberg (1995).
Transcripts with a potential role in HCO3− influx that were significantly enriched in AMsh cells compared with control cells were abts-4, the ClC-type chloride channels clh-1 and clh-3, the sulfate permease sulp-3, the putative bestrophin channel best-9, and the ATP-binding cassette transporters mrp-3 and mrp-4, which share homology with the HCO3−-permeable channel CFTR (Table 2). Of the transcripts enriched in the abts, sulp, best, mrp, and clh families, clh-1 showed the highest reads in AMsh cell pools (144.70 ± 39.27 FPKM; Table 2), which could be indicative of high levels of CLH-1 in these cells, whereas clh-3 showed the highest fold enrichment (3.90 fold vs control; Table 2). Interestingly as well, counts for abts-1 and abts-3 transcripts in AMsh cell pools were significantly lower than in control pools, whereas reads for abts-2 were quite low and not significantly different in AMsh cells versus the other C. elegans cells (Table 2). As stated above, ABTS-1 is an Na+-driven Cl−/HCO3− exchanger, whereas ABTS-2 and ABTS-3 share homology to the mammalian Na+-coupled anion transporter SLC4A11 (Sherman et al., 2005). Thus, our RNA sequencing data correlate well with our pH imaging data from Figure 2, showing that removal of Na+ has no effect on HCO3− entry in AMsh cells, confirming that indeed these types of transporters are not operative in these cells.
Table 2.
Expression levels of transcripts for putative HCO3− transport proteins in AMsh glia as measured by RNA sequencinga
| Gene class | Gene | Sequence | AMsh expression (FKPM) | Control expression (FKPM) | Fold AMsh Enrichment | p value |
|---|---|---|---|---|---|---|
| Anion/HCO3− transporter | abts-1 | F52B5.1b | 25.12 ± 5.21b | 55.27 ± 11.56b | 0.45b | 0.0113 |
| abts-2 | F52D10.1 | 1.42 ± 0.40 | 1.11 ± 0.11 | 1.28 | 0.3324 | |
| abts-3 | F57F10.1 | 13.91 ± 0.48 | 56.23 ± 22.67 | 0.25 | 0.0337 | |
| abts-4 | R03E9.3b | 33.85 ± 5.97b | 12.48 ± 3.77b | 2.71b | 0.0187 | |
| Sulfate permease | sulp-1 | C55B7.6 | 18.63 ± 4.88 | 15.97 ± 5.19 | 1.17 | 0.5087 |
| sulp-2 | F14D12.5 | 0.90 ± 0.75 | 0.57 ± 0.24 | 1.56 | 0.9922 | |
| sulp-3 | F41D9.5b | 2.55 ± 0.52b | 1.44 ± 0.35b | 1.77b | 0.0356 | |
| sulp-4 | K12G11.1 | 0.20 ± 0.18 | 0.85 ± 0.36 | 0.24 | 0.0559 | |
| sulp-5 | K12G11.2 | 1.20 ± 1.85 | 0.31 ± 0.15 | 3.85 | 0.8517 | |
| sulp-6 | W01B11.2 | 1.13 ± 0.98 | 1.00 ± 0.72 | 1.14 | 0.8037 | |
| sulp-7 | W04G3.6 | 2.21 ± 3.12 | 1.51 ± 1.02 | 1.46 | 0.8622 | |
| sulp-8 | ZK287.2 | 5.22 ± 3.07 | 6.57 ± 1.16 | 0.79 | 0.4647 | |
| CLC-type Cl− channel | clh-1 | T27D12.2b | 144.70 ± 39.27b | 53.46 ± 4.17b | 2.71b | 0.0165 |
| clh-2 | B0491.8 | 2.67 ± 1.59 | 7.51 ± 3.77 | 0.36 | 0.1154 | |
| clh-3 | E04F6.11b | 9.15 ± 1.10b | 2.35 ± 0.27b | 3.90b | 0.0002 | |
| clh-4 | T06F4.2 | 0.27 ± 0.14 | 0.67 ± 0.53 | 0.39 | 0.3418 | |
| clh-5 | C07H4.2 | 3.99 ± 6.67 | 0.54 ± 0.44 | 7.33 | 0.8601 | |
| clh-6 | R07B7.1 | 4.91 ± 1.86 | 6.59 ± 0.36 | 0.75 | 0.2974 | |
| ATP-binding cassette transporters | mrp-1 | F57C12.5 | 21.21 ± 7.14 | 42.92 ± 13.84 | 0.49 | 0.0762 |
| mrp-2 | F57C12.4 | 315.54 ± 71.14 | 255.76 ± 53.12 | 1.23 | 0.3054 | |
| mrp-3 | E03G2.2b | 10.08 ± 2.65b | 4.93 ± 1.34b | 2.04b | 0.0319b | |
| mrp-4 | F21G4.2b | 3.77 ± 1.16b | 1.81 ± 0.49b | 2.08b | 0.0402b | |
| mrp-5 | F14F4.3 | 48.28 ± 8.15 | 38.15 ± 6.69 | 1.27 | 0.1764 | |
| mrp-6 | F20B6.3b | 6.09 ± 1.63b | 11.11 ± 1.91b | 0.55b | 0.0370b | |
| mrp-7 | Y43F8C0.12 | 7.63 ± 1.62 | 21.56 ± 9.66 | 0.35 | 0.0779 | |
| mrp-8 | Y75B8A0.26 | 1.72 ± 1.42 | 10.46 ± 1.04 | 0.16 | 0.1466 | |
| Bestrophin channels | best-1 | B0564.3 | 0.59 ± 0.54 | 1.72 ± 1.04 | 0.34 | 0.1871 |
| best-2 | B0564.4 | 1.11 ± 1.75 | 0.72 ± 0.39 | 1.54 | 0.6142 | |
| best-3 | C01B12.3 | 0.73 ± 0.62 | 0.55 ± 0.35 | 1.34 | 0.9841 | |
| best-4 | C01B12.5 | NF | NF | |||
| best-5 | C07A9.8 | 10.90 ± 17.42 | 0.45 ± 0.25 | 24.02 | 0.3918 | |
| best-6 | C09B9.3 | NF | NF | |||
| best-7 | C29F4.2b | 0.15 ± 0.05b | 0.90 ± 0.23b | 0.17b | 0.0031b | |
| best-8 | C37A5.1b | 0.10 ± 0b | 0.47 ± 0.07b | 0.21b | 0.0039b | |
| best-9 | C43G2.4b | 7.40 ± 3.54b | 1.67 ± 0.64b | 4.42b | 0.0129b | |
| best-10 | C49A1.2 | 2.15 ± 3.55 | 0.45 ± 0.11 | 4.82 | 0.9473 | |
| best-11 | C49A1.3 | NF | NF | |||
| best-12 | F14H3.2 | NF | NF | |||
| Bestrophin channels | best-13 | F32B6.9 | 16.25 ± 8.20 | 6.46 ± 2.23 | 2.52 | 0.1094 |
| best-14 | F32G8.4 | 1.89 ± 2.49 | 4.14 ± 1.97 | 0.46 | 0.2731 | |
| best-15 | R13.3b | 0.18 ± 0.12b | 0.77 ± 0.28b | 0.24b | 0.0319b | |
| best-16 | R13A1.9 | NF | NF | |||
| best-17 | T19C3.1 | NF | NF | |||
| best-18 | T20G5.4 | 3.35 ± 2.37 | 6.38 ± 0.62 | 0.52 | 0.2674 | |
| best-19 | T21D12.15b | 2.20 ± 0.66b | 9.22 ± 1.70b | 0.24b | 0.0029b | |
| best-20 | Y37E11AR0.1 | 1.36 ± 2.18 | 0.81 ± 0.42 | 1.67 | 0.5876 | |
| best-21 | Y73F8A0.11 | NF | NF | |||
| best-22 | ZC518.1b | 1.97 ± 0.84b | 10.48 ± 4.20b | 0.19b | 0.0080b | |
| best-23 | ZK675.3b | 0.68 ± 0.67b | 10.35 ± 6.53b | 0.07b | 0.0389b | |
| best-24 | ZK688.2 | 0.54 ± 0.40 | 1.44 ± 0.77 | 0.38 | 0.1590 | |
| best-25 | ZK849.4 | NF | NF | |||
| best-26 | ZK849.5 | NF | NF |
aFKPM, Fragments per kilobase of gene per million fragments mapped, expressed as mean ± SD; NF, transcript not found in RNAseq data. n = 3 independent pools of AMsh and control cells.
bTranscripts from AMsh cells showing significant (p < 0.05) enrichment or reduction compared with control cells (determined using Student's t test).
Because clh-1 and clh-3 were highly enriched and seemed promising candidates for HCO3− influx into AMsh cells, and abts-4 is a member of a known HCO3− transporter family, we crossed our pT02B11.3;pHlourin strain with clh-1(ok658), clh-3(ok763), and abts-4(ok953) knock-out strains and looked at HCO3− entry at baseline pH into the AMsh cells. Clh-1(ok658) has a 1031 bp deletion and 1 bp insertion (T) resulting in a 3 exon deletion near the N terminus and a subsequent frameshift mutation of CLH-1. Clh-3 (ok763) has a 1495 bp deletion resulting in the loss of 4 or 6 central exons (depending on the predicted splice variants) of CLH-3. Abts-4(ok953) has a 1196 bp deletion causing the disruption of 4 exons of ABTS-4. Although the clh-3 and abts-4 knock-outs had no effect on HCO3− entry (Fig. 2B,C; ΔF/F = 0.24 ± 0.05, n = 5 cells and 0.26 ± 0.07, n = 4 cells, respectively, vs 0.30 ± 0.05), knock-out of clh-1 significantly reduced the extent of HCO3− entry in AMsh cells (Fig. 2B,C; ΔF/F = 0.08 ± 0.05, n = 11 cells). As there was still residual HCO3− entry in AMsh cells in the absence of CLH-1, we surmised that CLH-3 may have had a smaller role in HCO3− entry that was only obvious upon knock-out of CLH-1. However, clh-1;clh-3 double knock-out AMsh cells did not have any further deficits in HCO3− entry beyond that of clh-1 alone (Fig. 2C; ΔF/F = 0.10 ± 0.09, n = 4 cells). Finally, we rescued AMsh CLH-1 expression in clh-1 knock-out animals by creating a transgenic clh-1;pT02B11.3::pHlourin line that also expressed the clh-1 genomic sequence under the control of the pT02B11.3 promoter sequence. The AMsh cells of these rescue animals showed a significant increase in HCO3− uptake compared with the AMsh cells of clh-1 knock-out animals (Fig. 2D,E; 0.73 ± 0.23, n = 8 cells). The mean of the total HCO3− uptake in the clh-1 rescue animals was also larger than the mean of HCO3− uptake in wild-type animals, although not significantly so (Fig. 2D,E). We also noted that the variability of HCO3− uptake in these rescue animals was higher than the variability of HCO3− uptake in wild-type and clh-1 knock-out animals. This suggests that there may be considerable variability in clh-1 expression from one rescue animal to another. Together, these data support a major role of ClC type Cl− channel CLH-1, but not of ABTS-4 or CLH-3, in HCO3−-dependent pH buffering in these glial cells.
Another method to examine the role of anion transporters in HCO3−-mediated pH regulation is by replacement of extracellular Cl− with gluconate− in HCO3−-buffered solution. This maneuver increases the driving force for Cl− efflux, which in turn causes more rapid HCO3− uptake. This is generally thought to be indicative of Cl−/HCO3− exchanger activity. (Buckler et al., 1991; Taylor et al., 2006; Ishiguro et al., 2007; Damkier et al., 2010). AMsh cells did have an increase in pH upon removal of Cl− in HCO3−-buffered solution, suggesting the presence of this type of mechanism in these cells (Fig. 2F,G; ΔF/F = 0.30 ± 0.03, n = 16 cells). In clh-1 animals however, this alkalization upon Cl− removal was significantly decreased (Fig. 2F,G; ΔF/F = 0.15 ± 0.03 n = 8 cells) and was restored to wild-type levels in clh-1;pT02B11.3:clh-1 rescue animals (Fig. 2F,G; ΔF/F = 0.39 ± 0.10, n = 8 cells). While certain types of ClC Cl− channels have been shown to function as antiporters, their antiport activity involves the exchange of protons not bicarbonate for a Cl− ion. Thus, we speculate that this instead may be due to increased driving force for HCO3− entry into the cell through the CLH-1 channel due to changes in membrane potential caused by efflux of Cl−, which would result in a more depolarized (positive) membrane potential. Alternatively, removal of Cl− may reduce the anomalous mole fraction effect caused by Cl− on HCO3− permeation and allow for more entry of HCO3− ions (Rychkov et al., 1998). Finally, this may be also due to changes in membrane potential caused by the loss of Cl− flux through CLH-1, reducing uptake of HCO3− through another channel or transporter.
