Abstract
Tsetse flies survive in a variety of environments across tropical Africa, often rising to large numbers, despite their low birth rate of one offspring every seven to nine days. They use olfactory receptors to process chemical signals in their environments to find food, escape from predators, and locate suitable larviposition sites. We discuss the identification of odorant and gustatory receptors in Glossina morsitans morsitans and the role genomics could play in management of nuisance insects.
Keywords: chemosensory proteins, odorant receptors, gustatory receptors, tsetse flies, ecology, genomics
The genome content of the Odorant Receptors (Ors) and Gustatory Receptors (Grs) of Glossina morsitans morsitans was published recently [1,2]. These receptors belong to the chemoreceptor family that mediates olfaction. Obiero et al. [1] found 46 Ors and 14 Grs in G. m. morsitans. Odorant receptors are expressed in the olfactory sensory neurons (OSNs), where they receive olfactory molecules – small molecules transported from the environment – in the initial step of processing of odors [3]. Grs are taste receptors, for example, distinguishing bitter and sweet. The number of Ors and Grs is less in tsetse flies compared to Drosophila melanogaster as well as vectors of human pathogens such as Anopheles gambiae and Aedes aegypti. Six Ors (GmmOr41-46) were identified as homologs of a single D. melanogaster Or (DMOr67d) that serves as the receptor for cis-vaccenyl acetate, a chemical known for mate seeking [4]. Because tsetse flies do not swarm and often rest singly, finding mates is probably reliant more on chemical than visual signals. Hence, this could explain the need for multiple Or copies to provide more of the receptor protein. This study also found that tsetse flies have lost all Grs for sweet taste that are present in Drosophila, but those that detect carbon dioxide, which is produced by animals, are retained and expanded. This is perhaps not surprising because tsetse flies feed only on blood, and both sexes are vectors of trypanosomes; unlike most vectors of important diseases, they do not feed on plant sugars in addition to blood. For instance, male and female Anopheles mosquitoes feed on both blood and sugars, with blood needed to support egg development. Data from the tsetse fly genome shows that insects invest heavily in critical functions such as finding sources of food. Exploitation of these findings will be exceedingly beneficial for developing vector control strategies.
Chemosensory proteins process olfactory and visual signals for critical survival functions, such as seeking food, resting sites, and suitable places to lay their larvae, as well as to escape from predators; Ors and Grs are central to this. Although the identification of the genes that provide the molecular basis of these functions is new knowledge, such observations have their foundation in experiments under-taken more than a century ago. Investigations on tsetse attractants date back at least a century, with skin secretions used in experiments by Balfour in 1913 (reviewed in [5]). Over the past century, the utility of various compounds, particularly those that are volatile, were established as attractants for tsetse. The identification and synthesis of odor attractants for tsetse in the 1970s and 1980s is acknowledged as a landmark development in tsetse fly control and considerably impacted trap and target based control strategies. Studies in Zimbabwe (e.g., [6]) showed the effect of animal odors on tsetse fly attraction. Tsetse show aversive responses to odors from some animals, such as the Water-buck [7], indicating that some level of chemically defined host choice by tsetse is at play.
Since the publication of the human genome, there has been considerable progress in sequencing technologies, resulting in a virtually continuous rollout of genome and transcription data. Among the beneficiaries of this sequencing revolution, driven largely by next generation sequencing (NGS) technologies and associated analyses, is the field of the biology of infectious diseases and their vectors. One of the most ambitious projects is the effort to generate genome sequences of 5000 insects from diverse groups, known as the i5k Insect and other Arthropod Sequencing Initiative (http://www.arthropodgenomes.org/wiki/i5K) [8], the growing assemblage of genomes of disease vectors (www.vectorbase.org), and multiple transcriptomes of insects (http://www.1kite.org). The authors of i5k state that the project will sequence insect genomes of representatives from all branches of insect phylogeny, including those used as model organisms and all known species important to medicine, worldwide agriculture, food safety, and energy production.
The availability of diverse genome sequences has huge potential for increasing understanding of vector biology and for providing solutions to insect vectored diseases and pests. When the human genome project was completed in 2001 (www.genome.gov), data acquisition was a bottleneck. The burden has now shifted to translation of these masses of data into knowledge and tools that can make a difference to health.
There is considerable optimism that vector genomics will spawn novel strategies for vector-based disease control [3]. Knowledge obtained recently from the genome of G. m. morsitans confirms the genetic basis of many of the decades old ecological observations, indicating the validity of genome-derived hypotheses. For example, bioassays to determine the attractiveness of some colors showed that blue was the most likely color to attract tsetse flies to a trap [9],confirming the validity of trap designs by Challier and colleagues [10]. The genome sequence also revealed the presence of the receptor gene for opsin Rh5, which indicates the likely presence of blue-sensitive R8p photoreceptors [2].
The G. m. morsitans genome provides opportunities for functional analyses to identify chemicals to which these receptors respond, using a panel of available approaches (Box 1). Assays such as the Drosophila ‘empty neuron approach’ have been used to identify odors and also define which ones are activated by single or multiple odors [3].
Box 1. Approaches for functional analysis of chemosensory proteins.