Acid load recovery in AMsh does not require CLH-1
We also wanted to determine whether CLH-1, in addition to HCO3− uptake at baseline pH, was involved in HCO3−-mediated recovery from an acid load. In contrast to HCO3− transport at baseline pH (Fig. 2A,C), here we found that replacement of Na+ in the extracellular buffer with NMDG+ (control ΔF/F/s = 7.16 × 10−4 ± 7.53 × 10−5, n = 12 cells vs NMDG+ = 4.60 × 10−4 ± 8.69 × 10−5, n = 12 cells) and incubation of the AMsh cell with DIDS (250 μm, control ΔF/F/s = 9.24 × 10−4 ± 7.31 × 10−5, n = 8 cells vs DIDS ΔF/F/s = 5.02 × 10−4 ± 5.33 × 10−5, n = 8 cells) caused a decrease in the rate of pH recovery to baseline after an acid load (Fig. 4A–D). We also found that Cl− replacement with gluconate caused a significant decrease in the rate of recovery to neutral pH after an acid load (Fig. 4E,F; control ΔF/F/s = 6.44 × 10−4 ± 6.90 × 10−5, n = 10 cells vs gluconate ΔF/F/s = 4.56 × 10−4 ± 5.00 × 10−5 n = 10 cells). These data suggest a different mechanism than that involved in CLH-1-mediated HCO3− influx seen at baseline pH. Indeed, knock-out of CLH-1 had no effect on acid load recovery (Fig. 4G,H). To conclude, CLH-1 is mostly required for HCO3− influx at baseline pH, and another mechanism mediates pH recovery after an acid load.
Figure 4.
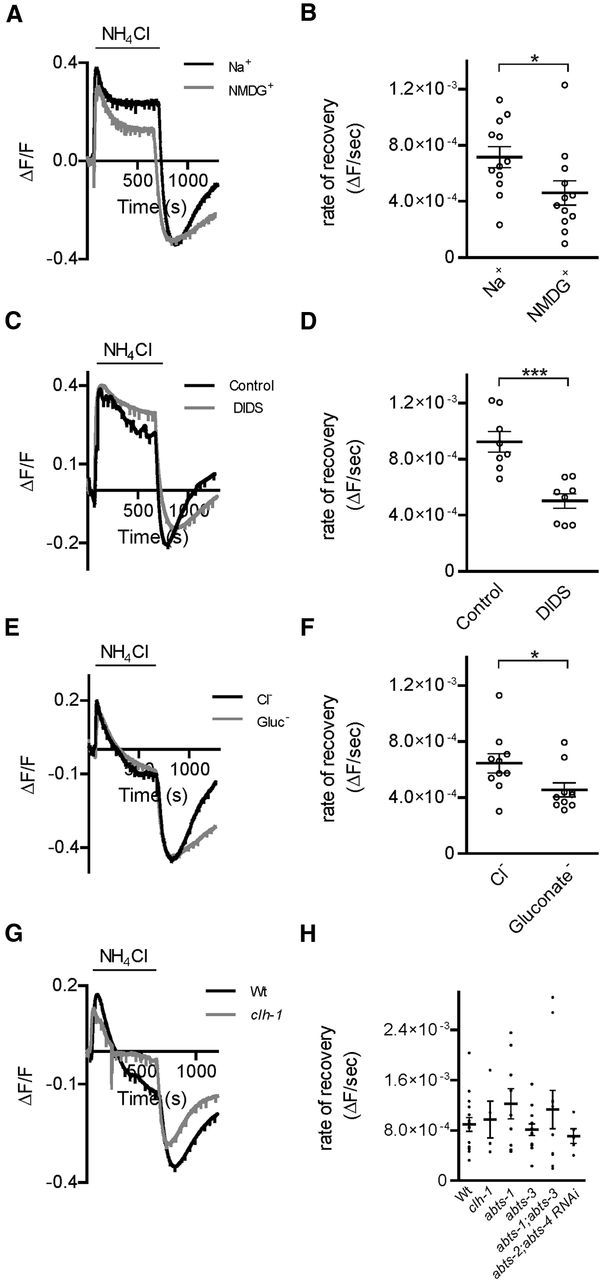
Acid load recovery in AMsh cells involves a Na+, Cl−, and DIDS-sensitive mechanism independent of CLH-1. A, Representative traces from NH4Cl acid pulse experiments in extracellular buffer with 20 mm HCO3−, in the presence (Na+, black trace) and in the absence (NMDG+, gray trace) of extracellular Na+. B, The rate of recovery after the NH4Cl-mediated acid load is significantly decreased in the absence of Na+ (NMDG+, ΔF/F/s = 4.60 × 10−4 ± 8.69 × 10−5, n = 12 cells) compared with in the presence of Na+ (ΔF/F/s = 7.16 × 10−4 ± 7.53 × 10−5, n = 12 cells). C, Same as in A, in the absence (control, black trace) and in the presence of DIDS (250 μm, DIDS, gray trace). D, The average rate of recovery from an acid load after an NH4Cl acid pulse in 20 mm HCO3− is significantly decreased in the presence of DIDS (DIDS, ΔF/F/s = 5.02 × 10−4 ± 5.33 × 10−5, n = 8 cells) compared with control conditions (control, ΔF/F/s = 9.24 × 10−4 ± 7.31 × 10−5, n = 8 cells). E, Representative traces from NH4Cl acid pulse experiments in extracellular buffer with 20 mm HCO3, in both the presence (Cl−, black trace) and absence (gluconate−, gray trace) of Cl−. F, The average rate of recovery from acid load after an NH4Cl pulse is significantly decreased in the absence of Cl− (gluconate−, ΔF/F/s = 4.56 × 10−4 ± 5.00 × 10−5 n = 10 cells) compared with control (Cl−, ΔF/F/s = 6.44 × 10−4 ± 6.90 × 10−5, n = 10 cells). *p < 0.05 (Student's t test). G, Representative traces from NH4Cl acid pulse experiments in extracellular buffer with 20 mm HCO3, in glia from wild-type (Wt, black trace) and clh-1 (gray trace) animals. H, The average rate of recovery from an acid load after an NH4Cl acid pulse in 20 mm HCO3− is not significantly different between wild-type animals (Wt, ΔF/F/s = 8.93 × 10−4 ± 1.09 × 10−4, n = 16 cells), clh-1 (ΔF/F/s = 9.72 × 10−4 ± 2.94 × 10−4, n = 4 cells), abts-1 (ΔF/F/s = 1.22 × 10−3 ± 2.40 × 10−4, n = 9 cells), abts-3 (ΔF/F/s = 8.11 × 10−4 ± 9.13 × 10−5, n = 13 cells), abts-1;abts-3 (ΔF/F/s = 1.13 × 10−3 ± 3.06 × 10−4, n = 10 cells) knock-out animals, and pT02B11.3:abts-2;abts-4 RNAi animals (abts-2;abts-4 RNAi, ΔF/F/s = 7.09 × 10−4 ± 1.17 × 10−4, n = 5 cells). p = 0.4916 (one-way ANOVA). ***p < 0.001, Student's t test. Data are mean ± SEM.
The above results suggested the involvement of Na+-driven Cl−/HCO3− exchange in acid load recovery in AMsh cells, which is typically mediated by genes homologous to the C. elegans abts family (Romero et al., 2013). However, rates of recovery in AMsh glia from clh-1 (Fig. 4H; ΔF/F/s = 9.72 × 10−4 ± 2.94 × 10−4, n = 4 cells), abts-1 (Fig. 4H; ΔF/F/s = 1.22 × 10−3 ± 2.40 × 10−4, n = 9 cells), abts-3 (Fig. 4H; ΔF/F/s = 8.11 × 10−4 ± 9.13 × 10−5, n = 13 cells), and abts-1;abts-3 (Fig. 4H; ΔF/F/s = 1.13 × 10−3 ± 3.06 × 10−4, n = 10 cells) knock-out animals, and animals expressing a construct that causes AMsh-specific knockdown of abts-2 and abts-4 (Fig. 4H; ΔF/F/s = 7.09 × 10−4 ± 1.17 × 10−4, n = 5 cells) transcripts were similar to wild-type (Fig. 4H; ΔF/F/s = 8.93 × 10−4 ± 1.09 × 10−4, n = 16 cells). This is consistent with our RNA sequencing data showing little to no expression of ABTS-1, -2, and -3. ABTS-4 is expressed in AMsh cells as shown in the RNA sequencing data (Table 2); however, it shares the most homology with Na+-independent Cl−/HCO3− exchangers, which generally function physiologically to mediate intracellular HCO3− efflux rather than influx, but may also show a reversal of ion transport under conditions favoring Cl− efflux rather than influx (Romero et al., 2013). However, under our experimental conditions of high extracellular [Cl−], ABTS-4 is not likely to function to alkalize the cytoplasm, but rather acidify it. These data suggest that AMsh glia have a mechanism of recovery from acid load that does not require CLH-1 and is not mediated by known transporters that are usually involved in this phenomenon.
A CLH-1 GFP reporter is expressed in AMsh cells
CLH-1 is most homologous to plasma membrane bound ClC channels (ClC-0, ClC-1, ClC-2, ClC-Ka, and ClC-Kb; Fig. 5A), sharing the most identity with ClC-2 (37%). ClC anion channels form a structure of two-pore homodimers (Fig. 5B). As a further confirmation of CLH-1 being expressed in AMsh cells, we examined transgenic animals expressing a GFP reporter construct under the control of the clh-1 promoter (Nehrke et al., 2000). Fluorescent images of the head of the animals revealed GFP expression in the AMsh cell body (Fig. 5C). Images of the tip of the nose of the animals revealed GFP reporter expression in the distinctive AMsh processes (Fig. 5D), which envelop the dendrites of sensory neurons and form the pore through which the cilia of some of these neurons gain access to the outside environment (Oikonomou and Shaham, 2011). In addition to AMsh glia, we observed expression of GFP under the control of the clh-1 promoter in other cells in the head, including 3 sets of neurons located posterior to the C. elegans nerve ring (data not shown), as was previously reported (Nehrke et al., 2000).
Figure 5.
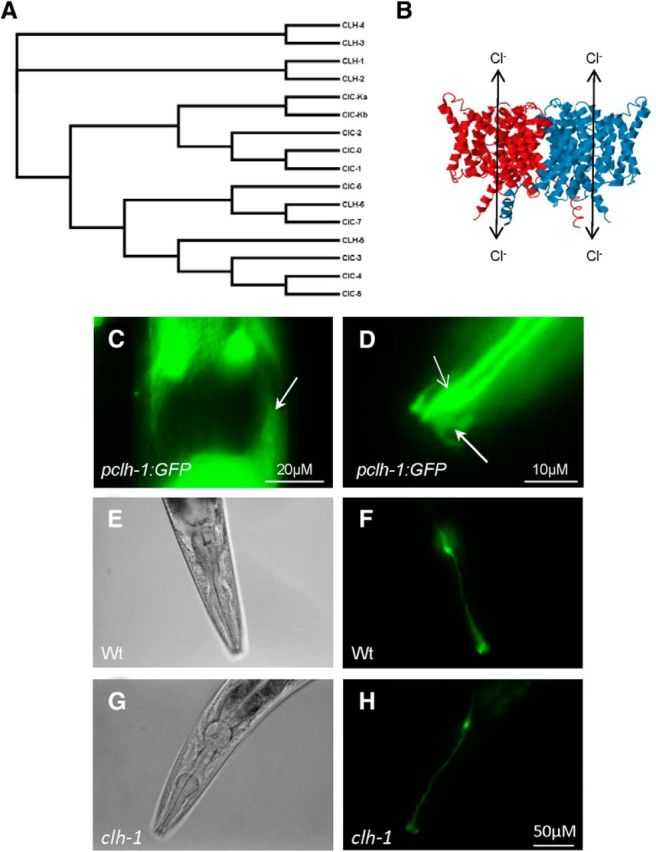
CLH-1 is expressed in AMsh glia of C. elegans and does not affect AMsh development. A, Phylogenetic relationship between the ClC family of chloride channels and the C. elegans CLH family. Multiple sequence alignment of the protein sequences of these channels was performed using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) to generate the phylogenic tree data. FigTree version 1.4.2 was used to assemble the tree. B, Diagram of the general structure of ClC-type chloride channels, generated using the E. coli ClC channel 3D structure 1KPK from RCSB protein databank. Cl− (and other anions) permeate through the two pores of ClC channel homodimers. C, Example of pCLH-1-driven GFP expression in a cell resembling the AMsh cell body (white arrow), along with high levels of pCLH-1 driven GFP expression from surrounding cells. D, Example of pCLH-1 driven GFP expression in the AMsh process at the tip of the nose of C. elegans (thick arrow, dimmer fluorescent structure). Expression is also apparent in a neuron sensory dendritic process that extends to the nose as well (thin arrow, brighter fluorescent structure). E–H, Bright-field (left panels) and fluorescent (right panels) images of wild-type (Wt, E, F) and clh-1 (G, H) animals that express GFP under the control of the AMsh-specific promoter pT02B11.3 reveal no major structure abnormalities of the AMsh cell of clh-1 animals compared with wild-type animals.
One mechanism by which AMsh HCO3− uptake is disrupted in clh-1 knock-out animals could be due to improper development of these cells, which could be independent from any possible HCO3− uptake via CLH-1 channels. To determine whether knock-out of clh-1 affects the development and/or structure of AMsh glia, we compared the morphology of AMsh glia in wild-type and clh-1 animals that expressed GFP under the control of the AMsh-specific promoter pT02B11.3. We observed no obvious differences in the structure of AMsh cell bodies and processes between wild-type (Fig. 5E,F) and clh-1 (Fig. 5G,H) animals. ClC channels are known to also be involved in cellular volume regulation (Sardini et al., 2003). However, we did not observe significant differences in the areas of wild-type and clh-1 AMsh glia cell bodies (753 ± 266 μm2 vs 716 ± 171 μm2, respectively, mean ± SD, n = 5). These observations indicate that the effects of clh-1 knock-out on HCO3− flux are likely not due to improper development, volume changes, or general sickness of these cells. It is important to note, though, that fluorescent microscopy cannot resolve subcellular structures.
CLH-1 is a pH-sensitive anion channel that is permeable to HCO3−
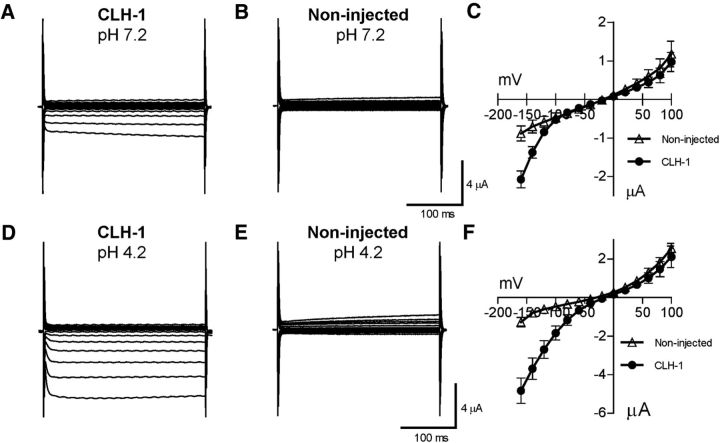
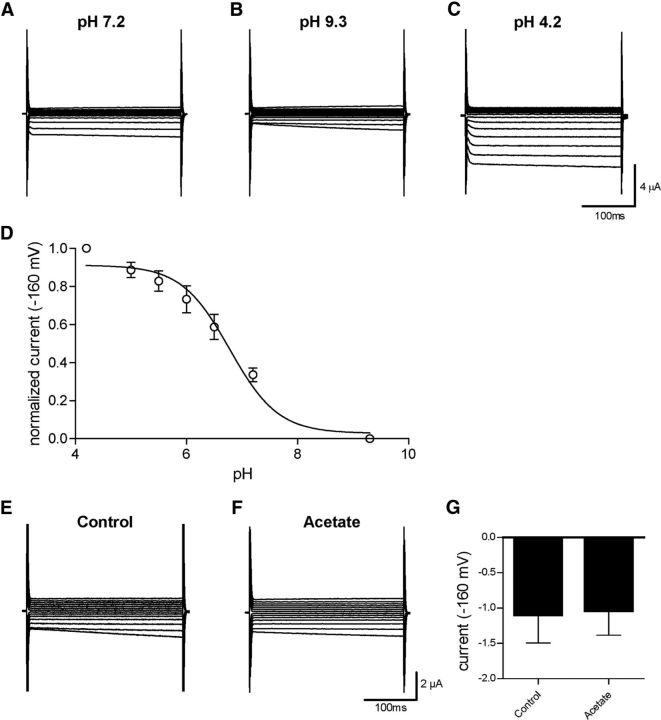
Because CLH-1 is homologous to mammalian ClC chloride channels, especially ClC-2 (37% identity), and has previously been shown to mediate chloride flux (Nehrke et al., 2000), we wished to further define the properties of this channel and how it might be involved in pH regulation through HCO3− transport. We thus expressed CLH-1 heterologously in Xenopus oocytes and measured membrane currents through two-electrode voltage clamp. Oocytes injected with CLH-1 RNA showed a small, inwardly rectifying current when clamped at voltages ranging from −160 mV to 100 mV (Fig. 6A–C). As ClC-type chloride channels have previously been shown to be activated by decreases in extracellular pH (Jentsch, 2008), we then measured membrane currents in extracellular solution of pH 4.2 in CLH-1-expressing oocytes. Here, currents at negative voltages were greatly increased compared with currents measured at pH 7.2 and noninjected oocytes at pH 4.2 (Fig. 6D–F). We next wanted to characterize the pH dependence of CLH-1 by measuring currents at extracellular pH ranging from 4.2 to 9.3. We have previously shown that these changes of pH in extracellular solution do not affect the intracellular pH of Xenopus oocytes (Wang and Bianchi, 2009). As it has been observed in other ClC channels (Gradogna and Pusch, 2013), alkaline pH inhibited CLH-1 while lowering the pH-activated CLH-1 (Fig. 7A–D). We plotted the sigmoidal pH-activation curve at −160 mV for CLH-1 using pH 4.2 as the maximum activation and pH 9.3 as the minimum, resulting in a LogEC50 of pH 6.778 ± 0.1050 for extracellular pH activation of CLH-1 (Fig. 7D; n = 7 cells). To test whether intracellular acidification had an effect on CLH-1 currents, we incubated CLH-1-expressing oocytes in physiological solution containing 20 mm acetate for 1–2 h. Incubation of cells in similar concentrations of acetate is known to cause sustained intracellular acidification without having to change the extracellular pH (Thomas, 1984; Zampighi et al., 1988) and was previously used in our laboratory to measure the intracellular pH sensitivity of the DEG/ENaC channel ACD-1 (Wang et al., 2008). However, we observed no difference in currents from CLH-1-expressing oocytes incubated in 20 mm acetate versus control buffer (Fig. 7E–G; average current at −160 mV for control = −1.10 ± 0.40 μA, n = 3, and for acetate = −1.04 ± 0.35 μA, n = 4, p = 0.9154 Student's t test). Thus, although CLH-1 is sensitive to changes in extracellular pH, we found no evidence that CLH-1 is sensitive to changes in intracellular pH.
Figure 6.
CLH-1 is a pH-sensitive inwardly rectifying anion channel. A, B, Example traces of current from a CLH-1 (A) and a noninjected (B) oocyte at an extracellular pH of 7.2, clamped at voltages ranging from −160 mV to 100 mV. C, The averaged current–voltage relationships of CLH-1 (dark circles, n = 10) and noninjected (open triangles, n = 9) oocytes at pH 7.2. D–F, Same as in A–C for pH 4.2 (CLH-1, n = 11; noninjected, n = 6).
Figure 7.
CLH-1 is sensitive to extracellular pH but insensitive to intracellular pH. A–C, CLH-1 currents in an oocyte that was perfused with solutions at pH 7.2 (A), pH 9.3 (B), and pH 4.2 (C), using the same voltage protocol as in Figure 6. D, Sigmoidal dose–response curve of currents from CLH-1-expressing oocytes (n = 7) at −160 mV, normalized to pH 4.2 as maximum and pH 9.3 as minimum activation. LogEC50 = pH 6.778 ± 0.1050 for extracellular pH activation of CLH-1. Data are mean ± SEM. E, F, Example traces of currents from CLH-1-expressing oocytes that had been preincubated for 1–2 h in control buffer (E) or 20 mm acetate (F) clamped at voltages ranging from −160 mV to 100 mV. G, There was no significant difference in CLH-1 currents between control (−1.10 ± 0.40 μA, n = 3) or acetate-incubated (−1.04 ± 0.35 μA, n = 4) oocytes at a voltage of −160 mV. Data are mean ± SEM. p = 0.9154 (t test).
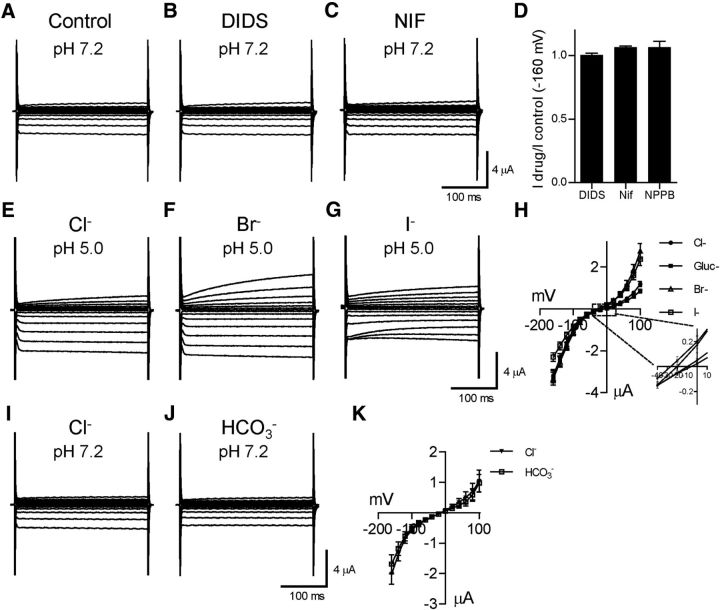
Next, we characterized the effect of chloride channel inhibitors on CLH-1. Surprisingly, CLH-1 currents (Fig. 8A) were not significantly affected by the known chloride channel blockers DIDS (1 mm, Fig. 8B,D; fraction of control current at −160 mV = 1.01 ± 0.02, n = 4 cells), niflumic acid (1 mm; Fig. 8C,D; 1.07 ± 0.01, n = 4 cells), or 5-nitro-2-(3-phenylpropylamino)-benzoate (500 μm; Fig. 8D; 1.07 ± 0.05, n = 5 cells). These data correlate well with our pH imaging data shown in Figure 2 that DIDS and niflumic acid did not block HCO3− entry into AMsh cells. We also characterized the permeability of CLH-1 to other anions relative to Cl−. We replaced 85 mm chloride from the extracellular buffer with an equimolar amount of gluconate, bromide, or iodide (leaving in 20 mm Cl−), and examined the change in current and reversal potential. These experiments were done at an external pH of 5.5 to activate CLH-1 and increase currents compared with the relatively small currents seen at neutral pH (Figs. 6, 7). The I-V curve for gluconate (Fig. 8H) was shifted to the right compared with chloride (Fig. 8E,H), whereas the I-V curves for bromide (Fig. 8F,H) and iodide (Fig. 8G,H) were shifted to the left. The reversal potentials for chloride (−10.36 ± 2.75 mV), gluconate (−7.72 ± 3.60 mV), bromide (−19.45 ± 3.02 mV), and iodide (−24.99 ± 2.80 mV, n = 10 for each condition) gave relative permeabilities to Cl− of 0.90, 1.44, and 1.80 for gluconate−, Br−, and I−, respectively, and a permeability sequence for CLH-1 of I− > Br− > Cl− > gluconate−. Importantly, the change in reversal potential upon lowering the [Cl−] and replacing it with the less permeant gluconate− proves that the inward current seen in CLH-1-injected oocytes is indeed mediated by outward movement of Cl− ions.