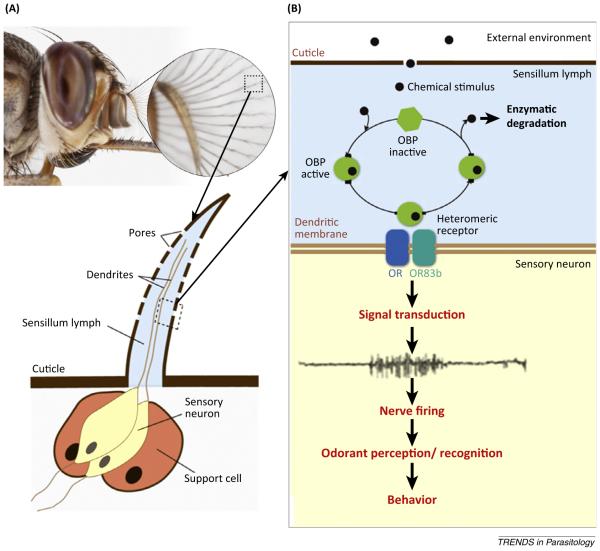
The main events in sensing chemical cues in the environment include (i) uptake of signal molecules from the external environment; (ii) transport through the sensory hair; and (iii) interaction with the chemoreceptor, which in turn activates the cascade of events leading to spike activity in sensory neurons (Figure I) [12]. Many functional studies aim to determine the specificity of volatile chemicals for olfactory receptors, and many tests exploit the discovery that the voltage between the base and tip of an insect’s antenna changes measurably when exposed to volatile chemicals of biological importance. The procedure, known as coupled gas chromatography-electroantennographic detection (GC-EAD) enables identification of compounds that stimulate olfactory sensilla. Compounds identified in this way can be further analyzed in bioassays involving Y-tube olfactometry (http://www.science.gov/topicpages/y/y-tube+olfactometer+bioassays.html), in which arthropods are given a choice of odors to determine their response (attraction or repellency). Other techniques include: (i) single cell recordings (SSR) via ‘surface contact’ technique and (ii) the Drosophila empty neuron system, a null mutant for one receptor gene, which allows heterologous expression of a foreign receptor (e.g., from tsetse) in the ‘empty space’. It integrates the Gal4-UAS system [3] and RNAi that has been used to demonstrate reduced electrophysiological responses when odorant binding proteins were targeted for knock-down [3].
Determining the chemical compounds to which insects respond has profound implications for describing the language of communication between insects and their environments, as attractants or repellents, which can lead to novel tools for control of disease vectors and other nuisance insects. Additional genome sequences from other species of Glossina, and other insects, as well as their transcriptomes, give us an opportunity to generate hypotheses about insect behaviour and test them to develop tools for disease management more efficiently and rapidly. More data that enable comparative evolution across species gives a unique opportunity to understand the function and anatomy of the nervous system of insects [11]. New synergy between field ecology, genomics and bioinformatics, and chemistry, should be exploited to advance the health of humans and the environment we occupy.
Figure I.
Schematic representation of insect olfaction. (A) shows the antennae of tsetse, with extensive branching to which sensilla (little hairs) are attached. Sensilla details are shown, with the structure depicting pores through which odorants enter. (B) shows how odorants are transported through sensillum fluid to the olfactory receptors in the neurons. Two olfactory receptors are necessary for activation – a specific receptor and a common receptor. Odorant binding triggers a signal transduction cascade that results in neuronal firing and brain-centered perception of the odorant, which triggers the appropriate behavioral response. For additional details, see [12]. Photo of tsetse fly head courtesy of Geoff Attardo, Yale School of Public Health; graphics by Brian Mwashi, icipe.
References
- 1.Obiero GFO, et al. Odorant and gustatory receptors in the tsetse fly Glossina morsitans morsitans. PLoS Negl. Trop. Dis. 2014;8:e2663. doi: 10.1371/journal.pntd.0002663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International Glossina Genome Initiative Genome sequence of the tsetse fly (Glossina morsitans): vector of African trypanosomiasis. Science. 2014;344:380–386. doi: 10.1126/science.1249656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carey AF, Carlson JR. Insect olfaction from model systems to disease control. PNAS. 2011;108:2987–12995. doi: 10.1073/pnas.1103472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ha TS, Smith DP. A pheromone receptor mediates 11-cis-vaccenyl acetate-induced responses in Drosophila. J. Neurosci. 2006;26:8727–8733. doi: 10.1523/JNEUROSCI.0876-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.International Livestock Research Institute ILRI 1998: Linking Livestock and Natural Resource Management. 1999 ILRI (aka ILCA and ILRAD) [Google Scholar]
- 6.Hargrove JW, Vale GA. The effect of host odour concentration on catches of tsetse flies (Glossinidae) and other Diptera in the field. Bull. Ent. Res. 1978;68:607–612. [Google Scholar]
- 7.Saini RK, Hassanali A. A 4-alkyl-substituted analogue of guaiacol shows greater repellency to savannah tsetse (Glossina spp.) J. Chem. Ecol. 2007;33:985–995. doi: 10.1007/s10886-007-9272-7. [DOI] [PubMed] [Google Scholar]
- 8.Robinson, Gene E, et al. Creating a buzz about insect genomes. Science. 2011;331:1386. doi: 10.1126/science.331.6023.1386. [DOI] [PubMed] [Google Scholar]
- 9.Green CH, Flint S. An analysis of colour effects in the performance of the F2 trap against Glossina pallidipes Austen and G. morsitans morsitans Westwood (Diptera: Glossinidae) Bull. Ent. Res. 1986;76:409–418. [Google Scholar]
- 10.Challier, et al. Amelioration du rendement du piege biconique pour glossines (Diptera, Glossinidae) par l’emploi d’un cone inferieur bleu. Can. ORSTOM Ser. Entomol. Med. Parasitol. 1977;15:283–286. [Google Scholar]
- 11.Hannson BS, Stensmyr MC. Evolution of Insect olfaction. Neuron. 2011;72:698–711. doi: 10.1016/j.neuron.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Sánchez-Gracia A, et al. Molecular evolution of the major chemosensory gene families in insects. Heredity. 2009;103:208–216. doi: 10.1038/hdy.2009.55. [DOI] [PubMed] [Google Scholar]