Figure 8.
The chloride channel blocker sensitivity and the anion permeability of CLH-1. A–D, Example traces of currents from a CLH-1-expressing oocyte clamped at voltages ranging from −160 mV to 100 mV at an extracellular pH of 7.2 in control (A), DIDS (B, 1 mm), and niflumic acid (C, 1 mm) containing solution. D, The relative current amplitudes at −160 mV of CLH-1-expressing oocytes in the presence of DIDS (1.01 ± 0.02, n = 4), niflumic acid (1.07 ± 0.01, n = 4), and 5-nitro-2-(3-phenylpropylamino)-benzoate (500 μm, 1.07 ± 0.05, n = 5) compared with the same oocyte in control solution. E–G, Example traces of currents from a CLH-1-expressing oocyte clamped at voltages ranging from −160 mV to 100 mV at an extracellular pH of 5.5 with Cl− (E), Br− (F), or I− (G) as the dominant anion. H, Current–voltage relationship of CLH-1 at pH 5.5 in the presence of extracellular Cl− (dark circles), gluconate− (dark squares), Br− (open triangles), and I− (open squares). n = 10 cells. Inset, Magnification of the area of the graph that contains the reversal potentials for the anions. The reversal potentials for chloride (−10.36 ± 2.75 mV), gluconate (−7.72 ± 3.60 mV), bromide (−19.45 ± 3.02 mV), and iodide (−24.99 ± 2.80 mV) gave a permeability sequence for CLH-1 of I− > Br− > Cl− > gluconate−. I, J, Example traces of currents from a CLH-1-expressing oocyte clamped at voltages ranging from −160 mV to 100 mV at extracellular pH of 7.2 with Cl− (I) or HCO3− (J) as the dominant anion. K, Current–voltage relationship of CLH-1 in the presence of extracellular Cl− (dark triangles, n = 7) or HCO3− (open squares, n = 7). The reversal potentials of CLH-1 were −14.01 ± 2.70 mV in Cl− solution and −13.26 ± 2.16 mV in HCO3− solution (p = 0.83, Student's t test). Data are mean ± SEM.
Finally, we wished to characterize the permeability of CLH-1 to HCO3−. We substituted 85 mm HCO3− for Cl− in the extracellular solution and compared CLH-1 currents under this condition with the currents at pH 7.2. We did not perform these experiments at pH 5.5 as with the others as an acidic pH will move the HCO3−-buffering reaction more toward the conversion of HCO3− to CO2, resulting in loss of HCO3−. There was no significant difference in the reversal potential in HCO3− solution (Fig. 8J,K; −13.26 ± 2.16 mV) compared with regular physiological solution (Fig. 8I,K; −14.01 ± 2.70 mV, p = 0.83). The relative permeability of HCO3− to Cl− was 0.97. Thus, CLH-1 appears to be equally permeable to both Cl− and HCO3−.
Discussion
In this paper, we have demonstrated that C. elegans AMsh glia regulate intracellular pH in an HCO3−-dependent manner via HCO3− flux through the ClC-type Cl− channel CLH-1. Although ClC channels are generally well characterized in terms of Cl− flux (Jentsch, 2008), to our knowledge this is the first study to demonstrate a ClC channel that regulates pH via HCO3− flux. Bolívar et al. (2014) characterized an anion conductance in rat inner medullary collecting duct cells that was highly permeable to HCO3−, which shared similar properties to ClCk1 channels; however, the molecular identity of this channel was not apparent. Human and Xenopus ClC-5 was shown to be conductive to HCO3− when expressed in a heterologous expression system (Mo et al., 1999), but ClC-5 is localized only to endosomal membranes (Zifarelli, 2015), so it likely does not have a role in HCO3− flux from the extracellular environment.
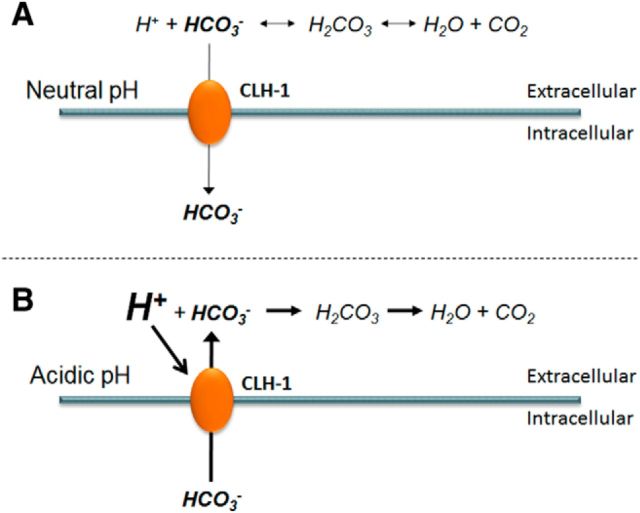
CLH-1 is most homologous to mammalian CLC-2 (37% identity). The inward rectification of CLH-1 would suggest that its main function would be extrude anions, such as Cl− and HCO3−. However, two recent studies have shown that the CLH-1 homolog ClC-2 contributes significantly to Cl− influx in neuronal cells, likely due to incomplete rectification of ClC-2 and conditions favoring Cl− influx at neuronal resting potentials (Rinke et al., 2010; Ratté and Prescott, 2011). Indeed, in C. elegans, another CLC-2 homolog, CLH-3, was shown to reduce excitability in neurons mediating egg-laying behavior via Cl− influx (Branicky et al., 2014). Thus, ClC-2 channels and homologs appear to be able to mediate both Cl− influx and efflux, depending on neuronal resting potential and driving force for Cl−. One can envision a similar role for CLH-1 in HCO3−-mediated pH regulation. HCO3− may accumulate through CLH-1 into AMsh glia under basal conditions. Upon extracellular acidification, which would tend to deplete extracellular HCO3− via conversion to CO2 and H2O, CLH-1 may be activated and mediate HCO3− efflux to neutralize the extracellular space (Fig. 9). This would help to prevent any changes in glial or neuronal synaptic functions that might occur due to sensitivity to changes in pH. Thus, in addition to Cl− homeostasis, CLH-1 may be a homeostatic regulator of pH.
Figure 9.
Proposed role of glial-expressed CLH-1 in intracellular and extracellular pH buffering. A, At neutral steady-state baseline pH, AMsh maintain intracellular buffering capacity via HCO3− leak (thin arrow) through CLH-1 down the HCO3− concentration gradient. B, Acidification of the extracellular space would shift the HCO3−-buffering reaction to the right, resulting in depletion of extracellular HCO3−. The excess protons in the extracellular space then activate CLH-1, which opens to allow the accumulated intracellular HCO3− to flow into the extracellular space down the reversed concentration gradient (thick arrow).
Knock-out of CLH-1 did not affect the rate of ammonium acid load recovery in AMsh glia, however. This, coupled with the fact that HCO3− flux was not completely eliminated in CLH-1 knock-outs, suggests an alternate unknown mechanism of HCO3− flux in AMsh glia. In contrast to the HCO3− addition experiments, ammonium acid load recovery in AMsh was sensitive to removal of Na+ and Cl− and inhibited by the presence of DIDS. Because knock-out/knockdown of abts family members had no effect on this recovery, we can eliminate the classical Na+-coupled HCO3− transporters from mediating this phenomenon, much like in the HCO3− addition experiments. Another possible candidate is the bestrophin channel BEST-9, which was significantly enriched in AMsh transcripts analyzed by RNAseq. Because no knock-out of BEST-9 currently exists, RNAi knockdown or generation of a knock-out strain will be needed to test its role in AMsh HCO3− flux.
ClC-2 is broadly expressed in many mammalian cell types, including neurons and glia (Thiemann et al., 1992). Knock-out of ClC-2 in mice leads to retinal and testicular degeneration, as well as leukoencephalopathy, a vacuolization of brain and spinal cord white matter (Jentsch, 2008). In addition to Cl− homeostasis and regulation of neuronal excitability (Rinke et al., 2010; Ratté and Prescott, 2011), ClC-2 is theorized to have a role in cellular volume regulation as well (Furukawa et al., 1998). The role of mammalian ClC-2 in pH regulation has not been studied, but our results with the ClC-2 homolog CLH-1 suggest that these channels may also regulate pH by HCO3− conductance.
Our results also demonstrate the power of RNA sequencing to identify candidate genes for unique mechanisms as our initial pH imaging experiments eliminated all classical transporters as possible HCO3− influx mediators in AMsh glia. HCO3− flux has been demonstrated in channels, such as CFTR and bestrophins (Qu and Hartzell, 2008; Ishiguro et al., 2009; Yu et al., 2010). However, the fact that C. elegans has 6 ClC-type anion channel genes (clh-1 to clh-6), 8 ATP-binding cassette genes bearing homology to CFTR (mrp-1 to mrp-8), and 26 bestrophin genes (best-1 to best-26) makes knock-out or RNAi knockdown experiments to examine the effects of all of these genes on AMsh pH regulation prohibitively laborious. GFP-reporter constructs under the control of the promoters for these genes is a possibility to determine whether they are expressed in the AMsh glia; however, the loss of transcriptional elements found in gene introns and in other genomic areas decreases the reliability of expression of these reporters (Wenick and Hobert, 2004). Thus, the lack of reporter GFP expression does not necessarily mean lack of expression of that particular gene product. By performing RNA sequencing of AMsh glial transcripts, we were able to identify strong candidate anion channel genes expressed in these cells that could mediate HCO3− flux.
Our RNA sequencing data of AMsh transcripts matches well with AMsh microarray data published by Bacaj et al. (2008). A total of 97 of the 297 transcripts listed in their study as being enriched in AMsh were also found to be significantly enriched in our RNA sequencing data (Table 3). There were, however, 16 transcripts from the microarray data that were significantly depleted in our AMsh RNA sequencing data, and 15 transcripts from the microarray data that were not detected at all in our data. The remaining 169 genes were not significantly enriched or depleted in our data. Differences between our data and data published by Bacaj et al. (2008) could be explained by the limitations of microarray technology, which include high levels of background due to issues with cross-hybridization of probes or saturation of signal due to the lower dynamic range of microarrays (Wang and Bianchi, 2009).
Table 3.
Comparison of AMsh microarray versus RNAseq transcriptome analysisa
| Associated gene | Type | Microarray enrichment | RNAseq enrichment | RNAseq p value |
|---|---|---|---|---|
| F16F9.3 | Contains EF-hand domain pair | 416 | 26.55b | 9.26E-03b |
| F14D7.7 | Unknown function | 329 | NF | |
| T02B11.4 | Unknown function | 307 | 65.68b | 4.41E-04b |
| T02B11.3 | Unknown function | 222 | 62.72b | 1.52E-04b |
| F53F4.13 | Unknown function | 177 | 104.81b | 1.66E-03b |
| fig-1 | Predicted extracellular protein | 163 | 3.54b | 2.61E-02b |
| R05A10.3 | Unknown function | 134 | 16.59b | 2.46E-03b |
| R05A10.4 | Unknown function | 132 | 30.30b | 8.58E-04b |
| R13D7.2 | Unknown function | 101 | 10.63 | 3.07E-01 |
| F15D4.7 | Transposon | 96.7 | NF | |
| F52E1.2 | Similar to C-type lectin domain family | 91.1 | 11.15b | 4.26E-02b |
| C33G8.4 | Unknown function | 83.8 | 8.71 | 8.65E-02 |
| F07C6.3 | Unknown function | 83.6 | 20.29b | 1.51E-02b |
| R07A4.4 | Contains PAN-1 domain | 68.7 | 29.28b | 3.10E-04b |
| C08F11.13 | Unknown function | 68.2 | 3.65b | 1.05E-02b |
| tag-209 | Unknown function | 64.7 | 6.04 | 7.28E-01 |
| Y54G2A0.10 | Unknown function | 63.2 | 56.43b | 2.77E-04 |
| ZK822.4 | Unknown function | 61.8 | 15.35b | 3.53E-02 |
| vap-1 | Similar to venom allergen-like protein | 61.7 | 11.41b | 2.07E-02 |
| C44H4.11 | ncRNA | 58.6 | 0.06c | 4.05E-02c |
| R05A10.2 | Similar to yeast nucleolar protein NSR1 | 56.6 | 15.59b | 4.46E-04b |
| F35B12.9 | Unknown function | 55.7 | 26.41b | 9.81E-05b |
| Y69H2.3 | Similar to human IgGFc-binding protein | 53.4 | 0.93 | 6.59E-01 |
| Y23H5B0.3 | Unknown function | 52.8 | NF | |
| C05B5.9 | Pseudogene | 46.1 | 9.57b | 1.58E-02b |
| Y46B2A0.2 | redicted DNA-helicase | 44.3 | 2.29b | 5.00E-02b |
| F11C7.2 | Contains thrombospondin, Type 1 repeat domain | 43.6 | 20.85 | 4.01E-01 |
| F20A1.1 | Unknown function | 43.3 | 9.39b | 3.18E-02b |
| F59A7.2 | Unknown function | 38.7 | 4.91 | 4.68E-01 |
| K09B3.1 | Similar to mouse keratin-associated protein 6–2 | 37.3 | 8.27b | 3.34E-02b |
| Y69A2AR0.22 | Unknown function | 37.2 | 15.25b | 1.45E-02b |
| nas-32 | Astacin family zinc metalloprotease | 36.6 | NF | |
| Y38H6A0.3 | Contains cysteine-rich repeat domain | 35.8 | 9.11b | 3.28E-03b |
| K06A9.1 | Similar to human TCOF1 | 34.8 | 6.54b | 1.42E-02b |
| nas-31 | Astacin family zinc metalloprotease | 34.7 | 19.70b | 0.01373769b |
| F14D2.7 | Unknown function | 30.6 | NF | |
| K11E4.1 | Contains fungal lipase-like domain | 29.8 | 10.33b | 1.27E-03b |
| F35E2.5 | Similar to human MUCIN-2 | 28.6 | 0.22c | 3.36E-03c |
| F40F8.4 | Unknown function | 27.5 | 4.85 | 8.48E-02 |
| C25E10.11 | Unknown function | 26.9 | 6.21 | 5.78E-02 |
| ceh-37 | Homeobox protein | 26.3 | 12.53b | 2.27E-03b |
| Y105C5B0.5 | Unknown function | 25 | 0.25 | 1.49E-01 |
| F59A7.5 | Unknown function | 24.5 | 3.19b | 2.95E-02b |
| C56E10.3 | Similar to human myosin-18A | 24.2 | 3.67b | 2.14E-02b |
| B0285.6 | Putative sodium-coupled carboxylate transporter | 23.6 | 1.98 | 7.94E-01 |
| C56C10.4 | Ortholog of human FAM46 family | 23.5 | 13.53b | 1.02E-03b |
| C12D8.9 | Unknown function | 23.4 | 0.92 | 9.86E-01 |
| C05D12.1 | Ortholog of human ferric-chelate reductase 1 | 22.3 | 12.12b | 9.22E-03b |
| F13B6.3 | Similar to human mucin-5AC (fragment) | 21.4 | 0.86 | 9.11E-01 |
| tre-5 | Similar to human trehalase (brush-border membrane glycoprotein) | 20.8 | 2.59 | 4.70E-01 |
| F07F6.7 | Ortholog of human apolipoprotein L family | 20.5 | 1.44 | 3.44E-01 |
| K02E11.3 | Contains UDP-glucuronosyl/UDP-glucosyltransferase domain | 20.3 | 7.49b | 3.82E-02b |
| H22K11.11 | ncRNA | 20 | 1.91 | 6.71E-01 |
| K02E11.5 | Unknown function | 18.3 | 10.57 | 4.12E-01 |
| C41C4.16 | ncRNA | 18.3 | 0.68 | 4.27E-01 |
| C06E2.2 | Unknown function | 16.9 | 19.51 | 4.31E-01 |
| F16H6.10 | Involved in innate immune response. | 16.8 | 9.64b | 1.36E-04b |
| Y71G12B0.17 | Ortholog of human phosphatidylinositol transfer proteins | 16.5 | 4.86b | 4.47E-02b |
| Y39H10A0.1 | Unknown function | 14.9 | 11.06 | 1.54E-01 |
| T05C1.1 | Unknown function | 14.7 | 18.67b | 1.73E-03b |
| K02E11.4 | Contains UDP-glucuronosyl/UDP-glucosyltransferase domain | 14.6 | 0.18 | 1.84E-01 |
| Y73B6A0.3 | Contains UDP-glucuronosyl/UDP-glucosyltransferase domain | 14.6 | 39.55b | 2.20E-02b |
| F36H2.3 | Ortholog of human complement component receptor 2 | 14.5 | 5.00b | 6.30E-03b |
| cnp-3 | Unknown function | 13.5 | 12.87b | 9.78E-04b |
| M01G12.14 | Ortholog of human decapping exoribonuclease | 13.4 | 1.19 | 8.68E-01 |
| E02H9.3 | Unknown function | 13 | 1.01 | 8.39E-01 |
| C40H5.2 | Contains epidermal growth factor-like domain | 12.4 | 8.89b | 2.04E-02b |
| F49F1.7 | Similar to human MUCIN-2 | 12.3 | 0.20c | 9.22E-03c |
| H11L12.1 | Unknown function | 12.1 | 0.61 | 3.01E-01 |
| lec-9 | Predicted lectin | 12 | 34.88b | 6.41E-03b |
| F59E11.2 | Ortholog of human dehydrogenase/reductase member 1 | 12 | 7.39 | 1.39E-01 |
| hsp-70 | Heat shock protein | 11.8 | 0.68 | 2.85E-01 |
| daf-6 | Patched-related protein, required for amphid channel morphogenesis | 11.6 | 6.09b | 7.68E-03b |
| C27C7.1 | Unknown function | 11.5 | 0.87 | 5.73E-01 |
| nhr-158 | Nuclear hormone receptor | 11.5 | 13.55b | 9.44E-04b |
| F08G5.6 | CUB-like domain-containing, involved in innate immunity | 11.3 | 20.28b | 2.34E-04b |
| T19H12.3 | Contains UDP-glucuronosyl/UDP-glucosyltransferase domain | 11.3 | NF | |
| M03E7.2 | Ortholog of human prostaglandin E synthase 3 | 11.1 | 1.44 | 7.16E-01 |
| Y54F10BM0.3 | Ortholog of human (protein tyrosine phosphatase, nonreceptor Type 9) | 10.8 | 2.91 | 6.45E-01 |
| C38D9.2 | Unknown function | 10.6 | 0.57 | 7.94E-02 |
| F59D6.7 | Ortholog of human calcineurin-like EF-hand protein | 10.4 | 2.50b | 3.71E-02b |
| F59D12.3 | Contains sulfotransferase domain | 10.4 | 3.82 | 5.14E-01 |
| T13B5.9 | Ortholog of human LMLN metalloendopeptidase | 10.3 | 2.91 | 7.72E-01 |
| C25F9.2 | Probable DNA polymerase | 9.9 | 1.39 | 3.83E-01 |
| F10A3.16 | Contains 7TM GPCR, serpentine receptor class domain | 9.8 | 0.53 | 6.44E-01 |
| kcc-3 | Potassium/chloride transporter | 9.7 | 9.24b | 1.12E-02b |
| fkh-10 | Forkhead transcription factor | 9.7 | 0.77 | 4.66E-01 |
| lec-10 | Soluble galactose-binding lectin | 9.6 | 33.23b | 1.42E-03b |
| W06D11.3 | Similar to yeast EBNA1-binding protein | 9.6 | 2.06 | 7.51E-01 |
| C11H1.9 | Similar to humanSEC14-like protein 3 | 9.4 | 8.51b | 4.88E-02b |
| hsp-43 | Heat sock protein | 9.4 | 2.31 | 9.83E-02 |
| Y38H6C0.7 | Pseudogene | 9.3 | 2.20 | 1.39E-01 |
| ptc-2 | Similar to human isoform S of protein patched homolog 1 | 9.3 | 0.97 | 8.34E-01 |
| ceh-26 | Similar to human Prospero homeobox protein 1 | 9.3 | 3.93b | 5.93E-04b |
| K10C2.3 | Aspartyl protease | 9.3 | 2.21b | 1.06E-02b |
| C14F11.6 | Contains dTDP-4-dehydrorhamnose 3,5-epimerase domain | 9.1 | 0.85 | 9.96E-01 |
| T04A11.14 | Pseudogene | 9.1 | 0.15 | 1.24E-01 |
| Y55B1BL0.1 | Unknown function | 9.1 | 2.78 | 9.06E-01 |
| rdy-2 | Involved in regulation of amount of matrix material secreted by sheath cells | 9.1 | 3.13 | 7.41E-01 |
| pqn-8 | Predicted prion-like-(Q/N-rich)- domain-bearing protein | 8.9 | 4.53b | 3.05E-02b |
| C38D9.5 | Pseudogene | 8.9 | 2.01 | 6.90E-01 |
| C56C10.6 | Ortholog of human tau tubulin kinase family | 8.8 | 2.97b | 1.86E-02b |
| gba-1 | Ortholog of human GBA (glucosidase, beta, acid) | 8.5 | 4.92 | 5.55E-01 |
| clec-174 | Similar to human MUCIN-2 | 8.4 | 0.31 | 8.20E-02 |
| R05D7.5 | Contains cytosolic motility protein domain | 8.3 | 13.09b | 8.77E-03b |
| lipl-6 | Ortholog of human lipase family | 8.2 | 17.11b | 1.01E-02b |
| Y6E2A0.4 | Unknown function | 8.1 | 9.27 | 7.07E-01 |
| deps-1 | P-granule-associated protein involved in germline development | 8 | 0.10 | 5.43E-02 |
| Y110A2AL0.4 | Unknown function | 8 | 0.41 | 2.89E-01 |
| F32B5.4 | Unknown function | 8 | 1.09 | 5.91E-01 |
| col-111 | Structural constituent of cuticle | 7.9 | 0.57 | 2.98E-01 |
| Y69H2.10 | Unknown function | 7.9 | 0.62 | 3.05E-01 |
| C05B5.8 | Unknown function | 7.8 | 21.07b | 2.29E-03b |
| bath-29 | Similar to human Kelch repeat and BTB domain-containing protein 2 | 7.8 | 0.32 | 8.44E-02 |
| T13C2.2 | Unknown function | 7.6 | 5.22b | 1.10E-02b |
| C17C3.5 | Unknown function | 7.6 | 2.02 | 3.72E-01 |
| R09H3.3 | Unknown function | 7.5 | 0.83 | 3.86E-01 |
| W09B6.5 | Unknown function | 7.4 | 1.14 | 5.73E-01 |
| ZC449.2 | Contains PAN-1 domain | 7.4 | 1.37 | 8.21E-01 |
| cyp-33A1 | Similar to human cytochrome P450 2C9 | 7.4 | 4.23 | 7.58E-02 |
| C17E7.9 | Unknown function | 7.3 | 4.88 | 3.65E-01 |
| rnh-1.3 | RNase H ribonuclease | 7.3 | 9.96 | 1.33E-01 |
| Y105C5A0.25 | Pseudogene | 7.3 | 2.75 | 6.78E-01 |
| F59H6.5 | Putative DNA helicase | 7.3 | 2.10 | 1.35E-01 |
| C34D4.10 | Similar to human glycosyltransferase MGAT1 | 7.2 | 1.08 | 8.97E-01 |
| C49A9.4 | Ortholog of human cystatin B | 7.1 | 22.24b | 9.54E-03b |
| kca-1 | Kinesin cargo adaptor | 7.1 | 0.45 | 3.11E-01 |
| T04G9.7 | Unknown function | 7.1 | NF | |
| tdo-2 | Tryptophan 2,3-dioxygenase | 7 | 2.55 | 1.08E-01 |
| Y38H6C0.8 | Ortholog of human regenerating islet-derived (REG) family | 7 | 2.14 | 3.28E-01 |
| T14G8.4 | Unknown function | 7 | 1.53 | 6.18E-01 |
| F59B1.2 | Unknown function | 7 | 7.39b | 3.78E-03b |
| inft-1 | Similar to human FH2 domain-containing protein 1 | 7 | 3.62b | 7.03E-03b |
| Y65B4BR0.8 | Ortholog of human DNA replication complex protein GINS3 | 6.8 | 0.07 | 8.76E-03 |
| nspe-3 | Unknown function | 6.8 | 10.79 | 4.84E-01 |
| dmd-3 | DNA-binding Zn finger DM domain transcription factor | 6.8 | 0.03 | 5.22E-03 |
| F40G12.5 | Unknown function | 6.7 | 9.19 | 3.44E-01 |
| T05F1.7 | Similar to yeast vesicle-mediated ER to Golgi transport protein USO1 | 6.7 | 1.65 | 9.36E-01 |
| F20A1.10 | Unknown function | 6.7 | 2.12 | 2.12E-01 |
| F53A9.8 | Involved in defense response to Gram-positive bacterium | 6.7 | 33.73b | 2.15E-03b |
| gly-5 | UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase | 6.7 | 1.58 | 1.22E-01 |
| zip-10 | bZIP transcription factor, involved in innate immune response | 6.7 | 172.52b | 2.59E-03b |
| Y57G7A0.5 | Unknown function | 6.7 | 0.85 | 4.83E-01 |
| F20A1.9 | Similar to human ATP-dependent RNA helicase DDX1 | 6.6 | 2.15 | 2.59E-01 |
| T16G1.5 | Predicted transferase | 6.5 | 0.61 | 3.67E-01 |
| fbxa-197 | Unknown function | 6.5 | 1.24 | 8.20E-01 |
| C17H12.12 | Contains BTB/POZ domain | 6.5 | 0.14c | 1.98E-02c |
| col-63 | Similar to human collagen alpha-1(I) chain | 6.5 | 0.10c | 2.72E-02c |
| T05E11.8 | Unknown function | 6.5 | 1.19 | 9.34E-01 |
| C04B4.2 | Unknown function | 6.3 | 5.60b | 7.10E-04b |
| H06H21.11 | Unknown function | 6.3 | 2.35b | 2.30E-03b |
| glr-2 | AMPA-type ionotropic glutamate receptor subunit | 6.3 | 0.85 | 0.50002708 |
| unc-25 | GABA neurotransmitter biosynthetic enzyme, glutamic acid decarboxylase | 6.3 | 0.43 | 2.69E-01 |
| Y43F8B0.9 | Unknown function | 6.3 | 3.26b | 4.02E-02b |
| glh-1 | Putative DEAD-box RNA helicase | 6.2 | 0.19 | 8.43E-02 |
| F26A1.9 | Unknown function | 6.2 | 2.05 | 9.94E-01 |
| Y39A3B0.3 | Unknown function | 6.2 | 0.95 | 6.50E-01 |
| C05D2.8 | Unknown function | 6.2 | 9.69b | 1.99E-02b |
| B0034.4 | Similar to O. cuniculus triadin, Involved in regulation of ER Ca2+ release | 6.1 | 0.05c | 5.66E-03c |
| inx-13 | Innexin channel | 6 | 1.37 | 7.82E-01 |
| ifb-1 | Essential intermediate filament protein | 6 | 3.21b | 1.94E-02b |
| F14B4.1 | Ortholog of human low density lipoprotein receptor-related protein family | 6 | 0.38 | 3.22E-02 |
| cbd-1 | Contains chitin-binding peritrophin-A domains | 5.9 | 0.28 | 8.27E-02 |
| eak-4 | Acts in parallel to akt-1 to regulate insulin-like signaling | 5.9 | 0.53 | 3.38E-01 |
| zig-3 | Member of the immunoglobulin superfamily of proteins | 5.8 | 3.39b | 1.54E-02b |
| srx-11 | Pseudogene | 5.8 | 0.44 | 2.46E-01 |
| ptr-11 | Member of the sterol sensing domain protein family | 5.7 | 2.45 | 1.12E-01 |
| M01G12.9 | Ortholog of human decapping exoribonuclease | 5.7 | 1.96b | 5.89E-03b |
| itr-1 | Putative inositol (1,4,5) trisphosphate receptor | 5.7 | 4.17b | 1.30E-02b |
| gly-6 | Ortholog of human polypeptide N-acetylgalactosaminyltransferase | 5.7 | 1.35 | 1.54E-01 |
| R09H3.2 | Unknown function | 5.7 | 15.78 | 7.94E-01 |
| rrn-3.56 | Pseudogene | 5.6 | NF | |
| F47C12.8 | Unknown function | 5.6 | 3.34b | 4.68E-02b |
| hpk-1 | Predicted homeodomain interacting protein kinase | 5.6 | 0.92 | 6.29E-01 |
| nhr-247 | Predicted nuclear hormone receptor | 5.6 | 6.29b | 5.70E-03b |
| F57A8.1 | Unknown function | 5.5 | 0.42 | 1.47E-01 |
| kbp-4 | Kinetochore null binding protein, involved in reproduction | 5.5 | 0.82 | 4.11E-01 |
| Y67A10A0.9 | Homologous to integral membrane protein claudin | 5.5 | 2.24 | 8.36E-02 |
| bath-12 | pseudogene | 5.5 | 4.89 | 6.57E-02 |
| T08G5.3 | Unknown function | 5.5 | 4.95 | 9.56E-01 |
| mnk-1 | Ortholog of the Mnk MAP kinase-interacting kinase | 5.5 | 0.55c | 1.81E-02c |
| F21H7.2 | Similar to human high mobility group nucleosome-binding domain-containing protein 5 | 5.4 | 11.93b | 2.96E-03b |
| ZK669.5 | Contains small GTPase superfamily domain | 5.4 | 3.10 | 9.55E-01 |
| C39B5.2 | Unknown function | 5.4 | 1.37 | 4.88E-01 |
| nspe-7 | Unknown function | 5.4 | 0.30 | 2.41E-01 |
| C05D10.4 | Unknown function | 5.4 | 4.70b | 2.09E-02b |
| F35E12.6 | CUB domain containing protein involved in innate immunity | 5.3 | 2.47 | 1.59E-01 |
| hsp-16.41 | Heat shock protein | 5.3 | 1.21 | 7.63E-01 |
| kin-6 | Predicted tyrosine protein kinase | 5.3 | 0.33 | 2.41E-01 |
| nhr-90 | Nuclear hormone receptor | 5.3 | 6.87b | 2.45E-02b |
| Y59E1B0.1 | Unknown function | 5.3 | 4.34 | 1.53E-01 |
| ttr-43 | Contains transthyretin-like domain | 5.3 | 12.06 | 5.16E-02 |
| duox-2 | Similar to human isoform 1 of dual oxidase 1 | 5.3 | 1.20 | 7.87E-01 |
| C01B10.3 | Similar to Type I inositol-1,4,5-trisphosphate 5-phosphatase | 5.3 | 3.24b | 2.17E-02b |
| W03A5.4 | Similar to human disks large-associated protein 1 | 5.2 | 4.33b | 2.80E-02b |
| F49C5.8 | Transposon | 5.2 | NF | |
| Y75B12B0.3 | Unknown function | 5.2 | 1.59 | 2.46E-01 |
| F17C8.5 | TWK potassium channel subunit | 5.2 | NF | |
| nhr-204 | Nuclear hormone receptor | 5.2 | 2.45 | 4.68E-01 |
| rab-11.2 | Ortholog of human RAB11 small GTPase family | 5.2 | 13.36b | 1.83E-03b |
| mua-3 | Similar to human fibrillin-1 | 5.1 | 0.43c | 1.05E-02c |
| lec-8 | Galectin family member | 5.1 | 40.08b | 3.55E-04b |
| F40F9.3 | Contains PDZ domain found in signaling proteins | 5.1 | 2.66 | 2.78E-01 |
| pgp-8 | Member of the P-glycoprotein subclass of the ATP-binding cassette transporter superfamily | 5.1 | 1.29b | 4.13E-02b |
| ZK355.5 | Insulin/EGF-receptor L domain protein | 5 | NF | |
| F25H5.8 | Similar to yeast vacuolar intralumenal vesicle intergral membrane protein SNA3 | 5 | 9.30 | 1.08E-01 |
| twk-45 | Ortholog of human KCNK potassium channels | 5 | 0.12 | 8.84E-02 |
| K04H4.2 | Predicted secreted chitin binding domain containing protein | 5 | 0.50c | 1.38E-02c |
| fbxa-170 | Contains protein-protein interaction mediating F-box domain | 5 | 3.71b | 1.07E-02b |
| R10H10.4 | Unknown function | 5 | 1.59 | 5.58E-01 |
| Y57G11C0.31 | Contains nucleotide-diphospho-sugar transferase | 5 | 1.39 | 2.53E-01 |
| hlh-3 | Helix-loop-helix transcription factor | 4.9 | 12.70b | 6.59E-03b |
| srt-42 | 7TM GPCR, serpentine receptor Class T | 4.9 | 0.62 | 2.47E-01 |
| W02D7.3 | Unknown function | 4.9 | 22.87b | 4.85E-03b |
| R03H10.6 | Ortholog of human replication protein A1 | 4.8 | 1.86b | 3.80E-02b |
| sri-43 | 7TM GPCR, serpentine chemoreceptor Class I | 4.8 | 0.41 | 3.18E-01 |
| ZK470.12 | ncRNA | 4.8 | 11.78 | 3.77E-01 |
| ifa-4 | Intermediate filament protein | 4.8 | 12.22b | 6.66E-03b |
| T28F4.6 | Unknown function | 4.8 | 1.84 | 2.28E-01 |
| T25F10.3 | Contains epidermal growth factor-like domain | 4.8 | 3.34b | 6.16E-03b |
| inx-12 | Innexin channel | 4.8 | 1.57 | 1.59E-01 |
| such-1 | Component of the anaphase promoting complex/cyclosome | 4.8 | 0.22c | 3.48E-02c |
| itx-1 | Member of neurexin superfamily | 4.8 | 3.37b | 8.07E-03b |
| F55D1.2 | Unknown function | 4.7 | 0.99 | 9.61E-01 |
| B0281.4 | Ortholog of several human BTB/POZ domain-containing proteins | 4.7 | 0.59 | 2.92E-01 |
| gei-17 | SUMO E3 protein ligase | 4.7 | 1.19 | 2.19E-01 |
| F30F8.10 | Ortholog of human (N(alpha)-acetyltransferase 60 | 4.7 | 0.56 | 2.83E-01 |
| ugt-45 | Ortholog of human UDP glycosyltransferase 3 family | 4.7 | 4.17 | 6.22E-02 |
| apm-1 | Similar to human AP-1 complex subunit mu-1 | 4.7 | 3.40b | 3.51E-03b |
| magu-2 | Similar to the human membrane-associated guanylate kinase family | 4.7 | 2.00 | 5.50E-02 |
| nspd-4 | Unknown function | 4.6 | 0.30 | 1.94E-01 |
| F20A1.6 | Unknown function | 4.6 | 2.79 | 1.83E-01 |
| ceh-27 | NK-2 class homeodomain protein | 4.6 | 0.59 | 2.02E-01 |
| C03G6.17 | Contains acyl-CoA N-acyltransferase domain | 4.6 | 1.22 | 9.26E-01 |
| C24H12.5 | Ortholog of human downstream neighbor of SON | 4.6 | 0.04c | 1.52E-04c |
| D1086.3 | Unknown function | 4.6 | 1.30 | 4.79E-01 |
| T19D7.5 | Unknown function | 4.6 | 0.17 | 1.34E-01 |
| Y42G9A0.2 | Unknown function | 4.5 | NF | |
| cysl-3 | Ortholog of human cystathionine-beta-synthase | 4.5 | 2.14 | 3.89E-01 |
| ins-37 | Insulin-like peptide | 4.5 | 0.13 | 9.41E-02 |
| fbxa-211 | Contains protein-protein interaction mediating F-box domain | 4.5 | 1.49 | 2.19E-01 |
| clec-216 | Contains C-type lectin-like domain | 4.5 | 0.39 | 2.31E-01 |
| clec-223 | Contains C-type lectin-like domain | 4.5 | 5.85b | 4.23E-02b |
| fasn-1 | Fatty acid synthase | 4.5 | 0.83 | 5.48E-01 |
| srv-15 | 7TM GPCR, serpentine receptor Class V | 4.5 | 0.04c | 9.94E-04c |
| C28C12.4 | Similar to yeast GAR1, which is involved in modification and cleavage of 18S pre-rRNA | 4.5 | 9.96 | 1.05E-01 |
| tyr-4 | Similar to human tyrosinase | 4.4 | 2.33 | 8.57E-02 |
| C16B8.3 | Similar to yeast RNA-binding protein NAB3 | 4.4 | 21.97b | 2.31E-03b |
| pat-12 | Similar to mouse serine/arginine repetitive matrix protein 2 | 4.4 | 2.60b | 7.04E-03b |
| T10E9.3 | Similar to yeast GPI-anchored cell surface glycoprotein (flocculin) | 4.4 | 1.80b | 1.71E-02b |
| nsy-4 | Member of PMP22/EMP/claudin/calcium channel gamma subunit family | 4.4 | 3.69b | 4.00E-02b |
| C27B7.5 | Predicted nucleic acid and zinc ion binding activity | 4.4 | 0.87 | 7.46E-01 |
| F36H5.10 | Unknown function | 4.4 | 0.07c | 4.62E-02c |
| ceh-32 | Six/sine oculis-type homeodomain protein | 4.4 | 0.64 | 2.05E-01 |
| F56H6.2 | Unknown function | 4.4 | 0.09c | 3.92E-04c |
| C01B7.5 | PDZ domain-containing protein | 4.4 | 0.30c | 2.09E-02c |
| sek-1 | Encodes a mitogen-activated protein kinase kinase | 4.4 | 3.75b | 1.57E-03b |
| gei-14 | Similar to human cylicin-1 | 4.4 | 0.48 | 2.75E-01 |
| C06A8.3 | Similar to O. volvulus Ov17 hypodermal antigen | 4.4 | 1.07 | 8.38E-01 |
| H08J19.1 | Unknown function | 4.3 | 0.67 | 2.83E-01 |
| T01D1.1 | Declared dead | 4.3 | NF | |
| lec-11 | Galectin family member | 4.3 | 20.86b | 6.74E-05b |
| B0353.1 | Similar to yeast GPI-anchored cell surface glycoprotein (flocculin) | 4.3 | 8.14b | 1.83E-03b |
| Y50D4B0.2 | Ortholog of human saccharopine dehydrogenase | 4.3 | 0.57 | 3.98E-01 |
| F21A9.2 | Contains zinc finger, C2H2-like domain | 4.3 | 1.57 | 5.97E-01 |
| F52H3.5 | Ortholog of human tetratricopeptide repeat domain 36 | 4.3 | 5.72 | 2.68E-01 |
| lfe-2 | Inositol (1,4,5) triphosphate-3-kinase | 4.3 | 4.37b | 6.55E-03b |
| Y22D7AR0.3 | Unknown function | 4.3 | 0.07 | 1.89E-01 |
| fbxa-140 | Contains protein-protein interaction mediating F-box domain | 4.3 | 2.35 | 2.54E-01 |
| vps-4 | Predicted ATP binding protein | 4.3 | 1.22 | 5.67E-01 |
| dnj-12 | Similar to human DnaJ homolog subfamily A member 1 | 4.2 | 1.00 | 8.91E-01 |
| hsp-16.2 | Heat shock protein | 4.2 | 1.17 | 8.44E-01 |
| F26A1.3 | Predicted protein kinase and ATP binding protein | 4.2 | 3.20 | 6.93E-01 |
| crh-2 | Predicted DNA binding transcription factor activity | 4.2 | 2.54b | 9.33E-03b |
| ZC239.2 | Contains potassium channel tetramerization-type BTB domain | 4.2 | 0.40 | 2.27E-01 |
| gst-36 | Similar to yeast RNA-binding protein NAB3 | 4.2 | 7.06b | 4.02E-03b |
| Y55B1AL0.1 | Similar to mouse serine/arginine repetitive matrix protein 2 | 4.2 | 7.23b | 4.45E-04b |
| C33C12.4 | Similar to yeast GPI-anchored cell surface glycoprotein (flocculin) | 4.2 | NF | |
| ram-2 | Member of PMP22/EMP/claudin/calcium channel gamma subunit family | 4.2 | 0.20 | 1.49E-01 |
| C04F12.11 | Predicted nucleic acid and zinc ion binding activity | 4.2 | NF | |
| rps-20 | Unknown function | 4.1 | 0.94 | 6.47E-01 |
| tts-1 | Six/sine oculis-type homeodomain protein | 4.1 | 22.99b | 5.39E-04b |
| F19B10.10 | Unknown function | 4.1 | 1.40 | 2.19E-01 |
| C17G1.5 | PDZ domain-containing protein | 4.1 | 3.37b | 1.98E-03b |
| ser-2 | Encodes a mitogen-activated protein kinase kinase | 4.1 | 0.98 | 4.77E-01 |
| F53A9.6 | Similar to human cylicin-1 | 4.1 | 46.43b | 3.80E-03b |
| C30G12.6 | Similar to O. volvulus Ov17 hypodermal antigen | 4.1 | 1.09 | 7.32E-01 |
| F31A3.3 | Unknown function | 4.1 | 23.69b | 4.63E-03b |
| K09F6.6 | Declared dead | 4.1 | 0.90 | 5.93E-01 |
| F54E7.6 | Galectin family member | 4 | 0.50 | 3.38E-01 |
| H25P19.1 | Similar to yeast GPI-anchored cell surface glycoprotein (flocculin) | 4 | 1.06 | 8.97E-01 |
| nhr-145 | Ortholog of human saccharopine dehydrogenase | 4 | 0.17 | 7.86E-02 |
| ttr-9 | Contains zinc finger, C2H2-like domain | 4 | 0.07c | 3.80E-03c |
| rom-2 | Ortholog of human tetratricopeptide repeat domain 36 | 4 | 8.30b | 1.78E-03b |
| eps-8 | Inositol (1,4,5) triphosphate-3-kinase | 4 | 4.49b | 3.84E-03b |
| R07B7.10 | Unknown function | 4 | 1.17 | 6.40E-01 |
| dct-18 | Contains protein-protein interaction mediating F-box domain | 4 | 7.23b | 3.96E-02b |
| ptc-1 | Predicted ATP binding protein | 4 | 1.20 | 3.65E-01 |
aRNAseq fold enrichment and p values for transcripts enriched ≥4-fold from Bacaj et al. (2008) microarray analysis. NF, Transcript not found in RNAseq data. Gene descriptions were derived from Wormbase (www.wormbase.org).
bValues from transcripts significantly enriched in RNAseq.
cValues from transcripts significantly depleted.
Because both intracellular and extracellular pH changes can affect so many aspects of synaptic function, it will also be interesting to examine in future experiments how disruption of pH regulation due to loss of CLH-1 in these glia affects sensory behavior in C. elegans. Loss of AMsh glia is known to affect the structure and/or function of several associated amphid sensory neurons (Bacaj et al., 2008), and our laboratory has previously shown that knock-out of the AMsh glia specific DEG/ENaC channel exacerbated deficits in sensory behavior (Wang et al., 2008, 2012). Thus, it is conceivable that disruption of glial pH regulation by loss of HCO3− flux through CLH-1 may have a multitude of consequences to glial and associated sensory neuronal function and sensory behaviors.
Footnotes
This work was supported by National Institutes of Health Grant RO1NS070969 to L.B. We thank the C. elegans Genetic Consortium and Keith Nehrke for providing the C. elegans strains; Keith Nehrke for the gift of CLH-1 cDNA; Diana Lopez for helping with the RNA sequencing data analysis; Rachele Sangaletti for helping with the cell culture; Peter H. Larsson and Stephen Roper for critical reading of the manuscript; and Shai Shaham and David Miller III for helpful discussions.
The authors declare no competing financial interests.
References
- Allaman I, Bélanger M, Magistretti PJ. Astrocyte-neuron metabolic relationships: for better and for worse. Trends Neurosci. 2011;34:76–87. doi: 10.1016/j.tins.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Bacaj T, Tevlin M, Lu Y, Shaham S. Glia are essential for sensory organ function in C. elegans. Science. 2008;322:744–747. doi: 10.1126/science.1163074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI. Chemosensation in C. elegans. WormBook: the online review of C elegans biology. 2006. pp. 1–29. www.wormbase.org. [DOI] [PMC free article] [PubMed]
- Bargmann CI, Horvitz HR. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991;7:729–742. doi: 10.1016/0896-6273(91)90276-6. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-H. [DOI] [PubMed] [Google Scholar]
- Bellemer A, Hirata T, Romero MF, Koelle MR. Two types of chloride transporters are required for GABA(A) receptor-mediated inhibition in C. elegans. EMBO J. 2011;30:1852–1863. doi: 10.1038/emboj.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple Testing. J R Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Bergles DE, Dzubay JA, Jahr CE. Glutamate transporter currents in bergmann glial cells follow the time course of extrasynaptic glutamate. Proc Natl Acad Sci U S A. 1997;94:14821–14825. doi: 10.1073/pnas.94.26.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevensee MO, Weed RA, Boron WF. Intracellular pH regulation in cultured astrocytes from rat hippocampus: I. Role Of HCO3. J Gen Physiol. 1997;110:453–465. doi: 10.1085/jgp.110.4.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolívar JJ, Lara-Figueroa CO, Martínez-Mayorquin RH, Monroy-Romero F, Arenas G. A chloride conductance exhibiting bicarbonate conductivity in renal inner medullary collecting duct cells. Gen Physiol Biophys. 2014;33:321–334. doi: 10.4149/gpb_2014006. [DOI] [PubMed] [Google Scholar]
- Bormann J, Hamill OP, Sakmann B. Mechanism of anion permeation through channels gated by glycine and gamma-aminobutyric acid in mouse cultured spinal neurones. J Physiol. 1987;385:243–286. doi: 10.1113/jphysiol.1987.sp016493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boron WF. Regulation of intracellular pH. Adv Physiol Educ. 2004;28:160–179. doi: 10.1152/advan.00045.2004. [DOI] [PubMed] [Google Scholar]
- Boron WF, De Weer P. Intracellular pH transients in squid giant axons caused by CO2, NH3, and metabolic inhibitors. J Gen Physiol. 1976;67:91–112. doi: 10.1085/jgp.67.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branicky R, Miyazaki H, Strange K, Schafer WR. The voltage-gated anion channels encoded by clh-3 regulate egg laying in C. elegans by modulating motor neuron excitability. J Neurosci. 2014;34:764–775. doi: 10.1523/JNEUROSCI.3112-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler KJ, Vaughan-Jones RD, Peers C, Nye PC. Intracellular pH and its regulation in isolated type I carotid body cells of the neonatal rat. J Physiol. 1991;436:107–129. doi: 10.1113/jphysiol.1991.sp018542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt AM, Kalsi A. Inwardly rectifying potassium channels (Kir) in central nervous system glia: a special role for Kir4.1 in glial functions. J Cell Mol Med. 2006;10:33–44. doi: 10.1111/j.1582-4934.2006.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HC, Shi QX, Zhou CX, Wang XF, Xu WM, Chen WY, Chen AJ, Ni Y, Yuan YY. Critical role of CFTR in uterine bicarbonate secretion and the fertilizing capacity of sperm. Mol Cell Endocrinol. 2006;250:106–113. doi: 10.1016/j.mce.2005.12.032. [DOI] [PubMed] [Google Scholar]
- Chen MH, Chen H, Zhou Z, Ruan YC, Wong HY, Lu YC, Guo JH, Chung YW, Huang PB, Huang HF, Zhou WL, Chan HC. Involvement of CFTR in oviductal HCO3− secretion and its effect on soluble adenylate cyclase-dependent early embryo development. Hum Reprod. 2010;25:1744–1754. doi: 10.1093/humrep/deq094. [DOI] [PubMed] [Google Scholar]
- Coulter DA, Eid T. Astrocytic regulation of glutamate homeostasis in epilepsy. Glia. 2012;60:1215–1226. doi: 10.1002/glia.22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damkier HH, Aalkjaer C, Praetorius J. Na+-dependent HCO3− import by the slc4a10 gene product involves Cl− export. J Biol Chem. 2010;285:26998–27007. doi: 10.1074/jbc.M110.108712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Di Schiavi E, Bergamasco C, Bazzicalupo P. Efficient and cell specific knock-down of gene function in targeted C. elegans neurons. Gene. 2007;395:170–176. doi: 10.1016/j.gene.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Fasseas MK, Tsikou D, Flemetakis E, Katinakis P. Molecular and biochemical analysis of the beta class carbonic anhydrases in Caenorhabditis elegans. Mol Biol Rep. 2010;37:2941–2950. doi: 10.1007/s11033-009-9857-z. [DOI] [PubMed] [Google Scholar]
- Fasseas MK, Tsikou D, Flemetakis E, Katinakis P. Molecular and biochemical analysis of the alpha class carbonic anhydrases in Caenorhabditis elegans. Mol Biol Rep. 2011;38:1777–1785. doi: 10.1007/s11033-010-0292-y. [DOI] [PubMed] [Google Scholar]
- Fatima-Shad K, Barry PH. Anion permeation in GABA- and glycine-gated channels of mammalian cultured hippocampal neurons. Proc Biol Sci R Soc. 1993;253:69–75. doi: 10.1098/rspb.1993.0083. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Ogura T, Katayama Y, Hiraoka M. Characteristics of rabbit ClC-2 current expressed in Xenopus oocytes and its contribution to volume regulation. Am J Physiol. 1998;274:C500–C512. doi: 10.1152/ajpcell.1998.274.2.C500. [DOI] [PubMed] [Google Scholar]
- Gradogna A, Pusch M. Alkaline pH block of CLC-K kidney chloride channels mediated by a pore lysine residue. Biophys J. 2013;105:80–90. doi: 10.1016/j.bpj.2013.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Wang Y, Sangaletti R, D'Urso G, Lu Y, Shaham S, Bianchi L. Two novel DEG/ENaC channel subunits expressed in glia are needed for nose-touch sensitivity in Caenorhabditis elegans. J Neurosci. 2013;33:936–949. doi: 10.1523/JNEUROSCI.2749-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson E, Eriksson P, Nilsson M. Amino acid and monoamine transport in primary astroglial cultures from defined brain regions. Neurochem Res. 1985;10:1335–1341. doi: 10.1007/BF00964976. [DOI] [PubMed] [Google Scholar]
- Harpur RP. Haemolymph gases and buffers in Ascaris lumbricoides. Comp Biochem Physiol A Comp Physiol. 1974;48:133–143. doi: 10.1016/0300-9629(74)90861-5. [DOI] [PubMed] [Google Scholar]
- Hille B. Ion channels of excitable membranes. Ed 3. Sunderland, MA: Sinauer; 2001. Selective permeability: Independence; p. xviii. [Google Scholar]
- Hug MJ, Tamada T, Bridges RJ. CFTR and bicarbonate secretion by [correction of to] epithelial cells. News Physiol Sci. 2003;18:38–42. doi: 10.1152/nips.01412.2002. [DOI] [PubMed] [Google Scholar]
- Ishiguro H, Namkung W, Yamamoto A, Wang Z, Worrell RT, Xu J, Lee MG, Soleimani M. Effect of Slc26a6 deletion on apical Cl−/HCO3− exchanger activity and cAMP-stimulated bicarbonate secretion in pancreatic duct. Am J Physiol Gastrointest Liver Physiol. 2007;292:G447–G455. doi: 10.1152/ajpgi.00286.2006. [DOI] [PubMed] [Google Scholar]
- Ishiguro H, Steward MC, Naruse S, Ko SB, Goto H, Case RM, Kondo T, Yamamoto A. CFTR functions as a bicarbonate channel in pancreatic duct cells. J Gen Physiol. 2009;133:315–326. doi: 10.1085/jgp.200810122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch TJ. CLC chloride channels and transporters: from genes to protein structure, pathology and physiology. Crit Rev Biochem Mol Biol. 2008;43:3–36. doi: 10.1080/10409230701829110. [DOI] [PubMed] [Google Scholar]
- Kaplan JM, Horvitz HR. A dual mechanosensory and chemosensory neuron in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1993;90:2227–2231. doi: 10.1073/pnas.90.6.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressin K, Kuprijanova E, Jabs R, Seifert G, Steinhäuser C. Developmental regulation of Na+ and K+ conductances in glial cells of mouse hippocampal brain slices. Glia. 1995;15:173–187. doi: 10.1002/glia.440150210. [DOI] [PubMed] [Google Scholar]
- Loria PM, Hodgkin J, Hobert O. A conserved postsynaptic transmembrane protein affecting neuromuscular signaling in Caenorhabditis elegans. J Neurosci. 2004;24:2191–2201. doi: 10.1523/JNEUROSCI.5462-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo L, Hellmich HL, Fong P, Wood T, Embesi J, Wills NK. Comparison of amphibian and human ClC-5: similarity of functional properties and inhibition by external pH. J Membr Biol. 1999;168:253–264. doi: 10.1007/s002329900514. [DOI] [PubMed] [Google Scholar]
- Mori I, Ohshima Y. Neural regulation of thermotaxis in Caenorhabditis elegans. Nature. 1995;376:344–348. doi: 10.1038/376344a0. [DOI] [PubMed] [Google Scholar]
- Nehrke K, Begenisich T, Pilato J, Melvin JE. Into ion channel and transporter function: Caenorhabditis elegans ClC-type chloride channels: novel variants and functional expression. Am J Physiol Cell Physiol. 2000;279:C2052–C2066. doi: 10.1152/ajpcell.2000.279.6.C2052. [DOI] [PubMed] [Google Scholar]
- Oikonomou G, Shaham S. The glia of Caenorhabditis elegans. Glia. 2011;59:1253–1263. doi: 10.1002/glia.21084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MD, Boron WF. The divergence, actions, roles, and relatives of sodium-coupled bicarbonate transporters. Physiol Rev. 2013;93:803–959. doi: 10.1152/physrev.00023.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen JH, Fischer H, Illek B, Machen TE. Bicarbonate conductance and pH regulatory capability of cystic fibrosis transmembrane conductance regulator. Proc Natl Acad Sci U S A. 1994;91:5340–5344. doi: 10.1073/pnas.91.12.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C, Lu Y, Shaham S. Glia delimit shape changes of sensory neuron receptive endings in C. elegans. Development. 2011;138:1371–1381. doi: 10.1242/dev.058305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z, Hartzell HC. Bestrophin Cl− channels are highly permeable to HCO3. Am J Physiol Cell Physiol. 2008;294:C1371–C1377. doi: 10.1152/ajpcell.00398.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratté S, Prescott SA. ClC-2 channels regulate neuronal excitability, not intracellular chloride levels. J Neurosci. 2011;31:15838–15843. doi: 10.1523/JNEUROSCI.2748-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke I, Artmann J, Stein V. ClC-2 voltage-gated channels constitute part of the background conductance and assist chloride extrusion. J Neurosci. 2010;30:4776–4786. doi: 10.1523/JNEUROSCI.6299-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero MF, Chen AP, Parker MD, Boron WF. The SLC4 family of bicarbonate (HCO(3) (−)) transporters. Mol Aspects Med. 2013;34:159–182. doi: 10.1016/j.mam.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychkov GY, Pusch M, Roberts ML, Jentsch TJ, Bretag AH. Permeation and block of the skeletal muscle chloride channel, ClC-1, by foreign anions. J Gen Physiol. 1998;111:653–665. doi: 10.1085/jgp.111.5.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangaletti R, Bianchi L. A method for culturing embryonic C. elegans cells. J Vis Exp. 2013;70:e50649. doi: 10.3791/50649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanan S, De Angelis D, Rothman JE, Ryan TA. The use of pHluorins for optical measurements of presynaptic activity. Biophys J. 2000;79:2199–2208. doi: 10.1016/S0006-3495(00)76468-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santello M, Calì C, Bezzi P. Gliotransmission and the tripartite synapse. Adv Exp Med Biol. 2012;970:307–331. doi: 10.1007/978-3-7091-0932-8_14. [DOI] [PubMed] [Google Scholar]
- Sardini A, Amey JS, Weylandt KH, Nobles M, Valverde MA, Higgins CF. Cell volume regulation and swelling-activated chloride channels. Biochim Biophys Acta. 2003;1618:153–162. doi: 10.1016/j.bbamem.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Sherman T, Chernova MN, Clark JS, Jiang L, Alper SL, Nehrke K. The abts and sulp families of anion transporters from Caenorhabditis elegans. Am J Physiol Cell Physiol. 2005;289:C341–C351. doi: 10.1152/ajpcell.00071.2005. [DOI] [PubMed] [Google Scholar]
- Sontheimer H. Voltage-dependent ion channels in glial cells. Glia. 1994;11:156–172. doi: 10.1002/glia.440110210. [DOI] [PubMed] [Google Scholar]
- Taylor CJ, Nicola PA, Wang S, Barrand MA, Hladky SB. Transporters involved in regulation of intracellular pH in primary cultured rat brain endothelial cells. J Physiol. 2006;576:769–785. doi: 10.1113/jphysiol.2006.117374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiemann A, Gründer S, Pusch M, Jentsch TJ. A chloride channel widely expressed in epithelial and non-epithelial cells. Nature. 1992;356:57–60. doi: 10.1038/356057a0. [DOI] [PubMed] [Google Scholar]
- Thomas RC. Experimental displacement of intracellular pH and the mechanism of its subsequent recovery. J Physiol. 1984;354:3P–22P. doi: 10.1113/jphysiol.1984.sp015397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bianchi L. Insights into the molecular determinants of proton inhibition in an acid-inactivated degenerins and mammalian epithelial Na(+) channel. Biochemistry. 2009;48:10005–10013. doi: 10.1021/bi9014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Apicella A, Jr, Lee SK, Ezcurra M, Slone RD, Goldmit M, Schafer WR, Shaham S, Driscoll M, Bianchi L. A glial DEG/ENaC channel functions with neuronal channel DEG-1 to mediate specific sensory functions in C. elegans. EMBO J. 2008;27:2388–2399. doi: 10.1038/emboj.2008.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, D'Urso G, Bianchi L. Knock-out of glial channel ACD-1 exacerbates sensory deficits in a C. elegans mutant by regulating calcium levels of sensory neurons. J Neurophysiol. 2012;107:148–158. doi: 10.1152/jn.00299.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenick AS, Hobert O. Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev Cell. 2004;6:757–770. doi: 10.1016/j.devcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- White MM, Aylwin M. Niflumic and flufenamic acids are potent reversible blockers of Ca2(+)-activated Cl− channels in Xenopus oocytes. Mol Pharmacol. 1990;37:720–724. [PubMed] [Google Scholar]
- Wulff H. New light on the “old” chloride channel blocker DIDS. ACS Chem Biol. 2008;3:399–401. doi: 10.1021/cb800140m. [DOI] [PubMed] [Google Scholar]
- Yu K, Lujan R, Marmorstein A, Gabriel S, Hartzell HC. Bestrophin-2 mediates bicarbonate transport by goblet cells in mouse colon. J Clin Invest. 2010;120:1722–1735. doi: 10.1172/JCI41129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampighi G, Kreman M, Ramón F, Moreno AL, Simon SA. Structural characteristics of gap junctions: I. Channel number in coupled and uncoupled conditions. J Cell Biol. 1988;106:1667–1678. doi: 10.1083/jcb.106.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zifarelli G. A tale of two CLCs: biophysical insights toward understanding ClC-5 and ClC-7 function in endosomes and lysosomes. J Physiol. 2015;593:4139–4150. doi: 10.1113/JP270604. [DOI] [PMC free article] [PubMed] [Google Scholar]